Abstract
It was suggested that the most effective way to improve rice grain yield is to increase the grain number per panicle (GN) through the breeding practice in recent decades. GN is a representative quantitative trait affected by multiple genetic and environmental factors. Understanding the mechanisms controlling GN has become an important research field in rice biotechnology and breeding. The regulation of rice GN is coordinately controlled by panicle architecture and branch differentiation, and many GN-associated genes showed pleiotropic effect in regulating tillering, grain size, flowering time, and other domestication-related traits. It is also revealed that GN determination is closely related to vascular development and the metabolism of some phytohormones. In this review, we summarize the recent findings in rice GN determination and discuss the genetic and molecular mechanisms of GN regulators.
Keywords: grain number, panicle architecture, branch differentiation, vascular development, rice
Introduction
Rice (Oryza sativa L.), one of the most widely consumed food crops, feeds over a half of world population, and provides more than 21% of the dietary calories worldwide (Fitzgerald et al., 2009; Muthayya et al., 2014; Ito, 2019). The total milled rice consumption is 490 million tons in 2018, and is predicted to be 590 million tons in the year 2040 (Ito, 2019). Therefore, increasing the rice grain yield is an essential task for the fulfillment of global food security.
Rice grain yield is a complex quantitative trait determined by three major yield components, panicle number (PN), grain number per panicle (GN), and grain weight (GW; Xing and Zhang, 2010; Zuo and Li, 2014). Among them, GN is suggested to be the critical trait in increasing grain yield in the breeding practice (Huo et al., 2017; Gouda et al., 2020b; Wu et al., 2021). According to the ideal plant architecture model, low tillering and large panicle (200–250 grains per panicle) are the future targets for the breeding of rice (Khush, 2005). In a survey of yield traits covering 200 japonica rice cultivars bred in central China, a significant increase can be observed for GN during the past 30 years, and many GN-associated genes experienced artificial selection during the breeding process (Xiao et al., 2021), suggestive potential pivot of GN in genetic improvement of rice yield.
GN is mainly determined by the panicle architecture and branch differentiation, which are closely associated with the phytohormone pathways and vascular development (Terao et al., 2010; Duan et al., 2019; Deveshwar et al., 2020). It has been shown that several important genes associated with these processes have significant potential in improving GN and rice grain yield. Gn1a (Grain number 1a)/CKX2 (Cytokinin oxidase 2) is the first isolated GN-associated gene, which was identified through map-based cloning (Ashikari et al., 2005). Favorable allele of Gn1a increased rice grain yield by up to 11.9% when introgressed into an elite japonica rice cultivar Kongyu 131 (Feng et al., 2017), suggestive great potential of GN-associated genes in the breeding practice of rice yield improvement. Another GN-associated QTL, GN4-1, caused a 14.3% increase of grain yield when introgressed into an elite indica cultivar (Zhou et al., 2018). Similarly, introduction of a GN-associated gene NOG1 (Number of Grains 1) into a deficient rice cultivar increased the grain yield by 25.8% (Huo et al., 2017). Moreover, a major quantitative trait loci (QTL) of GN, qPE9-1 (QTL panicle erect 9–1)/DEP1 (Dense and Erect Panicle 1), which control the dense and erect panicle architecture, has been applied in breeding since the 1980s, and has occupied a dominant position among japonica rice cultivars in northern China, long before the gene was isolated (Yan et al., 2007). It can be inferred that the beneficial alleles of GN-associated genes or QTLs confer high-yield potential in rice breeding. Identification and mechanic studies of genes associated with GN would provide valuable gene resources for rice yield improvement. In this review, we summarized current progress on the molecular and genetic basis under the control of GN, and raise future perspectives on the approaches for improving GN in rice breeding.
Genetic characteristics and influencing factors of GN in rice
The rice panicle consists of main rachis, rachis branches (including primary and secondary branches) and spikelets. Spikelets are the basic units of the inflorescence, which are attached to the branches through pedicels, while branches were arise from the nodes of main rachis (Wang and Li, 2005; Xing and Zhang, 2010). As a canonical quantitative trait, GN is controlled by multiple genes, and can be affected by various environmental factors (Yin et al., 2021; Li et al., 2022). The broad-sense heritability of GN is relatively high, which ranged from about 70 to 90% in different studies (Tuhina-Khatun et al., 2015; Roy and Shil, 2020), suggesting that genetic factors are the major determinant of GN. Furthermore, GN is positively correlated with panicle length, number of primary and secondary branches (Rebolledo et al., 2016; Li et al., 2021b).
The spikelet fertility is influenced by environmental conditions, such as temperature, nutrition and water supply. Hence, the environment conditions and cultivation methods also have significant effect on GN. High temperatures occurring at flowering and young microspore stages cause decrease of GN in rice due to spikelet degeneration and decreased pollen viability (Hu et al., 2021; Park et al., 2021); while chilling stress also results in reduced GN through impairing pollen germination and reducing spikelet fertility (Zeng et al., 2017; Hussain et al., 2018; Ali et al., 2021). Water stress during the meiosis stage leads to a severe reduction (40–45%) in GN due to pre-flowering spikelet abortion (Kato et al., 2008; Yang et al., 2019). Cultivation methods also affect GN through regulating the light and nutrition availability. Evidence revealed that high density planting causes decreased GN due to limited light and nutrition supply, while application of nitrogen fertilizer has a positive effect on GN (Jiang et al., 2021; Ju et al., 2021; Wang et al., 2022b). Both the genetic background and the cultivation and environmental influences are essential for the determination of GN.
Regulation of GN by panicle architecture
The panicle architecture, including panicle length, shape, and the number and arrangement of primary and secondary branches, is an important factor for determining GN. G-protein signal pathway is widely involved in controlling plant growth and morphogenesis (Perfus-Barbeoch et al., 2004; Ghusinga et al., 2022). qPE9-1 (QTL panicle erect 9–1)/DEP1 (Dense and Erect Panicle 1) is characterized as a major QTL for rice grain yield through regulating panicle architecture. qPE9-1/DEP1 encodes the γ subunit of a G-protein, and a deletion mutation of the cysteine-rich domain enhanced the transmission of G-protein signaling, which leads to an erect and compact panicle shape, shortened panicle length, enhanced cell division and increased number of branches (Huang et al., 2009b; Zhou et al., 2009; Xu et al., 2016). The favorable allele of qPE9-1/DEP1 has been widely applied in the practice of rice breeding for high yield in China (Sun et al., 2014; Liu et al., 2018).
In addition to the G-protein signal pathway, many other genes were associated with the panicle architecture regulation. The SP1 (Short Panicle 1) gene encodes a peptide transporter family protein, which is predominantly expressed in the branch vascular bundle. Enhanced expression of SP1 in young panicles can promote the elongation of the cob, increase the panicle size, and increase GN (Li et al., 2009). The sped1-D gene, encoding a pentatricopeptide repeat protein, affects panicle structure through blocking the action of GID1L2 (GA-INSENSITIVE DWARF1-L2), RFL (RICE FLORICAULA) and WOX3 (WUSCHEL-related homeobox 3), and caused shortened pedicels and decreased GN (Jiang et al., 2014). Additionally, mutation of DEP3 (Qiao et al., 2011) and EP3 (Erect Panicle 3; Piao et al., 2009) were also associated with erect panicle shape, condensed branches, and elevated GN, for which the regulatory mechanisms are currently not clear.
Transcription factors were also major regulators of panicle development through targeting different genes and affecting their expression. RGN1 (REGULATOR OF GRAIN NUMBER1) encodes an R2R3 MYB family protein, which regulates panicle architecture through activating the expression of LOG (LONELY GUY) and promote cytokinin biosynthesis (Li et al., 2022). Mutation of RGN1 resulted in loss of lateral grains on secondary branches (Li et al., 2022). IPA1 (Ideal Plant Architecture 1)/WFP (WEALTHY FARMER’S PANICLE) encodes a transcription factor OsSPL14 (SQUAMOSA promoter binding protein-like 14), which directly targets DEP1 to promote its expression and thus increase GN (Jiao et al., 2010; Miura et al., 2010). A point mutation in IPA1 perturbs the targeting of OsmiR156, resulting in reduced tiller number, increased panicle size and GN, and improved yield potential (Jiao et al., 2010). In addition, the transcription factor OsSHI1 (SHORT INTERNODES1) was suggested to physically interact with IPA1, and to represses its transcriptional activation activity (Duan et al., 2019). Another SPL family transcription factor OsSPL18 also regulates panicle architecture and GN through activating the expression of DEP1, while the expression of OsSPL18 is further regulated by microRNAs miR156k and miRNA529 (Yuan et al., 2019; Yan et al., 2021).
Regulation of GN by branch differentiation of panicle
The reproductive growth of rice begins with the transition of shoot apical meristem (SAM) into inflorescence meristem (IM). Then, the branch meristem (BM) and spikelet meristem (SM) were sequentially generated from IM (Ikeda et al., 2004; Tanaka et al., 2013; Sreenivasulu et al., 2021). Determination of the meristem identity, regulation of meristem activity and phase transition are crucial factors affecting branch differentiation and GN.
Increasing the activity of IM usually results in elevated branch number and GN. The Gn1a (Grain number 1a)/CKX2 (Cytokinin oxidase 2) gene is the first major QTL implicated to GN, which encodes an enzyme involved in cytokinin (CK) degradation (Ashikari et al., 2005). Down-regulation of Gn1a leads to the increase of CK level in the IM, thereby enhancing IM activity, branch number, and GN (Ashikari et al., 2005). PAP2 (PANICLE PHYTOMER 2) encodes a MADS-box protein which belongs to the SEPALLATA (SEP) subfamily (Kobayashi et al., 2010). PAP2 is exclusively expressed during the primary stage of panicle development, and functions in specifying the IM identity, and promote branch differentiation through controlling the expression of TFL1-like (TERMINAL FLOWER1-like) genes (Gao et al., 2010; Kobayashi et al., 2010; Liu et al., 2013; Lin et al., 2014). LAX1 (LAX PANICLE 1) and LAX2/GNP4 (GRAIN NUMBER PER PANICLE 4) jointly regulate the initiation and maintenance of BM, thereby regulating branch differentiation and GN (Oikawa and Kyozuka, 2009; Tabuchi et al., 2011; Sreenivasulu and Schnurbusch, 2012). NOG1 (NUMBER OF GRAINS 1) encodes an enoyl-CoA hydratase, which is highly expressed in the BM, and positively regulates GN and grain yield (Huo et al., 2017).
The maintenance of BM identity and the phase transition from BM to SM are regulated by the well-characterized APO1/APO2-LARGE2 module. APO1 (Aberrant Panicle Organization 1)/SCM2 (STRONG CULM 2) encodes an F-box-containing protein, which functions in controlling meristem cell proliferation (Ikeda-Kawakatsu et al., 2009). APO1 can promote the expression of class-C floral homeotic genes, and suppresses the precocious transition from BM into SM, thereby positively regulate the number of branches and GN (Huang et al., 2021a). APO2/RFL (Rice FLORICAULA) gene encodes a homolog protein of Arabidopsis LEAFY, which interacts and cooperates with APO1 in regulating the phase transition (Ikeda-Kawakatsu et al., 2012). Apo1 and apo2 mutants both showed decreased panicle size and primary branch number, which is caused by mis-regulation of floral meristem identity (Ikeda-Kawakatsu et al., 2012). LARGE2 encodes an E3 ubiquitin ligase, which is predominantly expressed in the developing inflorescence, and negatively regulates the stability of APO1 and APO2, thereby represses the maintenance of BM activity, decrease panicle size and GN (Huang et al., 2021a). In addition, FZP (FRIZZY PANICLE) encodes an AP2/ERF family transcription factor, which negatively regulates GN through repressing the expression of APO2, and promote the BM to SM transition and establishment of SM identity (Komatsu et al., 2003; Bai et al., 2016). The protein abundance of FZP is further controlled by NAL1 (NARROW LEAF 1), which encodes a serine and cysteine protease and interacts with FZP to promote its degradation (Huang et al., 2018). Furthermore, TAW1 (TAWAWA 1) suppresses the phase transition of BM to SM, thereby prolong the branch extension, and increase the secondary branch number (Yuan et al., 2021). RCN1 (RICE CENTRORADIALIS 1) and RCN2 also negatively regulate the transition of BM to SM, and overexpression of these two genes resulted in more high order branches and increased GN (Nakagawa et al., 2002).
The genes regulating the SM activity also have the potential to increase GN (Ren et al., 2020). FON4 (Floral organ number 4) controls the activity of SM (Chu et al., 2006; Ren et al., 2019), while MFS1 (MULTI-FLORET SPIKELET 1) and MFS2 are involved in the transition from SM to floral organ (Ren et al., 2013; Li et al., 2020). Mutants of these genes resulted in multi-floret spikelets and increased GN.
Regulation of GN by phytohormone
Phytohormones are ubiquitously involved in plant growth, development, and stress responses. The biosynthesis, metabolism, and signal transduction of phytohormones have significant impact on GN, through controlling both the panicle architecture and the branch differentiation processes (Deveshwar et al., 2020).
Cytokinin (CK) is an evolutionary conserved regulator of cell division and meristem activity in plants, which play crucial roles in the floral organ development (Kieber and Schaller, 2018; Rashotte, 2021), and is recognized as a key driver of grain yield (Jameson and Song, 2016). The level of CK in the IM is positively associated with floral organ number through promoting the activity of meristem (Ashikari et al., 2005; Kurakawa et al., 2007). LOG (LONELY GUY) encodes a phosphoribohydrolase, which catalyzes the conversion of inactive CK into its active form, and its mutation resulted in reduced number of branches and decreased GN (Kurakawa et al., 2007). An2 (Awn-2) also encodes a LOG family protein, which positively regulates GN through promoting CK biosynthesis (Gu et al., 2015). On the contrary, CKX (cytokinin oxidase) genes are negative regulators of GN through degradation of CK. In addition to the previously described GN1a/CKX2 (Ashikari et al., 2005), some other CKX genes were also found to negatively regulate GN, including CKX9 (Huang et al., 2021b) and CXK11 (Zhang et al., 2021a). Furthermore, the expression level of the genes related to CK biosynthesis and metabolism are under the control of transcription factors and MAPK signal cascade. DST encodes a zinc-finger transcription factor, which negatively regulates GN through promoting the expression of Gn1a (Li et al., 2013), while OsMPK6 can directly phosphorylate DST and enhance its transcriptional activation activity (Guo et al., 2020b). GSN1 (GRAIN SIZE AND NUMBER1)/GLA1 (GRAIN LENGTH AND AWN1)/LARGE8 encodes a MAPK phosphatase, which regulates CK metabolism through directly dephosphorylating OsMPK6, and inactivating the MAPK signal (Guo et al., 2018; Xu et al., 2018; Wang et al., 2019; Zhang et al., 2019a). ERECTA1 (OsER1) acts upstream of GSN1, and negatively regulates GN through promoting CK metabolism (Guo et al., 2020a,b). Moreover, the mediator protein OsMED25 physically interacts with DST and functions as a coactivator through recruiting RNA polymerase II to the promoter of OsCKX2 and promote its transcription (Lin et al., 2022). These genes coordinately regulate panicle architecture and GN through integrative control of CK homeostasis.
Gibberellin (GA) is known as a positive regulator of cell division and elongation in vegetative organs (Binenbaum et al., 2018). However, GA play negative roles in regulating the IM activity (Kwon and Paek, 2016; Su et al., 2021). Previous studies have shown that the OsCYP71D8L (CYTOCHROME P450-71 D8L) gene negatively regulates GN and panicle length through GA biosynthesis (Gao and Chu, 2020; Zhou et al., 2020). GA signal transduction depends on the GID family genes. It was suggested that Sped1-D can repress the expression of several GID1L2 genes, and promote the elongation of pedicels and secondary branches, thereby increase GN (Jiang et al., 2014). Furthermore, a crosstalk between GA and cytokinin was implicated in GN regulation. GNP1 (grain number per panicle1) encodes a GA20 oxidase, and increased expression of GNP1 caused a feed-back regulation of GA catabolism genes, reduced GA accumulation and enhanced CK level in panicle meristems to increase GN (Bessho-Uehara et al., 2016).
Auxin is recognized as a negative regulator of IM activity (He et al., 2018; Goetz et al., 2021). The dynamic efflux of auxin conducted by PIN (PIN-FORMED) protein family are essential for the establishment of axillary meristems (Deveshwar et al., 2020). PAY1 (PLANT ARCHITECTURE AND YIELD1) improves GN and plant architecture through influencing polar auxin transport and shifting auxin distribution (Zhao et al., 2015). NAL1 also functions in the polar transport of auxin, and overexpressing NAL1 can promote panicle branching and GN (Qi et al., 2008).
Brassinosteroid (BR) is an essential regulator of cell expansion and grain size (Li et al., 2018a; Fan and Li, 2019). It is revealed that BR signal is also involved in the meristem differentiation during panicle development. The QTLs CPB1 (CLUSTERED PRIMARY BRANCH1), GNS4 (grain number and size on chromosome 4) and PMM1 (Panicle Morphology Mutant1), were independently identified to regulate GN and grain size through affecting spikelet meristem differentiation as well as panicle architecture (Wu et al., 2016; Zhou et al., 2017; Li et al., 2018c). Further analysis identified them as multiple alleles of D11 (DWARF11), a cytochrome P450 encoding gene involved in BR biosynthesis pathway (Tanabe et al., 2005). In addition, it has been revealed that ABA and ethylene negatively regulate GN (Hirose et al., 2007; Wuriyanghan et al., 2009). It is worth noting that GN is a complex agronomic trait, and its regulation is usually the result of the synergistic effect of multiple phytohormones. Dissecting the genetic architecture and molecular mechanism of these genes would facilitate the genetic improvement of grain production in rice and other crops.
Regulation of GN by vascular development
The vascular system connects the entire plant body and conducts the long-distance transport of water, inorganic salts, nutrients and assimilates, which are crucial for plant growth and grain yield (De Rybel et al., 2016; Agusti and Blazquez, 2020). The vascular bundles in the stem internode consists of the large vascular bundles (lvbs) arranged in the inner side of the cortex, and the small vascular bundles (svbs) arranged around the outer side of stem. Each vascular bundle consists of the phloem and the xylem. The vascular bundles in panicle neck determines the transport efficiency of photoassimilates from “source” leaf to “sink” grain. Furthermore, the lvbs in panicle neck are directly connected to the primary branches of the panicle (Liao et al., 2021). Therefore, the number of lvbs in the panicle neck is positively correlated with the number of branches and GN (Zhai et al., 2018; Fei et al., 2019; Liao et al., 2021).
Emerging evidence have revealed an association between vascular development and GN. Ghd7 gene is highly expressed in the vascular tissue, and its elite allele shows an improved vascular system and increased GN (Xue et al., 2008). Genome-wide association analysis also revealed that Ghd7 is a key locus affecting vascular development (Liao et al., 2021). The NAL1 (Narrow Leaf 1) gene, which is associated with leaf development, has a significant effect on GN in both indica and japonica populations (Chen et al., 2012; Wang et al., 2020c). Recent research shows that NAL1 can also significantly affect the vascular bundle morphology in leaves and panicle neck (Liao et al., 2021). The qPE9-1/DEP1 and EP2 genes associated with erect panicle shape were also found to regulate the patterning and development of large vascular bundles in the panicle neck (Zhu et al., 2010; Xu et al., 2015b). APO1 is predominantly expressed in developing vascular tissues, and promotes the translocation of photoassimilates (Terao et al., 2010). SP1 is also expressed in the vascular bundle of developing panicles, where it functions as a nitrate transporter (Li et al., 2009).
Studies on vascular patterning and development revealed key genes which possess the potential to increase GN. The transaldolase gene TAL is a key regulator of vascular development in rice, which also has a positive effect on GN and grain yield (Yang et al., 2015). Knock down of OsTAL reduced the number and area of stem lvbs, and significantly decreased GN and grain yield (Yang et al., 2015). The OsCOMT (caffeic acid O-methyl transferase) gene, which encodes the rate-limiting enzyme in melatonin biosynthesis, positively regulates GN and grain yield through promoting vascular development and delaying leaf senescence (Huangfu et al., 2022). Overexpression of OsCOMT can significantly improve the vascular bundle size and number, and increase GN (Huangfu et al., 2022). It is suggested that the improved vascular system (flow) may promote the translocation of photoassimilates from source to sink organs (Li et al., 2018b). These observations also suggested that the vascular development associated genes may be useful in improving GN and rice grain yield.
Pleiotropy of the genes regulating GN
Many genes associated with GN have been revealed to possess pleiotropic effects in other important agronomic traits, such as tiller number, grain shape, grain weight, heading date and plant architecture. Tillering and panicle branching are both controlled by the activity of axillary meristem (Liang et al., 2014). Therefore, the genes involved in axillary meristem establishment and maintenance often exhibit co-regulation of GN and tiller number. MOC1 (MONOCULM 1)/GNP6 (grain number per panicle 6) encodes a GRAS (GAI, RGA and SCR) family protein, which is an essential regulator for the initiation of axillary meristem, and its null mutation resulted in almost complete loss of tillers and arrested branch growth (Li et al., 2003; Shao et al., 2019). LAX1, LAX2, APO1 and APO2 were required for the maintenance of axillary meristems (Oikawa and Kyozuka, 2009; Tabuchi et al., 2011), therefore, mutation of these genes also significantly reduced tiller number and GN (Zhang et al., 2021b). However, GN and tiller number can sometimes show opposite regulations. PAY1 encodes a peptidase S64 domain protein, which is associated with auxin transport (Zhao et al., 2015). Enhanced expression of PAY1 increases the number of secondary branches, but reduces the number of tillers (Zhao et al., 2015). Moreover, IPA1 also increases GN but reduces PN through directly binding to the promoter of TB1 (Teosinte Branched 1) and DEP1 to promote their transcription, thereby suppress rice tillering, promote branching, and regulate rice plant architecture (Miura et al., 2010; Lu et al., 2013).
The regulation of GN and grain size (GS) or grain weight (GW) often show an antagonistic relationship (Fan and Li, 2019). Down-regulation of GS3 (GRAIN SIZE/SHAPE 3; Fan et al., 2006; Mao et al., 2010), GSN1 (GRAIN SIZE AND NUMBER1; Zhang et al., 2019a), and GW2 (GRAIN WIDTH 2; Song et al., 2007) resulted in increased GW but reduced GN; while down-regulation of GW10 (GRAIN WIDTH 10) decreased GW but increased GN (Zhan et al., 2021). On the other hand, down-regulation of GN-associated genes DEP1 (Huang et al., 2009b; Yi et al., 2011; Li et al., 2019), GAD1 (grain number, grain length and awn development 1; Bessho-Uehara et al., 2016; Jin et al., 2016), and FZP (Bai et al., 2016; Fujishiro et al., 2018) increased GN but reduced GW. This antagonistic relationship between GN and GW can be attributed to the competition effects for photoassimilates (Fan and Li, 2019). The balancing between GW and GN would be an important target in the future improvement of rice gain yield.
Some genes regulating heading date were also found to be associated with GN. Ghd7 (Grain Number, Plant Height, and Heading Date 7) gene encodes a CCT domain protein, and its overexpression under long-day condition delays the heading date and increased GN (Weng et al., 2014). Ghd8 gene can simultaneously regulate the heading date, tiller number, plant height and the number of branches, thereby affect GN (Yan et al., 2011). Such synergistic regulation of GN and heading date has also been observed in studies on other genes including APO2/RFL (Ikeda-Kawakatsu et al., 2012), RCN1 (RICE CENTRORADIALIS 1; Nakagawa et al., 2002; Wang et al., 2020b), RCN2 (Nakagawa et al., 2002), and OsCOL13 (CONSTANS-LIKE 13; Sheng et al., 2016). However, the regulatory mechanism of the association between GN and heading date remains elusive.
It was suggested that GN-associated genes can also affect other important agronomic traits, such as nutrient metabolism, plant architecture, and stress response. For instance, qPE9-1/DEP1 has been revealed as a multifunctional regulator of nitrogen use efficiency (NUE; Dong et al., 2022), root elongation and phosphorus uptake (Wang et al., 2021), as well as drought stress response (Zhang et al., 2015). In addition, DEP1 can interact with RGA1 (Rice G-protein Alpha subunit1) and RGB1 (Rice G-protein Beta subunit1) to increase nitrogen absorption and utilization, and ultimately increase the plant biomass and grain yield (Sun et al., 2014). OsEBS (ENHANCING BIOMASS AND SPIKELET NUMBER) positively regulates the plant height, leaf size and biomass in addition to GN (Dong et al., 2013). DST functions in rice drought and salt tolerance through modulating stomatal aperture, and also positively regulates GN (Huang et al., 2009a). Moreover, loss of function of Gn1a/OsCKX2 not only increased GN, but also enhanced lodging resistance through accelerating root development and increasing the culm diameter (Tu et al., 2022). Further studies would be expected for elucidating the relationship between GN and these traits, and revealing the application value of these pleiotropic genes in rice breeding.
GN is a common domestication syndrome trait in cereal crops. Interestingly, many GN-associated genes were involved in the regulation of other domestication related traits. PROG1 (PROSTRATE GROWTH 1) is a key gene in the process of rice domestication, which controls the critical transition from prostrate to erect growth, and changes the plant architecture (Tan et al., 2008; Huang et al., 2020). PROG1 is predominantly expressed in the axillary meristems, and promotes GN through increasing the number of primary and secondary branches (Jin et al., 2008). GAD1/RAE2 (regulator of awn elongation2)/ GLA (Grain Length and Awn Development) encodes a secreted peptide, and the loss-of-function of GAD1 resulted in decreased grain length, increased GN, and loss of awn, suggestive important role of GAD1 in the domestication of rice (Bessho-Uehara et al., 2016; Jin et al., 2016; Zhang et al., 2019b; Xu and Sun, 2021). The mutants of An-1 (Awn-1) and An-2 showed reduced awn length, increased GN and GW, and these genes have experienced artificial selection during rice domestication (Luo et al., 2013; Gu et al., 2015). Furthermore, recent study revealed that OsKRN2 (Kernel Row Number 2), which negatively regulates GN, has undergone convergent selection with its maize ortholog KRN2 during the domestication of rice and maize (Chen et al., 2022). These findings partially revealed the genetic basis and molecular mechanisms underlying the selective forces of GN and other domestication related traits. Understanding the molecular mechanism of the co-regulation of these traits would provide novel insights for the improvement of grain yield in crop genetic improvement.
Conclusions and future perspectives
During the past two decades, substantial progresses have been made in understanding the genetic and molecular factors in determining GN in rice (Figure 1; Table 1). GN is mainly determined by panicle architecture and branch differentiation. Panicle architecture consists of the panicle length and the number and arrangement of rachis branches, while branch differentiation is controlled by the establishment, maintaining, phase transition, and differentiation of IM, BM, and SM. These processes are regulated by various phytohormones and G-protein signal pathways, and are closely associated with the vascular development. Furthermore, the GN-associated genes play pleiotropic roles in regulating PN, GW, flowering time and domestication related traits. However, rice GN is a complex quantitative trait which is regulated by multiple factors. In addition to these major aspects, some other factors, including spikelet sterility (Heng et al., 2018; Sekhar et al., 2021), nitrogen allocation (Guo et al., 2020a), sugar transport (Seki et al., 2015; Xu et al., 2019), and circadian clock regulation (Wang et al., 2020a), might also participate in the regulation of rice GN.
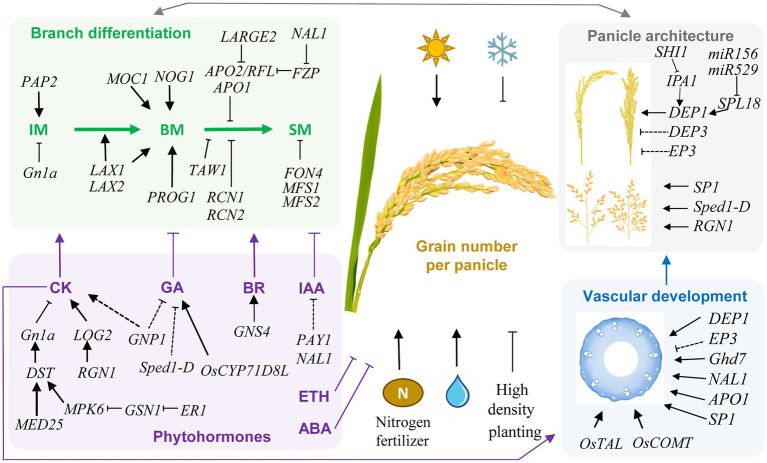
Figure 1.
Schematic representation of genetic and environmental factors controlling grain number per panicle (GN) in rice. Key genes and regulatory pathways in controlling GN are indicated. Arrows indicate positive regulation, while T lines indicate negative regulation. Dashed lines indicate indirect regulations. Rice GN is determined by the panicle architecture and branch differentiation, which are associated with phytohormones and vascular development. These regulatory pathways are interconnected in regulating GN. In the determination of panicle architecture, the DEP1, DEP3, and EP3 genes control the dense and erect panicle type, while SP1, Sped1-D, and RGN1 regulate panicle length and branch number. Transcription factors IPA1 and SPL18 positively regulates the expression of DEP1. SHI1 interacts with IPA1 and inhibit its activity, while microRNAs miR156 and miR529 regulate the expression level of SPL18. The branch differentiation involves the activity and phase transition of inflorescence meristem (IM), branch meristem (BM), and spikelet meristem (SM). Increasing the meristem activity and delaying the transition from BM to SM have a positive effect on GN. Phytohormones were also crucial regulators of GN through manipulating branch differentiation, among which cytokinin (CK) and brassinosteroid (BR) are positive regulators, while indole 3-acetic acid (IAA), gibberellin (GA), abscisic acid (ABA), and ethylene (ETH) are negative regulators. The key GN regulator GN1a encodes a CKX, which negatively regulate CK level, and is regulated by the ER1-GSN1-MPK6-DST signal cascade, while MED25 functions as a coactivator with DST to promote expression of GN1a. Vascular development is closely associated with GN through affecting the panicle architecture and translocation capability for water and nutrients. Many GN-associated genes also function in regulating vascular development, while the key vascular development regulators OsTAL and OsCOMT both have significant positive effects on GN. Moreover, vascular development is positively regulated by CK. In addition, light, water, and nitrogen availability positively regulate GN, while low temperature and high planting density negatively control GN. These genetic and non-genetic factors together determine the GN of rice.
Table 1.
List of the genes involved in rice grain number per panicle regulation.
| Gene name | Locus | Protein | Function | References |
|---|---|---|---|---|
| APO1 | LOC_Os06g45460 | F-box protein | Control meristem cell proliferation; enhance the formation of vascular bundle systems | Ikeda-Kawakatsu et al. (2009), Terao et al., (2010) |
| APO2/RFL | LOC_Os04g51000 | Transcription factor RFL | Control inflorescence and flower development | Kyozuka et al. (1998), Ikeda-Kawakatsu et al. (2012) |
| COMT | LOC_Os08g06100 | Caffeic acid O-methyltransferase | Promote GN through regulating vascular development | Huangfu et al. (2022) |
| DEP1/qPE9-1 | LOC_Os09g26999 | G-protein gamma subunit | Regulate GN, panicle length, and grain weight | Huang et al. (2009b), Zhou et al. (2009), Sun et al. (2014), Xu et al. (2015b) |
| DEP2 | LOC_Os07g42410 | Unknown plant-specific protein | Control panicle outgrowth and elongation | Li et al. (2010) |
| DEP3 | LOC_Os06g46350 | Patatin-related phospholipase A | Regulate the formation of vascular bundles | Qiao et al. (2011) |
| DST | LOC_Os03g57240 | Zinc-finger transcription factor | Directly regulate OsCKX2 expression in the reproductive meristem | Li et al. (2013) |
| EBS | LOC_Os05g51360 | Similar to the N-terminal conserved ATPase domain of Hsp70 | Enhancing biomass and spikelet number | Dong et al. (2013) |
| EP3 | LOC_Os02g15950 | F-box protein | Regulates panicle architecture and vascular development | Piao et al. (2009) |
| ER1 | LOC_Os06g10230 | receptor-like protein kinase | Regulate CK metabolism through the MAPK signal cascade | Guo et al. (2020b) |
| FON4 | LOC_Os11g38270 | Receptor-like protein kinase | Prevents the multi-floret spikelet through controlling SM identity | Ren et al. (2019) |
| FZP | LOC_Os07g47330 | ERF transcription factor | Promote GN through establishing floral organ identity | Komatsu et al. (2003), Bai et al. (2016) |
| GAD1/RAE2 | LOC_Os08g37890 | Cysteine-rich secretory peptide | Regulates GN, grain length, and awn development | Bessho-Uehara et al. (2016), Jin et al. (2016) |
| Gn1a/CKX2 | LOC_Os01g10110 | Cytokinin oxidase CKX2 | Reduce GN through cytokinin metabolism | Ashikari et al. (2005), Gouda et al. (2020a) |
| Ghd7 | LOC_Os07g15770 | CCT (CO, CO-LIKE and TIMING OF CAB1) domain protein | Regulates grain number, plant height, and heading date; promote vascular development | Xue et al. (2008), Weng et al. (2014) |
| Ghd7.1 | LOC_Os07g49460 | pseudo-response regulator (PRR) protein | Delays rice heading and enhances grain productivity | Luo et al. (2013), Yan et al. (2013) |
| Ghd8 | LOC_Os08g07740 | HAP3 subunit of the HAP | Regulate grain number, plant height, and heading date | Yan et al. (2011) |
| GNP1 | LOC_Os03g63970 | Gibberellin biosynthesis enzyme GA20ox1 | Promote gibberellin biosynthesis | Bessho-Uehara et al. (2016) |
| GNS4 | LOC_Os04g39430 | Cytochrome P450 protein | Positively regulate GN and GS through the BR pathway | Zhou et al. (2017) |
| GSN1/GLA1/ LARGE8 | LOC_Os05g02500 | MAPK phosphatase | Regulate CK metabolism through inactivating MAPK signal cascade | Guo et al. (2018), Xu et al. (2018), Wang et al. (2019), Zhang et al. (2019a) |
| IPA1/OsSPL14 | LOC_Os08g39890 | SOUAMOSA PROMOTER BINDING PROTEIN-LIKE transcription factor | Promote shoot branching through transcriptional activation of DEP1 | Jiao et al. (2010), Miura et al. (2010), Lu et al. (2013) |
| LARGE2 | LOC_Os12g24080 | HECT-domain E3 ubiquitin ligase | Negatively regulate GN through affecting the stability of APO1 and APO2 | Huang et al. (2021a) |
| LAX1 | LOC_Os01g61480 | bHLH transcription factor | Regulate axillary meristems formation | Oikawa and Kyozuka (2009) |
| LAX2/GNP4 | LOC_Os04g32510 | Nuclear protein with a plant-specific conserved domain | Interact with LAX1; regulate axillary meristem formation and lateral branching | Tabuchi et al. (2011), Zhang et al. (2018) |
| MED25 | LOC_Os09g13610 | Mediator protein | Interact with DST to promote expression of Gn1a | Lin et al. (2022) |
| MFS1 | LOC_Os05g41760 | AP2 domain containing protein | Repress SM determinacy and floral organ identity | Ren et al. (2013) |
| MFS2 | LOC_Os04g47890 | MYB transcription factor | Repress SM determinacy and floral organ identity | Li et al. (2020) |
| MOC1/GNP6 | LOC_Os06g40780 | GRAS-family nuclear protein | Promote axillary meristem initiation | Zhang et al. (2021b) |
| NAL1/qFLW4 | LOC_Os04g52479 | Trypsin-like serine and cysteine protease | Promote degradation of FZP; positively regulate leaf and vascular development | Qi et al. (2008), Fujita et al. (2013), Xu et al. (2015a), Huang et al. (2018), Lin et al. (2019), Wang et al. (2020c) |
| NOG1 | LOC_Os01g54860 | Enoyl-CoA hydratase/isomerase | Promote GN without affecting other yield traits | Huo et al. (2017) |
| PAP2/MADS34 | LOC_Os03g54170 | SEP-like MADS box transcription factor | Positively control spikelet meristem identity | Gao et al. (2010), Kobayashi et al. (2010), Lin et al. (2014) |
| PAY1 | LOC_Os08g31470 | Trypsin-like serine and cysteine protease | Improve plant architecture through affecting polar auxin transport and endogenous IAA distribution | Zhao et al. (2015) |
| PROG1 | LOC_Os07g05900 | Cys2-His2 zinc-finger protein | Regulate erect growth, promote GN and grain yield | Tan et al. (2008) |
| RCN1 | LOC_Os03g17350 | White-brown complex homolog protein | Promote branching through delaying the phase transition | Nakagawa et al. (2002) |
| RCN2 | LOC_Os02g32950 | Phosphatidylethanolamine-binding protein | Promote branching through delaying the phase transition | Nakagawa et al. (2002) |
| RGN1 | LOC_Os01g49160 | R2R3 MYB transcription factor | Promote GN through regulating LOG expression | Li et al. (2022) |
| RLB | LOC_Os07g03770 | KNOX type homebox protein | Promote GN through epigenetic silencing of OsCKX4 | Wang et al. (2022a) |
| SH1 | LOC_Os09g36160 | Transcription factor | Interacts with IPA1 and represses the transcriptional activation ability | Duan et al. (2019) |
| SP1 | LOC_Os11g12740 | Putative peptide transporter (PTR) family protein | Regulate panicle architecture through nitrate transport | Li et al. (2009) |
| SPL18 | LOC_Os09g32944 | SOUAMOSA PROMOTER BINDING PROTEIN-LIKE transcription factor | Promote expression of DEP1 | Yuan et al. (2019) |
| sped1-D | LOC_Os06g39650 | Pentatricopeptide repeat protein | Prompt the shortening of pedicels and secondary branches through repressing the GA signal transduction | Jiang et al. (2014) |
| TAL | LOC_Os01g70170 | Transaldolase | Promote vascular development | Yang et al. (2015) |
| TAW1 | LOC_Os10g33780 | Unknown nuclear protein | Promote panicle development | Yuan et al. (2021) |
Identification of GN-associated genes are of vital importance both for understanding the regulatory network of GN and for the improvement of rice yield. In addition to the traditional genetic mapping approach, GWAS (genome-wide association studies) also provides an effective tool in unraveling the genetic basis of GN and other yield traits (Rebolledo et al., 2016; Xiao et al., 2017). Moreover, the advent of third-generation long-range genome sequencing and pangenomes have greatly enriched the genomic information and expanded genetic diversity of rice and other crops (Zhao et al., 2018; Alonge et al., 2020). Crop pangenome studies highlighted structural variants and their association with important agronomic traits (Gabur et al., 2019), which would have great potential in GN-associated gene mining and yield improvement. On the other hand, innovation of rice germplasm populations with rich genetic and phenotypic variations are essential for mining novel genes and QTLs. Construction of multi parent populations (MPPs), including MAGIC (multiparent advanced generation inter-cross) and MCC-NAM (mini-core collection nested association mapping) populations, provide effective tools for the identification of novel genes controlling complex traits (Scott et al., 2020). Compared with traditional bi-parent populations, MPPs effectively expanded genetic diversity and increased genetic recombination. The application of these MPPs provide higher mapping power and resolution in exploring genetic architecture of yield traits in rice (Zaw et al., 2019; Han et al., 2020; Ayaad et al., 2021; Huerta et al., 2021), and would be effective strategies in the cloning of GN-associated genes and rice breeding. In addition, current studies also shed light on the roles of epigenetic modification related genes in GN regulation (Zhang et al., 2017; Wang et al., 2022a). Despite the increasing attention on the importance of epigenetic regulations in plant growth, stress response, and crop yield (Lu et al., 2018, 2020), the significance and mechanisms of epigenetic modification related genes in GN regulation remains largely elusive. Mining the epigenetic genes and mechanisms underlying GN regulation through bisulfite sequencing, chromatin immunoprecipitation (ChIP) assay, and epigenome editing based on RNA-dependent DNA methylation (Wakasa et al., 2018), will provide novel insight into the regulations on GN. The identification and mechanic revelation of genes controlling GN will further guide the molecular design breeding of rice.
The application of GN-associated genes in rice breeding is a pivotal task in the future genetic improvement. Gene editing technologies, including transcription activator-like effector nucleases (TALENs), zinc-finger nucleases (ZFNs), and clustered regularly interspaced short palindromic repeats (CRISPR-Cas9) system, are promising tools for reshaping crop breeding. These technologies enabled flexibility in improving target traits through precise targeting of multiple genes (Zhu et al., 2020; Ganie et al., 2021; Lu et al., 2021; Mohd Saad et al., 2022). It is suggested that CRISPR-Cas9 editing of GN-associated genes, including GN1a, DEP1, and IPA1, can significantly increase GN and rice grain yield (Li et al., 2016, 2021a). Therefore, editing of GN-associated genes in elite cultivars through these approaches would be effective and promising to accelerate the utilization of these genes in the breeding process. Genomic selection (GS) estimates the effects of all markers in a training population, and use this information to predict the breeding value of genotyped individuals (Crossa et al., 2017; Xu et al., 2020). GS holds enormous potential in transferring the elite allele into breeding cultivars and accelerating the breeding process (Xu et al., 2021b), which has been successfully applied in rice breeding (Cui et al., 2020; Xiao et al., 2021). Furthermore, the development of machine learning, deep learning, and neural network strategies, has greatly improved the efficiency in phenotyping and analyzing environmental variables that affect phenotypes. These strategies, combined with high throughput plant phenotyping technique, provide efficient and effective solution for improving the predictive capability and trait improvement (Bayer and Edwards, 2021). Moreover, the recent developed multi-trait GS technology offers a powerful and efficient solution for improving the predictive ability for complex traits (Wang et al., 2017; Moeinizade et al., 2020; Xu et al., 2021a). Considering the pleiotropy of GN-associated genes, multi-trait GS would be favorable for the improvement of GN and other associated traits in rice breeding, such as GW, plant architecture, and improved vascular system. The integration of gene identification and molecular breeding strategies will benefit for the future improvement of GN and other agronomic traits in rice.
Author contributions
YL, ZY, and CX conceived the idea and wrote the manuscript. YL, MC, HW, RC, and TT collected the materials. YZ, YX, and PL prepared the figures. YZ and YY revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32100448, 32070558, 32061143030, 32170636, and 31970248), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Natural Science Foundation of Jiangsu Province (BK20210799), the Seed Industry Revitalization Project of Jiangsu Province [JBGS(2021)009], and the Project of Hainan Yazhou Bay Seed Laboratory (B21HJ0223).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Agusti J., Blazquez M. A. (2020). Plant vascular development: mechanisms and environmental regulation. Cell. Mol. Life Sci. 77, 3711–3728. doi: 10.1007/s00018-020-03496-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali I., Tang L., Dai J., Kang M., Mahmood A., Wang W., et al. (2021). Responses of grain yield and yield related parameters to post-heading low-temperature stress in japonica rice. Plants (Basel) 10, 1425. doi: 10.3390/plants10071425, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M., Wang X., Benoit M., Soyk S., Pereira L., Zhang L., et al. (2020). Major impacts of widespread structural variation on gene expression and crop improvement in tomato. Cell 182:e123. doi: 10.1016/j.cell.2020.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashikari M., Sakakibara H., Lin S., Yamamoto T., Takashi T., Nishimura A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745. doi: 10.1126/science.1113373 [DOI] [PubMed] [Google Scholar]
- Ayaad M., Han Z., Zheng K., Hu G., Abo-Yousef M., Sobeih S., et al. (2021). Bin-based genome-wide association studies reveal superior alleles for improvement of appearance quality using a 4-way MAGIC population in rice. J. Adv. Res. 28, 183–194. doi: 10.1016/j.jare.2020.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X., Huang Y., Mao D., Wen M., Zhang L., Xing Y. (2016). Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Sci. Rep. 6:19022. doi: 10.1038/srep19022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer P., Edwards D. (2021). Machine learning in agriculture: from silos to marketplaces. Plant Biotechnol. J. 19, 648–650. doi: 10.1111/pbi.13521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho-Uehara K., Wang D. R., Furuta T., Minami A., Nagai K., Gamuyao R., et al. (2016). Loss of function at RAE2, a previously unidentified EPFL, is required for awnlessness in cultivated Asian rice. Proc. Natl. Acad. Sci. U. S. A. 113, 8969–8974. doi: 10.1073/pnas.1604849113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binenbaum J., Weinstain R., Shani E. (2018). Gibberellin localization and transport in plants. Trends Plant Sci. 23, 410–421. doi: 10.1016/j.tplants.2018.02.005 [DOI] [PubMed] [Google Scholar]
- Chen W., Chen L., Zhang X., Yang N., Guo J., Wang M., et al. (2022). Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 375:eabg7985. doi: 10.1126/science.abg7985 [DOI] [PubMed] [Google Scholar]
- Chen M., Luo J., Shao G., Wei X., Tang S., Sheng Z., et al. (2012). Fine mapping of a major QTL for flag leaf width in rice, qFLW4, which might be caused by alternative splicing of NAL1. Plant Cell Rep. 31, 863–872. doi: 10.1007/s00299-011-1207-7 [DOI] [PubMed] [Google Scholar]
- Chu H., Qian Q., Liang W., Yin C., Tan H., Yao X., et al. (2006). The floral organ number4 gene encoding a putative ortholog of Arabidopsis CLAVATA3 regulates apical meristem size in rice. Plant Physiol. 142, 1039–1052. doi: 10.1104/pp.106.086736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J., Perez-Rodriguez P., Cuevas J., Montesinos-Lopez O., Jarquin D., De Los Campos G., et al. (2017). Genomic selection in plant breeding: methods, models, and perspectives. Trends Plant Sci. 22, 961–975. doi: 10.1016/j.tplants.2017.08.011 [DOI] [PubMed] [Google Scholar]
- Cui Y., Li R., Li G., Zhang F., Zhu T., Zhang Q., et al. (2020). Hybrid breeding of rice via genomic selection. Plant Biotechnol. J. 18, 57–67. doi: 10.1111/pbi.13170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B., Mahonen A. P., Helariutta Y., Weijers D. (2016). Plant vascular development: from early specification to differentiation. Nat. Rev. Mol. Cell Biol. 17, 30–40. doi: 10.1038/nrm.2015.6 [DOI] [PubMed] [Google Scholar]
- Deveshwar P., Prusty A., Sharma S., Tyagi A. K. (2020). Phytohormone-mediated molecular mechanisms involving multiple genes and QTL govern grain number in rice. Front. Genet. 11:586462. doi: 10.3389/fgene.2020.586462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Wang X., Zhang L., Yang Z., Xin X., Wu S., et al. (2013). Identification and characterization of OsEBS, a gene involved in enhanced plant biomass and spikelet number in rice. Plant Biotechnol. J. 11, 1044–1057. doi: 10.1111/pbi.12097 [DOI] [PubMed] [Google Scholar]
- Dong G., Zhou Y., Zhang J., Wang J., Zhou J., Chen C., et al. (2022). Introgression of qPE9-1/DEP1, a major QTL for rice panicle erectness,drastically improves nitrogen use efficiency under limited nitrogen supply. Eur. J. Agron. 133:126444. doi: 10.1016/j.eja.2021.126444 [DOI] [Google Scholar]
- Duan E., Wang Y., Li X., Lin Q., Zhang T., Wang Y., et al. (2019). OsSHI1 regulates plant architecture through modulating the transcriptional activity of IPA1 in rice. Plant Cell 31, 1026–1042. doi: 10.1105/tpc.19.00023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y., Li Y. (2019). Molecular, cellular and Yin-Yang regulation of grain size and number in rice. Mol. Breed. 39:163. doi: 10.1007/s11032-019-1078-0 [DOI] [Google Scholar]
- Fan C., Xing Y., Mao H., Lu T., Han B., Xu C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1 [DOI] [PubMed] [Google Scholar]
- Fei C., Geng X., Xu Z., Xu Q. (2019). Multiple areas investigation reveals the genes related to vascular bundles in rice. Rice (N Y) 12, 17. doi: 10.1186/s12284-019-0278-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X., Wang C., Nan J., Zhang X., Wang R., Jiang G., et al. (2017). Updating the elite rice variety Kongyu 131 by improving the Gn1a locus. Rice (N Y) 10:35. doi: 10.1186/s12284-017-0174-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Mccouch S., Hall R. (2009). Not just a grain of rice: the quest for quality. Trends Plant Sci. 14, 133–139. doi: 10.1016/j.tplants.2008.12.004 [DOI] [PubMed] [Google Scholar]
- Fujishiro Y., Agata A., Ota S., Ishihara R., Takeda Y., Kunishima T., et al. (2018). Comprehensive panicle phenotyping reveals that qSrn7/FZP influences higher-order branching. Sci. Rep. 8, 12511. doi: 10.1038/s41598-018-30395-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita D., Trijatmiko K., Tagle A., Sapasap M., Koide Y., Sasaki K., et al. (2013). NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. Proc. Natl. Acad. Sci. U. S. A. 110, 20431–20436. doi: 10.1073/pnas.1310790110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabur I., Chawla H. S., Snowdon R. J., Parkin I.a. P. (2019). Connecting genome structural variation with complex traits in crop plants. Theor. Appl. Genet. 132, 733–750. doi: 10.1007/s00122-018-3233-0 [DOI] [PubMed] [Google Scholar]
- Ganie S., Wani S., Henry R., Hensel G. (2021). Improving rice salt tolerance by precision breeding in a new era. Curr. Opin. Plant Biol. 60:101996. doi: 10.1016/j.pbi.2020.101996 [DOI] [PubMed] [Google Scholar]
- Gao S., Chu C. (2020). Gibberellin metabolism and Signaling: targets for improving agronomic performance of crops. Plant Cell Physiol. 61, 1902–1911. doi: 10.1093/pcp/pcaa104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X., Liang W., Yin C., Ji S., Wang H., Su X., et al. (2010). The SEPALLATA-like gene OsMADS34 is required for rice inflorescence and spikelet development. Plant Physiol. 153, 728–740. doi: 10.1104/pp.110.156711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghusinga K., Elston T., Jones A. (2022). Towards resolution of a paradox in plant G-protein signaling. Plant Physiol. 188, 807–815. doi: 10.1093/plphys/kiab534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M., Rabinovich M., Smith H. (2021). The role of auxin and sugar signaling in dominance inhibition of inflorescence growth by fruit load. Plant Physiol. 187, 1189–1201. doi: 10.1093/plphys/kiab237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouda G., Gupta M., Donde R., Kumar J., Parida M., Mohapatra T., et al. (2020a). Characterization of haplotypes and single nucleotide polymorphisms associated with Gn1a for high grain number formation in rice plant. Genomics 112, 2647–2657. doi: 10.1016/j.ygeno.2020.02.016 [DOI] [PubMed] [Google Scholar]
- Gouda G., Gupta M., Donde R., Mohapatra T., Vadde R., Behera L. (2020b). Marker-assisted selection for grain number and yield-related traits of rice (Oryza sativa L.). Physiol. Mol. Biol. Plants 26, 885–898. doi: 10.1007/s12298-020-00773-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu B., Zhou T., Luo J., Liu H., Wang Y., Shangguan Y., et al. (2015). An-2 encodes a Cytokinin synthesis enzyme that regulates awn length and grain production in Rice. Mol. Plant 8, 1635–1650. doi: 10.1016/j.molp.2015.08.001 [DOI] [PubMed] [Google Scholar]
- Guo T., Chen K., Dong N., Shi C., Ye W., Gao J., et al. (2018). GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 Cascade to coordinate the trade-off between grain NUMBER per panicle and grain size in Rice. Plant Cell 30, 871–888. doi: 10.1105/tpc.17.00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo N., Gu M., Hu J., Qu H., Xu G. (2020a). Rice OsLHT1 functions in leaf-to-panicle nitrogen allocation for grain yield and quality. Front. Plant Sci. 11:1150. doi: 10.3389/fpls.2020.01150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T., Lu Z., Shan J., Ye W., Dong N., Lin H. (2020b). ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 Cascade to control spikelet number by regulating Cytokinin metabolism in Rice. Plant Cell 32, 2763–2779. doi: 10.1105/tpc.20.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z., Hu G., Liu H., Liang F., Yang L., Zhao H., et al. (2020). Bin-based genome-wide association analyses improve power and resolution in QTL mapping and identify favorable alleles from multiple parents in a four-way MAGIC rice population. Theor. Appl. Genet. 133, 59–71. doi: 10.1007/s00122-019-03440-y [DOI] [PubMed] [Google Scholar]
- He Q., Yang L., Hu W., Zhang J., Xing Y. (2018). Overexpression of an auxin receptor OsAFB6 significantly enhanced grain yield by increasing cytokinin and decreasing auxin concentrations in rice panicle. Sci. Rep. 8:14051. doi: 10.1038/s41598-018-32450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng Y., Wu C., Long Y., Luo S., Ma J., Chen J., et al. (2018). OsALMT7 maintains panicle size and grain yield in Rice by mediating malate transport. Plant Cell 30, 889–906. doi: 10.1105/tpc.17.00998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N., Makita N., Kojima M., Kamada-Nobusada T., Sakakibara H. (2007). Overexpression of a type-A response regulator alters rice morphology and cytokinin metabolism. Plant Cell Physiol. 48, 523–539. doi: 10.1093/pcp/pcm022 [DOI] [PubMed] [Google Scholar]
- Hu Q., Wang W., Lu Q., Huang J., Peng S., Cui K. (2021). Abnormal anther development leads to lower spikelet fertility in rice (Oryza sativa L.) under high temperature during the panicle initiation stage. BMC Plant Biol. 21:428. doi: 10.1186/s12870-021-03209-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Chao D., Gao J., Zhu M., Shi M., Lin H. (2009a). A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes Dev. 23, 1805–1817. doi: 10.1101/gad.1812409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Hilscher J., Stoger E., Christou P., Zhu C. (2021b). Modification of cereal plant architecture by genome editing to improve yields. Plant Cell Rep. 40, 953–978. doi: 10.1007/s00299-021-02668-7 [DOI] [PubMed] [Google Scholar]
- Huang L., Hua K., Xu R., Zeng D., Wang R., Dong G., et al. (2021a). The LARGE2-APO1/APO2 regulatory module controls panicle size and grain number in rice. Plant Cell 33, 1212–1228. doi: 10.1093/plcell/koab041 [DOI] [PubMed] [Google Scholar]
- Huang L., Liu H., Wu J., Zhao R., Li Y., Melaku G., et al. (2020). Evolution of plant architecture in Oryza driven by the PROG1 locus. Front. Plant Sci. 11:876. doi: 10.3389/fpls.2020.00876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Qian Q., Liu Z., Sun H., He S., Luo D., et al. (2009b). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352 [DOI] [PubMed] [Google Scholar]
- Huang Y., Zhao S., Fu Y., Sun H., Ma X., Tan L., et al. (2018). Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. Plant J. 96, 716–733. doi: 10.1111/tpj.14062 [DOI] [PubMed] [Google Scholar]
- Huangfu L., Chen R., Lu Y., Zhang E., Miao J., Zuo Z., et al. (2022). OsCOMT, encoding a caffeic acid O-methyltransferase in melatonin biosynthesis, increases rice grain yield through dual regulation of leaf senescence and vascular development. Plant Biotechnol. J. 20, 1122–1139. doi: 10.1111/pbi.13794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta A., Delorean E., Bossa-Castro A., Tonnessen B., Raghavan C., Corral R., et al. (2021). Resistance and susceptibility QTL identified in a rice MAGIC population by screening with a minor-effect virulence factor from Xanthomonas oryzae pv. oryzae. Plant Biotechnol. J. 19, 51–63. doi: 10.1111/pbi.13438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo X., Wu S., Zhu Z., Liu F., Fu Y., Cai H., et al. (2017). NOG1 increases grain production in rice. Nat. Commun. 8, 1497. doi: 10.1038/s41467-017-01501-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain H., Hussain S., Khaliq A., Ashraf U., Anjum S., Men S., et al. (2018). Chilling and drought stresses in crop plants: implications, cross talk, and potential management opportunities. Front. Plant Sci. 9:393. doi: 10.3389/fpls.2018.00393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Sunohara H., Nagato Y. (2004). Developmental course of inflorescence and spikelet in rice. Breed. Sci. 54, 147–156. doi: 10.1270/jsbbs.54.147 [DOI] [Google Scholar]
- Ikeda-Kawakatsu K., Maekawa M., Izawa T., Itoh J., Nagato Y. (2012). ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. Plant J. 69, 168–180. doi: 10.1111/j.1365-313X.2011.04781.x [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K., Yasuno N., Oikawa T., Iida S., Nagato Y., Maekawa M., et al. (2009). Expression level of ABERRANT PANICLE ORGANIZATION1 determines rice inflorescence form through control of cell proliferation in the meristem. Plant Physiol. 150, 736–747. doi: 10.1104/pp.109.136739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S. (2019). Contemporary global Rice economies: structural changes of Rice production/consumption and trade. J. Nutr. Sci. Vitaminol. (Tokyo) 65, S23–S25. doi: 10.3177/jnsv.65.S23 [DOI] [PubMed] [Google Scholar]
- Jameson P., Song J. (2016). Cytokinin: a key driver of seed yield. J. Exp. Bot. 67, 593–606. doi: 10.1093/jxb/erv461 [DOI] [PubMed] [Google Scholar]
- Jiang H., Thobakgale T., Li Y., Liu L., Su Q., Cang B., et al. (2021). Construction of dominant rice population under dry cultivation by seeding rate and nitrogen rate interaction. Sci. Rep. 11, 7189. doi: 10.1038/s41598-021-86707-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang G., Xiang Y., Zhao J., Yin D., Zhao X., Zhu L., et al. (2014). Regulation of inflorescence branch development in Rice Through a novel pathway involving the Pentatricopeptide repeat protein sped1-D. Genetics 197, 1395–U1544. doi: 10.1534/genetics.114.163931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Wang Y., Xue D., Wang J., Yan M., Liu G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–U536. doi: 10.1038/ng.591 [DOI] [PubMed] [Google Scholar]
- Jin J., Hua L., Zhu Z., Tan L., Zhao X., Zhang W., et al. (2016). GAD1 encodes a secreted peptide That regulates grain number, grain length, and awn development in Rice domestication. Plant Cell 28, 2453–2463. doi: 10.1105/tpc.16.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Huang W., Gao J. P., Yang J., Shi M., Zhu M. Z., et al. (2008). Genetic control of rice plant architecture under domestication. Nat. Genet. 40, 1365–1369. doi: 10.1038/ng.247 [DOI] [PubMed] [Google Scholar]
- Ju C., Liu T., Sun C. (2021). Panicle nitrogen strategies for nitrogen-efficient Rice varieties at a moderate nitrogen application rate in the lower reaches of the Yangtze River. China Agron. Basel 11:192. doi: 10.3390/agronomy11020192 [DOI] [Google Scholar]
- Kato Y., Kamoshita A., Yamagishi J. (2008). Preflowering abortion reduces spikelet number in upland Rice (Oryza sativa L.) under water stress. Crop Sci. 48, 2389–2395. doi: 10.2135/cropsci2007.11.0627 [DOI] [Google Scholar]
- Khush G. (2005). What it will take to feed 5.0 billion Rice consumers in 2030. Plant Mol. Biol. 59, 1–6. doi: 10.1007/s11103-005-2159-5 [DOI] [PubMed] [Google Scholar]
- Kieber J., Schaller G. E. (2018). Cytokinin signaling in plant development. Development 145:dev149344. doi: 10.1242/dev.149344 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Maekawa M., Miyao A., Hirochika H., Kyozuka J. (2010). PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 51, 47–57. doi: 10.1093/pcp/pcp166, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Chujo A., Nagato Y., Shimamoto K., Kyozuka J. (2003). FRIZZY PANICLE is required to prevent the formation of axillary meristems and to establish floral meristem identity in rice spikelets. Development 130, 3841–3850. doi: 10.1242/dev.00564 [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., et al. (2007). Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445, 652–655. doi: 10.1038/nature05504 [DOI] [PubMed] [Google Scholar]
- Kwon C., Paek N. (2016). Gibberellic acid: A key Phytohormone for spikelet fertility in Rice grain production. Int. J. Mol. Sci. 17, 794. doi: 10.3390/ijms17050794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J., Konishi S., Nemoto K., Izawa T., Shimamoto K. (1998). Down-regulation of RFL, the FLO/LFY homolog of rice, accompanied with panicle branch initiation. Proc. Natl. Acad. Sci. U. S. A. 95, 1979–1982. doi: 10.1073/pnas.95.5.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Chang T., Chang S., Ouyang X., Qu M., Song Q., et al. (2018b). Systems model-guided rice yield improvements based on genes controlling source, sink, and flow. J. Integr. Plant Biol. 60, 1154–1180. doi: 10.1111/jipb.12738 [DOI] [PubMed] [Google Scholar]
- Li B., Du X., Fei Y., Wang F., Xu Y., Li X., et al. (2021a). Efficient breeding of early-maturing Rice cultivar by editing PHYC via CRISPR/Cas9. Rice (N Y) 14, 86. doi: 10.1186/s12284-021-00527-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Li X., Fu D., Wu C. (2018c). Panicle morphology mutant 1 (PMM1) determines the inflorescence architecture of rice by controlling brassinosteroid biosynthesis. BMC Plant Biol. 18:348. doi: 10.1186/s12870-018-1577-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Li X., Zhou Z., Wu P., Fang M., Pan X., et al. (2016). Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in Rice using a CRISPR/Cas9 system. Front. Plant Sci. 7:377. doi: 10.3389/fpls.2016.00377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Liu W., Tang J. Y., Chen J., Tong H., Hu B., et al. (2010). Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Res. 20, 838–849. doi: 10.1038/cr.2010.69 [DOI] [PubMed] [Google Scholar]
- Li X., Qian Q., Fu Z., Wang Y., Xiong G., Zeng D., et al. (2003). Control of tillering in rice. Nature 422, 618–621. doi: 10.1038/nature01518 [DOI] [PubMed] [Google Scholar]
- Li S., Qian Q., Fu M., Zeng L., Meng B., Kyozuka J., et al. (2009). Short panicle1 encodes a putative PTR family transporter and determines rice panicle size. Plant J. 58, 592–605. doi: 10.1111/j.1365-313X.2009.03799.x [DOI] [PubMed] [Google Scholar]
- Li G., Tang J., Zheng J., Chu C. (2021b). Exploration of rice yield potential: decoding agronomic and physiological traits. Crop J. 9, 577–589. doi: 10.1016/j.cj.2021.03.014 [DOI] [Google Scholar]
- Li X., Tao Q., Miao J., Yang Z., Gu M., Liang G., et al. (2019). Evaluation of differential qPE9-1/DEP1 protein domains in rice grain length and weight variation. Rice (N Y) 12, 5. doi: 10.1186/s12284-019-0263-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N., Xu R., Duan P., Li Y. (2018a). Control of grain size in rice. Plant Reprod. 31, 237–251. doi: 10.1007/s00497-018-0333-6 [DOI] [PubMed] [Google Scholar]
- Li G., Xu B., Zhang Y., Xu Y., Khan N., Xie J., et al. (2022). RGN1 controls grain number and shapes panicle architecture in rice. Plant Biotechnol. J. 20, 158–167. doi: 10.1111/pbi.13702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Zeng X., Li Y., Wang L., Zhuang H., Wang Y., et al. (2020). MULTI-FLORET SPIKELET 2, a MYB transcription factor, determines spikelet meristem fate and floral organ identity in Rice. Plant Physiol. 184, 988–1003. doi: 10.1104/pp.20.00743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Zhao B., Yuan D., Duan M., Qian Q., Tang L., et al. (2013). Rice zinc finger protein DST enhances grain production through controlling Gn1a/OsCKX2 expression. Proc. Natl. Acad. Sci. U. S. A. 110, 3167–3172. doi: 10.1073/pnas.1300359110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang W., Shang F., Lin Q., Lou C., Zhang J. (2014). Tillering and panicle branching genes in rice. Gene 537, 1–5. doi: 10.1016/j.gene.2013.11.058 [DOI] [PubMed] [Google Scholar]
- Liao S., Yan J., Xing H., Tu Y., Zhao H., Wang G. (2021). Genetic basis of vascular bundle variations in rice revealed by genome-wide association study. Plant Sci. 302:110715. doi: 10.1016/j.plantsci.2020.110715 [DOI] [PubMed] [Google Scholar]
- Lin L., Du M., Li S., Sun C., Wu F., Deng L., et al. (2022). Mediator complex subunit MED25 physically interacts with DST to regulate spikelet number in rice. J. Integr. Plant Biol. 64, 871–883. doi: 10.1111/jipb.13238 [DOI] [PubMed] [Google Scholar]
- Lin X., Wu F., Du X., Shi X., Liu Y., Liu S., et al. (2014). The pleiotropic SEPALLATA-like gene OsMADS34 reveals that the 'empty glumes' of rice (Oryza sativa) spikelets are in fact rudimentary lemmas. New Phytol. 202, 689–702. doi: 10.1111/nph.12657 [DOI] [PubMed] [Google Scholar]
- Lin L., Zhao Y., Liu F., Chen Q., Qi J. (2019). Narrow leaf 1 (NAL1) regulates leaf shape by affecting cell expansion in rice (Oryza sativa L.). Biochem. Biophys. Res. Commun. 516, 957–962. doi: 10.1016/j.bbrc.2019.06.142 [DOI] [PubMed] [Google Scholar]
- Liu Q., Han R., Wu K., Zhang J., Ye Y., Wang S., et al. (2018). G-protein betagamma subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 9:852. doi: 10.1038/s41467-018-03047-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Teo Z., Bi Y., Song S., Xi W., Yang X., et al. (2013). A conserved genetic pathway determines inflorescence architecture in Arabidopsis and rice. Dev. Cell 24, 612–622. doi: 10.1016/j.devcel.2013.02.013 [DOI] [PubMed] [Google Scholar]
- Lu Y., Wang J., Chen B., Mo S., Lian L., Luo Y., et al. (2021). A donor-DNA-free CRISPR/Cas-based approach to gene knock-up in rice. Nat. Plants 7, 1445–1452. doi: 10.1038/s41477-021-01019-4 [DOI] [PubMed] [Google Scholar]
- Lu Y., Xu Q., Liu Y., Yu Y., Cheng Z. Y., Zhao Y., et al. (2018). Dynamics and functional interplay of histone lysine butyrylation, crotonylation, and acetylation in rice under starvation and submergence. Genome Biol. 19:144. doi: 10.1186/s13059-018-1533-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Yu H., Xiong G., Wang J., Jiao Y., Liu G., et al. (2013). Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 25, 3743–3759. doi: 10.1105/tpc.113.113639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y., Zhou D. X., Zhao Y. (2020). Understanding epigenomics based on the rice model. Theor. Appl. Genet. 133, 1345–1363. doi: 10.1007/s00122-019-03518-7 [DOI] [PubMed] [Google Scholar]
- Luo J., Liu H., Zhou T., Gu B., Huang X., Shangguan Y., et al. (2013). An-1 encodes a basic helix-loop-helix protein that regulates awn development, grain size, and grain number in rice. Plant Cell 25, 3360–3376. doi: 10.1105/tpc.113.113589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H., Sun S., Yao J., Wang C., Yu S., Xu C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U. S. A. 107, 19579–19584. doi: 10.1073/pnas.1014419107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Ikeda M., Matsubara A., Song X. J., Ito M., Asano K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592 [DOI] [PubMed] [Google Scholar]
- Moeinizade S., Kusmec A., Hu G., Wang L., Schnable P. S. (2020). Multi-trait genomic selection methods for crop improvement. Genetics 215, 931–945. doi: 10.1534/genetics.120.303305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd Saad N., Neik T., Thomas W., Amas J., Cantila A., Craig R., et al. (2022). Advancing designer crops for climate resilience through an integrated genomics approach. Curr. Opin. Plant Biol. 67:102220. doi: 10.1016/j.pbi.2022.102220 [DOI] [PubMed] [Google Scholar]
- Muthayya S., Sugimoto J. D., Montgomery S., Maberly G. F. (2014). An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 1324, 7–14. doi: 10.1111/nyas.12540 [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Shimamoto K., Kyozuka J. (2002). Overexpression of RCN1 and RCN2, rice TERMINAL FLOWER 1/CENTRORADIALIS homologs, confers delay of phase transition and altered panicle morphology in rice. Plant J. 29, 743–750. doi: 10.1046/j.1365-313x.2002.01255.x [DOI] [PubMed] [Google Scholar]
- Oikawa T., Kyozuka J. (2009). Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in Rice. Plant Cell 21, 1095–1108. doi: 10.1105/tpc.108.065425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Kim E., Jang Y., Kim K. (2021). Screening and identification of genes affecting grain quality and spikelet fertility during high-temperature treatment in grain filling stage of rice. BMC Plant Biol. 21, 263. doi: 10.1186/s12870-021-03056-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfus-Barbeoch L., Jones A. M., Assmann S. M. (2004). Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Curr. Opin. Plant Biol. 7, 719–731. doi: 10.1016/j.pbi.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Piao R., Jiang W., Ham T. H., Choi M. S., Qiao Y., Chu S. H., et al. (2009). Map-based cloning of the ERECT PANICLE 3 gene in rice. Theor. Appl. Genet. 119, 1497–1506. doi: 10.1007/s00122-009-1151-x [DOI] [PubMed] [Google Scholar]
- Qi J., Qian Q., Bu Q., Li S., Chen Q., Sun J., et al. (2008). Mutation of the rice Narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 147, 1947–1959. doi: 10.1104/pp.108.118778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao Y., Piao R., Shi J., Lee S. I., Jiang W., Kim B. K., et al. (2011). Fine mapping and candidate gene analysis of dense and erect panicle 3, DEP3, which confers high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 122, 1439–1449. doi: 10.1007/s00122-011-1543-6 [DOI] [PubMed] [Google Scholar]
- Rashotte A. (2021). The evolution of cytokinin signaling and its role in development before angiosperms. Semin. Cell Dev. Biol. 109, 31–38. doi: 10.1016/j.semcdb.2020.06.010 [DOI] [PubMed] [Google Scholar]
- Rebolledo M. C., Pena A. L., Duitama J., Cruz D. F., Dingkuhn M., Grenier C., et al. (2016). Combining image analysis, genome wide association studies and different field trials to reveal stable genetic regions related to panicle architecture and the number of Spikelets per panicle in Rice. Front. Plant Sci. 7:1384. doi: 10.3389/fpls.2016.01384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Li Y., He G., Qian Q. (2020). Multifloret spikelet improves rice yield. New Phytol. 225, 2301–2306. doi: 10.1111/nph.16303 [DOI] [PubMed] [Google Scholar]
- Ren D., Li Y., Zhao F., Sang X., Shi J., Wang N., et al. (2013). MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines SPIKELET meristem fate and sterile lemma identity in rice. Plant Physiol. 162, 872–884. doi: 10.1104/pp.113.216044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D., Xu Q., Qiu Z., Cui Y., Zhou T., Zeng D., et al. (2019). FON4 prevents the multi-floret spikelet in rice. Plant Biotechnol. J. 17, 1007–1009. doi: 10.1111/pbi.13083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Shil P. (2020). Assessment of genetic heritability in Rice breeding lines based on morphological traits and caryopsis ultrastructure. Sci. Rep. 10:7830. doi: 10.1038/s41598-020-63976-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott M., Ladejobi O., Amer S., Bentley A., Biernaskie J., Boden S., et al. (2020). Multi-parent populations in crops: a toolbox integrating genomics and genetic mapping with breeding. Heredity (Edinb) 125, 396–416. doi: 10.1038/s41437-020-0336-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhar S., Kumar J., Mohanty S., Mohanty N., Panda R. S., Das S., et al. (2021). Identification of novel QTLs for grain fertility and associated traits to decipher poor grain filling of basal spikelets in dense panicle rice. Sci. Rep. 11:13617. doi: 10.1038/s41598-021-93134-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Feugier F. G., Song X. J., Ashikari M., Nakamura H., Ishiyama K., et al. (2015). A mathematical model of phloem sucrose transport as a new tool for designing Rice panicle structure for high grain yield. Plant Cell Physiol. 56, 605–619. doi: 10.1093/pcp/pcu191 [DOI] [PubMed] [Google Scholar]
- Shao G., Lu Z., Xiong J., Wang B., Jing Y., Meng X., et al. (2019). Tiller bud formation regulators MOC1 and MOC3 cooperatively promote tiller bud outgrowth by activating FON1 expression in Rice. Mol. Plant 12, 1090–1102. doi: 10.1016/j.molp.2019.04.008 [DOI] [PubMed] [Google Scholar]
- Sheng P., Wu F., Tan J., Zhang H., Ma W., Chen L., et al. (2016). A CONSTANS-like transcriptional activator, OsCOL13, functions as a negative regulator of flowering downstream of OsphyB and upstream of Ehd1 in rice. Plant Mol. Biol. 92, 209–222. doi: 10.1007/s11103-016-0506-3 [DOI] [PubMed] [Google Scholar]
- Song X., Huang W., Shi M., Zhu M., Lin H. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N., Pasion E., Kohli A. (2021). Idealizing inflorescence architecture to enhance rice yield potential for feeding nine billion people in 2050. Mol. Plant 14, 861–863. doi: 10.1016/j.molp.2021.05.003 [DOI] [PubMed] [Google Scholar]
- Sreenivasulu N., Schnurbusch T. (2012). A genetic playground for enhancing grain number in cereals. Trends Plant Sci. 17, 91–101. doi: 10.1016/j.tplants.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Su S., Hong J., Chen X., Zhang C., Chen M., Luo Z., et al. (2021). Gibberellins orchestrate panicle architecture mediated by DELLA-KNOX signalling in rice. Plant Biotechnol. J. 19, 2304–2318. doi: 10.1111/pbi.13661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Qian Q., Wu K., Luo J., Wang S., Zhang C., et al. (2014). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nat. Genet. 46, 652–656. doi: 10.1038/ng.2958 [DOI] [PubMed] [Google Scholar]
- Tabuchi H., Zhang Y., Hattori S., Omae M., Shimizu-Sato S., Oikawa T., et al. (2011). LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. Plant Cell 23, 3276–3287. doi: 10.1105/tpc.111.088765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L., Li X., Liu F., Sun X., Li C., Zhu Z., et al. (2008). Control of a key transition from prostrate to erect growth in rice domestication. Nat. Genet. 40, 1360–1364. doi: 10.1038/ng.197 [DOI] [PubMed] [Google Scholar]
- Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17, 776–790. doi: 10.1105/tpc.104.024950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka W., Pautler M., Jackson D., Hirano H. Y. (2013). Grass meristems II: inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 54, 313–324. doi: 10.1093/pcp/pct016 [DOI] [PubMed] [Google Scholar]
- Terao T., Nagata K., Morino K., Hirose T. (2010). A gene controlling the number of primary rachis branches also controls the vascular bundle formation and hence is responsible to increase the harvest index and grain yield in rice. Theor. Appl. Genet. 120, 875–893. doi: 10.1007/s00122-009-1218-8 [DOI] [PubMed] [Google Scholar]
- Tu B., Tao Z., Wang S., Zhou L., Zheng L., Zhang C., et al. (2022). Loss of Gn1a/OsCKX2 confers heavy-panicle rice with excellent lodging resistance. J. Integr. Plant Biol. 64, 23–38. doi: 10.1111/jipb.13185 [DOI] [PubMed] [Google Scholar]
- Tuhina-Khatun M., Hanafi M. M., Rafii Yusop M., Wong M. Y., Salleh F. M., Ferdous J. (2015). Genetic variation, heritability, and diversity analysis of upland Rice (Oryza sativa L.) genotypes based on quantitative traits. Biomed. Res. Int. 2015:290861. doi: 10.1155/2015/290861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasa Y., Kawakatsu T., Harada T., Takaiwa F. (2018). Transgene-independent heredity of RdDM-mediated transcriptional gene silencing of endogenous genes in rice. Plant Biotechnol. J. 16, 2007–2015. doi: 10.1111/pbi.12934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Han T., Song Q., Ye W., Song X., Chu J., et al. (2020a). The Rice circadian clock regulates tiller growth and panicle development through Strigolactone signaling and sugar sensing. Plant Cell 32, 3124–3138. doi: 10.1105/tpc.20.00289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li J. (2005). The plant architecture of rice (Oryza sativa). Plant Mol. Biol. 59, 75–84. doi: 10.1007/s11103-004-4038-x [DOI] [PubMed] [Google Scholar]
- Wang X., Li L., Yang Z., Zheng X., Yu S., Xu C., et al. (2017). Predicting rice hybrid performance using univariate and multivariate GBLUP models based on North Carolina mating design II. Heredity 118, 302–310. doi: 10.1038/hdy.2016.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Lu Y., Guo Z., Ding Y., Ding C. (2020b). RICE CENTRORADIALIS 1, a TFL1-like gene, responses to drought stress and regulates rice flowering transition. Rice (N Y) 13, 70. doi: 10.1186/s12284-020-00430-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Shen C., Xu Q., Zafar S., Du B., Xing D. (2022b). Grain yield, nitrogen use efficiency and antioxidant enzymes of Rice under different fertilizer N inputs and planting density. Agron. Basel 12:430. doi: 10.3390/agronomy12020430 [DOI] [Google Scholar]
- Wang H., Tong X., Tang L., Wang Y., Zhao J., Li Z., et al. (2022a). RLB (RICE LATERAL BRANCH) recruits PRC2-mediated H3K27 tri-methylation on OsCKX4 to regulate lateral branching. Plant Physiol. 188, 460–476. doi: 10.1093/plphys/kiab494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Xu F., Yuan W., Zhang D., Liu J., Sun L., et al. (2021). Rice G protein gamma subunit qPE9-1 modulates root elongation for phosphorus uptake by involving 14-3-3 protein OsGF14b and plasma membrane H(+) -ATPase. Plant J. 107, 1603–1615. doi: 10.1111/tpj.15402 [DOI] [PubMed] [Google Scholar]
- Wang Y., Zhai L., Chen K., Shen C., Liang Y., Wang C., et al. (2020c). Natural sequence variations and combinations of GNP1 and NAL1 determine the grain number per panicle in Rice. Rice (N Y) 13:14. doi: 10.1186/s12284-020-00374-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Zou T., He Z., Yuan G., Luo T., Zhu J., et al. (2019). GRAIN LENGTH AND AWN 1 negatively regulates grain size in rice. J. Integr. Plant Biol. 61, 1036–1042. doi: 10.1111/jipb.12736 [DOI] [PubMed] [Google Scholar]
- Weng X., Wang L., Wang J., Hu Y., Du H., Xu C., et al. (2014). Grain number, plant height, and heading date7 is a central regulator of growth, development, and stress response. Plant Physiol. 164, 735–747. doi: 10.1104/pp.113.231308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Fu Y., Zhao S., Gu P., Zhu Z., Sun C., et al. (2016). CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol. J. 14, 377–386. doi: 10.1111/pbi.12391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Liang Y., Gao H., Wang J., Zhao Y., Hua L., et al. (2021). Enhancing rice grain production by manipulating the naturally evolved cis-regulatory element-containing inverted repeat sequence of OsREM20. Mol. Plant 14, 997–1011. doi: 10.1016/j.molp.2021.03.016 [DOI] [PubMed] [Google Scholar]
- Wuriyanghan H., Zhang B., Cao W. H., Ma B., Lei G., Liu Y. F., et al. (2009). The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 21, 1473–1494. doi: 10.1105/tpc.108.065391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y., Liu H., Wu L., Warburton M., Yan J. (2017). Genome-wide association studies in maize: praise and stargaze. Mol. Plant 10, 359–374. doi: 10.1016/j.molp.2016.12.008 [DOI] [PubMed] [Google Scholar]
- Xiao N., Pan C. H., Li Y. H., Wu Y. Y., Cai Y., Lu Y., et al. (2021). Genomic insight into balancing high yield, good quality, and blast resistance of japonica rice. Genome Biol. 22:283. doi: 10.1186/s13059-021-02488-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Zhang Q. (2010). Genetic and molecular bases of rice yield. Annu. Rev. Plant Biol. 61, 421–442. doi: 10.1146/annurev-arplant-042809-112209 [DOI] [PubMed] [Google Scholar]
- Xu Q., Liu T., Bi W., Wang Y., Xu H., Tang L., et al. (2015b). Different effects of DEP1 on vascular bundle-and panicle-related traits under indica and japonica genetic backgrounds. Mol. Breed. 35, 173. doi: 10.1007/s11032-015-0364-8 [DOI] [Google Scholar]
- Xu Y., Liu X., Fu J., Wang H., Wang J., Huang C., et al. (2020). Enhancing genetic gain through genomic selection: From livestock to plants. Plant Commun. 1:100005. doi: 10.1016/j.xplc.2019.100005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Ma K. X., Zhao Y., Wang X., Zhou K., Yu G. N., et al. (2021a). Genomic selection: A breakthrough technology in rice breeding. Crop J. 9, 669–677. doi: 10.1016/j.cj.2021.03.008 [DOI] [Google Scholar]
- Xu R., Sun C. Q. (2021). What happened during domestication of wild to cultivated rice. Crop J. 9, 564–576. doi: 10.1016/j.cj.2021.02.005 [DOI] [Google Scholar]
- Xu F., Wang K., Yuan W., Xu W., Shuang L., Kronzucker H. J., et al. (2019). Overexpression of rice aquaporin OsPIP1;2 improves yield by enhancing mesophyll CO2 conductance and phloem sucrose transport. J. Exp. Bot. 70, 671–681. doi: 10.1093/jxb/ery386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wang Y., Zhang F., Wu Y., Zheng T., Wang Y., et al. (2015a). SS1 (NAL1)- and SS2-mediated genetic networks underlying source-sink and yield traits in Rice (Oryza sativa L.). PLoS One 10:e0132060. doi: 10.1371/journal.pone.0132060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R., Yu H., Wang J., Duan P., Zhang B., Li J., et al. (2018). A mitogen-activated protein kinase phosphatase influences grain size and weight in rice. Plant J. 95, 937–946. doi: 10.1111/tpj.13971 [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhao Y., Wang X., Ma Y., Li P. C., Yang Z. F., et al. (2021b). Incorporation of parental phenotypic data into multi-omic models improves prediction of yield-related traits in hybrid rice. Plant Biotechnol. J. 19, 261–272. doi: 10.1111/pbi.13458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Zhao M., Zhang Q., Xu Z., Xu Q. (2016). The DENSE AND ERECT PANICLE 1 (DEP1) gene offering the potential in the breeding of high-yielding rice. Breed. Sci. 66, 659–667. doi: 10.1270/jsbbs.16120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Xing Y., Weng X., Zhao Y., Tang W., Wang L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143 [DOI] [PubMed] [Google Scholar]
- Yan W., Liu H., Zhou X., Li Q., Zhang J., Lu L., et al. (2013). Natural variation in Ghd7.1 plays an important role in grain yield and adaptation in rice. Cell Res. 23, 969–971. doi: 10.1038/cr.2013.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W., Wang P., Chen H., Zhou H., Li Q., Wang C., et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. doi: 10.1093/mp/ssq070 [DOI] [PubMed] [Google Scholar]
- Yan Y., Wei M., Li Y., Tao H., Wu H., Chen Z., et al. (2021). MiR529a controls plant height, tiller number, panicle architecture and grain size by regulating SPL target genes in rice (Oryza sativa L.). Plant Sci. 302:110728. doi: 10.1016/j.plantsci.2020.110728 [DOI] [PubMed] [Google Scholar]
- Yan C., Zhou J., Yan S., Chen F., Yeboah M., Tang S., et al. (2007). Identification and characterization of a major QTL responsible for erect panicle trait in japonica rice (Oryza sativa L.). Theor. Appl. Genet. 115, 1093–1100. doi: 10.1007/s00122-007-0635-9 [DOI] [PubMed] [Google Scholar]
- Yang X., Wang B., Chen L., Li P., Cao C. (2019). The different influences of drought stress at the flowering stage on rice physiological traits, grain yield, and quality. Sci. Rep. 9, 3742. doi: 10.1038/s41598-019-40161-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Zhou Y., Huang J., Hu Y., Zhang E., Xie Z., et al. (2015). Ancient horizontal transfer of transaldolase-like protein gene and its role in plant vascular development. New Phytol. 206, 807–816. doi: 10.1111/nph.13183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Zhang Z., Zeng S., Tian C., Peng J., Li M., et al. (2011). Introgression of qPE9-1 allele, conferring the panicle erectness, leads to the decrease of grain yield per plant in japonica rice (Oryza sativa L.). J. Genet. Genom. 38, 217–223. doi: 10.1016/j.jgg.2011.03.011 [DOI] [PubMed] [Google Scholar]
- Yin C., Zhu Y., Li X., Lin Y. (2021). Molecular and genetic aspects of grain number determination in Rice (Oryza sativa L.). Int. J. Mol. Sci. 22, 728. doi: 10.3390/ijms22020728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H., Qin P., Hu L., Zhan S., Wang S., Gao P., et al. (2019). OsSPL18 controls grain weight and grain number in rice. J. Genet. Genom. 46, 41–51. doi: 10.1016/j.jgg.2019.01.003 [DOI] [PubMed] [Google Scholar]
- Yuan H., Xu Z. Y., Tan X. Q., Gao P., Jin M. Y., Song W. C., et al. (2021). A natural allele of TAW1 contributes to high grain number and grain yield in rice. Crop J. 9, 1060–1069. doi: 10.1016/j.cj.2020.11.004 [DOI] [Google Scholar]
- Zaw H., Raghavan C., Pocsedio A., Swamy B. P. M., Jubay M. L., Singh R. K., et al. (2019). Exploring genetic architecture of grain yield and quality traits in a 16-way indica by japonica rice MAGIC global population. Sci. Rep. 9, 19605. doi: 10.1038/s41598-019-55357-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y., Zhang Y., Xiang J., Uphoff N. T., Pan X., Zhu D. (2017). Effects of low temperature stress on spikelet-related parameters during Anthesis in Indica-japonica hybrid Rice. Front. Plant Sci. 8:1350. doi: 10.3389/fpls.2017.01350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai L., Zheng T., Wang X., Wang Y., Chen K., Wang S., et al. (2018). QTL mapping and candidate gene analysis of peduncle vascular bundle related traits in rice by genome-wide association study. Rice (N Y) 11, 13. doi: 10.1186/s12284-018-0204-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan P., Wei X., Xiao Z., Wang X., Ma S., Lin S., et al. (2021). GW10, a member of P450 subfamily regulates grain size and grain number in rice. Theor. Appl. Genet. 134, 3941–3950. doi: 10.1007/s00122-021-03939-3 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Li J., Tang Z., Sun X., Zhang H., Yu J., et al. (2018). Gnp4/LAX2, a RAWUL protein, interferes with the OsIAA3-OsARF25 interaction to regulate grain length via the auxin signaling pathway in rice. J. Exp. Bot. 69, 4723–4737. doi: 10.1093/jxb/ery256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Peng K., Cui F., Wang D., Zhao J., Zhang Y., et al. (2021a). Cytokinin oxidase/dehydrogenase OsCKX11 coordinates source and sink relationship in rice by simultaneous regulation of leaf senescence and grain number. Plant Biotechnol. J. 19, 335–350. doi: 10.1111/pbi.13467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Qin P., Peng Y., Ma B., Hu J., Fan S., et al. (2019a). A single nucleotide substitution at 5'-UTR of GSN1 represses its translation and leads to an increase of grain length in rice. J. Genet. Genom. 46, 105–108. doi: 10.1016/j.jgg.2019.02.001 [DOI] [PubMed] [Google Scholar]
- Zhang Z., Sun X., Ma X., Xu B., Zhao Y., Ma Z., et al. (2021b). GNP6, a novel allele of MOC1, regulates panicle and tiller development in rice. Crop J. 9, 57–67. doi: 10.1016/j.cj.2020.04.011 [DOI] [Google Scholar]
- Zhang L., Yu H., Ma B., Liu G., Wang J., Wang J., et al. (2017). A natural tandem array alleviates epigenetic repression of IPA1 and leads to superior yielding rice. Nat. Commun. 8, 14789. doi: 10.1038/ncomms14789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhang Z., Sun X., Zhu X., Li B., Li J., et al. (2019b). Natural alleles of GLA for grain length and awn development were differently domesticated in rice subspecies japonica and indica. Plant Biotechnol. J. 17, 1547–1559. doi: 10.1111/pbi.13080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Zhou Y., Yin J., Yan X., Lin S., Xu W., et al. (2015). Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation. J. Exp. Bot. 66, 6371–6384. doi: 10.1093/jxb/erv350 [DOI] [PubMed] [Google Scholar]
- Zhao Q., Feng Q., Lu H., Li Y., Wang A., Tian Q., et al. (2018). Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 50, 278–284. doi: 10.1038/s41588-018-0041-z [DOI] [PubMed] [Google Scholar]
- Zhao L., Tan L., Zhu Z., Xiao L., Xie D., Sun C. (2015). PAY1 improves plant architecture and enhances grain yield in rice. Plant J. 83, 528–536. doi: 10.1111/tpj.12905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li Z., Xiao G., Zhai M., Pan X., Huang R., et al. (2020). CYP71D8L is a key regulator involved in growth and stress responses by mediating gibberellin homeostasis in rice. J. Exp. Bot. 71, 1160–1170. doi: 10.1093/jxb/erz491 [DOI] [PubMed] [Google Scholar]
- Zhou Y., Tao Y., Yuan Y., Zhang Y., Miao J., Zhang R., et al. (2018). Characterisation of a novel quantitative trait locus, GN4-1, for grain number and yield in rice (Oryza sativa L.). Theor. Appl. Genet. 131, 637–648. doi: 10.1007/s00122-017-3025-y [DOI] [PubMed] [Google Scholar]
- Zhou Y., Tao Y., Zhu J., Miao J., Liu J., Liu Y., et al. (2017). GNS4, a novel allele of DWARF11, regulates grain number and grain size in a high-yield rice variety. Rice (N Y) 10, 34. doi: 10.1186/s12284-017-0171-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhu J., Li Z., Yi C., Liu J., Zhang H., et al. (2009). Deletion in a quantitative trait gene qPE9-1 associated with panicle erectness improves plant architecture during rice domestication. Genetics 183, 315–324. doi: 10.1534/genetics.109.102681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Li C., Gao C. (2020). Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 21, 661–677. doi: 10.1038/s41580-020-00288-9 [DOI] [PubMed] [Google Scholar]
- Zhu K., Tang D., Yan C., Chi Z., Yu H., Chen J., et al. (2010). ERECT PANICLE2 encodes a novel protein that regulates PANICLE erectness in Indica Rice. Genetics 184, 343–U354. doi: 10.1534/genetics.109.112045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J., Li J. (2014). Molecular genetic dissection of quantitative trait loci regulating rice grain size. Annu. Rev. Genet. 48, 99–118. doi: 10.1146/annurev-genet-120213-092138 [DOI] [PubMed] [Google Scholar]