Abstract
The trehalose content in Saccharomyces cerevisiae can be significantly manipulated by including trehalose at an appropriate level in the growth medium. Its uptake is largely dependent on the expression of AGT1, which encodes an α-glucoside transporter. The trehalose found in a tps1 mutant of trehalose synthase may therefore largely reflect its uptake from the enriched medium that was employed.
Most work on trehalose metabolism in yeast concerns factors governing its endogenous level (6, 14, 15). Trehalose synthase is a multimeric protein composed of four subunits encoded by TPS1, TPS2, TSL1, and TPS3 (2, 18), of which only Tps1p catalyzing the formation of trehalose-6-phosphate from UDP-Glc and glucose-6-phosphate is essential for growth on rapidly fermentable carbon sources like glucose and fructose (8, 22). The molecular mechanism underlying this defect is not yet understood (21). The deletion of TPS1 in principle results in the loss of trehalose accumulation. However, the existence of another functional pathway for trehalose synthesis in yeast has been postulated, based on data showing that trehalose accumulation is somehow related to maltose metabolism or constitutive maltose gene expression (5, 15) and on one report of a putative ADP-Glc-dependent trehalose synthase activity (17). However, most demonstrations in wild-type and tps1 mutant strains have employed an enriched medium such as yeast extract-peptone (YEP), and our analysis of yeast extract from various commercial sources (Difco and BIOKAR Diagnostic) by using ionic exchange chromatography with pulsed amperometric detection (4) shows it to contain ca. 1.5% trehalose by weight, which would result in a yield of 150 μg of free trehalose per ml of YEP medium. Since Saccharomyces cerevisiae is known to show both a high- and a low-affinity trehalose uptake (13, 19, 20), we speculated that the trehalose accumulation of tps1 mutant cells might be related to uptake from the medium, perhaps involving AGT1, a maltose-controlled gene encoding a transporter with broad specificity for α-glucosides, including trehalose (10).
Yeast mutant strains used in this work were derived from the prototrophic CEN.PK113-7D strain (the kind gift of K.-D. Entian and P. Kötter, Frankfurt, Germany), which possesses a MAL2-8c dominant mutation. Culturing was carried out at 30°C in YEP medium (10 g of yeast extract and 10 g of Bacto Peptone per liter) or in mineral medium (MIN) prepared according to the method described in reference 23 and buffered at pH 5.8 by the addition of 10 g of succinic acid and 6 g of NaOH per liter. The carbon source was added to the media at a final concentration of 10 g · liter−1, and auxotrophic requirements, when required, were added at 100 mg · liter−1. The deletion of TPS1 and AGT1 was performed according to the PCR and short homolog fragment procedure of Wach et al. (24) by using the pUG6 plasmid bearing the loxP-kanMX4-loxP module (9) for TPS1 and pUG6lacZ bearing lacZ-kanMX4 (3) for AGT1. The oligonucleotides used to construct the deletion cassette were as follows: d-TPS1 (5′-ATGACTACGGATAA CGCTAAGGCGCAACTGACCTCGTCTTCAGCTGAAGC TTCGTACGC-3′), containing the sequence from nucleotide +1 (A of the start codon) to +40 of the TPS1 open reading frame (ORF), and f-TPS1 (5′-TCAGTTTTTGGTGGCAGA GGAGCTTGTTGAGCTGATGATGCATAGGCCACTAG TGGATCTG-3′), containing the complementary sequence from nucleotide +1445 to +1488 of the TPS1 ORF, and S1-LAGT1 (5′-ATGAAAAATATCATTTCATTGGTAAGCAA GAAGAAGGCTGCCTCAAAATTCGTACGCTGCAGGT CGAC-3′), containing the sequence from nucleotide +1 (A of the start codon) to +48 of the AGT1 ORF, and S2-AGT1 (5′-TAATTCTCGCTGTTTTATGCTTGAGGACTGACT GATACTCTCATCAGCGCATAGGCCACTAGTGGATC TG-3′), containing the complementary sequence from nucleotide +1783 to +1830 of the AGT1 ORF.
Amplifications were carried out with Expand high-fidelity polymerase (Boehringer, Mannheim, Germany) according to standard procedures, and the amplified fragments (5 μg) were used for gene disruption as described in reference 9. For the deletion of ATH1, a 4.4-kb fragment bearing the full gene was amplified by using a pair of primers (ATH1-250 [5′-CGTATCACGACAAACCAACAGCC-3] and ATH1-500 [5′-CAAACCCTACTGACGAGAGAAG]) and genomic DNA from CEN.PK113-7D as a template. The PCR fragment cloned into the pGEM-T vector (Promega) was digested with EcoRV-HpaI, which was replaced by a 1.5-kb EcoRV-SmaI kanMX4 fragment from pFAkanMX4 (24). This construct was cut with ScaII and SpeI, and the 3.45-kb fragment was gel purified and used for transformation (7). For the deletion of NTH1, a 1.2-kb StuI-SnaBI of plasmid pTZ18RNTH1 (12) was replaced by a 1.4-kb EcoRV-SmaI kanMX module from pFAKanMX4 to yield pΔATH1. This plasmid was cut with PvuII-AlfII, and the 3.6-kb fragment was used for transformation. Gene disruption was verified either by PCR (24) or by Southern blotting. The Δagt1 Δtps1 and Δnth1 Δath1 double mutants were obtained by the crossing of haploid mutant strains. The correct Δagt1 Δtps1 mutant was characterized by its inability to grow on YEP-dextrose and its ability to turn dark blue on maltose due to the expression of AGT1-lacZ. The correct Δnth1 Δath1 mutant was obtained from a typical tetrad as the only one lacking both acid and neutral trehalase activity. The level of intracellular trehalose was determined by the procedure described previously (16).
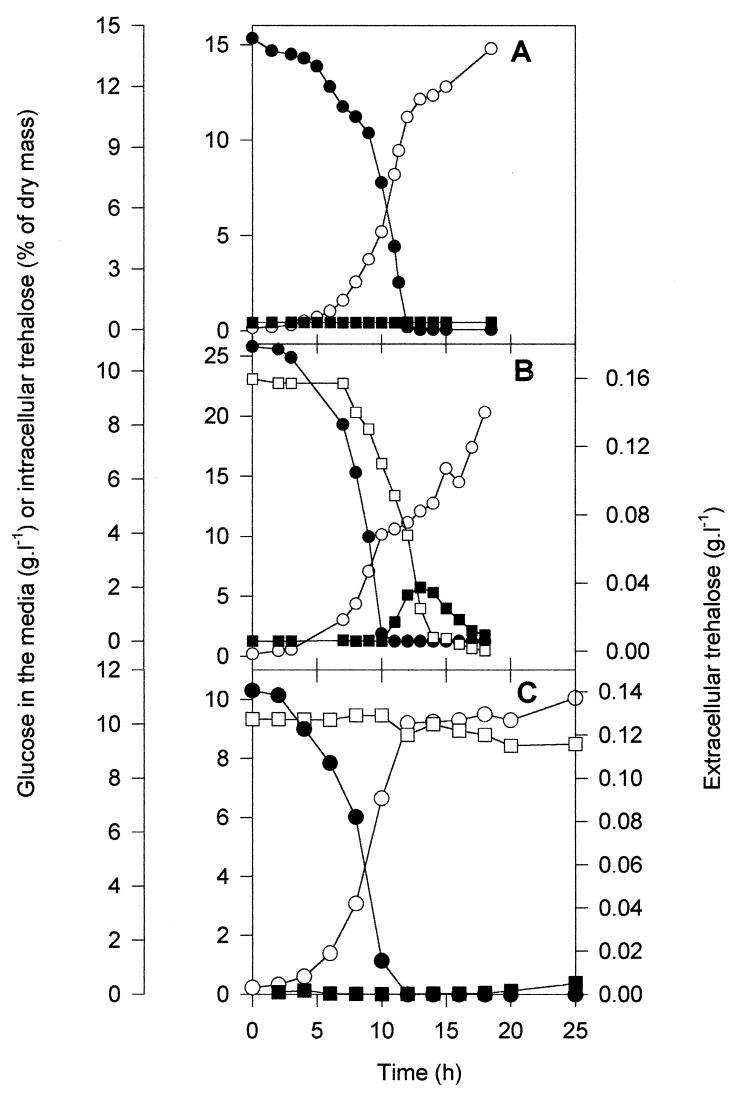
In agreement with the idea that the trehalose content can be manipulated by including trehalose in the growth medium, Fig. 1A shows that although there was a barely detectable level of trehalose in a wild-type strain (with a MALc mutation) cultivated on galactose-MIN, this level was significantly increased when the medium also contained 0.15 g of trehalose · liter−1 (Fig. 1B), but this accumulation was prevented by the deletion of AGT1 (Fig. 1C). Table 1 shows that the maximal content of trehalose during the growth of a wild-type strain on YEP-galactose was about 10% of the dry mass and that this level dropped to 3.2% in the tps1 isogenic strain. By contrast, in galactose-MIN, the content of the disaccharide in the wild-type strain reached ca. 1% and was not detectable in the tps1 mutant. Both strains, however, were able to accumulate about 2% trehalose when the medium was initially supplemented with 0.15 g of trehalose · liter−1.
FIG. 1.
Patterns of extracellular and intracellular trehalose levels during growth of wild-type CEN.PK113-7D (A and B) and RWY13 (Δagt1 [C]) on galactose-MIN without (A) or with 0.15 g of trehalose · liter−1 (B and C). Shown are the optical densities at 660 nm (○), the level of glucose in the medium (□), the level of extracellular trehalose (●), and the level of intracellular trehalose (■).
TABLE 1.
Levels of trehalose in wild-type and isogenic mutant strains with AGT1, TPS1, NTH1, or ATH1 deleted during growth on different media
| Growth medium | % Trehalose in
dry mass of yeast with genotypea
|
||||
|---|---|---|---|---|---|
| Wild type | AGT1 tps1 | agt1 TPS1 | agt1 tps1 | nth1 ath1 AGT1 | |
| YEP + galactose | 9.50 | 3.2 | ND | 0.0 | ND |
| MIN + galactose | 1.20 | BD | ND | 0.0 | ND |
| + trehalose (0.15 g · liter−1)b | 2.0 | 1.80 | 0.40 | 0.0 | ND |
| + trehalose (0.1 g · liter−1) | 1.28 | ND | 0.38 | 0.0 | ND |
| + trehalose (1.0 g · liter−1) | 8.80 | ND | 0.41 | 0.01 | ND |
| + trehalose (10 g · liter−1) | 12.9 | ND | 1.34 | 0.91 | ND |
| MIN + glucose | 0.39 | ND | 0.0 | ND | 2.0 |
| + trehalose (0.1 g · liter−1) | 1.28 | ND | 0.0 | ND | 3.80 |
| + trehalose (1.0 g · liter−1) | 8.80 | ND | ND | ND | 9.50 |
| + trehalose (4.0 g · liter−1) | 9.70 | ND | ND | ND | 11.10 |
Although the intracellular trehalose level was determined during growth, the only values reported were the maximal ones attained for each growth condition. BD, below detection; ND, not done.
Value of trehalose contained in YEP medium.
The use of varying exogenous trehalose concentrations shows that with 1 g · liter−1 (ca. 3 mM), a 9% intracellular concentration of trehalose can be attained even in galactose-MIN. For a cell sap of 2.4 ml per g of cell dry mass (1, 11), this content corresponds to 110 mM, suggesting an active uptake of trehalose. The accumulation of trehalose was largely abolished by the deletion of AGT1, with a residual internal trehalose concentration probably contributed by trehalose synthase activity, as indicated by its loss in the agt1 tps1 double mutant. However, at a much higher exogenous trehalose concentration (10 g · liter−1 or 30 mM), its endogenous level was on the order of 1% in both the single (agt1) and the double (agt1 tps1) mutants. This latter result is in agreement with previous work showing the existence of a nonconcentrative low-level Km uptake system for trehalose (13, 19). Since acid and neutral trehalases counteract the endogenous formation of trehalose (14), their influence on intracellular trehalose content was investigated by deleting the corresponding NTH1 and ATH1 genes. It is shown in Table 1 that the content of trehalose determined at the end of growth on glucose-MIN increased from 0.39% in a wild-type strain to 2% in cells lacking both trehalases. However, their absence had much less influence on the level of intracellular trehalose as its external concentration was increased, suggesting that yeast cells have a limiting capacity to store a maximum of 12 to 13% trehalose. Taken together, these results conclusively demonstrate that the accumulation of trehalose in yeast is mediated by at least two pathways: the first is via the endogenous UDP-Glc-linked trehalose synthase complex and the second is the uptake of exogenous trehalose via the high-affinity α-glucoside transporter encoded by AGT1.
Acknowledgments
Nathalie Dallies is acknowledged for her help in performing trehalose level determinations by high-performance liquid chromatography. We also thank the anonymous reviewer for his help in improving this paper.
This work was supported in part by the Commission of the European Union (Programme Cell Factories grant BIO.CT95.132 to J.F.). L.P. holds a fellowship from the Ministère de la Recherche et de l’Enseignement Supérieur (MENESR).
REFERENCES
- 1.Alexandre H, Plourde L, Charpentier C, François J. Lack of correlation between trehalose accumulation, cell viability and intracellular acidification as induced by various stresses in Saccharomyces cerevisiae. Microbiology. 1998;144:1103–1111. doi: 10.1099/00221287-144-4-1103. [DOI] [PubMed] [Google Scholar]
- 2.Bell W, Sun W, Hohmann S, Wera S, Reinders A, De Virgilio C, Wiemken A, Thevelein J M. Composition and functional analysis of the Saccharomyces cerevisiaetrehalose synthase complex. J Biol Chem. 1998;273:3311–3319. doi: 10.1074/jbc.273.50.33311. [DOI] [PubMed] [Google Scholar]
- 3.Boles E, de Jong-Gubbels P, Pronk J T. Identification and characterization of MAE1, the Saccharomyces cerevisiaestructural gene encoding mitochondrial malic enzyme. J Bacteriol. 1998;180:2875–2882. doi: 10.1128/jb.180.11.2875-2882.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dallies N, François J, Paquet V. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast. 1997;14:1297–1306. doi: 10.1002/(SICI)1097-0061(1998100)14:14<1297::AID-YEA310>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 5.Ferreira J C, Thevelein J M, Hohmann S, Paschoalin V M F, Tugo L C, Panek A D. Trehalose accumulation in mutants of Saccharomyces cerevisiaedeleted in the UDP-dependent trehalose synthase-phosphatase complex. Biochim Biophys Acta. 1997;1335:40–50. doi: 10.1016/s0304-4165(96)00127-4. [DOI] [PubMed] [Google Scholar]
- 6.François J, Blazquez M A, Arino J, Gancedo C. Storage carbohydrates in the yeast Saccharomyces cerevisiae. In: Zimmermann F K, Entian K-D, editors. Yeast sugar metabolism. Lancaster, Pa: Technomics Publishing Co., Inc.; 1997. pp. 285–311. [Google Scholar]
- 7.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 8.Gonzales M I, Stucka R, Blazquez M A, Feldmann H, Gancedo C. Molecular cloning of CIF1, a yeast gene necessary for growth on glucose. Yeast. 1992;8:183–192. doi: 10.1002/yea.320080304. [DOI] [PubMed] [Google Scholar]
- 9.Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann J H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;13:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han E-Y, Cotty F, Sottas C, Jiang H, Michels C A. Characterization of AGT1 encoding a general α-glucoside transporter from Saccharomyces cerevisiae. Mol Microbiol. 1995;17:1093–1107. doi: 10.1111/j.1365-2958.1995.mmi_17061093.x. [DOI] [PubMed] [Google Scholar]
- 11.Hottiger T, De Virgilio C, Hall M N, Boller T, Wiemken A. The role of trehalose synthesis for the acquisition of thermotolerance in yeast. II. Physiological concentrations of trehalose increase the thermal stability of proteins in vivo. Eur J Biochem. 1994;219:187–193. doi: 10.1111/j.1432-1033.1994.tb19929.x. [DOI] [PubMed] [Google Scholar]
- 12.Kopp M, Müller H, Holzer H. Molecular analysis of the neutral trehalase gene from Saccharomyces cerevisiae. J Biol Chem. 1993;268:4766–4774. [PubMed] [Google Scholar]
- 13.Kotyk A, Michaljanivova D. Uptake of trehalose by Saccharomyces cerevisiae. J Gen Microbiol. 1979;110:323–332. doi: 10.1099/00221287-110-2-323. [DOI] [PubMed] [Google Scholar]
- 14.Nwaka S, Holzer H. Molecular biology of trehalose and the trehalases in the yeast Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1998;58:197–237. doi: 10.1016/s0079-6603(08)60037-9. [DOI] [PubMed] [Google Scholar]
- 15.Panek A D, Panek A C. Metabolism and thermotolerance function of trehalose in Saccharomyces cerevisiae: a current perspective. J Biotechnol. 1990;14:229–238. [Google Scholar]
- 16.Parrou J L, François J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem. 1997;248:186–188. doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- 17.Paschoalin V M F, Silva J T, Panek A D. Identification of an ADPG-dependent trehalose synthase in Saccharomyces cerevisiae. Curr Genet. 1989;16:81–87. doi: 10.1007/BF00393399. [DOI] [PubMed] [Google Scholar]
- 18.Reinders A, Bürckert N, Hohmann S, Thevelein J M, Boller T, Wiemken A, DeVirgilio C. Structural analysis of the subunits of the trehalose-6-phosphate synthase/phosphatase complex in Saccharomyces cerevisiaeand their function during heat shock. Mol Microbiol. 1997;24:687–695. doi: 10.1046/j.1365-2958.1997.3861749.x. [DOI] [PubMed] [Google Scholar]
- 19.Stambuk B U, de Araujo P S, Panek A D, Serrano R. Kinetics and energetics of trehalose transport in Saccharomyces cerevisiae. Eur J Biochem. 1996;237:876–881. doi: 10.1111/j.1432-1033.1996.0876p.x. [DOI] [PubMed] [Google Scholar]
- 20.Stambuk B U, Panek A D, Crowe J H, Crowe L M, de Araujo P S. Expression of high-affinity trehalose-H+ symport in Saccharomyces cerevisiae. Biochim Biophys Acta. 1998;1374:118–128. doi: 10.1016/s0304-4165(97)00087-1. [DOI] [PubMed] [Google Scholar]
- 21.Thevelein J M, Hohmann S. Trehalose synthase: guard to the gate of glycolysis in yeast? Trends Biochem Sci. 1995;20:3–10. doi: 10.1016/s0968-0004(00)88938-0. [DOI] [PubMed] [Google Scholar]
- 22.Van Aelst L, Hohmann S, Bulaya B, de Koning W, Sierkstra L, et al. Molecular cloning of a gene involved in glucose sensing in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1993;8:927–943. doi: 10.1111/j.1365-2958.1993.tb01638.x. [DOI] [PubMed] [Google Scholar]
- 23.Verduyn C, Postma E, Scheffers A, Van Dijken J P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1991;8:501–516. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 24.Wach A, Brachat A, Pöhlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]