Abstract
The Human Cell Atlas (HCA) consortium aims to establish an atlas of all organs in the healthy human body at single-cell resolution to increase our understanding of basic biological processes that govern development, physiology and anatomy, and to accelerate diagnosis and treatment of disease. The Lung Biological Network of the HCA aims to generate the Human Lung Cell Atlas as a reference for the cellular repertoire, molecular cell states and phenotypes, and cell–cell interactions that characterise normal lung homeostasis in healthy lung tissue. Such a reference atlas of the healthy human lung will facilitate mapping the changes in the cellular landscape in disease. The discovAIR project is one of six pilot actions for the HCA funded by the European Commission in the context of the H2020 framework programme. discovAIR aims to establish the first draft of an integrated Human Lung Cell Atlas, combining single-cell transcriptional and epigenetic profiling with spatially resolving techniques on matched tissue samples, as well as including a number of chronic and infectious diseases of the lung. The integrated Human Lung Cell Atlas will be available as a resource for the wider respiratory community, including basic and translational scientists, clinical medicine, and the private sector, as well as for patients with lung disease and the interested lay public. We anticipate that the Human Lung Cell Atlas will be the founding stone for a more detailed understanding of the pathogenesis of lung diseases, guiding the design of novel diagnostics and preventive or curative interventions.
Short abstract
The discovAIR project contributes to the Human Cell Atlas Lung Biological Network by establishing a first draft of the Human Lung Cell Atlas, advancing our insight into the cellular complexity and spatial organisation of the lung in health and disease https://bit.ly/3zX4cad
Introduction
Lung diseases are leading causes of death worldwide [1], with incidences increasing at an alarming rate while curative interventions are lacking for most of these disorders. Research into lung diseases lags behind compared with cancer and cardiovascular diseases, the other two major causes of death [2]. The high anatomical complexity of lung tissue, with the bifurcating bronchial tree, facilitating transport of air, and the parenchyma containing the alveoli for gas exchange, as well as its extremely rich cellular heterogeneity, with more than 50 different cell types identified to date [3], are some of the obstacles hampering progress in understanding the mechanisms of the many different lung diseases. These discrete lung cell types are each defined by morphological features, as well as constitutive gene expression patterns that are essential for cell-type identity [4]. Each cell type can adopt various molecular cell states, defined by transient expression of additional, facultative gene modules to allow execution of specific functions, adaptations to environmental factors or stimuli, or transition to another cell type during differentiation or in disease pathogenesis [4]. Therefore, the exact molecular phenotype of lung cells in time and space is determined by their exact location within the highly complex, three-dimensional (3D) structure of the lung, their local interactions with other cells, with the lung matrix and with the external environment that is omnipresent in this organ. The rich cellular complexity and precise spatial organisation are critical for proper lung function in health and are often lost in disease. Hence, there is an urgent need to have a detailed description of this complexity in healthy lung tissue and propel basic, translational and clinical research in lung disease into a fast track for the development of precision diagnostics and therapeutics. To achieve this, we need a detailed understanding of the cells that make up the lung, their fixed and variable features, their interactions, and their organisation into local cellular neighbourhoods and higher-order structures that make up the macroscopic tissue architecture in health, and the deviations thereof in disease [4].
The central goal of the Human Cell Atlas (HCA; https://humancellatlas.org) is to establish such an atlas of all organs in the healthy human body [5]. Achieving this daunting task for the lung is the central goal of the Lung Biological Network of the HCA (HCA-Lung) [6], an open research community that encompasses a large number of research groups, as well as several research consortia, including discovAIR (https://discovAIR.org), CZI Seed Network for the Human Cell Atlas (http://bit.ly/HCALungCZI1), LungMAP (https://LungMAP.net) and HuBMAP (https://HuBMAPconsortium.org). The discovAIR consortium is one of six pilot actions for the HCA funded by the European Commission in the context of the H2020 framework programme.
discovAIR aims to contribute to establishing the first draft of an integrated Human Lung Cell Atlas. In this atlas, discovAIR will contribute significantly to the multimodal single-cell omics data from healthy human lung, to mapping the cellular heterogeneity observed in single-cell RNA sequencing (scRNA-seq) datasets onto the lung tissue architecture, and to identification of changes of molecular cell phenotypes and their interactions in lung disease cohorts such as asthma, chronic obstructive pulmonary disease (COPD), pulmonary arterial hypertension (PAH), coronavirus disease 2019 (COVID-19) and interstitial lung diseases (ILDs) such as interstitial pulmonary fibrosis, enabling accelerated translational and clinical research into lung diseases. The discovAIR results will facilitate progress in regenerative and precision medicine by identifying novel candidates for precision diagnostics and curative interventions in lung disease. Here, we present the contributions of the discovAIR project to the roadmap of the Human Lung Cell Atlas from HCA-Lung.
The Lung Biological Network of the HCA
The anatomy, physiology and cell-type composition of the lung have been studied in great detail using classical immunohistochemical and pathological approaches, with well-characterised relationships between cellular phenotype and function [7]. The application of scRNA-seq techniques to human lung tissue, however, showed that our knowledge of the cellular composition of the lung is incomplete and novel cell types such as the pulmonary ionocyte have been identified [8, 9]. Consequently, HCA-Lung aims to identify all cell types and their molecular states or activities present in healthy lung tissue, and their interactions and organisation into higher-order anatomical and/or functional units [4].
Lung tissue is exquisitely suitable for such a mapping effort, due to the presence of unambiguous tissue landmarks to relate local cellular neighbourhoods and larger-order cellular structures back to defined physical coordinates within the organ. Importantly, healthy lung tissue is available to a large number of research groups through lung resections in patients with lung disease (healthy lung tissue adjacent to well-defined regions of disease, such as lung cancer), but also from organ donation programmes and in some cases through research bronchoscopy programmes involving healthy control subjects. This combination of a highly ordered tissue architecture, facilitating the implementation of a common coordinate framework (CCF) [10], and good community-wide availability of tissue makes lung especially well-suited as a lead organ for the HCA to develop the infrastructure, workflows, platforms and computational approaches needed for a community-driven tissue mapping effort as laid down in the vision of the HCA consortium [5]. Consequently, atlases of both the airways and parenchymal lung tissue have been selected by the HCA consortium as a priority effort, with the Human Lung Cell Atlas having developed into one of the HCA Flagship projects [6]. The infrastructure, workflows, roadmap and platforms developed within HCA-Lung can serve as blueprints for other biological networks of the HCA community.
The roadmap of the Lung Biological Network
The identification of the pulmonary ionocyte as a previously unknown cell type present in healthy lung tissue [8, 9] sparked a number of studies mapping the cellular heterogeneity of healthy lung tissue with increasing spatial and cellular resolution, and taking into account smoking as one of the most important factors driving specific cell states in lung tissue [3, 11–13]. An overview of current lung tissue scRNA-seq datasets and their availability is provided in supplementary appendix S1. Progress in the Human Lung Cell Atlas from multiple sudies has also recently been summarised [14]. Notwithstanding the importance of these foundational datasets for the respiratory community, HCA-Lung aspires to move well beyond this current state-of-the-art that is hampered by a relatively low number of donors, poor coverage of different ethnicities and ancestral diversity, as well as age range of tissue donors, limited resolution across the CCF and lack of spatial mapping of the identified cell states onto the tissue architecture [4, 6].
Therefore, HCA-Lung aims to provide a true reference atlas of the lung that captures the heterogeneity of cell states associated with location within the organ, as well as the variation of cell states that can be observed within healthy lung tissue, as a function of genetic, demographic and geographical variables. One approach to establish such a reference atlas is to leverage recent advances in computational data integration techniques [15–19] to integrate the currently available scRNA-seq datasets into a single embedding, thereby establishing an integrated Human Lung Cell Atlas that captures most of the available data and identifies undersampled regions and cell populations, and efficiently corrects for batch effects. In addition to capturing the full heterogeneity in transcriptional cell states of lung cells, HCA-Lung aims to incorporate a much larger variation in ethnicity and ancestral diversity, age range and geographical location of tissue donors, as well as multiple layers of omics data into an advanced draft of the Human Lung Cell Atlas, including epigenetic features such as CpG methylation, chromatin accessibility and histone modifications, or proteomic features at single-cell resolution. Leveraging the recent advances in single-cell joint profiling protocols, paired modalities can enrich our RNA reference using transfer learning approaches that map the RNA measurements onto each other [20]. Finally, the lack of a systematic description of molecular cell phenotypes with spatially resolving methods is severely hampering the interpretation of the wealth of data generated by the single-cell omics approaches. Clearly, adding such spatial maps of healthy lung tissue is one of the current priorities of HCA-Lung. Merging spatial datasets with an integrated Human Lung Cell Atlas based on multiple layers of omics data will then establish a true Human Lung Cell Atlas of healthy lung tissue, which will be an invaluable resource for basic, translational and clinical research into lung and its diseases.
The discovAIR approach to establish the Human Lung Cell Atlas
The discovAIR project aims to contribute to the HCA-Lung goals by delivering the first version of the Human Lung Cell Atlas towards the summer of 2022. Since 2018, a number of studies have presented tissue atlases of both healthy and diseased human lung [3, 11, 13, 21–30], using scRNA-seq complemented with single-molecule fluorescent in situ hybridisation (smFISH) analyses for validation of the main results. While these studies provide a rich resource for mapping cell-type-specific gene expression patterns, as exemplified by the rapid description of the expression patterns of genes encoding the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cell entry factors by the HCA-Lung community during the COVID-19 pandemic [31], these are usually single-centre studies and limited by the relatively low number of biological replicates, the low resolution in spatial locations sampled, the annotation of “novel” cell-type labels lacking ontological or pathological context and the lack of detailed spatially resolving methods to accompany the scRNA-seq data.
To address this shortcoming, discovAIR will combine multimodal single-cell profiling of lung cells with multimodal spatial mapping of the lung cell types and their molecular states onto the lung tissue architecture (figure 1). Moreover, discovAIR partners will integrate multiple lung tissue single-cell datasets into a single embedding, establishing the first draft of an integrated Human Lung Cell Atlas, allowing cell-type-specific analysis of the transcriptomic variation explained by demographic covariates, as was recently piloted for a limited number of SARS-CoV-2 cell entry genes by a highly collaborative HCA-Lung effort [32]. We have recently benchmarked computational approaches for data integration of existing scRNA-seq datasets [19]. We will use this framework to identify the best-suited method for integration of available scRNA-seq datasets from healthy lung tissue. The resulting integrated dataset will be presented as the core reference of healthy lung tissue and can be used as the first draft of an integrated Human Lung Cell Atlas. This atlas can then be used as a reference to which newly generated datasets from either healthy or diseased lung tissue can be compared using transfer learning methods such as scArches [20]. Taking this approach is especially powerful, as it allows unified use of cell-type labels, direct comparisons of cellular composition across datasets and, in the case of diseased tissue, direct identification of unique, disease-associated cell states absent from the healthy reference. Moreover, the integrated Human Lung Cell Atlas can help to structure discussions around cell-type label harmonisation and mapping of the hierarchical cell-type labels used for data integration (figure 1) to existing ontologies of the cells of the lung [33].
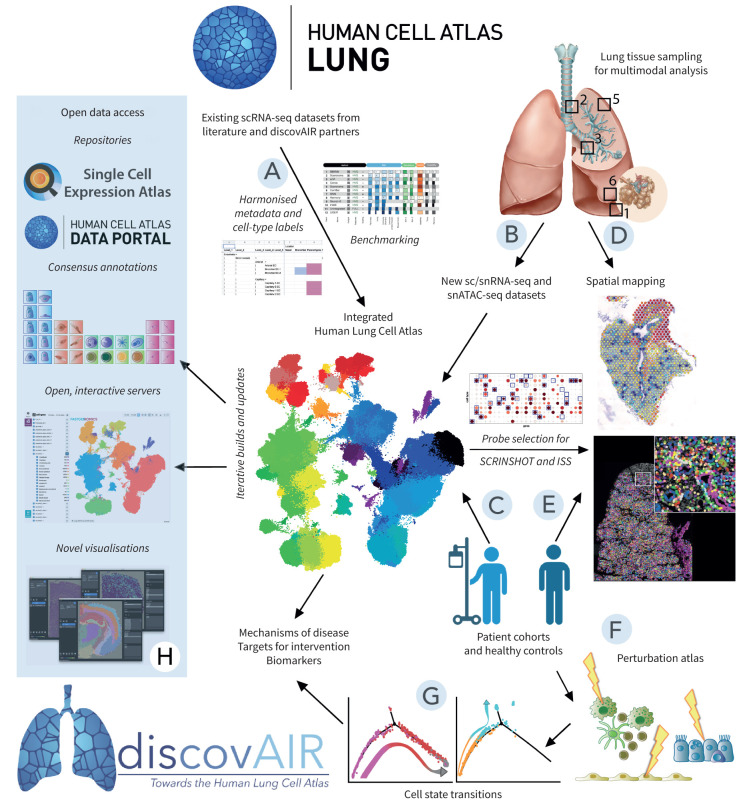
FIGURE 1.
discovAIR approach and workflow. The discovAIR consortium aims to establish a first draft of the Human Lung Cell Atlas by integrating existing single-cell RNA sequencing (scRNA-seq) datasets from the Lung Biological Network of the HCA (HCA-Lung) into a single embedding, allowing analyses on the integrated dataset (A). This dataset will be enriched by additional datasets generated by the discovAIR consortium: a multi-omics characterisation of lung tissue from five healthy donor lungs sampled at five locations (“deep dive”) (B) and a cohort of healthy controls (at least n=50) and patients with lung disease (at least n=50) (C). The “deep dive” tissue samples will also be used to generate spatially resolving datasets on matched tissue sections using Visium, in situ sequencing, SCRINSHOT (single-cell resolution in situ hybridisation on tissues) and laser capture microdissection (LCM)-guided single-nucleus RNA sequencing (snRNA-seq) analysis (D). Samples from the discovAIR cohort (patients and controls) will be used for spatial mapping using SCRINSHOT (E) and for establishment of the lung cell perturbation atlas (F). In the lung cell perturbation atlas, stimulations of ex vivo cultured primary cells or precision-cut lung tissue slices will be used to map cell state trajectories of healthy cells to diseased cell states (G). These novel datasets will be ingested into the next iterations of the integrated Human Lung Cell Atlas, used to establish consensus annotations for the different lung cell types and submitted to the appropriate data repositories (H) as open data using novel visualisation modalities for the spatial datasets. Finally, the integration of the Human Lung Cell Atlas (including datasets from lung disease) and the lung cell perturbation atlas will generate novel insights into mechanisms of disease, and help guide identification of drug targets and biomarkers for disease inception or treatment response. ISS: in situ sequencing; snATAC-seq: single-nucleus sequencing assay for transposase-accessible chromatin.
Towards the end of the project, discovAIR will establish a second draft of this integrated Lung Cell Atlas incorporating all newly generated data within the consortium, as well as data from the larger HCA-Lung community. The newly generated dataset from the discovAIR project will entail scRNA-seq data from at least 60 healthy controls spanning a large age range, both sexes and multiple ethnicities, and the detailed characterisation of the five-location dataset from healthy donor lung tissue encompassing multimodal data, as well as matching spatial datasets. It is important to bear in mind that any tissue sample obtained for building the reference atlas of healthy lung tissue will be derived from either deceased individuals whose lungs are suitable for organ transplant, lung resection programmes in routine clinical care (often as part of cancer treatment) or bronchoscopy studies performed for research purposes on healthy volunteers. Of these, only the latter source can be considered to reflect truly healthy, fresh lung tissue. The integrated Human Lung Cell Atlas will be made freely available to the respiratory and scientific community to ensure optimal impact of the discovAIR efforts. In addition to these integrated atlases of scRNA-seq data, discovAIR aims to develop innovative visualisations for spatial datasets, including 3D reconstruction of lung tissue architecture and molecular phenotyping of local cellular neighbourhoods by multiplexed smFISH or immunofluorescence analysis.
Most datasets with a large number of biological replicates sample only a limited number of locations within the organ and use a single-omics technique. To contribute to a more balanced coverage of multimodal sampling across the CCF in sufficient numbers of donors, discovAIR aims to establish a reference dataset offering an in-depth exploration of healthy lung tissue from five deceased transplant organ donors, each sampled at five discrete locations (figure 1) using both scRNA-seq and single-cell sequencing assay for transposase-accessible chromatin (scATAC-seq) to characterise their constant and variable features, as well as multimodal spatial profiling of lung cells on adjacent tissue sections to map their organisation in a 3D tissue architecture and their local and distant interactions. The generation of in-depth transcriptomic data will allow us to predict cell–cell interactions and the receptor–ligand pairs mediating these. The discovAIR approach will also allow validation of these predicted cell–cell interactions by spatial data that identifies, in a quantitative manner, cellular interactions.
The discovAIR project has selected four different spatially resolving methods. Of these, two are probe-based, allowing validation of the spatial expression pattern of a (limited) gene set, selected on the basis of the scRNA-seq data, by mapping these onto the tissue architecture. The other two methods are sequencing-based spatially resolving methods, allowing detection of spatial gene expression patterns without limiting the analysis to a prior selection of genes.
The two probe-based methods to map the transcriptional variation and cell-type heterogeneity observed in the single-cell datasets onto the spatial architecture of lung tissue differ in sensitivity and multiplexity. First, we will use a panel of 64 probes in the smFISH-based method SCRINSHOT (single-cell resolution in situ hybridisation on tissues) [34], which has high sensitivity but depends on sequential rounds of hybridisation and imaging of the same tissue section to achieve multiplexity. We have established two probe panels based on the transcriptional variation observed in a similar dataset covering these same five locations. One probeset was designed for airway wall-resident cell types and one for cell types of the lung parenchyma. In addition to SCRINSHOT, we will map the cell types and their molecular phenotypes in high detail with a panel of 150 probes using in situ sequencing (ISS), an alternative approach that has higher multiplexity but not the sensitivity of SCRINSHOT [35]. Probe selection for both SCRINSHOT and ISS was performed using a multi-objective computational approach, which optimises our ability to discern cell types of interest while recovering most of the transcriptional variation observed in these tissue samples with the selected probes (see supplementary appendix S2 for the current discovAIR smFISH gene lists for probe design in ISS and SCRINSHOT).
In addition to these probe-based spatial approaches, discovAIR has selected two unbiased, sequencing-based methods based on their capacity to generate spatially resolved transcriptomic data from the adjacent tissue sections from the same lung tissue samples as used in SCRINSHOT and ISS. In this pilot project for the Human Lung Cell Atlas, we have chosen to focus on spatially resolved transcriptome-based data only, while protein-based analyses will need to be integrated at a later stage. First, spatial transcriptomics using Visium [36] will be performed on 6.5 mm×6.5 mm×10 μm sections from the same lung tissue blocks also used for SCRINSHOT and ISS. The Visium spots are 55 μm in diameter with a 100 μm centre–centre distance and routinely yield more than 4000 genes per spot. Considering the architecture of the lung, we aim to profile pairs of adjacent sections (10 μm) followed by a gap of 100 μm so that a total of about 10 tissue sections will cover a tissue volume of over 1 cm3. A data-driven 3D model will be created based on the gene expression data. We have selected two out of the five “deep dive” locations to be included in the 3D transcriptomic approach: location 3 (third/fifth generation airway) and location 5 (lung parenchyma) (figure 1D), which will thus generate a first draft map of the transcriptome in two main sampling locations of the five deceased donor lungs. The 3D transcriptomic results will be integrated with the matching single-nucleus RNA sequencing (snRNA-seq) data generated on adjacent tissue sections to validate the spatial coordinates of the identified cell types. Moreover, SCRINSHOT and ISS data from adjacent sections will be available to guide further validation and higher-resolution mapping of the 3D transcriptomic map onto the tissue architecture. We will create probabilistic spatial cell maps of scRNA-seq-defined cell types using approaches like pciSeq [37] and Tangram [38].
The second sequencing-based approach with spatial resolution employs automated nuclear isolation using the laser capture microdissection (LCM) method [39], followed by snRNA-seq or snATAC-seq of the individual nuclei containing spatial coordinates. In this approach, a section of the lung tissue with a thickness of 10 μm and lateral sizes varying between a few millimetres and 1 cm is imaged in up to three colour channels. The nuclei of interest are detected by a machine learning method using the intensity information from the different colour channels. The detected nuclei are then sequentially cut and collected in a collector plate filled with the appropriate lysis buffer and further processed for single-nucleus sequencing (snRNA-seq or snATAC-seq). The spatial location of each nucleus is stored in a file by the LCM system. The RNA or ATAC sequencing data generated in this way will be correlated to the spatial location of the individual nuclei to provide insights on the distribution of particular cell populations across the human lung tissue. These datasets will be integrated with the matching snRNA-seq, as well as the SCRINSHOT and ISS data, and 3D Visium data for the two locations where this is available. Together, this will generate a spatial map of cell types and cell states of the healthy human lung, including 3D reconstruction of small airways and lung parenchyma.
Finally, discovAIR aims to provide a detailed profiling of the cellular trajectories of healthy adult lung cells towards the disease-associated cell states. To this end, discovAIR will study a (limited) number of chronic and/or inflammatory lung diseases: asthma, COPD, ILD, PAH and COVID-19. Moreover, discovAIR will establish a lung cell perturbation atlas to chart cell state transitions between health and disease (figure 1).
Lung diseases are highly heterogeneous and several phenotypes as well as endotypes can be observed for nearly all chronic lung diseases. The scope of discovAIR is to provide proof-of-principle data that a high-quality reference cell atlas of healthy lung can be used in combination with datasets from lung tissue of patients with lung disease to identify disease-associated cell states and cell–cell interactions, and the healthy-to-diseased cellular trajectories can be further characterised using the lung cell perturbation atlas. Therefore, discovAIR has chosen to include relatively small numbers of samples from patients with lung disease (10 tissue samples per disease condition). In addition, discovAIR has selected very specific disease groups, such as adult patients with (physician-diagnosed) childhood-onset asthma without a history of cigarette smoking, in an attempt to minimise the chance that heterogeneity of the disease obscures any reproducible disease-associated transcriptional cell states in the final dataset.
The discovAIR lung cell perturbation atlas will use primary epithelial, immune and endothelial cells, as well as precision-cut lung slices, all isolated from lung tissue of healthy donors and stimulated in advanced in vitro culture models [40, 41]. Time-series analysis of the ex vivo stimulated primary cells by scRNA-seq will allow the identification of transitional cell states in the cellular trajectories of healthy cells towards a potentially disease-associated transcriptional cell state. The discovAIR project focuses on testing this perturbation atlas concept in epithelial, endothelial and immune cells given the availability of well-developed cell culture models available to the consortium. However, other cell types such as mesenchymal cells including fibroblast subsets and smooth muscle cells are likely to play key roles in disease inception, progression and exacerbations, and will need to be studied in a similar approach in future follow-up projects if the concept of the perturbation atlas is validated. Integration of the datasets from ex vivo cultures of healthy primary lung cells and tissues with the scRNA-seq datasets acquired in lung tissue samples obtained from patients with lung disease is expected to allow the identification of the optimal in vitro proxy for the diseased cell states observed in vivo.
These studies are expected to further uncover cellular mechanisms of disease, guide identification of potential targets for preventive or therapeutic intervention and reveal biomarkers that can be used to detect the presence of these specific diseased cell states for use in diagnosis or treatment response monitoring. Given that discovAIR is a 2-year pilot project, the approach has a strong focus on transcriptomic data to establish the first draft of the Human Lung Cell Atlas in health and disease. Notwithstanding the significant progress beyond the state-of-the-art such an atlas would entail, these foundational transcriptomic datasets will need to be validated at the protein level, both for the Human Lung Cell Atlas describing the transcriptional heterogeneity in healthy lung tissue, as well as for the changes therein associated with chronic lung disease, to be able to achieve their full impact for the respiratory community and patients with lung disease.
Taken together, the multimodal healthy lung tissue datasets, the state-of-the-art integration methods, the spatial mapping of healthy lung tissue and the disease cohorts in combination with the lung cell perturbation atlas will allow discovAIR to generate a first draft of the Human Lung Cell Atlas (V1.0) as a standard reference for the respiratory community. This will not only encompass a healthy reference Human Lung Cell Atlas combining multimodal omics and spatial datasets, but also a comprehensive description of the changes thereof with disease and the cellular trajectories that might lead to the acquisition of the unique, disease-associated cell states. This will be a key asset for the respiratory community, and is expected to facilitate progress in regenerative and precision medicine for lung disease and to guide identification of novel candidates for precision diagnostics and curative interventions. All discovAIR methods and best practices will be openly shared through open access portals such as protocols.io (https://www.protocols.io/workspaces/hca) and GitHub (https://github.com/LungCellAtlas) (see supplementary appendix S3).
Dissemination, public engagement and outreach for the Human Lung Cell Atlas
To achieve full impact, discovAIR has the European Respiratory Society (ERS) and the European Lung Foundation (ELF) as project partners for dissemination of results and community involvement in the Human Lung Cell Atlas. ELF is a patient ambassador organisation, aiming to bring together patients and the public with respiratory professionals to positively influence lung health, and is essential to safeguard patient involvement, public engagement and outreach for discovAIR. ERS is the largest scientific and clinical organisation in respiratory medicine in Europe. Involvement of ERS in the Human Lung Cell Atlas initiative through discovAIR is instrumental to inform and involve the basic, translational and clinical respiratory scientific community, as well as diagnostic, regenerative medicine and pharmaceutical industries, with respect to the progress and achievements of the Human Lung Cell Atlas.
Needs from different user communities of a Human Lung Cell Atlas
A critical part of the discovAIR dissemination strategy is to involve the different user communities that will benefit from the Human Lung Cell Atlas early on. The discovAIR consortium organised a user group meeting for the Human Lung Cell Atlas at the 2020 ERS Lung Science Conference in Estoril, Portugal, together with the ELF. This meeting aimed to identify the needs of the different user groups with regard to content and design of the Human Lung Cell Atlas, as well as the best approach to ensure optimal use of the atlas. The user groups represented in this meeting were patients with different respiratory diseases, basic and translational scientists active in the respiratory field, clinical experts, and representatives of the diagnostic and pharmaceutical industry. The results from the user group meeting clearly indicate that the expectations and needs for the Human Lung Cell Atlas differ between the patient representatives and the lay public on the one hand, and experts including clinician, scientists and representatives from the private sector on the other hand.
Patient needs
The patient ambassadors clearly indicated that the added value of a Human Lung Cell Atlas for them is to better understand their lung disease. Patients with lung disease indicated that the Human Lung Cell Atlas might help them understand the structure and function of the lung, the changes in disease, as well as the identity and basic characteristics of the cells that make up the lung. Of special interest to the patient is the opportunity to understand how their disease condition affects the cells of the lung, how this is reflected in specific (diagnostic) test results, and how prescribed drugs can work to restore normal cell functions and interactions. As such, the Human Lung Cell Atlas can serve as a tool in interactions between patients and their doctors. Thus, information needs to be presented in the context of a specific disease.
In addition, the Human Lung Cell Atlas could be used as an educational tool, to explain the details of a lung disorder to relatives and the lay public, and might help to educate the next generation of patient advocates and respiratory scientists. Finally, the patient representatives also recognised the Human Lung Cell Atlas as a potential tool for scientists, clinicians and partners in the private sector to develop novel treatments for lung disease that can improve the quality of life for patients with lung disease.
Scientific, clinical and industry needs
The needs indicated by the scientific, clinical and industry representatives at the Human Lung Cell Atlas user group meeting were much more detailed, with open access to data being considered as one of the most important features of the Human Lung Cell Atlas. Data access could be facilitated either by querying the platform, by downloading the data or by obtaining contact details of the data guardians. Furthermore, the respiratory scientists would like to see details on the aspects of the atlas that are not well covered in the current draft of the Human Lung Cell Atlas, as well as opportunities to contribute their data to a next iteration of the atlas. Also, the Human Lung Cell Atlas should offer details on disease-induced molecular phenotypes of lung cells, as well as the cellular interactions causing disease or that are altered by disease, to increase its impact with the respiratory community.
Representatives from the pharmaceutical industry were highly interested to use the Human Lung Cell Atlas to map gene expression to specific locations in the bronchial tree or parenchyma and to guide design of drug delivery methods for targeted therapies. Furthermore, detailed insight into the cellular transcriptomes, behaviours and interactions in the different regions of the lung and in subtypes of disease could accelerate drug design for precision medicine. The industry representatives further stressed the importance of access to the raw data and future expansions of the Human Lung Cell Atlas with, for instance, single-cell epigenetic or proteomic datasets, as well as datasets from a large number of lung diseases.
Functionalities to meet the different needs
Given the divergent needs indicated by the patient representatives and the individuals representing respiratory science, medicine and industry, all stakeholders agreed that in designing a Human Lung Cell Atlas, two separate portals might need to be developed. All user groups indicated that curiosity about the biology of the lung and its disorders is an important incentive to access the Human Lung Cell Atlas. Patient representatives indicated that clear illustrations and a step-by-step introduction into the anatomy and physiology of the lung would be extremely helpful before accessing the individual cell types and their changes in disease, with complexity slowly increasing as the user gets to the more detailed parts of the atlas. Accessing the atlas through different mobile devices, with strong visual support and structured search functions, as well as the ability to leave feedback, were also of importance.
Interactive resources with stories from patients around specific regions or structures in the lung would greatly increase the attractiveness of the Human Lung Cell Atlas, especially when these are updated regularly and kept up-to-date and relevant (e.g. around cessation of cigarette smoking or the consequences of COVID-19). Finally, the Human Lung Cell Atlas would need to be available in different languages and hosted or mirrored by various local and national organisations for patients with lung disease, each in their own language, to make it truly accessible to patients and the public. The Human Lung Cell Atlas V1.0 may serve as a platform that could be expanded to accommodate these features in the future.
The individuals from academia, clinical medicine and industry indicated that the portal should allow maximal interaction with the data through a variety of analysis tools and the availability of the data for download to perform such analyses offline. Examples are analyses of gene coexpression networks, of trajectories along spatial, demographic or disease parameters, of DNA variant queries, of gene ontology, and of cell-type specificity regarding gene expression networks. An interactive analytical tool or browser, which could be used to analyse the data in such a way that its results could be presented in scientific publications or presentations, would clearly increase the impact and recognition of the Human Lung Cell Atlas as an authoritative reference tool. In addition, such functionalities would enable the Human Lung Cell Atlas to maximally contribute to open data and open science. These data analysis tools will be made available through a dedicated web portal at the Single-Cell Expression Atlas at the European Bioinformatics Institute [42], as well as through the FASTGenomics platform (https://fastgenomics.org).
Conclusions
The Human Lung Cell Atlas is a shared goal of several international consortia, all of which are represented in HCA-Lung. The discovAIR consortium is the main European consortium active within HCA-Lung, and is one of the six research and innovation actions funded by the European Commission in the H2020 framework programme. The discovAIR consortium aims to contribute to the goals and efforts of HCA-Lung by providing the multimodal characterisation of healthy lung tissue, including spatial mapping of cell types and cell states onto the tissue architecture. This will allow discovAIR to generate a first draft of the Human Lung Cell Atlas as a standard reference for the respiratory community.
In addition to the healthy reference Human Lung Cell Atlas combining multimodal omics and spatial datasets, discovAIR will also provide a first description of the changes thereof with several lung diseases and the cellular trajectories that might lead to the acquisition of the unique, disease-associated cell states. This will be a key asset for the respiratory community, and is expected to facilitate progress in regenerative and precision medicine for lung disease, and to guide identification of novel candidates for precision diagnostics and curative interventions. discovAIR will develop portals for data exploration and analysis suitable for academic and industrial end-users, as well as information portals for patients, their families and the interested lay audience in collaboration with the ELF, an ambassador organisation for patients with lung disease. All discovAIR methods, datasets and atlases will be made freely available and serve as a basis for further expansions and updates by HCA-Lung. Future iterations of the Human Lung Cell Atlas are expected to increase the genetic diversity of the atlas, to incorporate fetal and paediatric lung development, to expand the number of diseases and diseased samples included in the atlas, and to further develop the interactive and integrated features of the Human Lung Cell Atlas across transcriptomic, epigenomic, proteomic and spatial data modalities, evolving into a standard reference for the respiratory community.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Appendix 1: Overview of lung tissue scRNAseq studies ERJ-02057-2021.Appendix_1 (87.7KB, xlsx)
Appendix 2: discovAIR gene lists smFISH-based method ERJ-02057-2021.Appendix_2 (23.8KB, xlsx)
Appendix 3: Collection of open protocols and methods on Protocols.io and Githubl ERJ-02057-2021.Appendix_3 (128.3KB, pdf)
Shareable PDF
Footnotes
This publication is part of the Human Cell Atlas (www.humancellatlas.org/publications).
Conflict of interest: F.J. Theis reports personal fees and nonfinancial support from Cellarity and Dermagnostix, during the conduct of the study. J. Schniering was supported by an ERS/EU RESPIRE4 Marie Skłlodowska-Curie Postdoctoral Fellowship. J. Lundeberg reports personal fees from 10x Genomics, outside the submitted work. P. Powell and J. Denning are employees of the European Lung Foundation. W. Timens reports personal fees from Merck Sharp Dohme and Bristol-Myers-Squibb, outside the submitted work. M. Nilsson reports personal fees from 10x Genomics, outside the submitted work. G.H. Koppelman reports grants from the Netherlands Lung Foundation, GlaxoSmithKline, Vertex, TEVA, UBBO EMMIUS Foundation and European Union (H2020 grant), outside the submitted work, and has participated in advisory board meetings to GlaxoSmithKline and PURE-IMS, outside the submitted work. M. van den Berge reports research grants paid to UMCG from GlaxoSmithKline, Genentech, Roche and Novartis, outside the submitted work. M.C. Nawijn reports grants from the European Commission, Chan Zuckerberg Initiative and Netherlands Lung Foundation, during the conduct of the study; grants from GlaxoSmithKline, outside the submitted work. All other authors do not have a conflict of interest.
Support statement: This work is supported by the European Union's Horizon 2020 Research and Innovation Program under grant agreement 874656 (discovAIR) to S.A. Teichmann, K. Saeb-Parsy, S. Leroy, W. Timens, J. Lundeberg, M. van den Berge, M. Nilsson, P. Horváth, J. Denning, I. Papatheodorou, J.L. Schultze., H.B. Schiller, P. Barbry, M. von Papen, F.J. Theis, C. Samakovlis, K.B. Meyer and M.C. Nawijn, and a Seed Network grant from the Chan Zuckerberg Initiative to P. Barbry, H.B. Schiller, K.B. Meyer, A.V. Misharin, M.C. Nawijn and F.J. Theis. M. van den Berge and M.C. Nawijn are also supported by grants (5.1.14.020 and 4.1.18.226) from the Netherlands Lung Foundation. K.B. Meyer and S.A. Teichmann are also supported by Wellcome (WT211276/Z/18/Z and Sanger core grant WT206194). J. Schniering is also supported by an ERS/EU RESPIRE4 Marie Skłodowska-Curie Postdoctoral Fellowship (R4202007-00844). P. Barbry was also supported by Institut National contre le Cancer (PLBIO2018-156), FRM (DEQ20180339158), Inserm Cross-cutting Scientific Program HuDeCA 2018 and National Infrastructure France Génomique (Commissariat aux Grands Investissements, ANR-10-INBS-09-03, ANR-10-INBS-09-02). Funding information for this article has been deposited with the Crossref Funder Registry.
References
- 1.Gibson GJ, Loddenkemper R, Lundbäck B, et al. Respiratory health and disease in Europe: the new European Lung White Book. Eur Respir J 2013; 42: 559–563. doi: 10.1183/09031936.00105513 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization . Fact sheets. 2021. www.who.int/news-room/fact-sheets Date last accessed: 19 July 2021.
- 3.Travaglini KJ, Nabhan AN, Penland L, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020; 587: 619–625. doi: 10.1038/s41586-020-2922-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiller HB, Montoro DT, Simon LM, et al. The Human Lung Cell Atlas: a high-resolution reference map of the human lung in health and disease. Am J Respir Cell Mol Biol 2019; 61: 31–41. doi: 10.1165/rcmb.2018-0416TR [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Regev A, Teichmann SA, Lander ES, et al. The Human Cell Atlas. Elife 2017; 6: e27041. doi: 10.7554/eLife.27041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Regev A, Teichmann S, Rozenblatt-Rosen O, et al. The Human Cell Atlas White Paper. arXiv 2018; preprint [http://arxiv.org/abs/1810.05192]. [Google Scholar]
- 7.Franks TJ, Colby TV, Travis WD, et al. Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc 2008; 5: 763–766. doi: 10.1513/pats.200803-025HR [DOI] [PubMed] [Google Scholar]
- 8.Plasschaert LW, Žilionis R, Choo-Wing R, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 2018; 560: 377–381. doi: 10.1038/s41586-018-0394-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Montoro DT, Haber AL, Biton M, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 2018; 560: 319–324. doi: 10.1038/s41586-018-0393-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rood JE, Stuart T, Ghazanfar S, et al. Toward a common coordinate framework for the human body. Cell 2019; 179: 1455–1467. doi: 10.1016/j.cell.2019.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vieira Braga FA, Kar G, Berg M, et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat Med 2019; 25: 1153–1163. doi: 10.1038/s41591-019-0468-5 [DOI] [PubMed] [Google Scholar]
- 12.Goldfarbmuren KC, Jackson ND, Sajuthi SP, et al. Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium. Nat Commun 2020; 11: 2485. doi: 10.1038/s41467-020-16239-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deprez M, Zaragosi L-E, Truchi M, et al. A single-cell atlas of the human healthy airways. Am J Respir Crit Care Med 2020; 202: 1636–1645. doi: 10.1164/rccm.201911-2199OC [DOI] [PubMed] [Google Scholar]
- 14.Meyer KB, Wilbrey-Clark A, Nawijn M, et al. The Human Lung Cell Atlas: a transformational resource for cells of the respiratory system. In: Nikolić MZ, Hogan BLM, eds. Lung Stem Cells in Development, Health and Disease (ERS Monograph). Sheffield, European Respiratory Society, 2021; pp. 158–174. [Google Scholar]
- 15.Lotfollahi M, Wolf FA, Theis FJ. scGen predicts single-cell perturbation responses. Nat Methods 2019; 16: 715–721. doi: 10.1038/s41592-019-0494-8 [DOI] [PubMed] [Google Scholar]
- 16.Xu C, Lopez R, Mehlman E, et al. Probabilistic harmonization and annotation of single-cell transcriptomics data with deep generative models. Mol Syst Biol 2021; 17: e9620. doi: 10.15252/msb.20209620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez R, Regier J, Cole MB, et al. Deep generative modeling for single-cell transcriptomics. Nat Methods 2018; 15: 1053–1058. doi: 10.1038/s41592-018-0229-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell 2019; 177: 1888–1902. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luecken MD, Büttner M, Chaichoompu K, et al. Benchmarking atlas-level data integration in single-cell genomics. Nat Methods 2022; 19: 41–50. doi: 10.1038/s41592-021-01336-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lotfollahi M, Naghipourfar M, Luecken MD, et al. Mapping single-cell data to reference atlases by transfer learning. Nat Biotechnol 2022; 40: 121–130. doi: 10.1038/s41587-021-01001-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Habermann AC, Gutierrez AJ, Bui LT, et al. Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv 2020; 6: eaba1972. doi: 10.1126/sciadv.aba1972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reyfman PA, Walter JM, Joshi N, et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med 2018; 198: 440–446. doi: 10.1164/rccm.201801-0120PP [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morse C, Tabib T, Sembrat J, et al. Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J 2019; 54: 1802441. doi: 10.1183/13993003.02441-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valenzi E, Bulik M, Tabib T, et al. Single-cell analysis reveals fibroblast heterogeneity and myofibroblasts in systemic sclerosis-associated interstitial lung disease. Ann Rheum Dis 2019; 78: 1379–1387. doi: 10.1136/annrheumdis-2018-214865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adams TS, Schupp JC, Poli S, et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv 2020; 6: eaba1983. doi: 10.1126/sciadv.aba1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madissoon E, Wilbrey-Clark A, Miragaia RJ, et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol 2019; 21: 1. doi: 10.1186/s13059-019-1906-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grant RA, Morales-Nebreda L, Markov NS, et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 2021; 590: 635–641. doi: 10.1038/s41586-020-03148-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delorey TM, Ziegler CGK, Heimberg G, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature 2021; 595: 107–113. doi: 10.1038/s41586-021-03570-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chua RL, Lukassen S, Trump S, et al. COVID-19 severity correlates with airway epithelium–immune cell interactions identified by single-cell analysis. Nat Biotechnol 2020. 38: 970–979. doi: 10.1038/s41587-020-0602-4 [DOI] [PubMed] [Google Scholar]
- 30.Melms JC, Biermann J, Huang H, et al. A molecular single-cell lung atlas of lethal COVID-19. Nature 2021; 595: 114–119. doi: 10.1038/s41586-021-03569-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sungnak W, Huang N, Bécavin C, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med 2020; 26: 681–687. doi: 10.1038/s41591-020-0868-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muus C, Luecken MD, Eraslan G, et al. Single-cell meta-analysis of SARS-CoV-2 entry genes across tissues and demographics. Nat Med 2021; 27: 546–559. doi: 10.1038/s41591-020-01227-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan H, Deutsch GH, Wert SE, et al. Comprehensive anatomic ontologies for lung development: a comparison of alveolar formation and maturation within mouse and human lung. J Biomed Semantics 2019; 10: 18. doi: 10.1186/s13326-019-0209-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sountoulidis A, Liontos A, Nguyen HP, et al. SCRINSHOT enables spatial mapping of cell states in tissue sections with single-cell resolution. PLoS Biol 2020; 18: e3000675. doi: 10.1371/journal.pbio.3000675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gyllborg D, Langseth CM, Qian X, et al. Hybridization-based in situ sequencing (HybISS) for spatially resolved transcriptomics in human and mouse brain tissue. Nucleic Acids Res 2020; 48: e112. doi: 10.1093/nar/gkaa792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asp M, Bergenstråhle J, Lundeberg J. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays 2020; 42: e1900221. doi: 10.1002/bies.201900221 [DOI] [PubMed] [Google Scholar]
- 37.Qian X, Harris KD, Hauling T, et al. Probabilistic cell typing enables fine mapping of closely related cell types in situ. Nat Methods 2020; 17: 101–106. doi: 10.1038/s41592-019-0631-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Biancalani T, Scalia G, Buffoni L, et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat Methods 2021; 18: 1352–1362. doi: 10.1038/s41592-021-01264-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nichterwitz S, Chen G, Aguila Benitez J, et al. Laser capture microscopy coupled with Smart-seq2 for precise spatial transcriptomic profiling. Nat Commun 2016; 7: 12139. doi: 10.1038/ncomms12139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sachs N, Papaspyropoulos A, Zomer-van Ommen DD, et al. Long-term expanding human airway organoids for disease modeling. EMBO J 2019; 38: e100300. doi: 10.15252/embj.2018100300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruiz García S, Deprez M, Lebrigand K, et al. Novel dynamics of human mucociliary differentiation revealed by single-cell RNA sequencing of nasal epithelial cultures. Development 2019; 146: dev177428. doi: 10.1242/dev.174318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Athar A, Füllgrabe A, George N, et al. ArrayExpress update – from bulk to single-cell expression data. Nucleic Acids Res 2019; 47: D711–D715. doi: 10.1093/nar/gky964 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Appendix 1: Overview of lung tissue scRNAseq studies ERJ-02057-2021.Appendix_1 (87.7KB, xlsx)
Appendix 2: discovAIR gene lists smFISH-based method ERJ-02057-2021.Appendix_2 (23.8KB, xlsx)
Appendix 3: Collection of open protocols and methods on Protocols.io and Githubl ERJ-02057-2021.Appendix_3 (128.3KB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-02057-2021.Shareable (686.9KB, pdf)