Short abstract
The metabolome enables unprecedented insight into biochemistry, providing an integrated signature of the genome, transcriptome, proteome and exposome. Measurement requires rigorous protocols combined with specialised data analysis to achieve its promise. https://bit.ly/3yPiYkQ
Introduction
Obstructive lung diseases including asthma and COPD are heterogenous syndromes for which the molecular determinants of individual subtypes of pathogenesis remain unclear. This is a major barrier in understanding disease aetiology and in stratifying patients for treatment as well as in identifying actionable therapeutic targets. There is an unmet need to understand the dysregulated biochemical processes driven by the interaction between genetic and environmental factors [1]. While challenging to study, this complex intersection can be captured via the metabolome. Metabolic phenotyping (metabotyping) has demonstrated sufficient molecular resolution to identify phenotypes and endotypes of respiratory disease, including asthma [2–4], COPD [5, 6], respiratory syncytial virus bronchiolitis [7] and cystic fibrosis [8–10].
Although metabolomics is a valuable tool for investigating respiratory disease, it poses several challenges that result in metabolomics being a nonstandardised discipline for which there is no one-size-fits-all solution. To assist the respiratory community in asking the right questions and understanding the data generated by metabolomics, we herein discuss both challenges and recommendations for integrating metabolomics into respiratory research.
Background
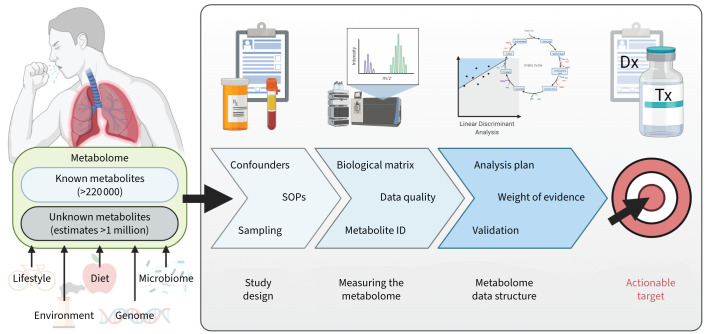
The metabolome is defined as the complement of small-molecule metabolites (generally <1500 Da) that arise from cellular metabolism. The human metabolome is complex, with size estimates varying widely and reaching upwards of 1 million molecules [11] compared to ∼21 000 protein-encoding genes and 15 000 gene products, which are determined by four bases or 20 amino acids, respectively. This chemical diversity renders the metabolome analytically challenging to measure relative to other omics technologies. The Human Metabolome Database currently contains 220 945 metabolite entries (https://hmdb.ca, accessed November 2021) [12]; however, only several thousand have been included in human metabolism models [13]. The utility of measuring the metabolome lies in its ability to provide insight into biochemical processes, essentially reflecting in real-time what the cell/tissue is doing at the moment of sampling, whereas genetics and transcriptomics reflect what the cell/tissue is potentially capable of doing or planning to do, respectively. The metabolome reflects the integrative effects of the genome, transcriptome, proteome and exposome together with other influencing factors such as diet and microbiome (figure 1). While the metabolome is the closest biological level to the phenotype, studying the metabolome poses its own unique challenges, discussed herein.
FIGURE 1.
Metabolomics overview illustrating the key components of a metabolomics experimental workflow. While the full complement of the human metabolome remains unknown, estimates range upwards of >1 million compounds. The utility of the metabolome lies in its ability to reflect the integrated metabolic signature of gene and environment (G×E) interactions combined with dietary and lifestyle influences. Measuring the metabolome requires a three-pronged approach that includes rigorous study protocols in combination with dedicated analytical methods and field-specific data analysis. When properly executed, the metabotype can inform clinical diagnosis and treatment as a component of precision medicine approaches. SOPs: standard operating procedures; ID: identification; Dx: diagnosis; Tx: treatment.
Metabolomics study design
Challenges
A strength and weakness of metabolomics is its inherent sensitivity to both internal and external stimuli, which has led to it being described as the canary of the genome [12]. First, it is necessary to control the study parameters to minimise physiological differences between study groups. For example, a systematic review reported 196 metabolites that significantly change their concentration in human biofluids 24 h post-exercise [14]. Second, the sampling and storage conditions of biosamples can also affect the observed metabolome; for example, haemolysis resulted in 30% of the measured blood metabolites significantly changing [15]. Third, given the sensitivity of the metabolome, it is vital to account for confounders (e.g. sex, age, body mass index, smoking, study site bias). External factors including diet and medication can exert strong effects on observed metabolite levels; for example, both inhaled and oral steroids exert metabolite-specific effects [3, 4, 16].
Recommendations
Prospectively plan sample collection, processing and biobanking to ensure integrity for metabolomics analysis. These efforts include standardised operating procedures for sample collection (e.g. collect samples at the same time of day, use the same collection tubes across all study centres), sample workup (e.g. same centrifugation speed, duration and temperature for preparation of plasma/serum) and sample storage (e.g. same temperature, defined biobanking protocols) [17, 18]. A comprehensive review of sampling aspects for metabolomics has been published [17]. Relevant clinical, lifestyle and treatment data should be recorded. When prospective planning is not possible, pilot studies are recommended to document potential bias associated with the sample handling conditions relevant for the study. It is useful to identify a priori metabolites or metabolic pathways that are central to the study to ensure that the selected method (e.g. choice of matrix, metabolomics method) will provide appropriate data. Multicentre recruitment sites can also exert bias on a metabolite-specific basis [3], and care must be taken to minimise site-related confounding factors. Failing to implement such recommendations can result in the introduction of statistical error (bias and variance), thus reducing the reliability in results and leaving the study vulnerable to irreproducibility.
Measuring the metabolome
Challenges
There are >350 000 chemicals registered for production and use [19], each of which can lead to multiple metabolites following intake to the human body. This chemical complexity, in combination with endogenous metabolism, requires specific considerations for measurement. First, the metabolome is physicochemically diverse. Metabolites range from small and polar molecules (e.g. amino acids) to large and nonpolar molecules (e.g. lipids). Second, the concentrations span several orders of magnitude [20], for example metabolite concentrations in blood range from picomolar (e.g. thromboxane B3) to millimolar (e.g. lactate) [21]. Third, the speciation and concentration of metabolites is biofluid- and tissue-specific. Fourth, many metabolites have short half-lives and/or low stability (e.g. thromboxane A2 has a half-life of ∼30 s [22]; glutathione oxidises easily and is highly reactive). Fifth, the full complement of metabolites in the metabolome is unknown.
Due to metabolite diversity, no single analytical platform can capture the metabolome in its entirety. For this reason, several analytical methods are used to increase coverage, each having their own benefits and limitations, with liquid chromatography-mass spectrometry (LC-MS) and gas chromatography-mass spectrometry (GC-MS) being the most common. Dunn et al. [23] published a comprehensive review of analytical platforms for metabolomics. These analytical techniques are designed to be discovery-based and are not developed in adherence to accreditation standards. Accordingly, alternative precision and quality-control measures are applied to ensure that high-quality data are achieved [24]. The most widely employed precision measure is the relative standard deviation (RSD) of biologically identical quality control samples. The dispersion ratio (D-ratio), the ratio of the standard deviation of quality-control samples to the standard deviation of biological test samples (i.e. the ratio of methodological variation to biological sample variation), has recently emerged as an alternative quality metric. For reliable statistical interpretation, it is recommended that the D-ratio should be <50% [24].
In LC-MS and GC-MS, metabolites are annotated and identified by comparing chromatographic and spectral properties to metabolite databases. Confident metabolite identification is required for accurate biological interpretation; however, this remains a key challenge and metabolites are often not all identified at the same confidence level [25, 26]. For reliable annotation, the chemical signal in the sample must match that of a chemical standard measured using the same method. However, for the majority of human metabolites, chemical standards are not available. In the absence of verification, mass spectral fragmentation patterns can be interpreted for metabolite identification but the identify is not considered confirmed, which can result in misidentification and subsequent misinterpretation.
Recommendations
Engage early with the metabolomics laboratory to discuss the specific study needs to choose methods that align with the study goals. It is essential to understand and report on the level of certainty in the metabolite identification [25] as well as the measurement precision of each metabolite (e.g. RSD, D-ratio). The reporting of unnamed metabolite features is discouraged. For pulmonary research, there is an unmet need to determine which matrices are most informative. Most metabolomics investigations have focused on blood and urine because the analytical methods are well established, and sampling is routine. Few studies have examined matrices that are potentially more insightful for respiratory medicine (e.g. bronchoalveolar lavage fluid [27], bronchial wash [28], exhaled breath [29], saliva [30], sputum [31]), with the exception of breathomics, which has been reviewed elsewhere [32]. Analysis of these matrices is challenging due to invasive or nonroutine sampling requirements, dilute metabolite concentrations and lack of standardised strategies for normalising intersample concentration differences.
Metabolome data structure
Challenges
Metabolism is interconnected and changes in metabolite concentration occur in combination with other metabolites. As a result, metabolomics data are highly covariate. Accordingly, data analysis methods used in other omics sciences are not always entirely relevant for metabolomics data. For example, univariate hypothesis testing followed by calculation of the false discovery rate (FDR) is often applied to omics data, but FDR methods assume that variables are independent [33, 34]. To model the covariance and assess the combinatorial effects of metabolites, data are routinely analysed using multivariate (clustering, dimensionality reduction) and machine learning (e.g. partial least squares discriminant analysis, support vector machines, random forests) methods [35–37]. In addition, as with other omics technologies, it is not straightforward to perform a power calculation. Pathway mapping approaches are sometimes used to aid biological interpretations; however, these should be used cautiously [38], because they are vulnerable to bias based upon choice of the background set (e.g. all the compounds identified in a particular assay versus reference pathway), differential metabolite selection methods (e.g. p-value versus q-value), and pathway database (e.g. Kyoto Encyclopedia of Genes and Genomes, Reactome, BioCyc). For example, the background set can be biased towards metabolites that are easily detectable (e.g. tryptophan metabolism) and pathway size can influence the findings with smaller pathways being more significant than larger pathways.
Recommendations
Develop a data analysis strategy prior to study initiation. In particular, determine which analysis methods are most appropriate to answer the study question; assess whether they are feasible to implement; and ensure they are performed in consultation with experts. A weight-of-evidence approach can be useful in interpreting results. A detailed protocol for statistical analysis of metabolomics data, including power calculations, has been published recently [39]. Metabolomics is a discovery science, and results should inform planning for future analysis and validation studies. Metabolomics data often require confirmation using targeted quantitative methods in the same way that transcriptomics data are confirmed with reverse transcriptase quantitative PCR. Another resource for data confirmation is to interrogate online repositories of metabolomics data (e.g. the COnsortium of METabolomics Studies [40]). A challenge for pulmonary research is that clinical cohorts are often small, resulting in decreased statistical power, sampling error and a paucity of data in online repositories. There is a need to collaborate to increase the size of studies in respiratory medicine, particularly in terms of longitudinal sampling, sampling in association with exacerbations, and intervention studies. In addition, there is a concomitant need for respiratory researchers to submit the results of their investigations into common online data repositories (e.g. MetaboLights [41], Metabolomics Workbench [42]).
Future of metabolomics in pulmonary medicine
Despite the challenges, metabolomics efforts have made important discoveries in pulmonary medicine [4, 43, 44]. The future applications lie in our ability to perform deep molecular phenotyping of disease to identify treatable endotypes (e.g. identify molecular markers to stratify patients for treatment with biologics). In addition, metabolomics is a useful component of precision medicine strategies for monitoring of individuals and can be readily adapted to home monitoring. For example, home microsampling can be paired with home spirometry and smartphone-based apps to provide increased understanding of the triggers of exacerbations as well as in identifying subgroups of individuals as unique responders and in determining appropriate treatment. It may be possible to link environmental and dietary exposures to specific metabotypes in an exposome-based approach to identify specific triggers of disease subphenotypes. There is increasing use of multiomics analysis, integrating data from metabolomics with other omics platforms to gain a more comprehensive understanding of molecular mechanisms [43]. The metabolome is dynamic and there is a need for longitudinal monitoring of individuals to enable temporal mapping and stability assessments. In addition, there is a need to develop clinically robust assays to be able to analyse data over long-term monitoring projects, particularly in regard to precision heath programmes. To realise its potential, the limitations of metabolomics outlined herein must be addressed. As the field continues to advance, it can be reasonably expected that metabolomics will become a routine tool; however, in its current phase, there is a need to understand the methods being used and in particular the limitations and biases of the chosen approach. This information is necessary to assess the data quality as well as ensure that the biological and clinical interpretations are accurate, which will enable extraction of the maximal information from a metabolomics experiment.
Shareable PDF
Footnotes
Conflict of interest: None declared.
Support statement: We acknowledge support from the Australian Government National Health and Medical Research Council (GNT1140234), Gunma University Initiative for Advanced Research (GIAR) and the Swedish Heart Lung Foundation (HLF 20210519, 20200693). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Wheelock CE, Rappaport SM. The role of gene-environment interactions in lung disease: the urgent need for the exposome. Eur Respir J 2020; 55: 1902064. doi: 10.1183/13993003.02064-2019 [DOI] [PubMed] [Google Scholar]
- 2.Kelly RS, Mendez KM, Huang M, et al. Metabo-endotypes of asthma reveal differences in lung function: discovery and validation in two TOPMed cohorts. Am J Respir Crit Care Med 2022; 205: 288–299. doi: 10.1164/rccm.202105-1268OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinke SN, Naz S, Chaleckis R, et al. Urinary metabotype of severe asthma evidences decreased carnitine metabolism independent of oral corticosteroid treatment in the U-BIOPRED study. Eur Respir J 2022; 59: 2101733. doi: 10.1183/13993003.01733-2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kachroo P, Stewart ID, Kelly RS, et al. Metabolomic profiling reveals extensive adrenal suppression due to inhaled corticosteroid therapy in asthma. Nat Med 2022; 28: 814–822. doi: 10.1038/s41591-022-01714-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowler RP, Jacobson S, Cruickshank C, et al. Plasma sphingolipids associated with chronic obstructive pulmonary disease phenotypes. Am J Respir Crit Care Med 2015; 191: 275–284. doi: 10.1164/rccm.201410-1771OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh N, Choudhury P, Kaushik SR, et al. Metabolomic fingerprinting and systemic inflammatory profiling of asthma COPD overlap (ACO). Respir Res 2020; 21: 126. doi: 10.1186/s12931-020-01390-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raita Y, Pérez-Losada M, Freishtat RJ, et al. Integrated omics endotyping of infants with respiratory syncytial virus bronchiolitis and risk of childhood asthma. Nat Commun 2021; 12: 3601. doi: 10.1038/s41467-021-23859-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler JD, Margaroli C, Horati H, et al. Myeloperoxidase oxidation of methionine associates with early cystic fibrosis lung disease. Eur Respir J 2018; 52: 1801118. doi: 10.1183/13993003.01118-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esther CR Jr, Turkovic L, Rosenow T, et al. Metabolomic biomarkers predictive of early structural lung disease in cystic fibrosis. Eur Respir J 2016; 48: 1612–1621. doi: 10.1183/13993003.00524-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raghuvanshi R, Vasco K, Vázquez-Baeza Y, et al. High-resolution longitudinal dynamics of the cystic fibrosis sputum microbiome and metabolome through antibiotic therapy. mSystems 2020; 5: e00292-20. doi: 10.1128/mSystems.00292-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uppal K, Walker DI, Liu K, et al. Computational metabolomics: a framework for the million metabolome. Chem Res Toxicol 2016; 29: 1956–1975. doi: 10.1021/acs.chemrestox.6b00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wishart D. Systems biology resources arising from the human metabolome project. In: Suhre K, ed. Genetics Meets Metabolomics: from Experiment to Systems Biology. New York, Springer, 2012; pp. 157–175. [Google Scholar]
- 13.Thiele I, Sahoo S, Heinken A, et al. Personalized whole-body models integrate metabolism, physiology, and the gut microbiome. Mol Syst Biol 2020; 16: e8982. doi: 10.15252/msb.20198982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schranner D, Kastenmüller G, Schönfelder M, et al. Metabolite concentration changes in humans after a bout of exercise: a systematic review of exercise metabolomics studies. Sports Med Open 2020; 6: 11. doi: 10.1186/s40798-020-0238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kamlage B, Maldonado SG, Bethan B, et al. Quality markers addressing preanalytical variations of blood and plasma processing identified by broad and targeted metabolite profiling. Clin Chem 2014; 60: 399–412. doi: 10.1373/clinchem.2013.211979 [DOI] [PubMed] [Google Scholar]
- 16.Reinke SN, Gallart-Ayala H, Gómez C, et al. Metabolomics analysis identifies different metabotypes of asthma severity. Eur Respir J 2017; 49: 1601740. doi: 10.1183/13993003.01740-2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirwan JA, Brennan L, Broadhurst D, et al. Preanalytical processing and biobanking procedures of biological samples for metabolomics research: a white paper, community perspective (for “Precision Medicine and Pharmacometabolomics Task Group” – the Metabolomics Society initiative). Clin Chem 2018; 64: 1158–1182. doi: 10.1373/clinchem.2018.287045 [DOI] [PubMed] [Google Scholar]
- 18.Elliott P, Peakman TC, UK Biobank . The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int J Epidemiol 2008; 37: 234–244. doi: 10.1093/ije/dym276 [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Walker GW, Muir DCG, et al. Toward a global understanding of chemical pollution: a first comprehensive analysis of national and regional chemical inventories. Environ Sci Technol 2020; 54: 2575–2584. doi: 10.1021/acs.est.9b06379 [DOI] [PubMed] [Google Scholar]
- 20.Rappaport SM, Barupal DK, Wishart D, et al. The blood exposome and its role in discovering causes of disease. Environ Health Perspect 2014; 122: 769–774. doi: 10.1289/ehp.1308015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Psychogios N, Hau DD, Peng J, et al. The human serum metabolome. PLoS One 2011; 6: e16957. doi: 10.1371/journal.pone.0016957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hamberg M, Svensson J, Samuelsson B. Thromboxanes: a new group of biologically active compounds derived from prostaglandin endoperoxides. Proc Natl Acad Sci USA 1975; 72: 2994–2998. doi: 10.1073/pnas.72.8.2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dunn WB, Broadhurst DI, Atherton HJ, et al. Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem Soc Rev 2011; 40: 387–426. doi: 10.1039/B906712B [DOI] [PubMed] [Google Scholar]
- 24.Broadhurst D, Goodacre R, Reinke SN, et al. Guidelines and considerations for the use of system suitability and quality control samples in mass spectrometry assays applied in untargeted clinical metabolomic studies. Metabolomics 2018; 14: 72. doi: 10.1007/s11306-018-1367-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis. Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007; 3: 211–221. doi: 10.1007/s11306-007-0082-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaleckis R, Meister I, Zhang P, et al. Challenges, progress and promises of metabolite annotation for LC-MS-based metabolomics. Curr Opin Biotechnol 2019; 55: 44–50. doi: 10.1016/j.copbio.2018.07.010 [DOI] [PubMed] [Google Scholar]
- 27.Walmsley S, Cruickshank-Quinn C, Quinn K, et al. A prototypic small molecule database for bronchoalveolar lavage-based metabolomics. Sci Data 2018; 5: 180060. doi: 10.1038/sdata.2018.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surowiec I, Karimpour M, Gouveia-Figueira S, et al. Multi-platform metabolomics assays for human lung lavage fluids in an air pollution exposure study. Anal Bioanal Chem 2016; 408: 4751–4764. doi: 10.1007/s00216-016-9566-0 [DOI] [PubMed] [Google Scholar]
- 29.Brinkman P, Wagener AH, Hekking PP, et al. Identification and prospective stability of electronic nose (eNose)-derived inflammatory phenotypes in patients with severe asthma. J Allergy Clin Immunol 2019; 143: 1811–1820. doi: 10.1016/j.jaci.2018.10.058 [DOI] [PubMed] [Google Scholar]
- 30.Cruickshank-Quinn C, Armstrong M, Powell R, et al. Determining the presence of asthma-related molecules and salivary contamination in exhaled breath condensate. Respir Res 2017; 18: 57. doi: 10.1186/s12931-017-0538-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brandsma J, Goss VM, Yang X, et al. Lipid phenotyping of lung epithelial lining fluid in healthy human volunteers. Metabolomics 2018; 14: 123. doi: 10.1007/s11306-018-1412-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ibrahim W, Carr L, Cordell R, et al. Breathomics for the clinician: the use of volatile organic compounds in respiratory diseases. Thorax 2021; 76: 514–521. doi: 10.1136/thoraxjnl-2020-215667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Methodol 1995; 57: 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 34.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA 2003; 100: 9440–9445. doi: 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Broadhurst DI, Kell DB. Statistical strategies for avoiding false discoveries in metabolomics and related experiments. Metabolomics 2006; 2: 171–196. doi: 10.1007/s11306-006-0037-z [DOI] [Google Scholar]
- 36.Gromski PS, Muhamadali H, Ellis DI, et al. A tutorial review: metabolomics and partial least squares-discriminant analysis – a marriage of convenience or a shotgun wedding. Anal Chim Acta 2015; 879: 10–23. doi: 10.1016/j.aca.2015.02.012 [DOI] [PubMed] [Google Scholar]
- 37.Mendez KM, Reinke SN, Broadhurst DI. A comparative evaluation of the generalised predictive ability of eight machine learning algorithms across ten clinical metabolomics data sets for binary classification. Metabolomics 2019; 15: 150. doi: 10.1007/s11306-019-1612-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wieder C, Frainay C, Poupin N, et al. Pathway analysis in metabolomics: recommendations for the use of over-representation analysis. PLoS Comput Biol 2021; 17: e1009105. doi: 10.1371/journal.pcbi.1009105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blaise BJ, Correia GDS, Haggart GA, et al. Statistical analysis in metabolic phenotyping. Nat Protoc 2021; 16: 4299–4326. doi: 10.1038/s41596-021-00579-1 [DOI] [PubMed] [Google Scholar]
- 40.Yu B, Zanetti KA, Temprosa M, et al. The Consortium of Metabolomics Studies (COMETS): metabolomics in 47 prospective cohort studies. Am J Epidemiol 2019; 188: 991–1012. doi: 10.1093/aje/kwz028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haug K, Cochrane K, Nainala VC, et al. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res 2020; 48: D440–D444. doi: 10.1093/nar/gkz1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sud M, Fahy E, Cotter D, et al. Metabolomics Workbench: an international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res 2016; 44: D463–D470. doi: 10.1093/nar/gkv1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kelly RS, Dahlin A, McGeachie MJ, et al. Asthma metabolomics and the potential for integrative omics in research and the clinic. Chest 2017; 151: 262–277. doi: 10.1016/j.chest.2016.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujimura KE, Sitarik AR, Havstad S, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med 2016; 22: 1187–1191. doi: 10.1038/nm.4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00102-2022.Shareable (376.5KB, pdf)