Abstract
Tumor-associated macrophages (TAMs) are an abundant tumor-promoting cell type in the tumor microenvironment (TME). Most TAMs exhibit a pro-tumor M2-like phenotype supportive of tumor growth, immune evasion, and metastasis. IL-4 and IL-13 are major cytokines that polarize macrophages to an M2 subset and share a common receptor, IL-4 receptor alpha (IL-4R alpha). Treatment of human ex vivo polarized M2 macrophages and M2 macrophage precursors with IL-4R alpha antagonist antibody Dupilumab (DupixentⓇ) reduces M2 macrophage features, including a shift in cell surface marker protein expression and gene expression. In animal models of prostate cancer, both pharmacologic inhibition of IL-4R alpha and genetic deletion of IL-4R alpha utilizing an Il4ra -/- mouse model result in decreased CD206 on TAMs. These data support IL-4R alpha as a target to reduce the pro-tumor, M2-like macrophage phenotype as a novel adjunct cancer therapy.
Keywords: Prostate cancer, Macrophage, IL-4R alpha, IL-4, Dupilumab, Dupixent
Introduction
Macrophages can comprise over 50% of solid tumors and play vital roles in cancer progression [1,2]. In patients, high infiltration of tumor-associated macrophages (TAMs) is correlated with poor prognosis [3], [4], [5], [6], [7]. While macrophages can be stimulated by their environment to adopt different phenotypes, the majority of TAMs are polarized towards a phenotype that promotes cancer progression and confers treatment resistance [1,6,[8], [9], [10], [11], [12], [13], [14]. In prostate cancer, higher pro-tumor macrophage infiltration in patient tumors is correlated with poorer prognosis [2]. Inflammatory disease research has developed successful strategies for macrophage targeting, but these strategies have yet to be tested as TAM-targeting agents in cancer [15].
Macrophages originate from either blood-circulating monocytes originating in the bone marrow or from proliferating tissue resident macrophages [16]. Intravasated monocytes that have differentiated into macrophages and tissue macrophages will adopt different phenotypes and functions to participate in various immune responses. It is recognized that macrophages exist on a continuous spectrum of differentiated phenotypes that are generally classified as anti-tumor (M1-like) or pro-tumor (M2-like). The phenotype that a macrophage adopts is determined by the stimulants in its local environment. Macrophages that encounter bacterial products (e.g. lipopolysaccharide) or inflammatory cytokines (e.g. IFNγ) are stimulated, or polarized, toward an M1 phenotype and participate in clearing intracellular infections [17]. Macrophages that encounter the cytokines interleukin 4 (IL-4) and interleukin 13 (IL-13) are polarized to a M2 phenotype and participate in processes such as wound healing [17], [18], [19]. M2 macrophages contribute to wound healing processes by remodeling the extracellular matrix, stimulating angiogenesis, and promoting cell growth and proliferation [17]. Unlike M1-like macrophages, M2-like macrophages do not employ free radical mechanisms to induce cell death of a target cell. Additionally, M2 macrophages prevent immune responses against the remodeling tissue by recruiting regulatory T cells and suppressing cytotoxic T cell function [20]. M2 macrophages contribute to many inflammatory diseases, such as asthma and atopic dermatitis, as well as pathogen immune evasion [21], [22], [23].
Most TAMs in solid tumors exhibit an M2-like phenotype and mirror the functions of M2 macrophages in normal tissue [1,8,9]. In the context of cancer, these functions contribute to uncontrolled proliferation of cancer cells and disease progression. Normal tissue remodeling signals produced by M2 macrophages are exploited to promote dysregulated tumor growth and survival of cancer cells. Additionally, the immunosuppressive signals produced by M2 macrophages prevent immune responses that would induce cancer cell death [20]. These functions also contribute to treatment resistance through various mechanisms [1,6,[10], [11], [12],24]. In particular, M2 TAMs counteract the effect of cytotoxic agents on cancer cells through secretion of survival signals and cathepsins [11,12]. Additionally, they counteract immunotherapies by secreting immunosuppressive cytokines, upregulating alternative immune checkpoint ligands, and sequestering checkpoint blockade agents [11], [12], [13],25]. The overwhelming evidence of M2 TAM contribution to cancer progression makes them a promising anti-cancer target [26,27].
The cytokines IL-4 and IL-13 are the predominant drivers of M2 polarization. These cytokines have significant overlap in signaling pathways and induce transcription of similar target genes [17,28]. The receptor complexes for both IL-4 and IL-13 include the IL-4 receptor alpha (IL-4R alpha) subunit and both receptor complexes are expressed on macrophages. Both pathways act through various JAK proteins (including JAK1, JAK2, and JAK3) and STAT6 to downregulate M1 genes, such as inflammatory cytokines IL1B and TNF, while upregulating transcription of M2 genes, such as MRC1 (CD206) and ARG1 [17,29]. Due to the involvement of IL-4 and IL-13 in inflammatory diseases, a number of therapeutic strategies have been developed to target these pathways [15]. A monoclonal antibody against IL-4R alpha, Dupilumab (DupixentⓇ), is FDA approved for the treatment of atopic dermatitis and asthma [15,30]. Studies have also implicated IL-4 and IL-13 signaling in pro-tumor mechanisms including direct signaling to cancer cells overexpressing IL-4 or IL-13 receptors [1,15,[31], [32], [33], [34], [35], [36]. However, the impact of these signaling pathways on macrophage function in the tumor remains unknown.
Despite the pro-tumor functions of M2 TAMs, evidence for pro-tumor effects of the IL-4R alpha pathway in cancer, and the success of IL-4R alpha-targeting strategies in inflammatory diseases, little research has described the effects of these strategies in M2 TAMs or tested them clinically against cancer. We found that disrupting IL-4R alpha signaling either pharmacologically with Dupilimab or with an Il4ra–/– mouse model reduces the pro-tumor M2-TAM phenotype. This data suggests that targeting IL-4R alpha is a promising strategy for undermining the tumor-promoting capabilities of M2-TAMs that may have utility in combination with conventional anti-cancer therapy.
Materials and methods
Human macrophage culture
Human peripheral blood mononuclear cells (PBMCs) were acquired from the New York Blood Center (New York, NY). Monocytes were isolated and M1 and M2 macrophages were generated using previously published methods [37]. Unpolarized macrophages (M0s) were generated using the same protocol except without addition of any cytokines. Prior to polarization or analysis, isolated monocytes were cryopreserved in 95% FBS (VWR) 5% DMSO.
Macrophage gene expression analysis
Expression levels of 770 immune-related mRNAs were assessed by human nCounter Myeloid Innate Immunity Panel and custom 30 gene Panel Plus (NanoString Technologies). Hybridization of human samples were performed using 75-100 ng of RNA. Hybridization of mouse samples were performed using 20 ng of RNA. Gene expression was analyzed with nSolver software 4.0 (NanoString Technologies). The expression levels of each gene were normalized to those of control genes. Heat maps and unsupervised hierarchical clustering were generated in nSolver with agglomerative cluster analysis using average Euclidean distance.
Dupilumab treatment of human macrophages
Three Dupilumab (DupixentⓇ, Sanofi and Regeneron Pharmaceuticals, Inc.) conditions were used: M2-polarized macrophages that were treated with Dupilumab at the monocyte stage (Dup-mono; 20 ug/ml Dupilumab when plated on Day 0 and when fresh media was supplied on Day 5 one hour prior to IL-4 and IL-13 cytokine addition), at the unpolarized macrophage stage (Dup-unpol; administered 20 ug/ml Dupilumab when fresh media was supplied on Day 5 one hour prior to IL-4 and IL-13 cytokine addition), and once polarized to M2 macrophages (Dup-M2; 20 ug/ml Dupilumab when fresh media without cytokines was supplied on Day 9 after full polarization). The “No IL-4/13” conditions followed M2 macrophage polarization protocol but were not given IL-4 or IL-13.
Immunoblot analysis
Cells were washed in PBS and centrifuged. Pellets were resuspended in Frackelton lysis buffer (2.5 mM Tris-HCl, 7.5 mM Na4P2O7 pH 7.1, 12.5 mM NaCl, 12.5 mM NaF, 0.25% Triton X-100 supplemented with 10 µg/mL leupeptin, 2 µg/mL aprotinin, 1 µg/mL pepstatin A, 1 mM phenylmethanesulfonyl fluoride, 0.5 mM Na3VO4, and 0.2 mM DTT) supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (78442, ThermoFisher). Protein concentration was determined by BCA assay (23225, ThermoFisher) and protein lysates were prepared for electrophoresis by adding 4x Laemmli Sample Buffer (161-047, BioRad) supplemented with fresh 2-β-mercaptoethanol (161-0710, BioRad) at 1:10. Samples were run on a 4-20% SDS-PAGE gel (456-1093, BioRad) and protein was transferred onto a nitrocellulose membrane (1704158, BioRad). Membranes were blocked with 1X Casein Blocking Buffer (B6429, Sigma-Aldrich) and incubated with STAT6 (9362S, Cell Signaling, diluted 1:1,000 in Casein) and β-actin (A5441, Sigma-Aldrich, diluted 1:5,000 in Casein) or p-STAT6 (Y641) (9361T, Cell Signaling, diluted 1:5,000 in Casein) and β-actin (A5441, Sigma-Aldrich, diluted 1:5,000 in Casein) antibodies overnight at 4°C under agitation. Membranes were washed in TBS with 0.1% Tween and incubated with HRP-conjugated anti-rabbit (7074P2, Cell Signaling, diluted 1:3,000 in Casein) and HRP-conjugated anti-mouse (7076, Cell Signaling, diluted 1:3,000 in Casein) antibodies for 1 hour at room temperature under agitation. Blots were imaged using SuperSignal™ West Dura Extended Duration Substrate (34076, ThermoFisher) and film processing.
Flow cytometric analysis of in vitro experiments
Cells were dissociated using enzyme-free Cell Dissociation Buffer (13151014, ThermoFisher) with scraping. In vitro human macrophages were stained with CD206-FITC (130-100-085, Miltenyi Biotec), CD163-PE-Cy7 (clone GHI/61, 333614, BioLegend), CD86-PE (305406, BioLegend), and propidium iodide (PI, 00-6990-50, eBioscience). Data was collected using a BioRad S3TM Cell Sorter and analysis was performed using FlowJoⓇ.
Cell line culture and MycCaP-luc cell line generation
HEK293T (CRL-3216™, ATCC) and MycCaP (CRL-3255™, ATCC) cells were maintained in DMEM (11995073, Gibco) supplemented with 10% FBS (97068-085, Avantor) and 1% Penicillin-Streptomycin (11995073, Gibco). For MycCaP-luc cell line generation, HEK293T cells were transfected with 0.5ug pQCX1B CMV/TO LUC, 0.444ug pUMCV3, 0.0556ug pMD2.G and 2uL X-tremeGENE™ HP (6366236001, Roche) in DMEM without supplements. pQCXIB CMV/TO LUC (w431-1) was a gift from Eric Campeau & Paul Kaufman (Addgene plasmid # 17475; http://n2t.net/addgene:17475; RRID:Addgene_17475) [38], pUMVC was a gift from Bob Weinberg (Addgene plasmid # 8449; http://n2t.net/addgene:8449; RRID:Addgene_8449) [39], and pMD2.G was a gift from Didier Trono (Addgene plasmid # 12259; http://n2t.net/addgene:12259; RRID:Addgene_12259). Virus containing media from HEK293T cells was transferred for MycCaP cells with polybrene (H9268, Sigma-Aldrich). Target cells were selected with Blasticidin (R21001, Invitrogen) and remained under selection until mock-transduced cells died. Luciferase expression in MycCaP-luc cells was validated with Luciferase Assay System (E1500, Promega). All cell lines were cultured at 37°C and 5% CO2. All cell lines were authenticated and tested for mycoplasma (Genetica).
Il4ra KO mouse model
The Johns Hopkins Institutional Animal Care and Use Committee approved all experiments involving mice (protocol # MO19M41). FVB/N mice were purchased from The Jackson Laboratory (Bar Harbor, ME). FVB/N Il4raem1/em1 (Il4ra KO) mice were created by The Jackson Laboratory (Stock No. 037518) by whole animal gene knockout using CRISPR/Cas9 to remove exon 4 of Il4ra. Genotyping was performed by tail snip DNA extraction and PCR using forward primer 5’- AGCCTGAGCCGTACAGATTG-3’ (common) and reverse primers 5’-ACAGAACGGCCAGATCAGTG-3’ (WT) and 5’- TAACAGAACGCAGGGTCATC-3’ (Mutant). To confirm IL-4R alpha protein knockout, mouse spleen cells were stained with anti-mouse CD124-PE (clone mIL4R-M1, 561695, BD Biosciences) or isotype rat IgG2a,κ-PE (553930, BD Biosciences) and PI. Data was collected using a BioRad S3TM Cell Sorter and analysis was performed using FlowJoⓇ.
In vivo tumor models
Male FVB/N mice (6-8 weeks old) were injected subcutaneous in the right flank with 1 million MycCaP cells in 100uL Matrigel Matrix Basement Membrane (Corning 35623) diluted in 100uL sterile Hank's Balanced Salt Solution (H6648, Sigma-Aldrich). Twice per week, mice were injected IP with either 0.2 mg anti-mouse IL-4R alpha clone MIL4R-M1(BD 624094) or 0.2 mg Rat IgG2a,k isotype control clone R35-95 (BD 624094), and either 0.5 mg InVivoMAb anti-mouse IL-4 (BioXCell BE0045) or 0.5 mg InVivoMAb rat IgG1 isotype control, anti-horseradish peroxidase (BioXCell BE0088). All antibodies were diluted in InVivoPure Dilution Buffer (BioXCell IP0070) to a final volume of 200µL per mouse. Caliper measurements were used to monitor tumor volume, calculated as 0.5 x L x W2 with L measured as the largest tumor diameter and W as perpendicular. Mice were euthanized at 24 days post-inoculation and tumors were removed and stained with the flow cytometry panels (Myeloid panel Version 1, Supplementary Table 2; Lymphocyte panel, Supplementary Table 3).
Male Il4ra KO and Il4ra WT (6-8 weeks old) were injected subcutaneous in the right flank with 1 million MycCaP-luc cells in 100uL Matrigel Matrix Basement Membrane (Corning 35623) diluted in 100uL sterile Hank's Balanced Salt Solution (H6648, Sigma-Aldrich). Mice were injected IP with 100uL of 30 mg/mL luciferin (360222, Regis) in DPBS, anesthetized, and were imaged 5 minutes later using the IVIS Spectrum In Vivo Imaging System (PerkinElmer). Whole body total flux (photons/sec) was quantified using Living ImageⓇ 4.4. Tumor volume was calculated as 0.5 x L x W2 with L measured as the largest tumor diameter and W as perpendicular via caliper measurements. Mice were euthanized at 20 days post-inoculation and tumors were removed and stained with flow cytometry panels (Myeloid panel Version 2, Supplementary Table 2; Lymphocyte panel, Supplementary Table 3).
Flow-cytometric immune cell analysis
Tumor tissue was subjected to single cell dissociation using the MACS Mouse Tumor Dissociation Kit protocol and gentleMACS Dissociator (Miltenyi). Suspended cells were blocked with rat serum (012-000-120, Jackson ImmunoResearch), stained with FVS570 viability dye (1 ul/ml, 564995, BD Biosciences) or LIVE/DEAD Fixable Yellow (1 ul/ml, L34959, Thermo Fisher Scientific) in the dark for 15 minutes at room temperature. Samples were washed with PBS and incubated with Myeloid extracellular antibody panel (Supplementary Table 2), Lymphocyte extracellular antibody panel (Supplementary Table 3), or corresponding isotype panels diluted in Brilliant Stain Buffer (566349, BD Biosciences) in the dark for 30 minutes at 4°C. Cells were washed with FACS buffer (1x PBS, 1% BSA, 2mM EDTA), fixed with 1x Fixation Buffer (420801, BioLegend) in the dark for 20 minutes at room temperature, and stored overnight in Flow Buffer at 4°C. Samples were incubated in 1x FoxP3 Fix/Perm Solution (421401, BioLegend) in the dark for 20 minutes at room temperature and washed with 1x FoxP3 Perm Buffer (421402, BioLegend). Cells were resuspended with Myeloid intracellular antibody panel (Supplementary Table 2), Lymphocyte intracellular antibody panel (Supplementary Table 3), or corresponding isotype panels diluted in Flow Buffer in the dark for 30 minutes at room temperature under gentle agitation. Cell suspensions were washed with Flow Buffer and analyzed with a Gallios flow cytometer (Beckman Coulter Life Sciences) or Attune NxT flow cytometer (Thermo Fisher Scientific) and analysis was performed with Kaluza Analysis Software (Beckman Coulter). Immune cell population markers are defined in Supplementary Table 4.
Statistical Analysis
Differentially expressed gene analyses were performed in nSolver software 4.0 (NanoString Technologies) using the Differential Expression Call Error Model. Outliers were identified by Grubbs’ test with a false discovery rate (q) = 0.05. All results are expressed as means ± SD. Data were analyzed using t-test, one- or two-way ANOVA as specified. Differences were considered significant at p < 0.05. Figures denote statistical significance of p < 0.05 as *, p < 0.01 as **, p < 0.001 as ***, and p < 0.0001 as ****.
Results
Pharmacologic blockade of IL-4Ra during M2 macrophage differentiation skews human macrophages away from the M2 phenotype
To assess the role of IL-4R alpha in initiating the pro-tumor M2-like macrophage phenotype, cultures were treated with IL-4R alpha antagonist antibody Dupilumab at multiple stages of macrophage differentiation and M2 polarization. Dupilumab was added at time of monocyte seeding and continued until M2 polarization was complete (Dup-mono) or at initiation of M2 polarization (Dup-unpol). Following M2 polarization without addition of Dupilumab, STAT6 phosphorylation indicated activation and signaling through IL-4R alpha (Figure 1A). In all Dupilumab-treated conditions, STAT6 phosphorylation was undetectable indicating that the signaling pathway was not activated, phenocopying cultures without addition of IL-4R alpha ligands IL-4/13.
Figure 1.
Effects of Dupilumab on monocyte, M0, M1, and M2 signaling and protein expression. (A) A representative immunoblot of M2 macrophages, macrophages polarized with the M2 protocol except without IL-4 or IL13 (“No IL-4/13”), and Dupilumab-treated conditions. (B) Flow cytometry and (C) corresponding delta median fluorescence intensities (MFIs) of Dupilumab-treated M2 precursors (Dup-mono and Dup-unol). (D) Flow cytometry and (E) corresponding delta median fluorescence intensities (MFIs) of Dupilumab-treated M2 macrophages (Dup-M2).
Canonical pro-tumor M2 macrophage markers and anti-tumor M1 macrophage markers were assessed to assess the impact of Dupilumab blockade of IL-4R alpha during M2 polarization. Flow cytometry analyses showed a decrease in pro-tumor M2 marker CD206, an increase in pro-tumor CD163, and a decrease in anti-tumor CD86 protein expression in Dupilumab treated conditions, consistent with the phenotype observed in the “No IL-4/13” conditions (Figure 1B-C).
To assess the impact of IL-4R alpha blockade on macrophage plasticity of polarized M2 macrophages, human macrophage cultures were treated with Dupilumab following M2 polarization (Dup-M2). IL-4R alpha blockade in treated cultures was confirmed by absence of STAT6 activating phosphorylation (Figure 1D). Classical markers of pro-tumor and anti-tumor macrophages were assessed to determine effects on macrophage plasticity. All Dupilumab-treated M2 macrophages mimicked the “No IL-4/13” conditions with decreased expression of both pro-tumor CD206 and anti-tumor CD86 expression (Figure 1E-F). CD163 expression was unchanged in the Dupilumab treated cultures, though it was elevated in macrophages cultured in the absence of IL4/13.
Dupilumab treatment skews macrophages away from a pro-tumor M2-like gene signature and towards an unpolarized M0 gene signature
Macrophage phenotype is often characterized by the expression of one or two genes, but this offers a limited view of actual macrophage biology and phenotype plasticity. To more holistically capture the phenotypic plasticity observed with IL-4R alpha blockade, gene expression of 800 genes was assessed using the nCounter technology. The Myeloid Innate Immunity panel supplemented with an additional custom 30 gene panel is sufficient to distinguish human monocytes, unpolarized M0 macrophages, M1 macrophages, and M2 macrophages with each cell type clustered independently from the other macrophage phenotypes, demonstrating a unique transcriptomic signature for each differentiation or polarization state (Supplementary Figure 1A). To investigate the expression of genes commonly used to define M1-like or M2-like tumor-associated macrophages, canonical pro-tumor genes and anti-tumor genes were interrogated (Supplementary Table 5). This limited gene set was sufficient to distinguish each macrophage subtype (Supplementary Figure 1B).
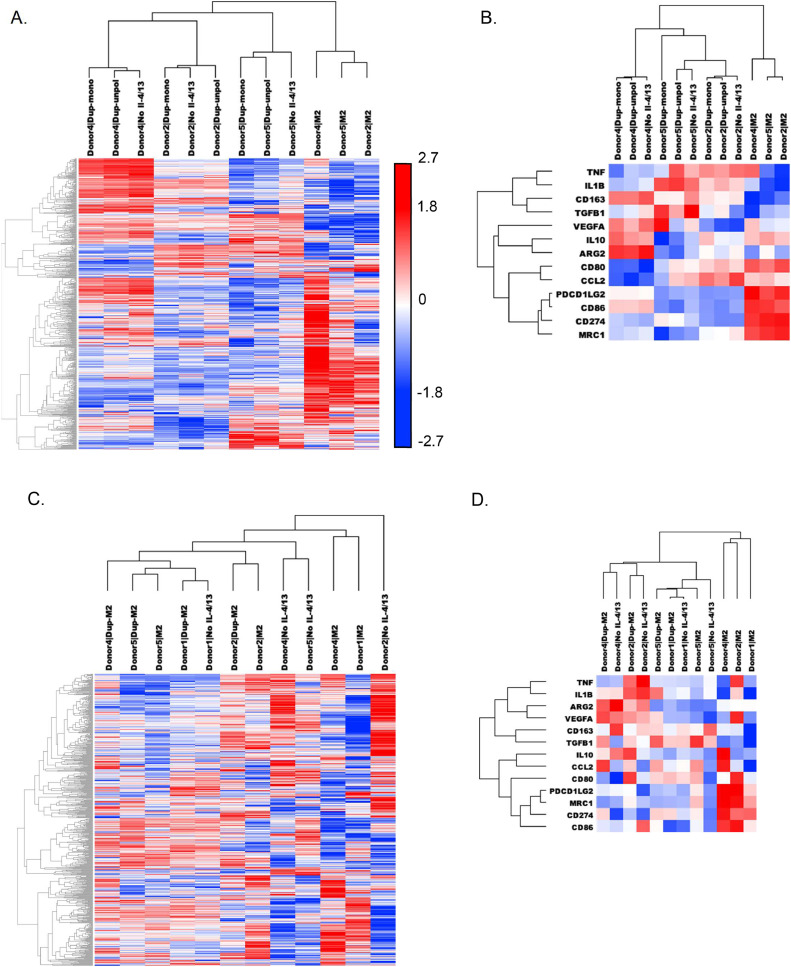
Gene expression patterns of human macrophage cultures treated with Dupilumab during macrophage differentiation (Dup-mono) and M2 polarization (Dup-unpol) and control cultures without IL-4/13 (“No IL-4/13”) and unmanipulated M2 culture conditions were assessed using unsupervised hierarchical clustering. Across all donors, M2 macrophages clustered independently from Dupilumab-treated and “No IL-4/13” conditions. In cultures in the absence of IL-4R alpha signaling, samples clustered independently by donor (Figure 2A). This pattern persisted when the analysis was limited to canonical pro- and anti-tumor macrophage genes (Supplementary Table 1), including genes encoding for proteins assessed by flow cytometry: CD163, CD86, and MRC1 (CD206) (Figure 2B).
Figure 2.
Gene expression changes with Dupilumab treatment. Dupliumab was added to M2 precursors prior to polarization: M2, No IL-4/13, Dupmono, and Dup-unpol conditions were prepared from 3 separate donors (Donors 2, 4, and 5). (A) Heat map and dendrograms comparing each condition across all expressed genes in the nCounter Myeloid Innate Immunity Panel. (B) Heat map and dendrograms comparing each comparing each condition across canonical pro- and anti-tumor genes. Dupliumab was added after M2 polarization: M2, No IL-4/13, and Dup-M2 conditions were prepared from 4 separate donors (Donors 1, 2, 4, and 5). (C) Heat map and dendrograms comparing each condition across all expressed genes in the nCounter Myeloid Innate Immunity Panel. (D) Heat map and dendrograms comparing each condition across canonical pro- and anti-tumor genes. (E) Heat map and dendrograms comparing Dupilumab treated M2 precursors and macrophages with M1s, M0s and monocytes across canonical pro- and anti-tumor genes. Heat maps were generated using unsupervised hierarchical clustering with centered Pearson Correlation.
When Dupilumab was introduced to polarized M2 macrophages (Dup-M2), the myeloid gene expression profiles taken as a whole did not cluster independently from the M2 controls (Figure 2C). Unlike the effects of Dupilumab on M2 precursors, the effects of Dupilumab on fully polarized M2s were more subject to donor variation. For Donors 1 and 4, Dup-M2 gene expression more closely resembled their respective “No IL-4/13” conditions. However, Dup-M2 from Donors 2 and 5 more closely resembled myeloid gene expression profiles of donor-matched M2s rather than donor-matched “No IL-4/13” conditions.
When limited to canonical pro- and anti-tumor macrophage genes, untreated M2 polarized macrophages from three donors clustered independently from the Dupliumab-treated and “No IL-4/13” conditions (Figure 2D). Gene expression of M2 polarized macrophages from Donor 5 clustered with the Dupilumab-treated and “No IL-4/13” cultures. In general, Dupilumab-treated cultures had a most similar gene expression profile to their matched “No IL-4/13” controls. Supplementary Figure 2 summarizes the key findings of significantly upregulated and downregulated genes across multiple Dupilumab-treated conditions. All significantly upregulated and downregulated genes with Dupilumab treatment from all experiments are listed in Supplementary Tables 6-11. Independent validation of changes in gene expression using immunological reagents are needed to strengthen these findings.
To assess how Dupilumab-treated macrophages compared other macrophage subtypes (i.e. monocytes, M0s, and M1s), gene expression data of canonical pro- and anti-tumor macrophage genes in these cell types was analyzed alongside data from the Dupilumab treatment experiments (Figure 2E). Polarized M1 macrophages and monocytes clustered independently by macrophage subtype, regardless of donor. In general, Dupilumab-treated M2 cultures (and the “No IL-4/13” conditions) were most similar to each other and to unpolarized M0 cultures with a high degree of relatedness within multiple subtypes/conditions each donor. The next closest relatedness was to M2 macrophage samples that in general clustered independently from M0 samples, Dupilumab-treated M2s samples, and “No IL-4/13” M2 samples (with the exception of Donor 2 M2 noted above).
Pharmacologic blockade of the IL-4R alpha pathway decreases pro-tumor TAM infiltrate in vivo
The effects of IL-4R alpha blockade in prostate cancer were investigated in vivo using a syngeneic prostate cancer tumor model. Mice were inoculated with MycCaP cells and treated with both IL-4 receptor and cytokine IL-4 targeting antibodies were used to ensure total IL-4R alpha signaling blockade. There was no change in tumor growth monitored via caliper measurements or in end tumor weight in antibody treated groups compared to isotype antibody controls (Figure 3A-B). To assess the immune cell infiltrate of the tumors, including TAMs, flow cytometry was performed. Although there was no change in total macrophage infiltration, IL-4R alpha inhibition decreased the percentage of M2-like CD206+ macrophages (Figure 3D) and decreased macrophage CD206 expression (Figure 3E). T cell, NK cell and B cell populations remained unchanged (Figure 3F-G, Supplementary Figure 3A-C).
Figure 3.
Tumor growth and immune characteristics with IL-4R alpha signaling inhibition. FVB/N mice were inoculated with subcutaneous MycCaP tumors and treated twice per week with combinations of anti-IL-4R alpha, anti-IL-4 antibodies, or corresponding isotype controls. (A) Tumor growth measured by caliper measures and (B) endpoint tumor weight. Immune cell populations were determined by flow cytometry for (C) macrophages, (D) CD206+ macrophages, (E) CD206 relative MFI on tumor macrophages, (F) CD8+ T cells, and (G) CD4+ T cells. Relative mean fluorescence intensity (MFI) was determined by setting the average MFI of the WT cohort to 1. Significance of bar graphs were determined by one-way ANOVA followed by pairwise comparison with * p < 0.05, ** p < 0.01 and *** p < 0.001.
Genetic deletion of Il4ra decreases pro-tumor TAM infiltrate in vivo
To further study the effects of IL-4R alpha inhibition, we generated an FVB/N Il4ra–/– mouse model. Flow cytometry of mouse spleens confirmed that the cells were null for IL-4R alpha (Figure 4A). Il4ra–/– (Il4ra KO) and wild type (WT) mice were injected with MycCaP-luc prostate cancer cells to generate a subcutaneous tumor. Cancer cell burden monitored by bioluminescent imaging and tumor size by caliper measurements. Il4ra KO mice had a lower BLI signal at the final timepoint on Day 18 (Figure 4C) but no significant difference in tumor volume assessed by caliper measurements (Figure 4D) or end tumor weight (Figure 4E).
Figure 4.
Il4ra KO versus WT tumor growth and immune characteristics. FVB/N Il4ra knockout (KO) and wild type (WT) mice were inoculated with subcutaneous MycCaP-luc tumors. (A) IL-4R alpha protein expression in FVB/N WT and FVB/N Il4ra KO spleens. (B) Tumor growth measured by BLI and (C) quantified at Day 18. (D) Tumor growth measured by caliper measurements. (E) Endpoint tumor weight. Immune cell populations were determined by flow cytometry for (F) macrophages, (G) CD206+ macrophages, (H) CD206 relative MFI on tumor macrophages, (I) CD8+ T cells, and (J) CD4+ T cells. Relative mean fluorescence intensity (MFI) was determined by setting the average MFI of the WT cohort to 1. Significance of bar graphs were determined by oneway ANOVA followed by pairwise comparison with * p < 0.05 and ** p < 0.01.
To assess the characteristics of immune cell infiltration flow cytometry was performed (Figure 4F-J, Supplementary Figure 3D-F). While there was no difference in total numbers of TAM infiltrate (Figure 4F), macrophage expression of pro-tumor M2-like marker CD206 was decreased in TAMs from Il4ra KO mice (Figure 4G-H). In the T cell compartment, cytotoxic CD8 T cell infiltrate was unchanged and there was an increase in percentage of CD4 T cells (Figure 4I-J). Regulatory T cells, NK cell and B cell infiltration did not change (Supplementary Figure 3D-F).
Discussion
Targeting TAMs and specifically the pro-tumor M2-like phenotype are emerging areas of interest for novel cancer therapeutics. Previously, we have shown that M2-like macrophages are increased in murine and human prostate cancer [2,40]. We investigated whether IL-4R alpha inhibition via Dupilumab treatment diminishes the M2-like macrophage phenotype in human macrophages in vitro. Overall, Dupilumab treatment skewed pre-polarized M2s and M2 precursors (monocytes and unpolarized macrophages) away from an M2 phenotype. With Dupilumab treatment, CD206 expression decreased and changes in cell surface markers CD206, CD163 and CD86 closely mimicked “No IL4/13” conditions. A greater effect was achieved when Dupilumab was introduced prior to polarization rather than to macrophage cultures already polarized to an M2-like state. This suggests that Dupilumab treatment may be more effective in altering actively infiltrating and polarizing macrophages rather than established pro-tumor TAMs.
To achieve a more holistic understanding of the role of IL-4R alpha in macrophage phenotype beyond canonical “M1-like” and “M2-like” markers, unbiased macrophage phenotype gene analysis was performed with the NanoString Human nCounter Myeloid Innate Immunity Panel and custom Panel Plus. This analysis showed that Dupilumab treatment in both M2 precursors and polarized M2s skewed macrophage characteristics away from a polarized M2-like phenotype and towards an unpolarized M0 phenotype, but did not induce adoption of an M1-like phenotype. Dupilumab-treated macrophages more closely resembled the gene expression of M1 macrophages than undifferentiated monocytes, suggesting that they share a common “macrophage differentiation” phenotype rather than that of a macrophage precursor.
Many canonical pro- and anti-tumor genes did not change as expected with Dupilumab treatment and exhibited high variation between donors (Figure 2E). For example, Donor 4 appears predisposed to high ARG2 expression and Donors 2 and 5 are predisposed to high CCL2 and TNF expression. While these genes are often accepted as strong macrophage subtype classifiers, these data show that they are subject to individual biological variation and uncontrolled confounding factors such as age, sex, genetic differences affecting immune responses, and environmental exposure to various pathogens.
Altogether, these in vitro data show that IL-4R alpha blockade with Dupilumab skews macrophages away from a pro-tumor subtype making it a promising strategy for altering M2-like TAMs. The drug did not repolarize macrophages towards an anti-tumor M1-like phenotype, however this is not surprising given that it does not involve an anti-tumor stimulant. By simply attenuating the pro-tumor capabilities of TAMs without amplifying the anti-tumor capabilities, we circumvent any anticipated autoimmune effects from systemic M1 stimulation while still weakening TAM tumor support. Therefore, targeting TAMs with Dupilumab in combination with a cytotoxic agent to target cancer cells shows promise as an effective cancer treatment strategy.
A tumor is in an incredibly complex and dynamic microenvironment that cannot be fully recapitulated in vitro. We investigated the effect of IL-4R alpha inhibition on tumor growth and immune characteristics in prostate cancer mouse models. In the MycCaP subcutaneous model, there was no difference in tumor growth with pharmacologic inhibition of IL-4R alpha. However, in the Il4ra KO mice, final tumor burden by BLI (a measure of only cancer cells) is decreased, suggesting that there may be a decrease cancer cell proliferation at later stages of tumor growth in this model. Differences in tumor growth between experiments with the MycCaP model may be due to cell line injected (luciferase versus no luciferase), IL-4R alpha inhibition on tumor cells in the antibody treatment model or mouse strain background. Additional model systems are needed to assess targeting IL-4R alpha to inhibit tumor growth.
IL-4R alpha inhibition by both antibody treatment and genetic deletion decreased CD206 expression in TAMs. CD206 is the most well characterized marker of the M2-like macrophage phenotype. These in vivo results support that findings from our in vitro studies showing that IL-4R alpha inhibition with Dupilumab skews macrophage phenotype away from a pro-tumor M2-like phenotype. In depth phenotype analyses of in vivo prostate cancer TAMs (expanded cell surface marker expression, gene expression) will investigate this further.
To better understand the effects of targeting macrophage IL-4R alpha on other immune cell compartments of the tumor, we investigated changes in CD4 and CD8 T cell infiltration. With genetic knockout of Il4ra there is an increase in CD4 T cells tumor infiltration (Figure 4J). It remains of interest whether there is an increase in infiltration of a CD4 T cell subset (Th1, Th2, etc.) or a global increase in all CD4 T cell subsets. Given the published evidence that M2-like macrophages inhibit T cells [41,42], we anticipated a decrease in CD8 T cells with IL-4Ra blockade. With inhibition of IL-4R alpha, there is no significant difference in CD8 T cell infiltration (Figures 3F and 4I). These data suggest that targeting macrophage IL-4R alpha is not sufficient for increasing CD8 T cell infiltration in these models. Future studies will combine IL-4R alpha inhibition with a T cell targeting agent (e.g. anti-PD1/PDL1) as well as looking at T cell activation and checkpoint markers (CD25, CD69, PD1, Lag3, etc.) to investigate targeting IL-4R alpha to inhibit tumor growth.
Conclusions
Despite the tumor-supporting role of M2-like TAMs, these cells have yet to be effectively targeted to improve cancer treatments. As an IL-4R alpha-targeting agent, Dupilumab both reduced the pro-tumor gene expression of M2 macrophages and also promoted key anti-tumor characteristics. Investigating the effects of IL-4R alpha signaling in M2-like TAMs in pre-clinical models has further elucidated the mechanism by which they enact their tumor-promoting functions and implicated IL-4R alpha as a candidate therapeutic target in cancer. Inhibiting IL-4R alpha in vivo demonstrates the promise of IL-4R alpha as a target for decreasing the pro-tumor M2-like phenotype. Further studies combining IL-4R alpha inhibition in combination with a cytotoxic therapy and/or immune checkpoint therapy (e.g. anti-PD1/PDL1 or anti-CTLA4) will investigate using Dupilumab as an adjunct therapy to fill an unmet need in prostate cancer treatments.
Author Contributions
Amber de Groot: Conceptualization, Methodology, Investigation, Writing - Original Draft, Writing - Review & Editing, Visualization. Kayla Myers: Conceptualization, Methodology, Investigation, Writing - Original Draft, Writing - Review & Editing, Visualization. Timothy Krueger: Methodology. W. Nathaniel Brennen: Methodology, Funding acquisition. Sarah Amend: Conceptualization, Writing - Review & Editing, Supervision, Funding acquisition. Kenneth Pienta: Conceptualization, Writing - Review & Editing, Supervision, Funding acquisition
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2022.100830.
Appendix. Supplementary materials
References
- 1.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Perspective. 2004;4:71. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 2.Zarif JC, Baena-Del Valle JA, Hicks JL, et al. Mannose receptor-positive macrophage infiltration correlates with prostate cancer onset and metastatic castration-resistant disease. Eur Urol Oncol. 2019;2(4):429–436. doi: 10.1016/j.euo.2018.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiao SL, Chu GC-Y, Chung LWK. Regulation of prostate cancer progression by the tumor microenvironment. Cancer Lett. 2016;380(1):340–348. doi: 10.1016/j.canlet.2015.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zarif JC, Taichman RS, Pienta KJ. TAM macrophages promote growth and metastasis within the cancer ecosystem. Oncoimmunology. 2014;3(7) doi: 10.4161/21624011.2014.941734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu W, Qian Y, Yu F, et al. Alternatively activated macrophages are associated with metastasis and poor prognosis in prostate adenocarcinoma. Oncol Lett. 2015;10(3):1390–1396. doi: 10.3892/ol.2015.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lanciotti M, Masieri L, Raspollini MR, et al. The role of M1 and M2 macrophages in prostate cancer in relation to extracapsular tumor extension and biochemical recurrence after radical prostatectomy. Biomed Res Int. 2014 doi: 10.1155/2014/486798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noy R, Pollard JW. Tumor-associated macrophages: from mechanisms to therapy. Immunity. 2014;41(1):49–61. doi: 10.1016/j.immuni.2014.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan A, Hsiao YJ, Chen HY, et al. Opposite effects of M1 and M2 macrophage subtypes on lung cancer progression. Sci Rep. Sep 24 2015;5:14273. doi: 10.1038/srep14273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang M, He Y, Sun X, et al. A high M1/M2 ratio of tumor-associated macrophages is associated with extended survival in ovarian cancer patients. J Ovarian Res. Feb 08 2014;7:19. doi: 10.1186/1757-2215-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yao W, Ba Q, Li X, et al. A Natural CCR2 Antagonist Relieves Tumor-associated Macrophage-mediated Immunosuppression to Produce a Therapeutic Effect for Liver Cancer. EBioMedicine. 2017;22:58–67. doi: 10.1016/j.ebiom.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi J, Gyamfi J, Jang H, Koo JS. The role of tumor-associated macrophage in breast cancer biology. Histol Histopathol. Jul 06 2017:11916. doi: 10.14670/HH-11-916. [DOI] [PubMed] [Google Scholar]
- 12.Engblom C, Pfirschke C, Pittet MJ. The role of myeloid cells in cancer therapies. Review. 2016;16:447. doi: 10.1038/nrc.2016.54. [DOI] [PubMed] [Google Scholar]
- 13.Arlauckas SP, Garris CS, Kohler RH, et al. In vivo imaging reveals a tumor-associated macrophage–mediated resistance pathway in anti–PD-1 therapy. Sci Transl Med. 2017;9(389) doi: 10.1126/scitranslmed.aal3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27(4):462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bankaitis KV, Fingleton B. Targeting IL4/IL4R for the treatment of epithelial cancer metastasis. Clin Exp Metastasis. 2015;32(8):847–856. doi: 10.1007/s10585-015-9747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sieweke MH, Allen JE. Beyond stem cells: self-renewal of differentiated macrophages. Science. 2013;342(6161) doi: 10.1126/science.1242974. [DOI] [PubMed] [Google Scholar]
- 17.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32(5):593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 18.McCormick SM, Heller NM. Regulation of macrophage, dendritic cell, and microglial phenotype and function by the SOCS proteins. Front Immunol. 2015;6:549. doi: 10.3389/fimmu.2015.00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luzina IG, Keegan AD, Heller NM, Rook GAW, Shea-Donohue T, Atamas SP. Regulation of inflammation by interleukin-4: a review of “alternatives. J Leukocyte Biol. 2012;92(4):753–764. doi: 10.1189/jlb.0412214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Review. 2010;11:889. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 21.Muraille E, Leo O, Moser M. Th1/Th2 Paradigm extended: macrophage polarization as an unappreciated pathogen-driven escape mechanism? Front Immunol. 2014;5:603. doi: 10.3389/fimmu.2014.00603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Z, Zhu L. Update on the role of alternatively activated macrophages in asthma. J Asthma Allergy. 2016;9:101–107. doi: 10.2147/JAA.S104508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasraie S, Werfel T. Role of macrophages in the pathogenesis of atopic dermatitis. Mediat Inflamm. 2013;2013 doi: 10.1155/2013/942375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Escamilla J, Schokrpur S, Liu C, et al. CSF1 receptor targeting in prostate cancer reverses macrophage-mediated resistance to androgen blockade therapy. Cancer Res. 2015;75(6):950–962. doi: 10.1158/0008-5472.CAN-14-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J, Ward JF, Pettaway CA, et al. VISTA is an inhibitory immune checkpoint that is increased after ipilimumab therapy in patients with prostate cancer. Brief Communication. Nat Med. 2017;23(5):551–555. doi: 10.1038/nm.4308. http://www.nature.com/nm/journal/v23/n5/abs/nm.4308.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cotechini T, Medler TR, Coussens LM. Myeloid cells as targets for therapy in solid tumors. Cancer J. 2015;21(4):343–350. doi: 10.1097/PPO.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Panni RZ, Linehan DC, DeNardo DG. Targeting tumor-infiltrating macrophages to combat cancer. Immunotherapy. 2013;5(10) doi: 10.2217/imt.13.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharjee A, Shukla M, Yakubenko VP, Mulya A, Kundu S, Cathcart MK. IL-4 and IL-13 Employ discrete signaling pathways for target gene expression in alternatively activated monocytes/macrophages. Free Radical Biol Med. 2013;54:1–16. doi: 10.1016/j.freeradbiomed.2012.10.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suzuki A, Leland P, Joshi BH, Puri RK. Targeting of IL-4 and IL-13 receptors for cancer therapy. Cytokine. 2015;75(1):79–88. doi: 10.1016/j.cyto.2015.05.026. [DOI] [PubMed] [Google Scholar]
- 30.Shirley M. Dupilumab: first global approval. Drugs. 2017;77(10):1115–1121. doi: 10.1007/s40265-017-0768-3. [DOI] [PubMed] [Google Scholar]
- 31.Venmar KT, Carter KJ, Hwang DG, Dozier EA, Fingleton B. IL4 Receptor ILR4α regulates metastatic colonization by mammary tumors through multiple signaling pathways. Cancer Res. 2014;74(16):4329. doi: 10.1158/0008-5472.CAN-14-0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeNardo DG, Barreto JB, Andreu P, et al. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009;16(2):91–102. doi: 10.1016/j.ccr.2009.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leen AM, Sukumaran S, Watanabe N, et al. Reversal of tumor immune inhibition using a chimeric cytokine receptor. Mol Ther. 2014;22(6):1211–1220. doi: 10.1038/mt.2014.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H-W, Joyce JA. Alternative activation of tumor-associated macrophages by IL-4: priming for protumoral functions. Cell Cycle. 2010;9(24):4824–4835. doi: 10.4161/cc.9.24.14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roth F, De La Fuente AC, Vella JL, Zoso A, Inverardi L, Serafini P. Aptamer-mediated blockade of IL4Rα triggers apoptosis of mdscs and limits tumor progression. Cancer Res. 2012;72(6):1373. doi: 10.1158/0008-5472.CAN-11-2772. [DOI] [PubMed] [Google Scholar]
- 36.Srabovic N, Mujagic Z, Mujanovic-Mustedanagic J, Muminovic Z, Cickusic E. Interleukin 13 expression in the primary breast cancer tumour tissue. Med Glas. 2011;8(1):109–115. [PubMed] [Google Scholar]
- 37.Zarif JC, Hernandez JR, Verdone JE, Campbell SP, Drake CG, Pienta KJ. A phased strategy to differentiate human CD14+monocytes into classically and alternatively activated macrophages and dendritic cells. BioTechniques. 2016;61(1):33–41. doi: 10.2144/000114435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campeau E, Ruhl VE, Rodier F, et al. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4(8):e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart SA, Dykxhoorn DM, Palliser D, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Groot AE, Myers KV, Krueger TEG, et al. Characterization of tumor-associated macrophages in prostate cancer transgenic mouse models. Prostate. 2021;81(10):629–647. doi: 10.1002/pros.24139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peranzoni E, Lemoine J, Vimeux L, et al. Macrophages impede CD8 T cells from reaching tumor cells and limit the efficacy of anti-PD-1 treatment. Proc Natl Acad Sci U S A. 2018;115(17):E4041–E4050. doi: 10.1073/pnas.1720948115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pu J, Xu Z, Nian J, et al. M2 macrophage-derived extracellular vesicles facilitate CD8+T cell exhaustion in hepatocellular carcinoma via the miR-21-5p/YOD1/YAP/beta-catenin pathway. Cell Death Discov. 2021;7(1):182. doi: 10.1038/s41420-021-00556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.