Abstract
Background
As the most widespread mRNAs modification, N6‐methyladenosine (m6A) is dynamically and reversibly modulated by methyltransferases and demethylases. ALKBH5 is a major demethylase, and plays vital roles in the progression of cancers. However, the role and mechanisms of ALKBH5 in colorectal cancer (CRC) is unclear.
Results
Herein, we discovered that in CRC, downregulated ALKBH5 was closely related to poor prognosis of CRC patients. Functionally, our results demonstrated that knockdown of ALKBH5 enhanced the proliferation, migration and invasion of LOVO and RKO in vitro, while overexpression of ALKBH5 inhibited the functions of these cells. The results also demonstrated that knockdown of ALKBH5 promoted subcutaneous tumorigenesis of LOVO in vivo, while overexpression of ALKBH5 suppressed this ability. Mechanistically, results from joint analyses of MeRIP‐seq and RNA‐seq indicated that PHF20 mRNA was a key molecule that was regulated by ALKBH5‐mediated m6A modification. Further experiments indicated that ALKBH5 may inhibit stability of PHF20 mRNA by removing the m6A modification of PHF20 mRNA 3′UTR.
Conclusions
ALKBH5 suppresses CRC progression by decreasing PHF20 mRNA methylation. ALKBH5‐mediated m6A modification of PHF20 mRNA can serve as a hopeful strategy for the intervention and treatment of CRC.
Keywords: ALKBH5, colorectal cancer, m6A modification, PHF20
ALKBH5 expression is down‐regulated in CRC and plays a crucial tumor suppressive role.
ALKBH5 inhibits the stability of PHF20 mRNA by removing the m6A modification.
Targeting the ALKBH5‐mediated m6A modification of PHF20 mRNA may be a promising therapeutic strategy for intervention and treatment of CRC.
1. INTRODUCTION
As one of the most common malignant tumours, colorectal cancer (CRC) ranks the third with regard to incidence and the second in terms of mortality worldwide. 1 It was reported that there would be about 2 million new CRC cases, and 0.9 million CRC‐associated deaths in 2020. 1 Although comprehensive treatment methods have improved, the prognosis of CRC remains poor. The molecular mechanisms of CRC progression need to be explored.
N6‐methyladenosine (m6A) is reported to be the most widespread mRNAs modification in eukaryotic cells. M6A modification of mRNAs regulates its metabolic process, including splicing, transport, stability and translation of mRNAs. 2 , 3 Recent studies have demonstrated that the m6A modification of mRNAs is dynamically and reversibly regulated by methyltransferases and demethylases. The methyltransferase complex consists of METTL3 (methyltransferase‐like 3), METTL14 (methyltransferase‐like 14) and additional adaptor molecules. 4 , 5 Demethylases include FTO (fat mass and obesity‐associated protein) and ALKBH5 (alkB homologue 5). 6 , 7 Lots of studies have demonstrated that methyltransferases and demethylases are frequently dysregulated among various cancers, making it vital roles in the progression of cancers. 8 , 9
ALKBH5 is one of two RNA demethylases, which is able to remove the m6A modification on RNAs. 7 ALKBH5 is dysregulated in many tumours, such as hepatocellular cancer, pancreatic cancer and gastric cancer. 10 With regard to CRC, ALKBH5 may promote cancer cell motility by demethylating the lncRNA NEAT1. 11 However, ALKBH5 has been reported to be downregulated in CRC tissues and positively associated with overall survival and disease‐free survival. 12 Therefore, the role and mechanisms of ALKBH5 in CRC need to be further studied.
Herein, we identified that downregulated ALKBH5 was closely associated with the poor prognosis of CRC patients. ALKBH5 significantly inhibited the proliferation, migration and invasion abilities of LOVO and RKO in vitro, and suppressed the subcutaneous tumourigenicity of LOVO in vivo. Mechanistically, PHF20 mRNA was a key downstream molecule of ALKBH5. ALKBH5 might inhibit the stability of PHF20 mRNA via removing the m6A modification of PHF20 mRNA 3′UTR, thereby suppressing the progression of CRC.
2. MATERIALS AND METHODS
2.1. Patient samples and cell lines
CRC tumour tissues and tumour‐adjacent normal tissues were collected from Peking University People's Hospital. All colorectal tissues were pathologically confirmed.
Six human colon cancer cell lines (RKO, SW480, HCT116, HCT8, LS174T, LOVO) were purchased from the Cell Resource Center of Peking Union Medical College (China). RKO, SW480, HCT116, HCT‐8, LS174T and LOVO were cultured in MEM, IMDM, IMDM, RPMI 1640, MEM and F12K, respectively. All the cell lines were cultivated in the corresponding medium containing 10% FBS (Gibco, USA) in 5% CO2 environment at 37°C.
2.2. Establishment of stable knock‐down and overexpression cells
Three siRNAs of ALKBH5 (siALKBH5‐1: ACAAGTACTTCTTCGGCGA, siALKBH5‐2: GCGCCGTCATCAACGACTA, siALKBH5‐3: CTGAGAACTACTGGCGCAA) and two siRNAs of human PHF20 (siPHF20‐1: CCCGAGAAATACACCTGTTAT, siPHF20‐2: ATTGTGCCACTGATGATAAAC) were synthesized by RiboBio (China). In addition, the two most efficient sequences for ALKBH5 were used to construct lentiviral shRNA plasmids. The lentiviral plasmid expressing shALKBH5 or shNC, overexpressing ALKBH5 or an empty vector were purchased from GeneCopoeia (USA). The packaging plasmid, envelope plasmid and target plasmid were transfected to the 293T cells to obtain the lentivirus using the Lenti‐Pac HIV Expression Packaging Kit (GeneCopoeia, USA). Then, the lentivirus was used to infect LOVO and RKO.
2.3. Immunohistochemical staining (IHC)
Immunohistochemical staining (IHC) was performed, as previously described. 13 ALKBH5 staining index score (0–12) was calculated using the staining intensity multiplying by the percentage of ALKBH5 positive staining. The staining intensity is divided into four grades (negative: 0; weak: 1; moderate: 2; strong: 3), and the percentage of positive staining is divided into five grades (<5%: 0; 5%–25%: 1; 26%–50%: 2; 51%–75%: 3; >75%: 4). Anti‐ALKBH5 (Sigma, USA) was used. All scores were independently evaluated by two pathologists.
2.4. RT‐qPCR
RT‐qPCR was performed, as previously described. 13 The primer sequences are listed in Table S1.
2.5. Western blot
Western blot assays were performed, as previously described. 13 Primary antibodies included anti‐ALKBH5 (Sigma, USA), anti‐PHF20 (CST, USA) and anti‐GAPDH (CST, USA).
2.6. Cell function experiments
To perform the cell proliferation assays, we seeded 3 × 103 cells into a 96‐well plate. Next, 10‐μl CCK8 solution was added into each well of the plate. After incubation for 2 h, the absorbance at 450 nm was detected to assess cell proliferation. Moreover, cell proliferation was assessed for five consecutive days after cells seeded.
To conduct the colony formation assays, we seeded 5 × 102 cells into a 6‐well plate. After cultivated for 10 days, the clones were fixed in 4% paraformaldehyde solution for 25 min. Then, the clones were stained with .1% crystal violet solution for 25 min. Last, the clones were counted after washed with PBS three times.
To perform the migration assays, we seeded 1 × 105 cells into the upper chambers (Corning, USA) and added medium with 10% FBS to the lower chambers. To perform the invasion assays, the upper chambers were coated with Matrigel (Sigma, USA). After cultivated for 48 h, the cells migrating below the chambers were fixed, stained and counted under a microscope.
2.7. Animal experiments
Female Balb/c nude mice (6–8 weeks) were purchased from the Experimental Animal Center of Military Medical Sciences (China). We injected 1 × 106 cells subcutaneously in the flanks of mice and measured the length (L) and width (W) of tumour every 2 days. Tumour volume (V) was calculated as follows: . The mice were euthanized at 16th days after the injection of cancer cells. Then, the subcutaneous tumours were removed and weighted.
2.8. RNA stability assays
Tumour cells were treated with actinomycin D (Sigma, USA) at 5 μg/ml. The cells were collected after incubated for 0, 3 or 6 h, and then RNA was extracted to perform RT‐qPCR assays as described earlier. The half‐life of mRNA was calculated to assess its stability.
2.9. mRNA m6A quantification
The mRNA was purified from total RNA using the GenElute mRNA Miniprep Kit (Sigma, USA). The m6A modification level of mRNA was evaluated utilizing the EpiQuik m6A RNA methylation quantification kit (EpiGentek, USA). In brief, 200‐ng mRNA was incubated with capture antibody and then incubated with detection antibody. The m6A modification level of mRNA was quantified colourimetrically by measuring the absorbance at 450 nm.
2.10. Luciferase reporter assays
The wild type and m6A sites mutated PHF20 were constructed into the luciferase reporter vector (GeneCopoeia, USA). The sequences of the wild type and m6A sites mutated PHF20 are listed in Table S2. Cells were transfected with siNC or siALKBH5. The cells were re‐seeded into a 24‐well plate after incubated with siNRA for 48 h and then were transfected with PHF20 3′UTR‐WT or PHF20 3′UTR‐MUT. The cells were lysed after cultivated for 24 h. Firefly and Renilla luciferase activities in cell lysates were analysed utilizing the Dual‐Glo Luciferase Assay system (Promega, USA).
2.11. RNA sequencing
Total RNA was isolated with a TRIzol reagent (Invitrogen, CA). A total of 1‐μg RNA was used to perform RNA sequencing. Sequencing libraries were generated using the NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA). Sequencing was performed based on the Illumina NovaSeq platform. Differentially expressed genes were identified utilizing the DESeq2 R package (|log2(fold change)|>0 and p < 0.05).
2.12. MeRIP‐qPCR
The m6A modification of mRNA was quantified using the Magna MeRIP m6A Kit (Millipore, Germany). In brief, 10% of mRNA purified from total RNA was saved as inputs. Magna ChIP Protein A/G Magnetic Beads were washed and then incubated with 5‐μg anti‐m6A antibody with rotation for 30 min at room temperature. The antibody‐beads mixed with mRNA were incubated at 4°C with RNase inhibitors for 2 h. Then, the methylated mRNAs were eluted and purified utilizing the RNeasy mini kit (QIAGEN, Germany). RT‐qPCR was used to determine m6A enrichment by normalizing to the input.
2.13. Statistical analysis
All results are presented as mean ± SD. The SPSS 17.0 was used for data analysis. The χ 2 test was utilized to analyse the correlation between ALKBH5 and clinicopathological features of patients. Kaplan–Meier analysis was used to construct survival curves, and the differences of different groups were estimated by log‐rank test. For all continuous variables, Student's t‐test or ANOVA was performed to evaluate the differences. A p value of .05 was considered statistically significant.
3. RESULTS
3.1. ALKBH5 was downregulated in CRC
To evaluate the expression of ALKBH5 in cancers, we initially detected mRNA expression of ALKBH5 in 23 solid cancers in TCGA (The Cancer Genome Atlas) datasets. In 12 solid cancers, ALKBH5 was significantly dysregulated in comparison to adjacent normal tissues (Figure 1A). As shown in Figure 1A, compared to adjacent normal tissues, ALKBH5 was significantly downregulated in seven solid cancers (CRC, GBM, KICH, KIRP, PRAD, THCA and UCEC) and upregulated in five solid cancers (CHOL, HNSC, KIRC, LIHC and LUSC). We discovered that, compared with adjacent normal tissues, ALKBH5 was significantly reduced in tumour tissues in TCGA‐CRC cohort. We also downloaded the RNA sequencing data of CRC from GEO (Gene Expression Omnibus) datasets. The results confirmed the reduction of ALKBH5 mRNA in CRC tissues (GSE39582 and GSE87211; Figure 1B). Furthermore, RT‐qPCR showed that ALKBH5 mRNA expression in CRC tissues was significantly lower than that in adjacent normal tissues (Figure 1C). We also evaluated ALKBH5 protein expression in CRC. IHC assays indicated that ALKBH5 protein expression was significantly decreased in CRC tissues (Figure 1D,E). What is more, the m6A quantification assays of mRNA implied that m6A modification levels of mRNAs were higher in CRC tissues (Figure 1F).
FIGURE 1.
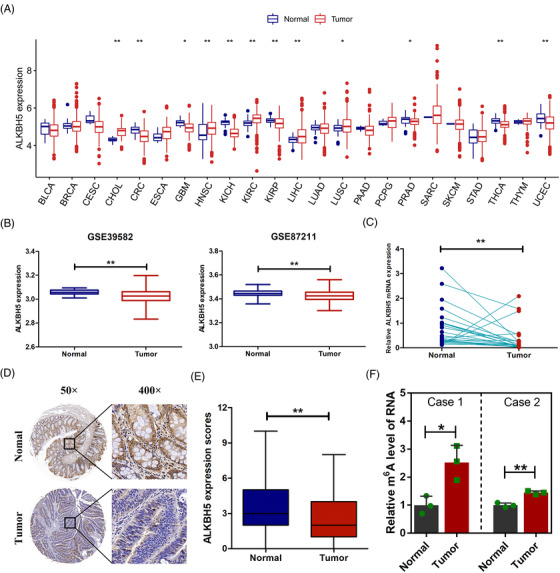
AlkB homologue 5 (ALKBH5) was downregulated in colorectal cancer (CRC). (A) ALKBH5 mRNA expression in 23 solid cancers in The Cancer Genome Atlas (TCGA) databases. (B) ALKBH5 mRNA expression in CRC in the Gene Expression Omnibus (GEO) databases. (C) RT‐qPCR analysis of ALKBH5 mRNA expression in 24 paired CRC tumours and adjacent normal tissues. (D) Representative immunohistochemical staining (IHC) images of ALKBH5 expression in CRC tumour and paired normal tissues. (E) Quantification of the IHC score of ALKBH5 in CRC tumour tissues (n = 114) and adjacent normal tissues (n = 107). (F) Relative quantitative analysis of global N6‐methyladenosine (m6A) levels of mRNA in CRC tumour tissues and adjacent normal tissues. Statistical significance was determined using Wilcoxon test (A, B, E), paired Wilcoxon test (C) and t‐test (F) (* p < .05, ** p < .01).
3.2. Loss of ALKBH5 predicted worse prognosis of CRC patients
In order to explore the clinical value of ALKBH5, we analysed the correlation between ALKBH5 and clinicopathological features of CRC patients at the mRNA and protein levels. The results indicated that, at the mRNA levels, downregulated ALKBH5 was significantly associated with distant metastasis (p = .025) and late clinical stage (p = .034) (Table 1; Figure 2A,B). Consistently, at the protein levels, downregulated ALKBH5 was significantly related to distant metastasis (p = .042) and late clinical stage (p = .024) (Table 2; Figure 2D,E). The Kaplan–Meier analysis indicated that CRC patients with a loss of ALKHB5 had shorter overall survival both at the mRNA (Figure 2C, p = .026) and protein (Figure 2F, p = .048) levels.
TABLE 1.
Clinical characteristics of 130 colorectal cancer (CRC) patients according to alkB homologue 5 (ALKBH5) mRNA levels
| ALKBH5 | ||||
|---|---|---|---|---|
| All cases | Low expression | High expression | p‐Value | |
| Gender | ||||
| Female | 61 | 33 | 28 | .38 |
| Male | 69 | 32 | 37 | |
| Age | ||||
| ≤60 | 46 | 22 | 24 | .714 |
| >60 | 84 | 43 | 41 | |
| Tumour size | ||||
| ≤5 | 65 | 30 | 35 | .384 |
| >5 | 65 | 35 | 30 | |
| Histological type | ||||
| Adenocarcinoma | 118 | 58 | 60 | .545 |
| Others | 12 | 7 | 5 | |
| Tumour differentiation | ||||
| Well and moderate | 104 | 53 | 51 | .661 |
| Poor | 26 | 12 | 14 | |
| Lymphovascular invasion | ||||
| Positive | 71 | 36 | 35 | .86 |
| Negative | 59 | 29 | 30 | |
| T stage | ||||
| T1 + T2 | 21 | 8 | 13 | .233 |
| T3 + T4 | 109 | 57 | 52 | |
| N stage | ||||
| N0 | 81 | 37 | 44 | .295 |
| N1 + N2 | 49 | 28 | 21 | |
| M stage | ||||
| M0 | 111 | 51 | 60 | .025* |
| M1 | 19 | 14 | 5 | |
| AJCC stage | ||||
| I + II | 74 | 31 | 43 | .034* |
| III + IV | 56 | 34 | 22 | |
p‐Value <.05.
FIGURE 2.
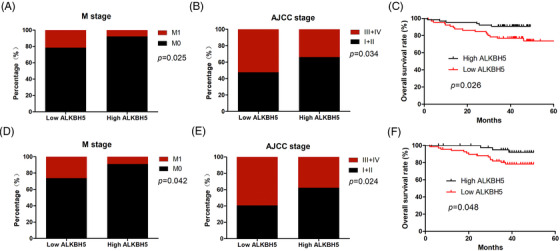
Loss of alkB homologue 5 (ALKBH5) predicted worse prognosis of colorectal cancer (CRC) patients. (A) The relationship between ALKBH5 mRNA expression and metastasis (M) stage. (B) The relationship between ALKBH5 mRNA expression and AJCC stage. (C) The Kaplan–Meier analysis of overall survival (OS) for CRC patients based on ALKBH5 mRNA expression. (D) The relationship between ALKBH5 protein expression and M stage. (E) The relationship between ALKBH5 protein expression and AJCC stage. (F) The Kaplan–Meier analysis of OS for CRC patients, based on ALKBH5 protein expression. Statistical significance was determined using χ 2 test (A, B, D and E) and log‐rank tests (C and F).
TABLE 2.
Clinical characteristics of 114 colorectal cancer (CRC) patients according to alkB homologue 5 (ALKBH5) protein levels
| ALKBH5 | ||||
|---|---|---|---|---|
| All cases | Low expression | High expression | p‐Value | |
| Gender | ||||
| Female | 47 | 26 | 21 | .341 |
| Male | 67 | 43 | 24 | |
| Age | ||||
| ≤60 | 41 | 30 | 11 | .038* |
| > 60 | 73 | 39 | 34 | |
| Tumour size | ||||
| ≤5 | 77 | 47 | 30 | .872 |
| > 5 | 37 | 22 | 15 | |
| Histological type | ||||
| Adenocarcinoma | 112 | 67 | 45 | .249 |
| Others | 2 | 2 | 0 | |
| Tumour differentiation | ||||
| Well and moderate | 93 | 58 | 35 | .398 |
| Poor | 21 | 11 | 10 | |
| Lymphovascular invasion | ||||
| Positive | 53 | 30 | 23 | .424 |
| Negative | 61 | 39 | 22 | |
| T stage | ||||
| T1 + T2 | 19 | 9 | 10 | .199 |
| T3 + T4 | 95 | 60 | 35 | |
| N stage | ||||
| N0 | 62 | 33 | 29 | .082 |
| N1 + N2 | 52 | 36 | 16 | |
| M stage | ||||
| M0 | 92 | 51 | 41 | .042* |
| M1 | 22 | 18 | 4 | |
| AJCC stage | ||||
| I + II | 56 | 28 | 28 | .024* |
| III + IV | 58 | 41 | 17 | |
p‐Value <.05.
3.3. Knock‐down of ALKBH5 enhanced growth and metastasis of colon cancer cells
Next, we explored the effect of ALKBH5 on colon cancer cell function. We initially identified ALKBH5 mRNA expression in five colon cancer cells, and noted that ALKBH5 was elevated in LOVO and RKO cells, compared to other colon cancer cell lines (Figure 3A). Three specific siRNAs were utilized to knock‐down ALKBH5 in LOVO and RKO cells (Figure 3B,C). Furthermore, two siRNAs (siALKBH5‐2, siALKBH5‐3) with high knock‐down efficiency were utilized to construct knock‐down plasmids (shALKBH5‐2, shALKBH5‐3) for subsequent experiments. We constructed two stable cell lines using two independent ALKBH5‐knock‐down lentiviruses. The knock‐down efficiency was validated both at the mRNA and protein levels (Figure 3D–F). Subsequently, the CCK‐8 assays revealed that the knock‐down of ALKBH5 markedly enhanced cellular growth, as well as the viability of LOVO and RKO cells (Figure 3G). It was confirmed that the knock‐down of ALKBH5 significantly increased the colony formation abilities of LOVO and RKO cells through the colony formation assays (Figure 3H). Additionally, we assessed migration and invasion abilities of LOVO and RKO cells using the transwell assays. The results indicated that the knock‐down of ALKBH5 drastically elevated cell migration and invasion abilities (Figure 3I,J).
FIGURE 3.
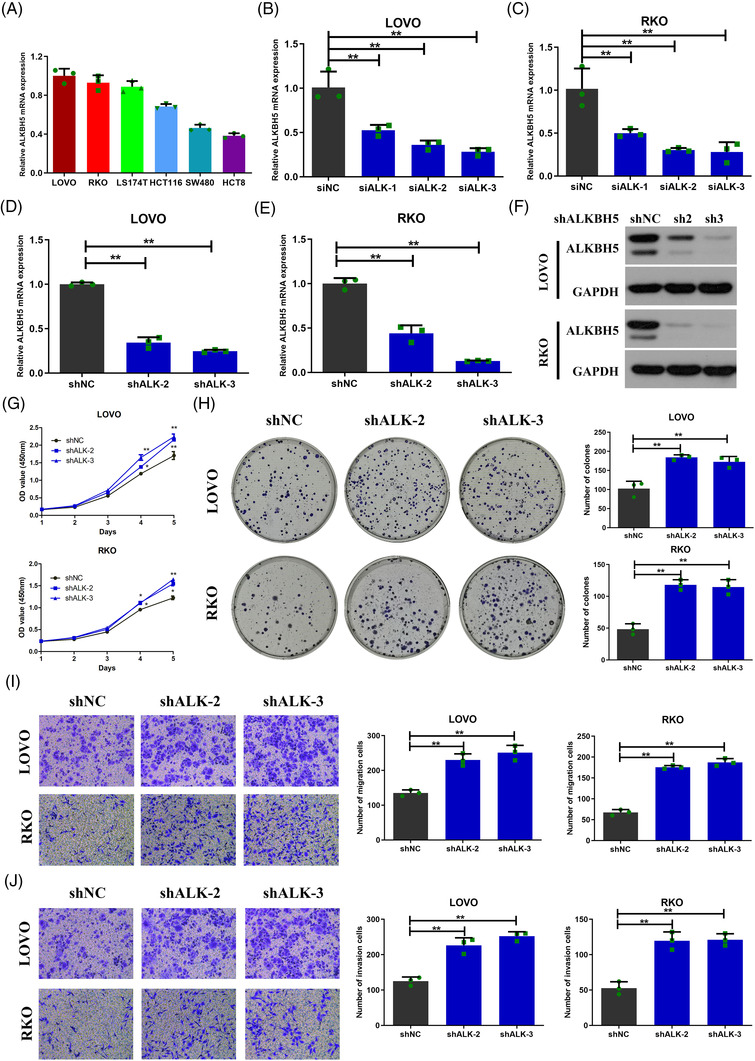
Knock‐down of alkB homologue 5 (ALKBH5) enhanced proliferation, migration and invasion of colon cancer cells in vitro. (A) ALKBH5 mRNA expression in five colon cancer cells. (B and C) ALKBH5 mRNA expression in LOVO (B) and RKO (C) cells infected with three independent siRNAs targeting ALKBH5. (D and E) ALKBH5 mRNA expression in LOVO (D) and RKO (E) cells infected with two independent ALKBH5‐knock‐down lentiviruses. (F) ALKBH5 protein expression in LOVO and RKO infected with two independent ALKBH5‐knock‐down lentiviruses. (G and H) The proliferation ability of LOVO and RKO cells with ALKBH5 knock‐down determined by CCK8 (G) and colony formation (H) assays. (I and J) The migration (I) and invasion (J) ability of LOVO and RKO cells with ALKBH5 knock‐down, as determined by transwell assays. Statistical significance was determined using ANOVA (* p < .05, ** p < .01).
To further determine whether ALKBH5 affects colon cancer cell function in vivo, we established tumour xenograft models. We found that tumour growth rate was faster (Figure 4C), and tumour volume and weight were increased (Figure 4A,B,D) when ALKBH5‐knock‐down LOVO cells were implanted (n = 5). Moreover, the expression of Ki‐67 was also increased in the shALKBH5 groups in comparison to the shNC groups (Figure S1).
FIGURE 4.
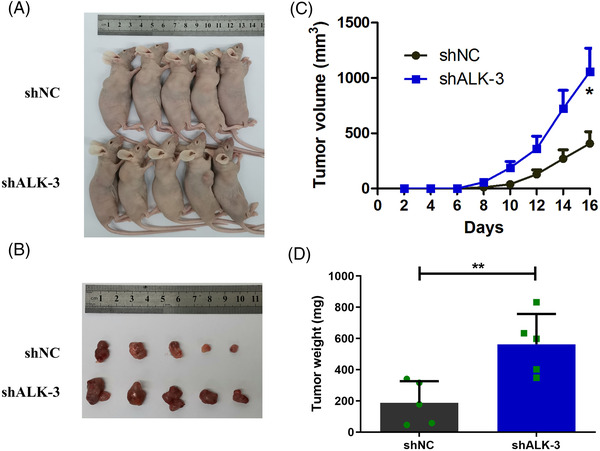
Knock‐down of alkB homologue 5 (ALKBH5) enhanced the proliferation of colon cancer cells in vivo. (A and B) Tumour xenograft models were constructed using ALKBH5‐knock‐down LOVO cells (n = 5). (C) Tumour formation and size in the tumour xenograft model were monitored every 2 days. (D) Tumour weights were measured from sacrificed mice. Statistical significance was determined using t‐test (* p < .05, ** p < .01).
3.4. Overexpression of ALKBH5 inhibited growth and metastasis of colon cancer cells
We also constructed stably transfected cell lines with ALKBH5 overexpression utilizing a lentivirus vector, and RT‐qPCR and Western blot assays validated the overexpression efficiency (Figure 5A–C). The CCK‐8 assays revealed that the overexpression of ALKBH5 markedly repressed cellular growth and the viability of LOVO and RKO cells (Figure 5D). The colony formation assays confirmed that the overexpression of ALKBH5 resulted in a significant decrease of the colony formation abilities of LOVO and RKO cells (Figure 5E). Additionally, the overexpression of ALKBH5 drastically weakened cell migration and invasion by the transwell assays (Figure 5F,G). Then, our results of tumour xenograft models indicated that tumour growth rate was slower (Figure 6C), and that tumour volume and weight were decreased (Figure 6A,B,D) when LOVO cells overexpressing ALKBH5 were implanted (n = 5).
FIGURE 5.
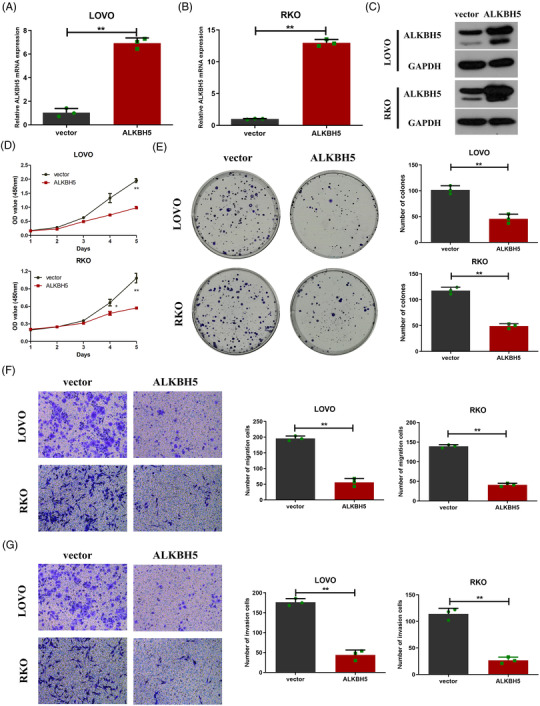
Overexpression of alkB homologue 5 (ALKBH5) inhibited proliferation, migration and invasion of colon cancer cells in vitro. (A and B) ALKBH5 mRNA expression in LOVO (A) and RKO (B) cells infected with ALKBH5‐overexpression or control vector lentiviruses. (C) ALKBH5 protein expression in LOVO and RKO cells infected with ALKBH5‐overexpression or control vector lentiviruses. (D and E) The proliferation ability of LOVO and RKO with ALKBH5 knock‐down, as determined by CCK8 (D) and colony formation (E) assays. (F and G) The migration (F) and invasion (G) ability of LOVO and RKO cells with ALKBH5 knock‐down, as determined by transwell assays. Statistical significance was determined using t‐test (* p < .05, ** p < .01).
FIGURE 6.
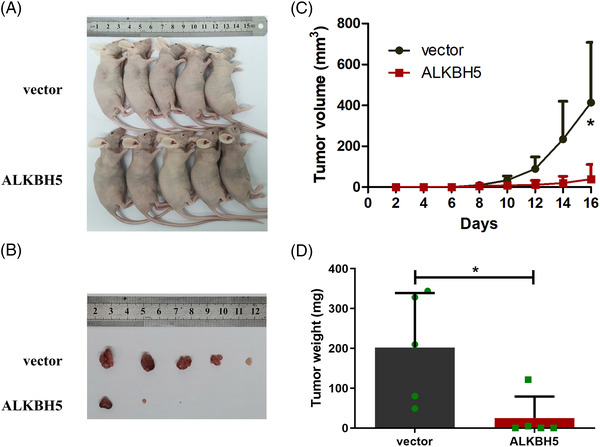
Overexpression of alkB homologue 5 (ALKBH5) inhibited the proliferation of colon cancer cells in vivo. (A and B) Tumour xenograft models were constructed using ALKBH5‐overexpression LOVO cells (n = 5). (C) Tumour size and formation in the tumour xenograft models were monitored every 2 days. (D) Tumour weights were measured from sacrificed mice. Statistical significance was determined using t‐test (* p < .05).
3.5. ALKBH5 targeted PHF20
To identify the m6A‐modified targets, we collected tumour tissues and normal tissues from six CRC patients to perform MeRIP‐seq. 14 We identified 1343 dysregulated m6A peaks in six pairs of tissues by MeRIP‐seEq (Figure 7A). Among them, 625 m6A peaks were hypermethylated in tumour tissues, whereas 718 m6A peaks were hypomethylated in tumour tissues (Figure 7A). Next, 625 hypermethylated m6A peaks were distributed among the transcripts of 265 genes, and 718 hypomethylated m6A peaks were distributed among the transcripts of 311 genes. Interestingly, we discovered that in the six pairs of tissues for MeRIP‐seq, the expression of ALKBH5 in tumour tissues was significantly lower than that in normal tissues (Figure 7B). We hypothesized that the transcripts of these 265 genes were regulated by ALKBH5. In order to further explore m6A modification targets regulated by ALKBH5, we performed RNA‐seq in ALKBH5‐knock‐down LOVO cells and control cells. The results demonstrated that after ALKBH5 knocked down, 679 genes were differentially expressed, including 362 upregulated genes and 317 downregulated genes (Figure 7C,D). Combining MeRIP‐seq and RNA‐seq, we found 19 genes regulated potentially by ALKBH5 through m6A modification, including 11 upregulated genes (PCDH7, QSOX1, LAMA5, CCDC69, LITAF, MDK, TGFB1I1, ATP2B4, DUSP3, PHF20 and PLIN3) and 8 downregulated genes (NPNT, KRT20, PCSK7, SAMD4A, NEDD4L, SLC35A3, SPTLC2 and DNM1L) after ALKBH5 knock‐down (Figure 7E).
FIGURE 7.
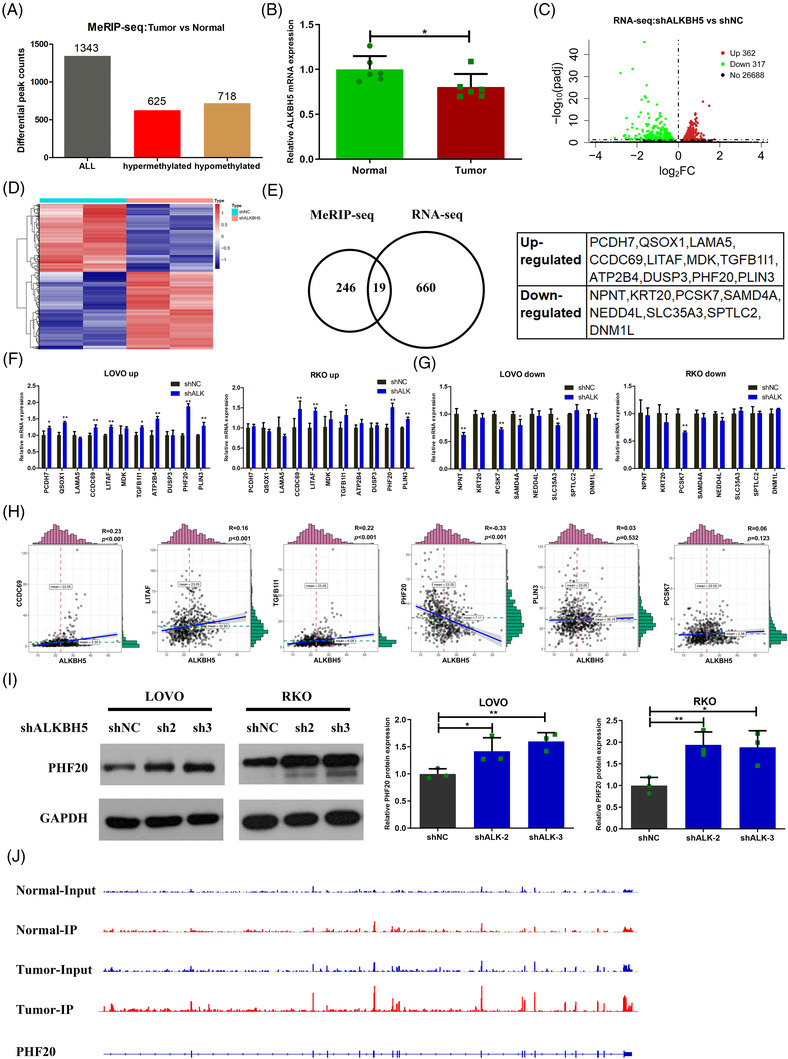
PHF20 was a downstream target of alkB homologue 5 (ALKBH5)‐mediated N6‐methyladenosine (m6A) modification. (A) MeRIP‐seq results of tumour tissues and tumour‐adjacent normal tissues showing the distribution of altered m6A peaks in six colorectal cancer (CRC) patients. (B) ALKBH5 mRNA expression in the six paired tumour tissues and tumour‐adjacent normal tissues. (C and D) Volcano plots (C) and heat map plots (D) indicating the differentially expressed genes in ALKBH5‐knock‐down cells, compared to control cells in RNA‐seq. (E) Conjoint analysis of MeRIP‐seq and RNA‐seq data. (F) Validation of upregulated genes in ALKBH5‐knock‐down cells, compared to the control group. (G) Validation of downregulated genes in ALKBH5‐knock‐down cells, compared to the control group. (H) The relationship between ALKBH5 and six candidate genes (CCDC69, LITAF, TGFB1I1, PHF20, PLIN3 and PCSK7) in The Cancer Genome Atlas (TCGA) database. (I) The protein expression of PHF20 in ALKBH5‐knock‐down and control cells. (J) Representative m6A peaks of PHF20 mRNA in CRC. Statistical significance was determined using ANOVA (I) and t‐test (B, F, G) (* p < .05, ** p < .01).
Then, we examined the expression of these 19 genes after ALKBH5 knock‐down. The results of RT‐qPCR displayed that among the 11 upregulated genes, 5 genes (CCDC69, LITAF, TGFB1I1, PHF20 and PLIN3) were upregulated both in ALKBH5‐knock‐down LOVO and RKO cells (Figure 7F). In addition, we discovered that among the eight downregulated genes, only PCSK7 was downregulated both in ALKBH5‐knock‐down LOVO and RKO cells (Figure 7G). Next, we examined the correlation between ALKBH5 and the six candidate genes in the TCGA database. Our results indicated that only PHF20 was negatively correlated with ALKBH5 in CRC (Figure 7H). We also verified elevated protein expression of PHF20 in ALKBH5‐knock‐down cells compared to control cells by Western blot assays (Figure 7I). Moreover, visualization analysis showed that compared to normal tissues, the m6A peaks of PHF20 mRNA were more abundant in tumour tissues (Figure 7J). Thus, PHF20 may be a key molecule regulated by ALKBH5 through m6A modification.
3.6. Loss of ALKBH5 increased the stability of PHF20 mRNA to facilitate CRC progression
As a demethylase, ALKBH5 is able to remove m6A modification from mRNA. The m6A quantification assays of mRNA indicated that ALKBH5 loss significantly increased the m6A modification level of mRNAs (Figure 8A). The MeRIP‐qPCR assays demonstrated that ALKBH5 loss significantly increased the m6A modification level of PHF20 mRNA (Figure 8B). Next, we predicted the m6A sites of PHF20 mRNA utilizing the SRAMP database (http://www.cuilab.cn/sramp) and BERMP database (http://www.bioinfogo.org/bermp). We discovered that the m6A methylation sites on PHF20 mRNA were mainly located in the 3′UTR, and that there were eight possible m6A methylation sites. According to the m6A methylation sites on PHF20 mRNA, we constructed a luciferase reporter vector (Figure 8C). The luciferase reporter assays illustrated that ALKBH5 loss significantly enhanced the expression of the wild‐type PHF20 3′UTR plasmid, but that there was no significant effect on PHF20 3′UTR plasmid with m6A mutation sites (Figure 8D).
FIGURE 8.
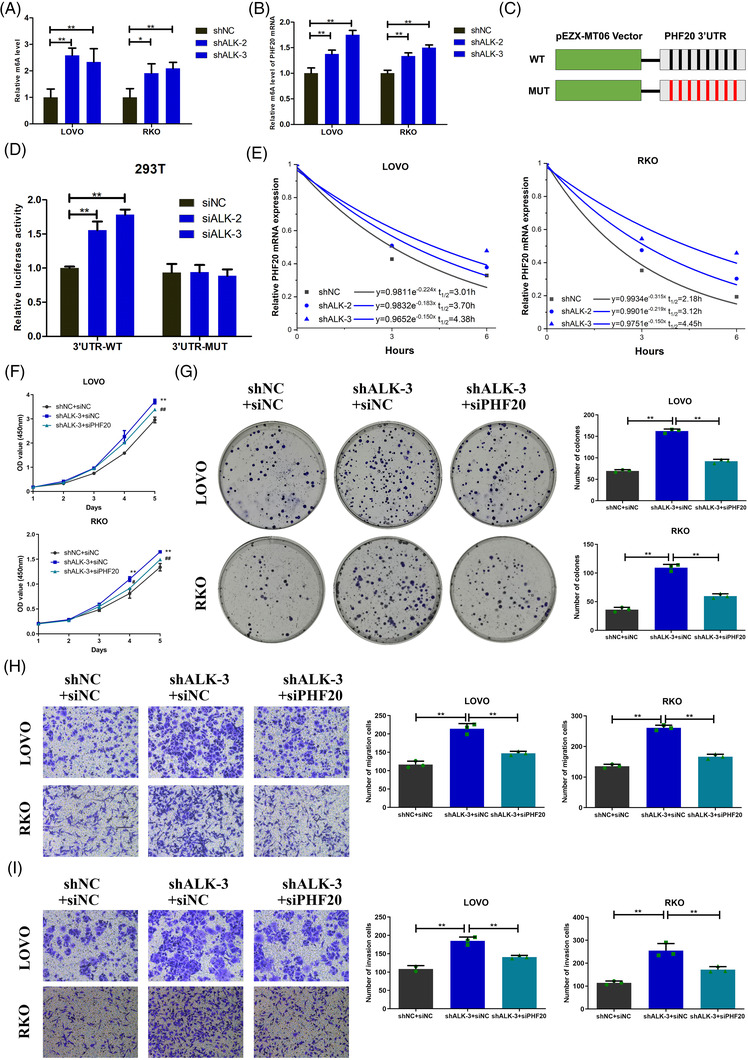
AlkB homologue 5 (ALKBH5) suppressed progression of colorectal cancer (CRC) by decreasing PHF20 mRNA methylation. (A) Relative quantitative analysis of global N6‐methyladenosine (m6A) levels of mRNA in ALKBH5‐knock‐down and control cells. (B) MeRIP‐qPCR analysis of alterations in the m6A levels of PHF20 mRNA in ALKBH5‐knock‐down and control cells. (C) Luciferase reporter constructs containing the human PHF20 3′UTR that have m6A motifs or mutants (A–T mutation) m6A sites. Black represents A, and red represents T. (D) Relative luciferase activities of 293T cells that were co‐transfected with plasmids containing wild‐type or mutant PHF20 3′UTR and siRNAs containing siNC or siALKBH5. (E) RT‐qPCR analysis of the decay rate of PHF20 mRNA after actinomycin D (5 μg/ml) treatment in LOVO or RKO cells after ALKBH5 inhibited. (F)–(I) The rescue experiment was utilized to determine whether ALKBH5 has an effect on proliferation (F and G), migration (H) and invasion (I) of LOVO and RKO cells by regulating PHF20. In (F), *: shALKBH5 + siNC versus shNC + siNC; #: shALKBH5 + siPHF20 versus shALKBH5 + siNC. Statistical significance was determined using ANOVA (* p < .05, ** p < .01, #p < .05, ##p < .01).
To assess the effect of ALKBH5 on PHF20 mRNA stability, we treated colon cancer cells with actinomycin D and found that ALKBH5 loss improved stability of PHF20 mRNA and prolonged its half‐life (Figure 8E). Furthermore, our data showed that PHF20 knock‐down contributed to a significant inhibition of the proliferation, colony formation, migration and invasion of LOVO (Figure S2). Then we carried out rescue experiments to determine whether ALKBH5 had an effect on the biological function of colon cancer cells by regulating PHF20. It showed that PHF20 knoc‐kdown inhibited the shALKBH5‐mediated enhancement of the proliferation, colony formation, migration and invasion of LOVO and RKO (Figure 8F–I).
4. DISCUSSION
M6A modification of RNA is a new research direction in epigenetics. Our previous study has first performed MeRIP‐seq in CRC and discovered dysregulated m6A peaks of transcripts. 14 Herein, the m6A quantification assays of mRNA revealed higher levels of mRNA m6A modification in CRC tumour tissues than that in adjacent normal tissues. It indicated that m6A modification regulated by methyltransferase and demethylase was dysregulated in CRC.
The discovery of demethylase is an important evidence of the reversible regulatory mechanism of m6A modification of RNA. So far, ALKBH5 is discovered as one of the two m6A demethylases. ALKBH5 is a highly conserved homologue of the AlkB family of dioxygenases dependent on α‐ketoglutaric acid and iron(II). 7 The AlkB family is a class of repair enzymes that can directly reverse an alkylation damage of DNA or RNA through oxidative dealkylation to remove alkyl adjunct on the base. 15 , 16 ALKBH5 specifically catalyses the removal of m6A modification on ssRNAs and is localized at the nuclear speckles, which is conducive to its direct interaction with mRNA substrates, regulating mRNA export, RNA metabolism and mRNA processing factor assembly in nuclear speckle. 17
It is reported that ALKBH5 plays a vital and specific role in cancer regulation. ALKBH5 functions as an oncogene in many cancers, including glioblastoma, breast cancer and esophageal squamous cell cancer. 18 , 19 , 20 ALKBH5 also functions as a suppressor gene in hepatocellular cancer, pancreatic cancer, bladder cancer and so on. 21 , 22 , 23 , 24 With regard to CRC, there are contrasting effects of ALKBH5 across different studies. Guo et al. indicated that ALKBH5 may promote cancer cell motility by demethylating the lncRNA NEAT1. 11 In contrast, Yang et al. illustrated that in CRC, ALKBH5 was downregulated and repressed tumour cell invasion in vitro and in vivo. 12 Previous studies were controversial, either drawing the opposite conclusion or with small samples validation. Therefore, a thorough and in‐depth study is necessary. In our research, the regulatory mechanism of ALKBH5 was studied in depth from clinical samples to cellular mechanisms. We demonstrated that ALKBH5 was downregulated in a large CRC cohort and closely related to the poor prognosis of CRC patients. Furthermore, ALKBH5 was found to significantly inhibit the growth and metastasis of colon cancer cells in vitro and in vivo. These suggested the suppressive role of ALKBH5 in CRC.
The expression of ALKBH5 is not fully consistent across different tumours, which indicates the tissue‐specific expression of ALKBH5. Therefore, it is necessary to explore downstream targets of ALKBH5. We comprehensively analysed the results of tissue sequencing and cell sequencing to screen out the downstream genes. Our data proved that PHF20 was an important downstream regulatory molecule of ALKBH5. ALKBH5 can remove the m6A modification of PHF20 mRNA 3′UTR, leading to the inhibition effect on the stability of PHF20 mRNA. IGF2BP1/2/3, as members of the m6A reading proteins, can recognize the m6A sites on 3′ UTR of mRNAs and maintain RNA stability. 25 We knocked down IGF2BP1, IGF2BP2 and IGF2BP3 in LOVO cells, respectively, and found that PHF20 was decreased both in the mRNA and protein levels only when IGF2BP3 was knocked down by siRNA (Figure S3). Therefore, IGF2BP3 may be a reader protein targeting PHF20 mRNA to reinforce its stability. We believe that after the m6A modification on PHF20 mRNA is removed by ALKBH5, the function of IGF2BP3 recognizing the m6A modigication and stabilizing PHF20 mRNA is weakened.
PHF20 is a component of the H4K16 histone acetyltransferase MOF complex, which binds to methylated lysine residues on histones. 26 , 27 Initially, PHF20 was identified as a tumour‐associated antigen that elicited an immune response in the serum of glioblastoma patients. 28 , 29 , 30 Furthermore, it was reported that PHF20 was highly expressed in many cancers, including nasopharyngeal cancer, lung cancer and CRC. 31 , 32 , 33 , 34 , 35 , 36 PHF20 was upregulated in CRC, particularly in invasive CRC. 35 PHF20 reduced p53 accumulation and inhibited p53 transcriptional activity to p21 and Bax in response to DNA damage in CRC. 36 We found that PHF20 knock‐down significantly suppressed the proliferation, colony formation, migration and invasion of LOVO. Moreover, our rescue experiments suggested that the knock‐down of PHF20 inhibited the shALKBH5‐mediated enhancement of colon cancer cellular functions in vitro. These results validate the tumour‐promoting role of PHF20 in CRC.
5. CONCLUSIONS
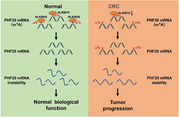
We determine that ALKBH5 expression is downregulated in CRC and plays a crucial tumour suppressive role. Mechanistically, ALKBH5 inhibits the stability of PHF20 mRNA by removing the m6A modification (Figure 9). Our study contributes novel insights into the anticancer roles of ALKBH5‐mediated m6A modification and suggests that targeting the ALKBH5‐mediated m6A modification of PHF20 mRNA can be a hopeful strategy for the intervention and treatment of CRC.
FIGURE 9.
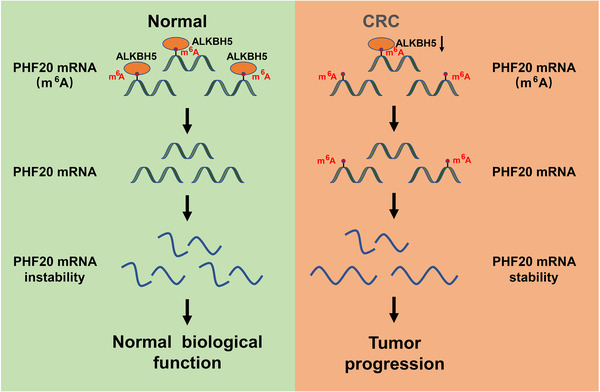
Schematic diagram of alkB homologue 5 (ALKBH5)‐mediated removal of N6‐methyladenosine (m6A) mRNA methylation, leading to the inhibition of PHF20 mRNA stability and suppression of colorectal cancer (CRC) progression
CONFLICT OF INTEREST
The authors declare no conflicts of interest for this study.
FUNDING INFORMATION
This work was supported by the National Natural Science Foundation of China (Grant no. 81972240) and the National Key Research and Development Program of China (Grant no. 2017YFC1308805).
Supporting information
Figure S1 Representative IHC images of ALKBH5, PHF20 and Ki67 expression in the tumour xenograft models (400×)
Figure S2 Knock‐down of PHF20 inhibited proliferation, migration and invasion of colon cancer cells in vitro.
Figure S3 Knock‐down of IGF2BP3 decreased the mRNA and protein level of PHF20.
Table S1 The primer sequences of genes
Table S2 The sequences of the wild type and m6A sites mutated PHF20
ACKNOWLEDGEMENTS
We thank the patients that are involved in this study, as well as our colleagues for their helpful suggestions.
Zhang Z, Wang L, Zhao L, et al. N6‐methyladenosine demethylase ALKBH5 suppresses colorectal cancer progression potentially by decreasing PHF20 mRNA methylation. Clin Transl Med. 2022;12:e940. 10.1002/ctm2.940
Zhen Zhang and Ling Wang contributed equally to this work.
REFERENCES
- 1. Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209‐249. [DOI] [PubMed] [Google Scholar]
- 2. Bi Z, Liu Y, Zhao Y, et al. A dynamic reversible RNA N(6) ‐methyladenosine modification: current status and perspectives. J Cell Physiol. 2019;234(6):7948‐7956. [DOI] [PubMed] [Google Scholar]
- 3. Yang Y, Hsu PJ, Chen YS, et al. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616‐624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tuck MT. Partial purification of a 6‐methyladenine mRNA methyltransferase which modifies internal adenine residues. Biochem J. 1992;288:233‐240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu J, Yue Y, Han D, et al. A METTL3‐METTL14 complex mediates mammalian nuclear RNA N6‐adenosine methylation. Nat Chem Biol. 2014;10(2):93‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jia G, Fu Y, Zhao X, et al. N6‐methyladenosine in nuclear RNA is a major substrate of the obesity‐associated FTO. Nat Chem Biol. 2011;7(12):885‐887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18‐29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li J, Liang L, Yang Y, et al. N(6)‐methyladenosine as a biological and clinical determinant in colorectal cancer: progression and future direction. Theranostics. 2021;11(6):2581‐2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang T, Kong S, Tao M, et al. The potential role of RNA N6‐methyladenosine in cancer progression. Mol Cancer. 2020;19(1):88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang J, Wang J, Gu Q, et al. The biological function of m6A demethylase ALKBH5 and its role in human disease. Cancer Cell Int. 2020;20:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guo T, Liu DF, Peng SH, et al. ALKBH5 promotes colon cancer progression by decreasing methylation of the lncRNA NEAT1. Am J Transl Res. 2020;12(8):4542‐4549. [PMC free article] [PubMed] [Google Scholar]
- 12. Yang P, Wang Q, Liu A, et al. ALKBH5 holds prognostic values and inhibits the metastasis of colon cancer. Pathol Oncol Res. 2020;26(3):1615‐1623. [DOI] [PubMed] [Google Scholar]
- 13. Yue B, Song C, Yang L, et al. METTL3‐mediated N6‐methyladenosine modification is critical for epithelial‐mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang Z, Wang Q, Zhang M, et al. Comprehensive analysis of the transcriptome‐wide m6A methylome in colorectal cancer by MeRIP sequencing. Epigenetics. 2021;16(4):425‐435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fedeles BI, Singh V, Delaney JC, et al. The AlkB family of Fe(II)/α‐ketoglutarate‐dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem. 2015;290(34):20734‐20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alemu EA, He C, Klungland A. ALKBHs‐facilitated RNA modifications and de‐modifications. DNA Repair (Amst). 2016;44:87‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aik W, Scotti JS, Choi H, et al. Structure of human RNA N⁶‐methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation. Nucleic Acids Res. 2014;42(7):4741‐4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang S, Zhao BS, Zhou A, et al. m(6)A Demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem‐like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31(4):591‐606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang C, Samanta D, Lu H, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF‐dependent and ALKBH5‐mediated m(6)A‐demethylation of NANOG mRNA. Proc Natl Acad Sci USA. 2016;113(14):E2047‐E2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nagaki Y, Motoyama S, Yamaguchi T, et al. m(6)A Demethylase ALKBH5 promotes proliferation of esophageal squamous cell carcinoma associated with poor prognosis. Genes Cells. 2020;25:547‐561. [DOI] [PubMed] [Google Scholar]
- 21. Chen Y, Zhao Y, Chen J, et al. ALKBH5 suppresses malignancy of hepatocellular carcinoma via m(6)A‐guided epigenetic inhibition of LYPD1. Mol Cancer. 2020;19(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tang B, Yang Y, Kang M, et al. m(6)A Demethylase ALKBH5 inhibits pancreatic cancer tumorigenesis by decreasing WIF‐1 RNA methylation and mediating Wnt signaling. Mol Cancer. 2020;19(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Guo X, Li K, Jiang W, et al. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A‐YTHDF2‐dependent manner. Mol Cancer. 2020;19(1):91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jin H, Ying X, Que B, et al. N(6)‐methyladenosine modification of ITGA6 mRNA promotes the development and progression of bladder cancer. EBioMedicine. 2019;47:195‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang H, Weng H, Sun W, et al. Recognition of RNA N(6)‐methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285‐295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li X, Wu L, Corsa CA, et al. Two mammalian MOF complexes regulate transcription activation by distinct mechanisms. Mol Cell. 2009;36(2):290‐301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dou Y, Milne TA, Tackett AJ, et al. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121(6):873‐885. [DOI] [PubMed] [Google Scholar]
- 28. Fischer U, Struss AK, Hemmer D, et al. Glioma‐expressed antigen 2 (GLEA2): a novel protein that can elicit immune responses in glioblastoma patients and some controls. Clin Exp Immunol. 2001;126(2):206‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pallasch CP, Struss AK, Munnia A, et al. Autoantibodies against GLEA2 and PHF3 in glioblastoma: tumor‐associated autoantibodies correlated with prolonged survival. Int J Cancer. 2005;117(3):456‐459. [DOI] [PubMed] [Google Scholar]
- 30. Heisel SM, Ketter R, Keller A, et al. Increased seroreactivity to glioma‐expressed antigen 2 in brain tumor patients under radiation. PLoS One. 2008;3(5):e2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaatar AM, Lim CR, Bong CW, et al. Whole blood transcriptome correlates with treatment response in nasopharyngeal carcinoma. J Exp Clin Cancer Res. 2012;31(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tang N, Ma L, Lin XY, et al. Expression of PHF20 protein contributes to good prognosis of NSCLC and is associated with Bax expression. Int J Clin Exp Pathol. 2015;8(10):12198‐12206. [PMC free article] [PubMed] [Google Scholar]
- 33. Bankovic J, Stojsic J, Jovanovic D, et al. Identification of genes associated with non‐small‐cell lung cancer promotion and progression. Lung Cancer. 2010;67(2):151‐159. [DOI] [PubMed] [Google Scholar]
- 34. Wang Y, Han KJ, Pang XW, et al. Large scale identification of human hepatocellular carcinoma‐associated antigens by autoantibodies. J Immunol. 2002;169(2):1102‐1109. [DOI] [PubMed] [Google Scholar]
- 35. Sugai T, Osakabe M, Sugimoto R, et al. A genome‐wide study of the relationship between chromosomal abnormalities and gene expression in colorectal tumors. Genes Chromosomes Cancer. 2021;60(4):250‐262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Y, Park J, Piao L, et al. PKB‐mediated PHF20 phosphorylation on Ser291 is required for p53 function in DNA damage. Cell Signal. 2013;25(1):74‐84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Representative IHC images of ALKBH5, PHF20 and Ki67 expression in the tumour xenograft models (400×)
Figure S2 Knock‐down of PHF20 inhibited proliferation, migration and invasion of colon cancer cells in vitro.
Figure S3 Knock‐down of IGF2BP3 decreased the mRNA and protein level of PHF20.
Table S1 The primer sequences of genes
Table S2 The sequences of the wild type and m6A sites mutated PHF20


