Abstract
The relA gene from Streptomyces antibioticus has been cloned and sequenced. The gene encodes a protein with an Mr of 93,653, which is 91% identical to the corresponding protein from Streptomyces coelicolor. Disruption of S. antibioticus relA produces a strain which grows significantly more slowly on actinomycin production medium than the wild type or a disruptant to which the intact relA gene was restored. Moreover, the disruptant was unable to accumulate ppGpp to the levels observed during the normal course of growth and actinomycin production in the wild type. The strain containing the disrupted relA gene did not produce actinomycin and contained significantly lower levels of the enzyme phenoxazinone synthase than the wild-type strain. Actinomycin synthetase I, a key enzyme in the actinomycin biosynthetic pathway, was undetectable in the relA disruptant. Growth of the disruptant on low-phosphate medium did not restore actinomycin production.
Guanosine tetraphosphate (ppGpp) mediates the stringent response to carbon and energy starvation in bacteria (2). In the case of amino acid starvation, the product of the relA gene, a (p)ppGpp synthetase (20), synthesizes ppGpp. Although considerable controversy exists regarding the mechanism of action of ppGpp, it is clear that one of the ultimate effects of the stringent response is to inhibit transcription from stringently controlled promoters, in particular the promoters responsible for the transcription of rRNA and tRNA genes.
While the details of the stringent response have been elucidated in studies performed with Escherichia coli, recent evidence indicates that similar phenomena occur in Streptomyces. The stringent response to amino acid starvation has been observed in many Streptomyces species (see, e.g., references 18 and 21), and the relA gene was recently cloned from Streptomyces coelicolor by Chakraburrty et al. (4). These workers also disrupted S. coelicolor relA and observed that the normal increase in ppGpp levels observed during the nutritional shiftdown of wild-type S. coelicolor was abolished by the disruption. In these experiments, relA disruption had no effect on the production of actinorhodin on either solid or liquid medium. However, these observations were compromised by the subsequent discovery that the relA disruptant was still capable of ppGpp production. In a subsequent study, Chakraburrty and Bibb (3) constructed a deletion within the coding region of the S. coelicolor relA gene and observed that the resulting strain was conditionally deficient in actinorhodin and undecylprodigiosin production under conditions of nitrogen limitation.
The stringent response has also been studied in Streptomyces antibioticus. Ochi isolated a relC mutant of S. antibioticus IMRU 3720 and demonstrated that the mutant mycelium contained significantly decreased levels of the enzyme phenoxazinone synthase compared with those in the wild-type strain (19). Moreover, the relC mutant did not produce actinomycin and was deficient in the production of a key enzyme in the biosynthetic pathway, actinomycin synthetase I (ACMSI) (16). Because relC is a ribosomal mutation, the possibility existed that the observed effects on the actinomycin pathway were due to impaired protein synthesis rather than to the decreased ppGpp levels observed in the mutant. To circumvent this potential problem and to gain further insight into the role of ppGpp in the regulation of antibiotic production, we have cloned and disrupted the relA gene from S. antibioticus. We demonstrate below that the relA disruptant is unable to produce actinomycin.
MATERIALS AND METHODS
Growth of organisms.
S. antibioticus IMRU 3720 was cultured on NZ-amine and galactose-glutamic acid (GGA) media as described previously (5). In some experiments, pseudomonic acid was added to GGA medium in concentrations up to 200 μg/ml. E. coli ET12567 (6) was grown in L broth containing chloramphenicol (25 μg/ml), kanamycin (50 μg/ml), and apramycin as needed.
Cloning and sequencing of S. antibioticus relA.
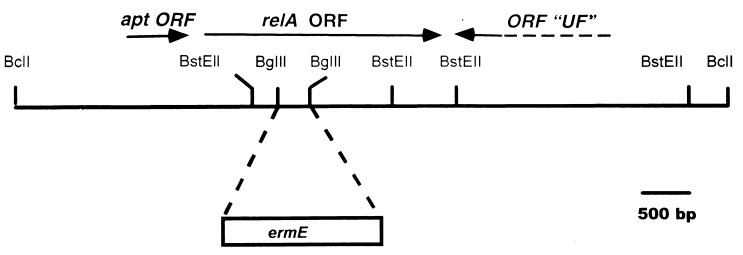
The S. antibioticus relA gene was identified by using a PCR fragment of ca. 400 bp, generously provided by Mervyn Bibb and Rekha Chakraburrty, representing a portion of the S. coelicolor relA gene. This PCR fragment was used to identify a ca. 4.5-kb BglII/BclI fragment in a Southern blot by using S. antibioticus chromosomal DNA as the target. BglII/BclI fragments of 3 to 6 kb were cloned into the BamHI site of pBluescript SK(+) (Stratagene), and the relevant transformant was identified by colony hybridization using the PCR fragment mentioned above as the probe. A plasmid containing an insert of ca. 4.5 kb was isolated from this clone and designated pJSE2000. Sequencing of the BglII/BclI fragment indicated that it contained only a portion of the relA open reading frame (ORF). Consequently, a ca. 7-kb BclI fragment, which overlapped the insert of pJSE2000, was identified by Southern blotting, and fragments of the appropriate size were cloned in pBluescript SK(+) as described above. The plasmid containing the 7-kb fragment was designated pJSE2001 (see Fig. 1). Nested deletions of the insert of pJSE2001 were constructed, and the DNA sequence of the relA ORF was obtained by the chain termination method using the TaqTrack system (United States Biochemicals).
FIG. 1.
Partial restriction map of the ca. 7-kb insert of pJSE2001. The approximate location of the relA ORF and the site of insertion of ermE are indicated.
Disruption of S. antibioticus relA.
In order to disrupt the relA ORF, the ca. 300-bp BglII fragment within the ORF was replaced with the ermE gene (22), excised as a BglII fragment from pIJ4642 (generously provided by Mervyn Bibb). The resulting plasmid was designated pJSE2012. The insert of pJSE2012 was excised as a ClaI/SpeI fragment, the ends of the fragment were filled in with Klenow polymerase, and the resulting blunt-ended fragment was cloned into the EcoRV site of pKC1132 (1) to produce pJSE2093. This plasmid was introduced by transformation into E. coli ET12567. pJSE2093 was transferred from ET12567 to S. antibioticus IMRU 3720 by conjugation as described previously (15). Exconjugants were selected by overlaying plates with 1 ml of distilled water containing 0.5 mg of nalidixic acid and 1 mg of apramycin. After about 7 days, melanin-producing colonies were observed on the conjugation plates. Several of these were streaked onto GGA agar without antibiotics. Once the colonies had sporulated, individual colonies were streaked again onto GGA agar without selection. When spores were again observed on the GGA agar plates, the process was repeated once more. Following the third round of sporulation, 25 colonies were transferred from the GGA agar plate to a similar plate containing either lincomycin at a concentration of 200 μg/ml or apramycin at a concentration of 50 μg/ml. Two colonies were observed to be lincomycin resistant and apramycin sensitive. One of these was chosen for further study and was designated 3720R9. Southern blotting confirmed the presence of the ermE gene within the relA ORF in the chromosome of 3720R9 (see Fig. 3).
FIG. 3.
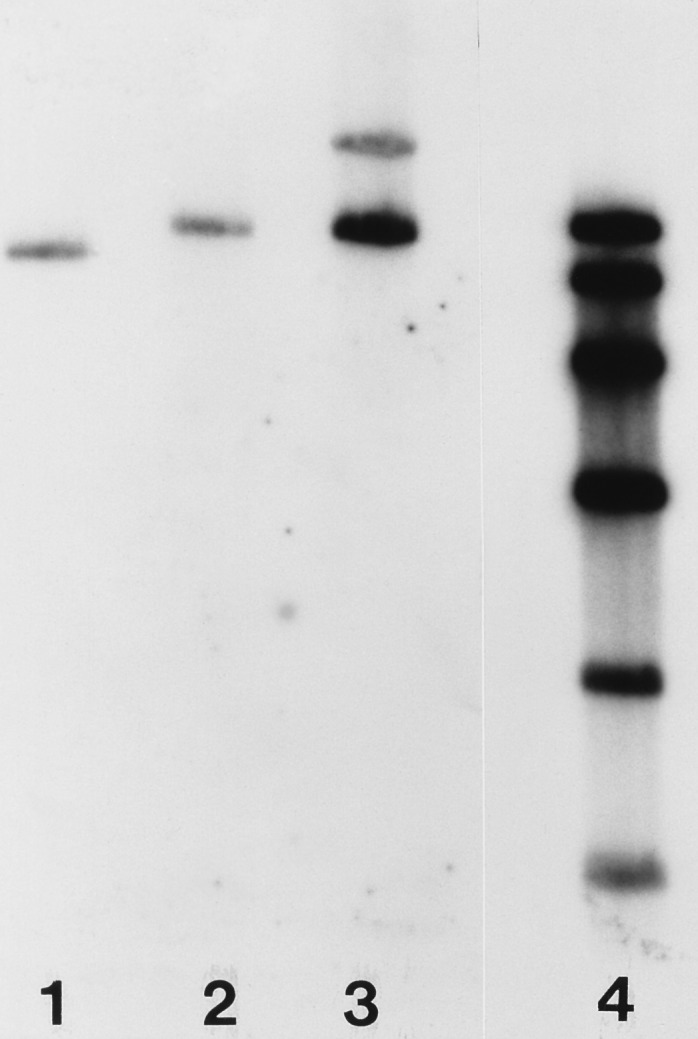
Southern blot of chromosomal DNA from strains 3720 (lane 1), 3720R9 (lane 2), and 3720R9.2 (lane 3). DNAs were digested with BclI. The radioactive probe used was the insert of pJSE2001 excised as a HindIII/XbaI fragment and labeled by random priming. The figure shows that 3720R9 contains a fragment that is larger than the insert of pJSE2001 by about 1.4 kb (the size of the ermE gene minus the size of the BglII fragment of pJSE2001) and that strain 3720R9.2 contains both this fragment and a second one whose size corresponds to that predicted for pJSE2089. That the band corresponding in size to pJSE2089 is less intense than the band below it is probably due to incomplete transfer of the former band to the Southern filter. Lane 4 contains a series of molecular weight markers of 9.3, 6.8, 4.4, 2.7, 1.5, and 0.85 kb.
The 7-kb insert of pJSE2001 was released as a HindIII/XbaI fragment whose ends were filled in with Klenow polymerase. The blunt-ended fragment was then cloned into the EcoRV site of pSET152 (1), and the resulting plasmid (pJSE2089) was transferred to S. antibioticus 3720R9 by conjugation from E. coli as described above. The resulting strain was designated 3720R9.2. Southern blotting confirmed the integration of pJSE2089 into the chromosome of 3720R9.2 (see Fig. 3).
Assays for growth, RNA synthesis, actinomycin production, and actinomycin enzymes.
Dry weights of mycelium were determined as described previously (14). The assays for phenoxazinone synthase and ACMSI were also performed as described previously (5, 10). Actinomycin synthesis was measured spectrophotometrically at 443 nm and also via the incorporation of [14C]valine into neutral ethyl acetate-extractable products. The data reported in Fig. 6 represent results obtained by extraction of the culture medium following removal of the mycelium by centrifugation. In other experiments, to eliminate the possibility that actinomycin was produced by strain 3720R9 but not excreted into the medium, culture samples were extracted without prior separation of the mycelium from the growth medium. Ethyl acetate extracts were examined by thin-layer chromatography as described previously (15). To measure RNA synthesis, cultures were grown for 13 h in 50 ml of GGA medium; then duplicate 1-ml portions were removed and incubated for 60 min with 0.2 μCi of [14C]uridine at a total uridine concentration of 50 μM. Incorporation was stopped by the addition of trichloroacetic acid to a final concentration of 10%, and mycelial pellets were collected by suction filtration and were examined by liquid scintillation counting.
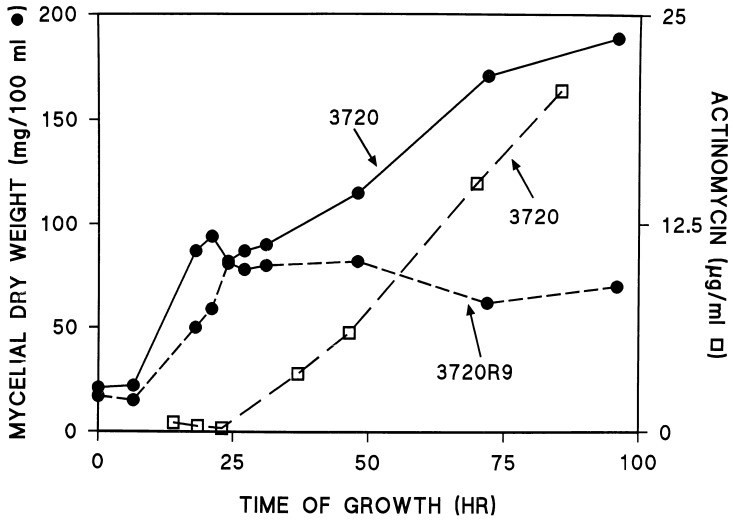
FIG. 6.
Growth and actinomycin production by strains 3720 and 3720R9. Details are provided in Materials and Methods.
Assay for ppGpp accumulation.
Relevant strains were grown in 1-liter GGA cultures for the times indicated in the Fig. 5. At those times, 110-ml portions were removed from each culture. Ten milliliters was used for the determination of the dry weight of the mycelium. The remaining 100 ml was collected rapidly by suction filtration, and the mycelium was scraped into tubes containing 3 ml of 2 M HCOOH. The HCOOH suspensions were stored frozen for at least 12 h. The mycelial residue was then removed by filtration through cotton in a 10-cc syringe, and the residue was washed in the syringe with 2 additional ml of 2 M HCOOH. The resulting extract was then passed through a 0.45-μm-pore-size filter and lyophilized. The dried material was dissolved in 500 μl of distilled water, and insoluble material was removed by centrifugation for 10 min at 13,000 rpm and 4°C in an Eppendorf centrifuge.
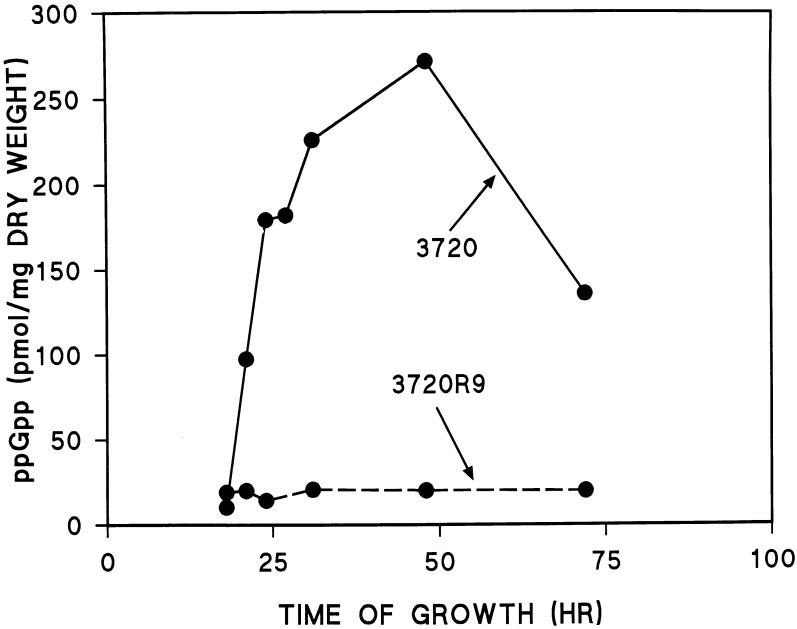
FIG. 5.
ppGpp levels in strains 3720 and 3720R9. Details are provided in Materials and Methods.
High-pressure liquid chromatography (HPLC) was used to determine ppGpp levels. Nucleotides were separated by using a Millenium HPLC system and a Spherisorb S5SAX column (both from Waters). A concave gradient was used with the buffer system described previously (18), and the flow rate was 1.5 ml/min. Authentic ppGpp was generously provided by Kozo Ochi. Results are expressed as picomoles of ppGpp per milligram (dry weight) of mycelium.
Nucleotide sequence accession number.
The DNA sequence of the S. antibioticus relA ORF has been deposited with GenBank under accession no. AF072829.
RESULTS
Cloning and sequencing of S. antibioticus relA.
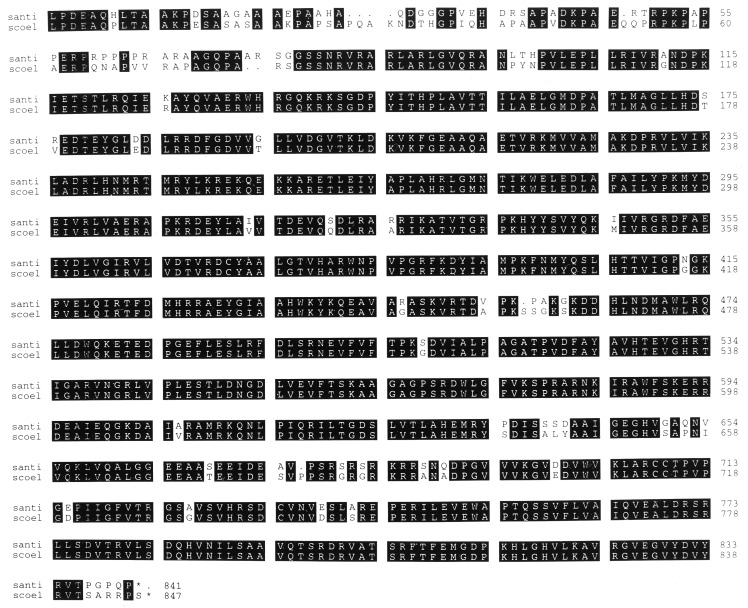
As described in Materials and Methods, the S. antibioticus relA gene was cloned as a ca. 7-kb BclI fragment by using a PCR product representing a portion of S. coelicolor relA as a probe in colony hybridization experiments. A partial restriction map of this fragment, indicating the position of the relA ORF, is shown in Fig. 1. The nucleotide sequence of the gene was obtained by using nested deletions constructed from pJSE2001, and the amino acid sequence of the protein product encoded by the ORF is shown in Fig. 2, in comparison with the sequence of S. coelicolor RelA. It is apparent that the two proteins are about 91% identical in their amino acid sequences. S. antibioticus relA has an Mr of 93,653 and an isoelectric point of 10.12. The corresponding values for S. coelicolor RelA are 94,162 and 10.14. Although the proteins are more than 90% identical in their overall amino acid sequences, they are considerably less similar at their amino-terminal ends. Over the first 115 amino acids, the degree of identity is only 68%. The functional significance of this difference, if there is any, is unknown.
FIG. 2.
Pileup comparison of the amino acid sequences of the RelA proteins from Streptomyces antibioticus (santi) and Streptomyces coelicolor (scoel). Identical residues are indicated by solid boxes.
Disruption of S. antibioticus relA.
The relA gene was disrupted by replacement of the ca. 300-bp BglII site within the ORF (Fig. 1) with the ermE gene from Saccharopolyspora erythrea (22). The insert from the resulting plasmid (pJSE2012) was transferred to the conjugative plasmid pKC1132 (1), and the plasmid obtained (pJSE2093) was introduced into S. antibioticus IMRU 3720 by conjugation from E. coli. After several rounds of sporulation without selection, two exconjugants that apparently contained the ermE gene (and were therefore resistant to lincomycin) but lacked the gene for apramycin resistance carried by pKC1132 were obtained. Southern blotting (Fig. 3, lane 2) demonstrated that one of these exconjugants, designated 3720R9, did indeed contain the disrupted relA gene, integrated into the S. antibioticus chromosome by homologous recombination from pJSE2093. An intact relA gene was restored to 3720R9 from pJSE2089 as described in Materials and Methods, and Southern blotting again confirmed the presence of the relevant construct in S. antibioticus following conjugation (Fig. 3, lane 3).
Disruption of relA leads to a relaxed response to amino acid starvation in S. antibioticus.
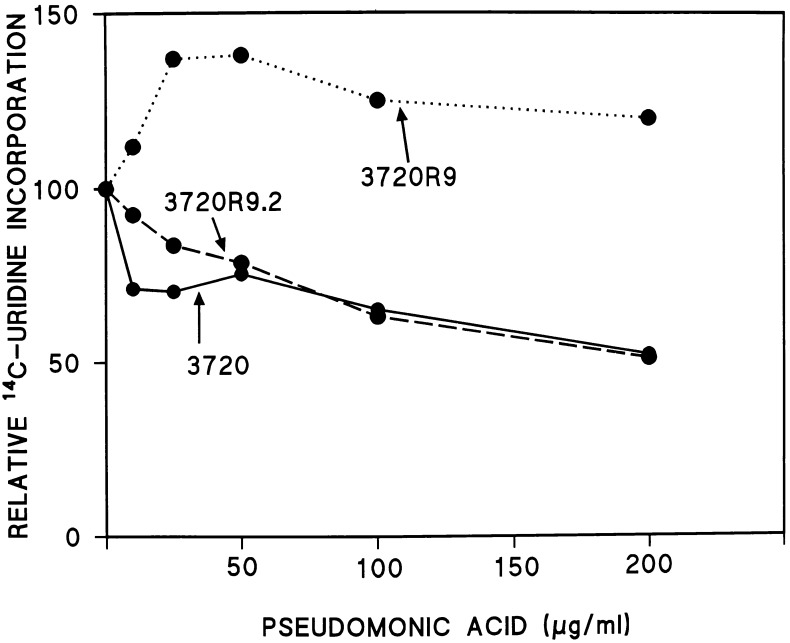
To verify that the cloned gene described herein was indeed S. antibioticus relA, the wild-type strain, 3720R9, and 3720R9.2 were subjected to amino acid starvation. The agent used to induce starvation in these experiments was pseudomonic acid, an inhibitor of isoleucyl-tRNA synthetase (9). As shown in Fig. 4, pseudomonic acid inhibited [14C]uridine incorporation by strains 3720 and 3720R9.2 (the wild-type strain and the disruptant with the relA gene restored, respectively) but did not inhibit incorporation by 3720R9. Thus, strain 3720R9 shows the classic relaxed response to amino acid starvation. We are unable to explain the increase in uridine incorporation observed in 3720R9 at the lowest concentrations of pseudomonic acid tested in the experiments of Fig. 4. It should be noted, however, that an increase in uridine incorporation in the early stages of the stringent response has been observed in other organisms (8).
FIG. 4.
Pseudomonic acid inhibition of [14C]uridine incorporation by strains 3720, 3720R9, and 3720R9.2. Results are expressed relative to the levels of incorporation in the absence of pseudomonic acid (12,000 cpm for 3720, 10,400 cpm for 3720R9, and 7,800 cpm for 3720R9.2), set at 100.
S. antibioticus 3720R9 accumulates only low levels of ppGpp.
Because the relaxed response to amino acid starvation in bacteria is the result of the inability of the mutant strains to produce appropriate levels of ppGpp, it seemed likely that strain 3720R9 would be deficient in ppGpp accumulation. The time course of changes in ppGpp levels in the wild-type strain and 3720R9 is shown in Fig. 5. As can be seen, the growth of 3720 in GGA medium is accompanied by an increase in ppGpp levels beginning at around 20 h postinoculation and continuing until about 50 h postinoculation. At this point, ppGpp levels had risen to approximately 280 pmol/mg (dry weight) of mycelium. Thereafter, ppGpp levels decreased to less than 150 pmol/mg and were maintained between 70 and 100 pmol/mg until at least 96 h postinoculation (data not shown). A similar pattern of ppGpp accumulation was observed for 3720R9.2 (data not shown). In contrast, 3720R9, the relaxed strain containing the disrupted relA gene, was unable to accumulate ppGpp to the levels observed for 3720 and 3720R9.2. ppGpp levels in 3720R9 were maintained at around 20 pmol/mg over the entire course of the experiment.
Effects of the disruption of relA on growth, actinomycin production, and actinomycin enzymes.
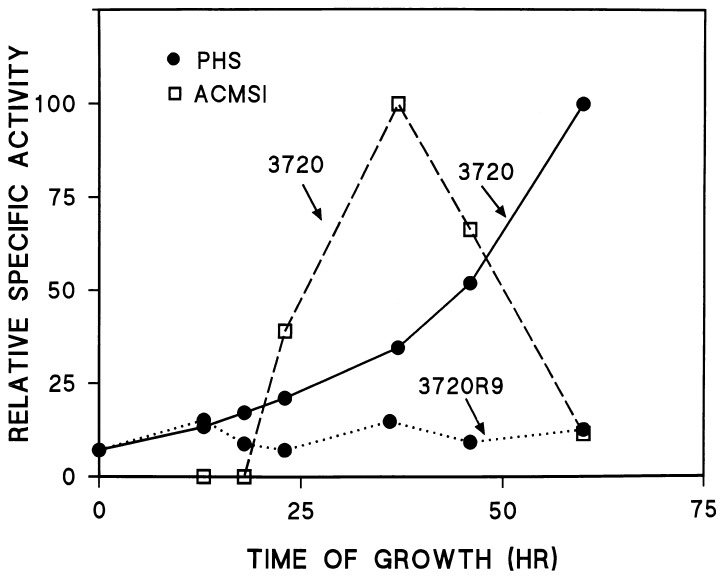
The time course of the change in the dry weight of the mycelium is shown for 3720 and 3720R9 in Fig. 6. The dry weight of the mycelium increased for 3720 throughout the course of the experiment shown and began to plateau around 95 h postinoculation. A similar pattern of growth was observed for 3720R9.2, with the exception that this strain grew somewhat more slowly than the wild type. At all time points after about 12 h postinoculation, the dry weights of the mycelium observed for 3720R9.2 were about 10% lower than those observed for 3720. Because these two strains were almost identical in physiological and biochemical properties, only data for 3720 are shown in Fig. 5, 6, and 8. In contrast to the results obtained for 3720 and 3720R9.2, strain 3720R9 grew significantly more slowly throughout the course of the experiment shown in Fig. 6. Moreover, this strain reached stationary phase very early in the growth process, after about 30 h of growth. Thereafter, there was a slight decrease in the dry weight of the mycelium in the 3720R9 culture.
FIG. 8.
Activities of phenoxazinone synthase (PHS) and ACMSI in strains 3720 and 3720R9. Phenoxazinone synthase activities are expressed relative to the activity observed at 60 h postinoculation (117 nmol of cinnabarinic acid formed per min per mg of protein) in extracts from 3720, and ACMSI activities are expressed relative to the activity observed at 37 h postinoculation (14.2 nmol of ATP formed per min per mg of protein) in 3720 extracts.
Actinomycin synthesis was measured spectrophotometrically after neutral ethyl acetate extraction of the antibiotic from portions of each culture. The increase in actinomycin concentration is depicted in Fig. 6 for 3720. A similar pattern of increase was observed for 3720R9.2 (data not shown). In dramatic contrast to these observations, no actinomycin was extractable from cultures of 3720R9 at any time during the experiments. These results were confirmed by the analysis of the incorporation of [14C]valine into ethyl acetate-extractable products and by thin layer-chromatography of those products. As shown in Fig. 7, 3720 and 3720R9.2 both incorporated valine into a product with the chromatographic mobility of authentic actinomycin. However, no such product was observed in the extracts obtained from the 3720R9 culture. Instead, valine was incorporated into products that moved with the solvent front in 3720R9.
FIG. 7.

Autoradiogram of a thin-layer chromatogram of neutral ethyl acetate-extractable products following incubation of strains 3720 (lane 1), 3720R9 (lane 2), and 3720R9.2 (lane 3) with [14C]valine. o, origin.
We also examined the effects of the disruption of relA on the enzymes phenoxazinone synthase and ACMSI. Figure 8 shows that for S. antibioticus 3720 (as for 3720R9.2 [data not shown]), the patterns of increase in the specific activities of these two enzymes were essentially identical to those previously reported (7, 16). Again, in dramatic contrast to these results, 3720R9 was shown to contain only low levels of phenoxazinone synthase at all time points at which the culture was sampled, and ACMSI was completely undetectable in 3720R9 at all times during the course of the experiment.
DISCUSSION
The results presented above identify the relA gene from S. antibioticus. That the cloned gene is a relA homologue is demonstrated not only by the sequence similarity of the proteins encoded by the S. antibioticus and S. coelicolor genes but also by the observation that S. antibioticus 3720R9, in which relA is disrupted, showed the relaxed response to amino acid starvation (Fig. 4). Strain 3720R9 was unable to accumulate ppGpp to the levels observed in the wild-type strain and in the disrupted strain to which an intact relA gene had been restored. Moreover, although strain 3720R9 sporulated normally and grew normally on NZ-amine medium, this disruptant grew significantly more slowly on GGA medium than did the wild type. Most significantly, strain 3720R9 was incapable of synthesizing actinomycin. No actinomycin could be detected in the medium or mycelium of 3720R9 spectrophotometrically, nor could that strain incorporate a radioactive precursor into actinomycin. Thus, the disruption of relA abolishes actinomycin synthesis in the disruptant strain. These results suggest that, as in S. coelicolor (3, 17), ppGpp plays a role in antibiotic production in S. antibioticus.
It might be argued that because the cloned fragment used in these experiments included sequences downstream of relA (Fig. 1), the effects of the disruption of relA might be due to polar effects on downstream genes rather than to a direct effect of relA disruption. To eliminate this possibility, we have sequenced the region downstream of relA and have determined that the sequence organization in that region in S. antibioticus is similar to that observed in S. coelicolor (4, 17). In the latter organism, the relA gene is followed by an intergenic region which separates relA from an ORF of unknown function (ORF UF in Fig. 1). That ORF is transcribed in the opposite direction to relA (4, 17). The intergenic region in S. antibioticus is 50% identical in sequence to the corresponding region from S. coelicolor. The end of the ORF of unknown function is situated 105 bases downstream of the relA stop codon in S. antibioticus. For the 3′-terminal 165 bases of this ORF, the sequence is 82% identical to the corresponding region from S. coelicolor (data not shown). These observations eliminate the possibility of polarity as the explanation for the effects of relA disruption. Thus, the likely explanation for the phenomena reported here is that the disruption of relA is directly responsible for the observed differences between the wild-type and disruptant strains of S. antibioticus.
While Chakraburrty and Bibb did not observe the marked effect on the growth of the S. coelicolor relA null mutant that we observed for the S. antibioticus relA disruptant, they did report that the relA null mutation affected the onset and extent of morphological differentiation in S. coelicolor (3). Moreover, these workers observed that the relA null mutation was conditional in S. coelicolor. In particular, they reported that a reduction in the phosphate concentration in the growth medium they used resulted in the restoration of both actinorhodin and undecylprodigiosin production (3). We also examined the effects of phosphate limitation on actinomycin production in strains 3720, 3720R9, and 3720R9.2. The phosphate concentration in GGA medium is 5.7 mM. We tested the three S. antibioticus strains at 1 and 0.1 mM phosphate. Although some differences in the growth rate were observed at the different phosphate concentrations (e.g., all grew more slowly at 0.1 mM phosphate than at 1 or 5.7 mM), 3720 and 3720R9.2 produced actinomycin at all phosphate concentrations tested, whereas 3720R9 did not produce actinomycin under any conditions tested (data not shown).
At least three unresolved issues remain with regard to the results presented above. First, as demonstrated in Fig. 6, the relA disruptant does not grow as vigorously as the wild-type S. antibioticus strain. Thus, it is possible that the effects of ppGpp on actinomycin production in the wild type are indirect, resulting from the impairment of vigor by ppGpp rather than from a direct effect on the regulation of actinomycin synthesis. Second, it has been reported that S. antibioticus contains at least one pathway to the production of (p)ppGpp that appears to be an alternative to the relA pathway (11, 12). That pathway involves an enzyme, designated guanosine pentaphosphate synthetase (GPSI), that apparently functions both as a pppGpp synthetase and as a polynucleotide phosphorylase (13). Since the ppGpp levels observed in 3720R9 were not zero, it seemed possible that GSPI was involved in the maintenance of the low levels observed in the studies reported here. Attempts to disrupt the gpsI gene have thus far been unsuccessful. It seems possible, therefore, that gpsI is an essential gene in S. antibioticus. The third question is the obvious and intriguing one: if ppGpp does directly affect the regulation of antibiotic production, what is the mechanism involved? It is tempting to speculate that ppGpp interacts with components of the transcriptional apparatus to regulate the transcription of promoters specifically required for antibiotic production. However, there is no direct evidence to support this hypothesis at this time. Nevertheless, the identification of a second antibiotic producer in which ppGpp is required for that production should facilitate studies designed to answer these important questions.
ACKNOWLEDGMENTS
This work was supported by grant 1 R01 GM52589 from the National Institutes of Health to G.H.J.
REFERENCES
- 1.Bierman R, Logan R, O’Brien K, Seno E T, Rao R N, Baltz R, Shoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomycesspp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 2.Cashel M, Gentry D R, Hernandez V J, Vinella D. The stringent response. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 3.Chakraburrty R, Bibb M J. The ppGpp synthetase gene (relA) of Streptomyces coelicolorA3(2) plays a conditional role in antibiotic production and morphological differentiation. J Bacteriol. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chakraburrty R, White J, Takano E, Bibb M J. Cloning, characterization and disruption of a (p)ppGpp synthetase gene (relA) of Streptomyces coelicolorA3(2) Mol Microbiol. 1996;19:357–368. doi: 10.1046/j.1365-2958.1996.390919.x. [DOI] [PubMed] [Google Scholar]
- 5.Choy H A, Jones G H. Phenoxazinone synthase from Streptomyces antibioticus: purification of the large and small enzyme forms. Arch Biochem Biophys. 1981;211:55–65. doi: 10.1016/0003-9861(81)90429-x. [DOI] [PubMed] [Google Scholar]
- 6.Flett F, Mersenias V, Smith C P. High efficiency intergenic conjugal transfer of plasmid DNA from Escherichia colito methyl DNA-restricting streptomycetes. FEMS Microbiol Lett. 1998;155:223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 7.Gallo M, Katz E. Regulation of secondary metabolite biosynthesis: catabolite repression of phenoxazinone synthase and actinomycin formation by glucose. J Bacteriol. 1972;109:659–667. doi: 10.1128/jb.109.2.659-667.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harris B Z, Kaiser D, Singer M. The guanosine nucleotide (p)ppGpp initiates development and A-factor production in Myxococcus xanthus. Genes Dev. 1998;12:1022–1035. doi: 10.1101/gad.12.7.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes J, Mellows G. Inhibition of isoleucyl-transfer ribonucleic acid synthetase in Escherichia coliby pseudomonic acid. Biochem J. 1978;176:305–318. doi: 10.1042/bj1760305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones G H. Combined purification of actinomycin synthetase I and 3-hydroxyanthranilic acid 4-methyltransferase from Streptomyces antibioticus. J Biol Chem. 1993;268:6831–6834. [PubMed] [Google Scholar]
- 11.Jones G H. Purification and properties of ATP-GTP 3′-pyrophosphotransferase (guanosine pentaphosphate synthetase) from Streptomyces antibioticus. J Bacteriol. 1994;176:1474–1481. doi: 10.1128/jb.176.5.1475-1481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones G H. Activation of ATP:GTP 3′-pyrophosphotransferase (guanosine pentaphosphate synthetase) in Streptomyces antibioticus. J Bacteriol. 1994;176:1482–1487. doi: 10.1128/jb.176.5.1482-1487.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones G H, Bibb M J. Guanosine pentaphosphate synthetase from Streptomyces antibioticusis also a polynucleotide phosphorylase. J Bacteriol. 1996;178:4281–4288. doi: 10.1128/jb.178.14.4281-4288.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones G H, Weissbach H. RNA metabolism in Streptomyces antibioticus: effect of 5-fluorouracil on the appearance of phenoxazinone synthetase. Arch Biochem Biophys. 1970;136:558–573. doi: 10.1016/0003-9861(70)90473-x. [DOI] [PubMed] [Google Scholar]
- 15.Jones G H, Paget M S B, Chamberlin L, Buttner M J. Sigma-E is required for the production of the antibiotic actinomycin in Streptomyces antibioticus. Mol Microbiol. 1997;23:169–178. doi: 10.1046/j.1365-2958.1997.2001566.x. [DOI] [PubMed] [Google Scholar]
- 16.Kelly K S, Ochi K, Jones G H. Pleiotropic effects of a relC mutation in Streptomyces antibioticus. J Bacteriol. 1991;173:2297–2300. doi: 10.1128/jb.173.7.2297-2300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martinez-Costa O H, Arias P, Romero N M, Parro V, Mellado R P, Malpartida F. A relA/spoT homologous gene from Streptomyces coelicolorA3(2) controls antibiotic biosynthetic genes. J Biol Chem. 1996;271:10627–10634. doi: 10.1074/jbc.271.18.10627. [DOI] [PubMed] [Google Scholar]
- 18.Ochi K. Occurrence of the stringent response in Streptomycesspp. and its significance for the inhibition of morphological and physiological differentiation. J Gen Microbiol. 1986;132:2621–2631. doi: 10.1099/00221287-132-9-2621. [DOI] [PubMed] [Google Scholar]
- 19.Ochi K. A rel mutation abolishes the enzyme induction needed for actinomycin synthesis by Streptomyces antibioticus. Agric Biol Chem. 1987;51:829–835. [Google Scholar]
- 20.Pedersen F S, Kjeldgaard N O. Analysis of the relA gene product of Escherichia coli. Eur J Biochem. 1977;76:91–97. doi: 10.1111/j.1432-1033.1977.tb11573.x. [DOI] [PubMed] [Google Scholar]
- 21.Strauch E, Takano E, Bayliss H A, Bibb M J. The stringent response in Streptomyces coelicolorA3(2) Mol Microbiol. 1991;5:289–298. doi: 10.1111/j.1365-2958.1991.tb02109.x. [DOI] [PubMed] [Google Scholar]
- 22.Thompson C J, Kieser T, Ward J M, Hopwood D A. Physical analysis of antibiotic resistance genes from Streptomycesand their use in vector construction. Gene. 1982;20:51–62. doi: 10.1016/0378-1119(82)90086-5. [DOI] [PubMed] [Google Scholar]