Abstract
Simultaneous inhibition of more than one target is considered to be a novel strategy in cancer therapy. Owing to the importance of histone deacetylases (HDACs) and p53-murine double minute 2 (MDM2) interaction in tumor development and their synergistic effects, a series of MDM2/HDAC bifunctional small-molecule inhibitors were rationally designed and synthesized by incorporating an HDAC pharmacophore into spirooxindole skeletons. These compounds exhibited good inhibitory activities against both targets. In particular, compound 11b was demonstrated to be most potent for MDM2 and HDAC, reaching the enzyme inhibition of 68% and 79%, respectively. Compound 11b also showed efficient antiproliferative activity towards MCF-7 cells with better potency than the reference drug SAHA and Nutlin-3. Furthermore, western blot analysis revealed that compound 11b increased the expression of p53 and Ac-H4 in MCF-7 cells in a dose-dependent manner. Our results indicate that dual inhibition of HDAC and MDM2 may provide a novel and efficient strategy for the discovery of antitumor drug in the future.
Keywords: multitarget drugs, histone deacetylase inhibitors, MDM2 inhibitors, spirooxindole, dual inhibitors, anticancer
1. Introduction
Over the past few decades, drug discovery always relies on the “single target single drug” model, and numerous drugs that act on a specific biological target with high potency and selectivity have been approved successfully (1–4). However, for the treatment of some heterogeneous, complex, and multigenic diseases such as cancer, single target chemotherapeutic agents might become less effective (5, 6). The main reason is that the tumor progression always involves the dysregulation of multiple signaling pathways due to their multifaceted and dynamic nature, which hinders the efficacy of single-target drugs, and even leads to drug resistance and toxic side effects after a period of treatment (7–12). Despite the fact that specific drug combination therapies are employed as an alternative method, they are often limited by many side effects, such as drug-drug interactions and poor patient compliance (3, 13). Compared with the traditional one-drug/one-target or drug combinations philosophy, multitarget antitumor agents could simultaneously regulate two or more relevant targets, thus enabling a synergic therapeutic efficacy and fewer side effects (3, 14, 15). In this context, the discovery of multitarget drugs that combine two distinct biologically active molecules or pharmacophores has drawn much more attention.
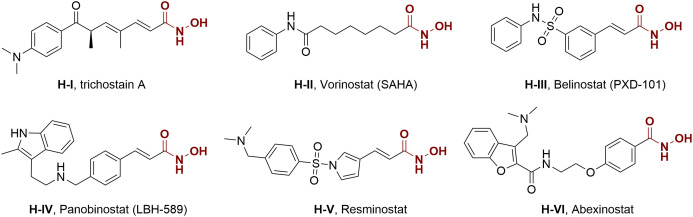
Actually, genetic and epigenetic aberrations have an evident influence on tumorigenesis and development, but the latter is more druggable because of its reversibility modulated by numerous enzymes (16, 17). Histone deacetylases (HDACs) represent a class of epigenetic enzymes that regulate the acetylation balance of histone proteins, are important regulatory factors in many crucial cellular functions, including angiogenesis, cell differentiation, cell proliferation, and cell apoptosis (18–23). The overexpression of HDACs can be observed in tumor initiation and proliferation, and thus they are regarded as potential epigenetic targets for cancer therapy. In 1990, Yoshida et al. reported the first potent HDAC inhibitor trichostatin A (H-I) (24). Since then, several HDAC inhibitors, such as vorinostat (SAHA, H-II), belinostat (PXD-101, H-III), and panobinostat (LBH-589, H-IV), have been approved for the treatment of hematological malignancy (25), while many others are currently in clinical trials for oncology ( Figure 1 , H-V and H-VI) (26–30). Despite the progress in hematologic tumor treatment, the therapeutic effects of some HDAC inhibitors and epigenetic agents are not significant against solid tumors. Considering the crucial role of HDAC in various cellular pathways and the above-mentioned advantages of multitarget antitumor drugs, intense interest is attracted to the discovery of HDAC inhibitor-based hybrid molecules (31–40).
Figure 1.
Structures of several representative HDAC inhibitors.
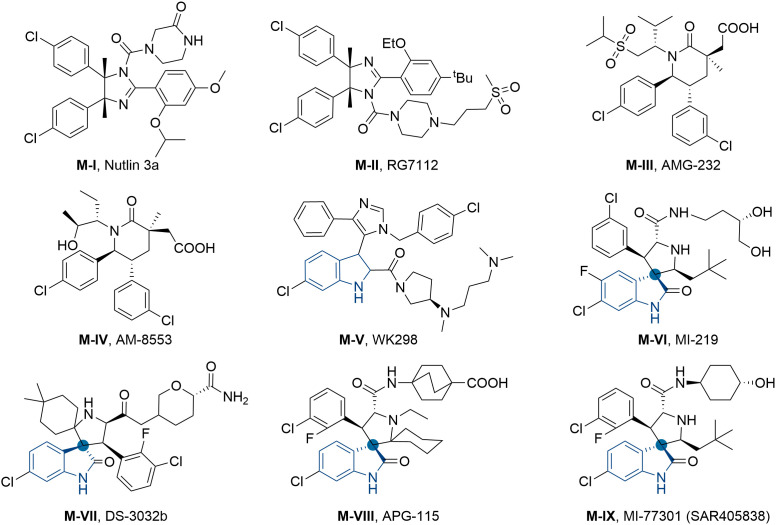
On the other hand, as a “guardian of the genome”, tumor suppressor protein p53 also played a pivotal role in regulating cell cascade reactions. Currently, approximately 50% of human tumors are related to the deletion or mutation of p53, whereas others exhibited wild-type status and lost p53 functions by negative regulation of murine double-minute 2 (MDM2) protein (41, 42). In general, MDM2 protein can directly bound to the N-terminal transactivation domain of p53 resulting in the disappearance of transcription function or proteasomal degradation (43–46). Therefore, inhibiting the interaction of p53-MDM2 can reactivate the p53 pathway and emerge as a prospective therapeutic method for cancers. Up to now, a number of small-molecule MDM2 inhibitors have been developed, and some of them even entered preclinical or clinical trials for the treatment of multiple tumors ( Figure 2 ), such as RG7122 (M-II), AMG-232 (M-III), APG-115 (M-VIII), MI-77301 (M-IX) and so on (47–52). However, the main side effect of p53-MDM2 inhibitors was thrombocytopenia and gastrointestinal toxicity, which was associated with dosage (53–55). In an effort to improve their efficacy, reducing the dose becomes an efficient way to avoid related toxicities. Therefore, modulation of multiple related targets to generate superior efficacy by hybrid molecule (single compound) is highly desirable. Our recent works and other studies have successfully demonstrated the effectiveness of MDM2-based dual inhibitors, like MDM2-GPX4, MDM2-CDK4, MDM2-XIAP, MDM2-MDM4, and so on (56–63).
Figure 2.
Structures of several representative MDM2 inhibitors.
Actually, p53 acetylation is one of the mechanisms for activating its function, previous studies have demonstrated that HDAC inhibitors can induce the acetylation of carboxyl-terminal lysine residue site Lys382 of p53 and activate p53 target genes (64). Therefore, inhibiting HDACs and MDM2 protein are both effective strategies to lead to the accumulation of activated p53. In addition, HDAC inhibitors could synergize with MDM2 inhibitors and enhance their antitumor activity by modifying the hyperacetylation of p53 (18, 65). These results indicated that rational design and synthesis of a single compound simultaneously inhibiting HDAC and MDM2 targets to exert desired biological functions would be an efficient strategy for developing novel antitumor agents.
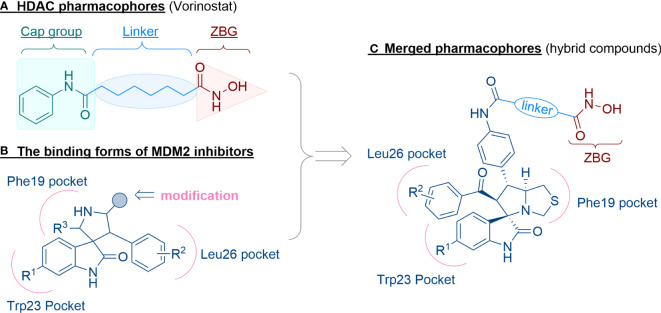
The general pharmacophore of most HDAC inhibitors, including Vorinostat, consists of three parts ( Figure 3A ): the hydrophobic cap group (CAP) that recognizes the protein surface and prevents other substrates from entering the active structure, a linker occupying the long tunnel of the active site, and the zinc-binding group (ZBG) that chelates with Zn2+ and forms hydrogen bonds in the catalytic site (66–68). More importantly, the cap group of HDAC inhibitors can be replaced by other chemical stents (or pharmacophore) to obtain new types of multi-targeted compounds. In 2018, Sheng and co-workers reported the first HDAC-MDM2 dual inhibitors with excellent anticancer activities in vitro and in vivo based on Nutlin-3a (M-I, MDM2 inhibitor) (69). Considering that spirooxindole skeletons have been well established as MDM2 inhibitors ( Figure 2 , M-VI−IX) (70–72) and encouraged by the pioneering work of Sheng’s group, we rationally designed new classes of hybrid compounds derived from spirooxindole scaffolds that could inhibit both HDAC as well as MDM2. According to the binding models of most spirooxindole derivatives and MDM2 target, three functional groups on the spirooxindole framework were assumed to occupy the Phe19, Trp23, and Leu26 pockets in p53, respectively ( Figure 3B ). As a result of the C3’ substitution in the spiro core of some inhibitors did not bury inside to MDM2 protein and was directly exposed to the solvent, this position may become appropriate to induce a hydrophobic linker and ZBG to generate HDAC-MDM2 dual inhibitors. As part of our continuing interest in assembling biologically important frameworks as lead compounds for drug discovery (73–78), we herein synthesized a series of spirooxindole-based dual HDAC-MDM2 small-molecule compounds, and evaluated their antitumor activity together with the mechanism of action ( Figure 3C ).
Figure 3.
Design of novel HDAC/MDM2 dual inhibitors via merging pharmacophores. (A) HDAC pharmacophores; (B) The binding forms of MDM2 inhibitor; (C) Merged pharmacophores.
2. Results and discussion
2.1. Chemistry
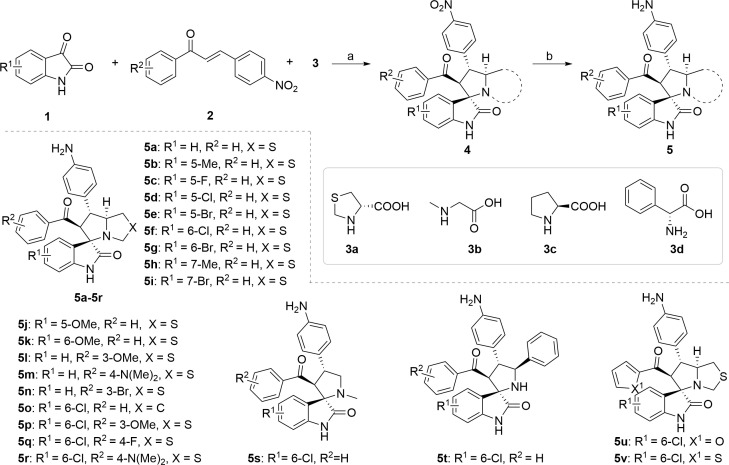
The synthetic methods of target compounds are depicted in Figures 4, 5. Compounds 5a-5v were prepared according to the procedure detailed in Figure 4. Using commercially available indirubin 1, 4-nitrochalcone derivatives 2 as the starting material to react with amino acids (thioproline 3a, sarcosine 3b, proline 3c, and phenylglycine 3d) in methanol at 65°C, a series of spirooxindole pyrrolidine, spirooxindole thiopyrrole, and their derivatives were obtained through dipolar cycloaddition reaction. Then compound 4a-4v was treated with iron powder, NH4Cl in MeOH/H2O at 60°C to produce intermediates 5a-5v.
Figure 4.
Synthesis of compounds 5a-5v. Reagents and conditions: (a) 1 (1.0 equiv), 2 (1.0 equiv), 3 (1.0 equiv), MeOH, 65 °C, 4 h; (b) Fe (10.0 equiv), NH4Cl (5.0 equiv), MeOH/H2O = 2:1, 60 °C, 6 h.
Figure 5.
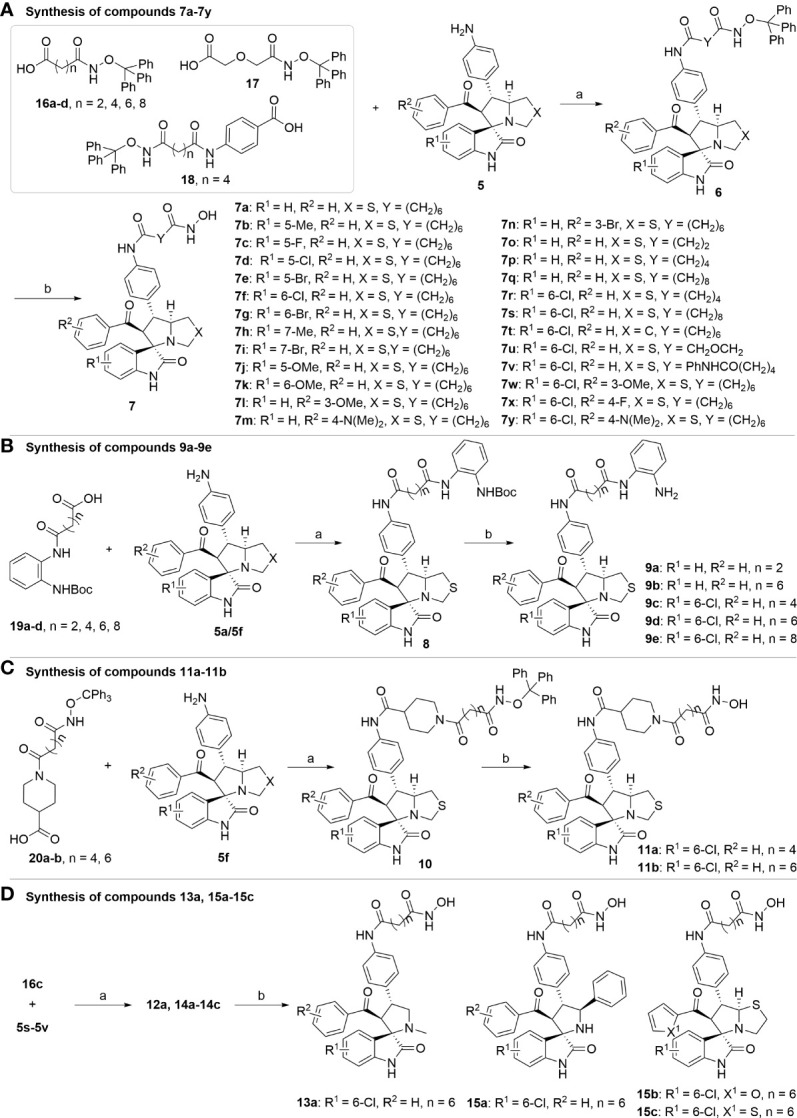
Synthesis of target compounds. (A) Synthesis of compounds 7a-7y; (B) Synthesis of compounds 9a-9e; (C) Synthesis of compounds 11a-11b; (D) Synthesis of compounds 13a, 15a-15c. Reagents and conditions: (a) HATU (1.2 equiv), DIPEA (2.0 equiv), DCM, rt; (b) TFA/DCM=1:3, rt.
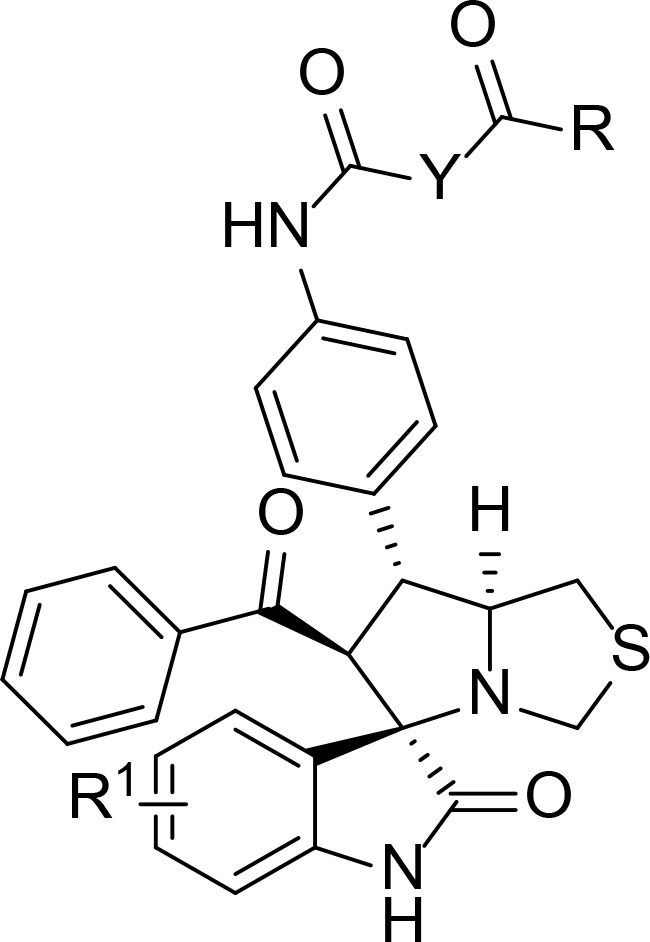
With the key spirooxindole intermediates 5a-5v in hand, we turned our attention to introduce HDAC pharmacophore. Thinking that 2-aminoaniline and hydroxamic acid are commonly used as Zn2+ binding groups (ZBG) in HDAC inhibitors, we subsequently introduce different linkers. Spirooxindole intermediates 5 were reacted with 16 to generate the corresponding derivatives 6a-6y (Figure 5A), which was followed by hydrolysis in the presence of triethylamine could finally afford the target HDAC-MDM2 dual inhibitors 7a-7y (Figure 5A). Target compounds 9a-9e, 11a-11b, 13a, 15a-15c were also prepared according to the above-mentioned methods (Figures 5B–D).
2.2. Biological Evaluation
2.2.1. Enzyme Inhibitory Activities
Having synthesized a series of novel spirooxindole-based derivatives, we turn to evaluate their inhibitory activities against HDAC and MDM2 at 1 μM, the results are summarized in Tables 1 – 4 . Given the fact that hydroxamic acid groups are prevalent in HDAC inhibitors, and can strongly chelate with zinc ions (79–84), most of the target compounds are fixed with hydroxamic acid as a ZBG to the side chain.
Table 1.
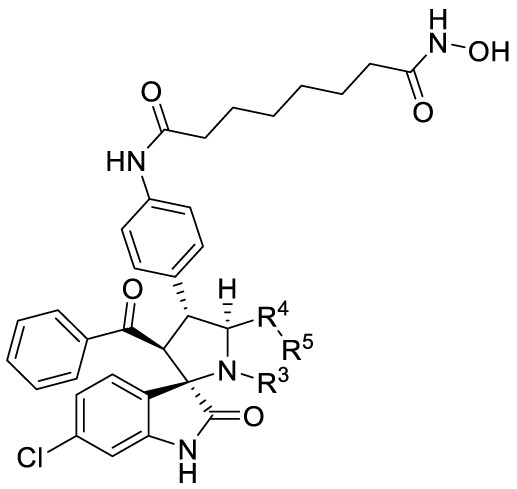
Effects of substituted group R1 and R2 on enzyme inhibition.
| ||||
|---|---|---|---|---|
| compound | R1 | R2 | % enzyme inhibition | |
| MDM2 | HDAC | |||
| 7a | H | Ph | 39 | 69 |
| 7b | 5-Me | Ph | 45 | 71 |
| 7c | 5-F | Ph | 51 | 68 |
| 7d | 5-Cl | Ph | 49 | 67 |
| 7e | 5-Br | Ph | 48 | 72 |
| 7f | 6-Cl | Ph | 65 | 64 |
| 7g | 6-Br | Ph | 59 | 68 |
| 7h | 7-Me | Ph | 23 | 71 |
| 7i | 7-Br | Ph | 30 | 67 |
| 7j | 5-OMe | Ph | 50 | 70 |
| 7k | 6-OMe | Ph | 54 | 70 |
| 7l | H | 3-OMe-C6H4 | 44 | 62 |
| 7m | H | 4-NMe2-C6H4 | 40 | 68 |
| 7n | H | 3-Br-C6H4 | 43 | 69 |
| 7w | 6-Cl | 3-OMe-C6H4 | 60 | 69 |
| 7x | 6-Cl | 4-F-C6H4 | 62 | 73 |
| 7y | 6-Cl | 4-NMe2-C6H4 | 54 | 71 |
| 15b | 6-Cl | 2-furyl | 48 | 70 |
| 15c | 6-Cl | 2-thienyl | 49 | 70 |
Table 4.
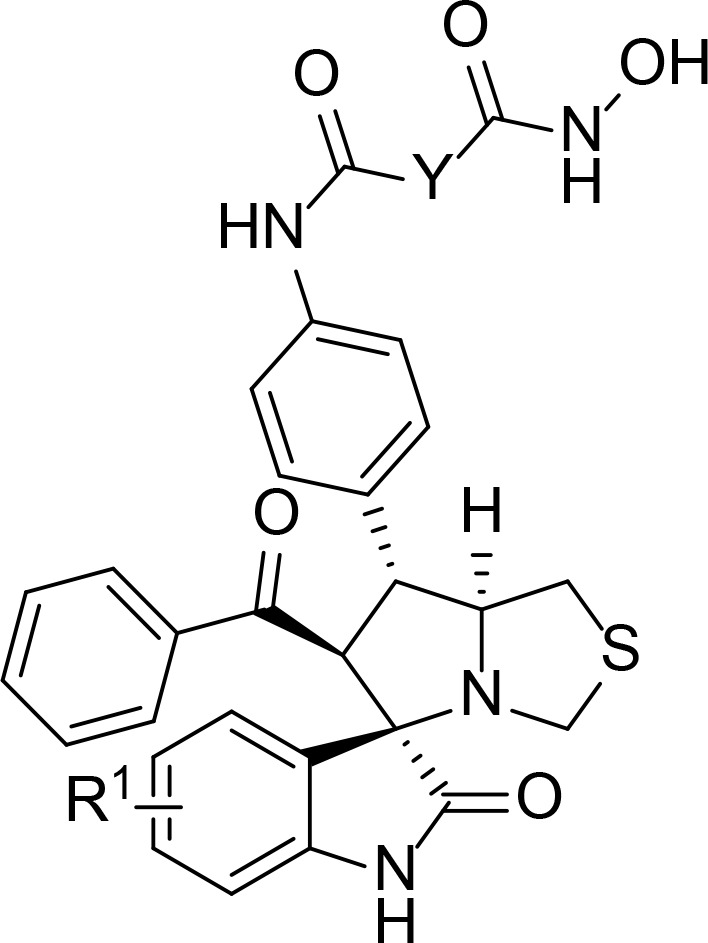
Effects of linkers on enzyme inhibition.
| ||||
|---|---|---|---|---|
| compound | R1 | Y | % enzyme inhibition | |
| MDM2 | HDAC | |||
| 7o | H | (CH2)2 | 43 | 54 |
| 7p | H | (CH2)4 | 44 | 60 |
| 7a | H | (CH2)6 | 39 | 69 |
| 7q | H | (CH2)8 | 42 | 71 |
| 7r | 6-Cl | (CH2)4 | 61 | 55 |
| 7f | 6-Cl | (CH2)6 | 65 | 64 |
| 7s | 6-Cl | (CH2)8 | 59 | 68 |
| 7u | 6-Cl |
|
61 | 54 |
| 7v | 6-Cl |
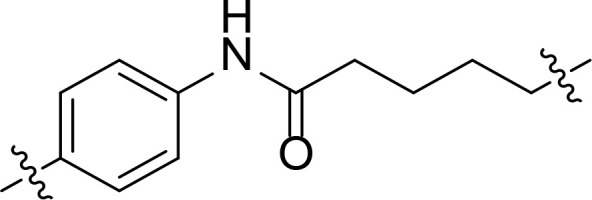
|
58 | 53 |
| 11a | 6-Cl |
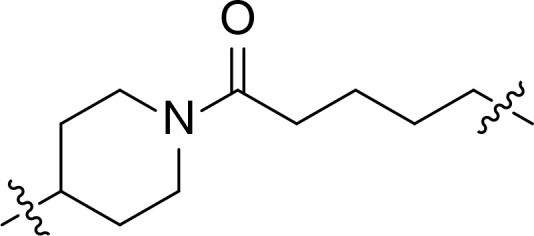
|
62 | 70 |
| 11b | 6-Cl |
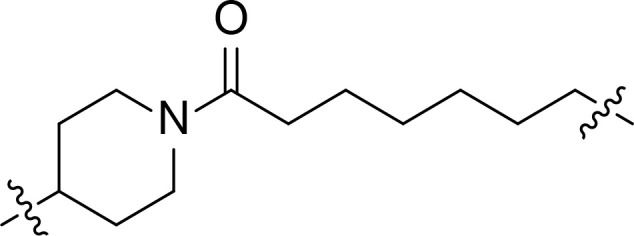
|
68 | 79 |
As shown in Table 1, we found that the variation of substituted groups at isatin scaffold has a strong influence on their MDM2 inhibition activity (7a-7k), with a chlorine atom at the C6 position exhibiting relatively good inhibitory activity against MDM2 (7f, 65%). On the other hand, various substituted group R2 on substrate 2 were also investigated, which did not enhance the inhibitory activity against MDM2 (7l-7y), some of them even decreased the corresponding activities on MDM2 target (15b and 15c). In fact, compounds 7w-7y, simultaneously substituted with chlorine moiety at the R1 position, exhibited better MDM2 inhibitory activity than those with R1 = H, such as 7l-7n. Moreover, the evaluation of amino acids demonstrated that thioproline-containing compound 7f exhibits the best activity, reaching the inhibition ratio of 65% and 64% toward MDM2 and HDAC, respectively (Table 2).
Table 2.
Effects of amino acids 3 on enzyme inhibition.
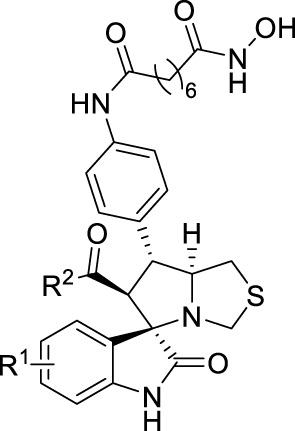
| |||||
|---|---|---|---|---|---|
| compound | R3 | R4 | R5 | % enzyme inhibition | |
| MDM2 | HDAC | ||||
| 7f | C | C | S | 65 | 64 |
| 7t | C | C | C | 61 | 61 |
| 13a | C | H | / | 52 | 65 |
| 15a | H | Ph | / | 67 | 61 |
We also introduced ortho-aminoanilide as ZBG, however, compounds 9a-9e caused a remarkable decrease of activity against HDAC compared to 7a, 7o, 7r, 7d, 7s fixed with hydroxamic acid as ZBG (Table 3). Considering the importance of the ZBG on dual inhibitory activity, a SAR study on linkers was performed, as shown in Table 4. In general, varying the linker length significantly influences the potency, with a longer alkyl chain leading to improved activity against HDAC. It is worth mentioning that introducing oxygen atom, and benzene ring to the alkyl chain led to a decrease in HDAC inhibition efficacy (7u and 7v), but further incorporation of the piperidine ring into the linker was beneficial for the HDAC inhibitory potency (11a and 11b).
Table 3.
Effects of ZBG on enzyme inhibition.
| |||||
|---|---|---|---|---|---|
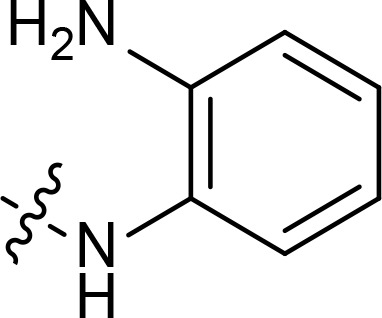
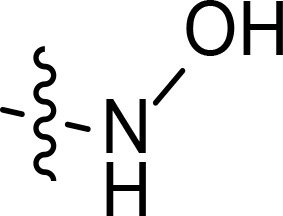
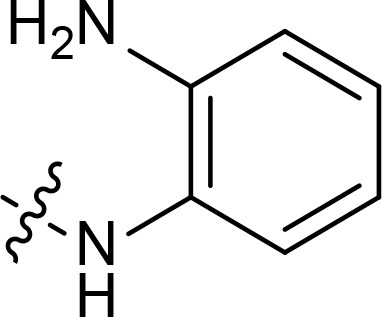
| compound | R | R1 | Y | % enzyme inhibition | |
| MDM2 | HDAC | ||||
| 7o |
|
H | (CH2)2 | 43 | 54 |
| 9a |
|
H | (CH2)2 | 50 | 37 |
| 7a |
|
H | (CH2)6 | 39 | 69 |
| 9b |
|
H | (CH2)6 | 45 | 45 |
| 7r |
|
6-Cl | (CH2)4 | 61 | 55 |
| 9c |
|
6-Cl | (CH2)4 | 57 | 40 |
| 7d |
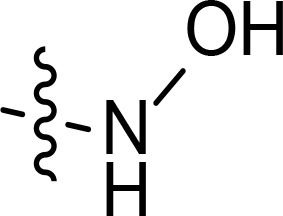
|
6-Cl | (CH2)6 | 49 | 67 |
| 9d |
|
6-Cl | (CH2)6 | 56 | 55 |
| 7s |
|
6-Cl | (CH2)8 | 59 | 68 |
| 9e |
|
6-Cl | (CH2)8 | 56 | 47 |
2.2.2. In Vitro Antiproliferative Assay
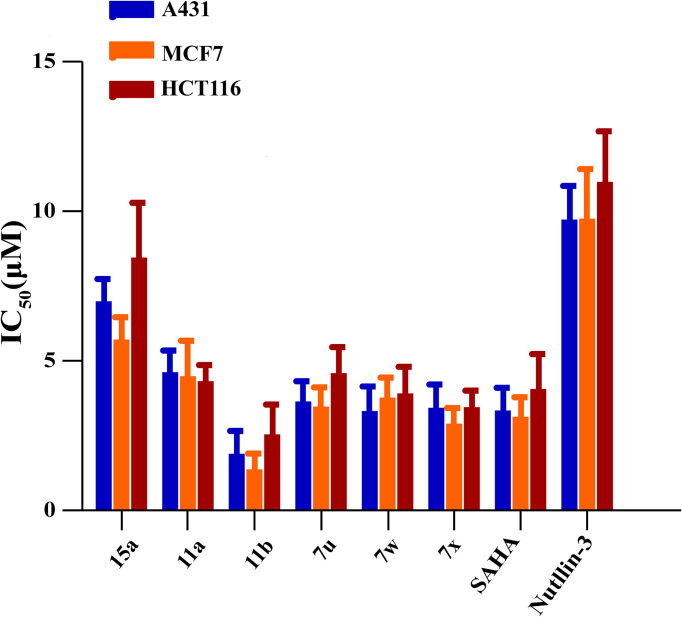
Considering their potent enzyme inhibitory potencies, six MDM2-HDAC dual inhibitors (7u, 7w, 7x, 11a, 11b, and 15a) were selected to evaluate for their antiproliferative activities against A431 (human epidermal carcinoma), MCF-7 (human breast cancer) and HCT116 (human colorectal cancer) cell lines by MTT assay, with the hydroxamic acid-based compound SAHA and Nutlin-3 as positive controls. According to the results in Table 5 and Figure 6, most compounds showed good inhibition activities against the three human cancer cells, they were more effective against the MCF-7 cells than A431 and HCT116 cells. Compound 11b was demonstrated to be the most potent against MCF-7 than both SAHA and Nutlin-3, whose IC50 value reached to 1.37 ± 0.45 μM. Therefore, considering the enzyme and cellular activities, the most active compound, 11b, was selected for further evaluation.
Table 5.
Antiproliferative activities of compounds 7u, 7w, 7x, 11a, 11b, and 15a.
| compound | IC50(μM) a | ||
|---|---|---|---|
| A431 | MCF-7 | HCT116 | |
| 7u | 3.64 ± 0.56 | 3.47 ± 0.92 | 4.59 ± 0.91 |
| 7w | 3.32 ± 0.68 | 3.77 ± 0.93 | 3.91 ± 0.84 |
| 7x | 3.43 ± 0.73 | 2.90 ± 0.56 | 3.45 ± 0.57 |
| 11a | 4.62 ± 0.78 | 4.48 ± 1.46 | 4.32± 0.90 |
| 11b | 1.89 ± 0.88 | 1.37 ± 0.45 | 2.54 ± 1.07 |
| 15a | 6.99 ± 0.92 | 5.71 ± 1.03 | 8.45± 2.44 |
| SAHA | 3.34 ± 0.95 | 3.13 ± 0.78 | 4.06 ± 1.36 |
| Nutlin-3 | 9.72 ± 1.66 | 9.75 ± 2.97 | 10.98 ± 2.42 |
IC50 = compound concentration required to inhibit tumor cell proliferation by 50%; Data displayed is the average of at least three independent replicates ± standard.
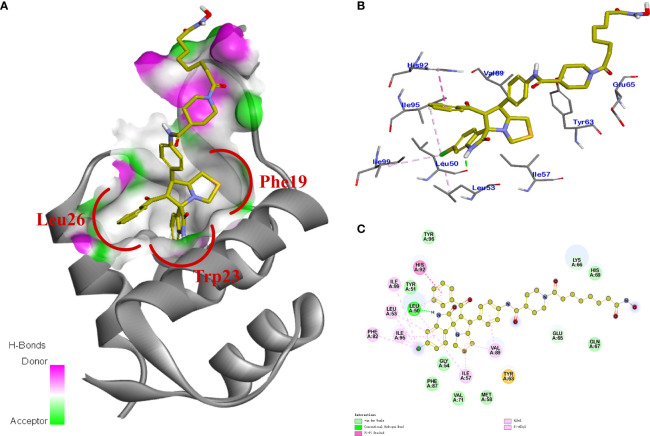
Figure 6.
Potential binding modes of 11b to HDAC. (A, B) the 3D diagram of the binding modes; (C) 2D diagram of the binding modes.
2.2.3. HDACs Isoform Selectivity of Compound 11b
Subsequently, to test the HDAC isoform selectivity profile, the representative compound 11b was subjected to estimate the enzyme inhibitory effects towards ten subtypes from the four respective HDAC subfamilies: HDAC1-3 (Class I), HDAC8 (Class I), HDAC4-5 (Class IIA), HDAC7 (Class IIA) and HDAC9 (Class IIA), HDAC6 (Class IIB), HDAC10 (Class IIB), and HDAC (Class IV). The result in Table 6 showed that compound 11b exhibited obvious selectivity for HDAC1 and HDAC2 (IC50 = 0.058 μM and 0.064 μM) compared with other isoforms. On the contrary, it showed submicromolar IC50 values variation against HDAC3 (IC50 = 0.115 μM), HDAC6 (IC50 = 0.120 μM), HDAC8 (IC50 = 0.313 μM). Moreover, similar to positive control SAHA, compound 11b was less active on HDAC10 (IC50 = 1.495 μM).
Table 6.
IC50 values of compound 11b against HDAC subtypes.
| Target | IC50(μM) a | Target | IC50(μM) a | ||
|---|---|---|---|---|---|
| 11b | SAHA | 11b | SAHA | ||
| HDAC1 | 0.058 ± 0.005 | 0.019 ± 0.004 | HDAC7 | >10 | >10 |
| HDAC2 | 0.064 ± 0.004 | 0.039 ± 0.007 | HDAC8 | 0.313 ± 0.023 | 0.183 ± 0.027 |
| HDAC3 | 0.115 ± 0.009 | 0.053 ± 0.006 | HDAC9 | >10 | >10 |
| HDAC4 | >10 | >10 | HDAC10 | 1.495 ± 0.037 | 0.379 ± 0.022 |
| HDAC5 | >10 | >10 | HDAC11 | >10 | >10 |
| HDAC6 | 0.120 ± 0.026 | 0.044 ± 0.005 | MDM2 | 0.339 ± 0.068 | ND |
IC50 = compound concentration required to inhibit tumor cell proliferation by 50%; Data displayed is the average of at least three independent replicates ± standard.
2.2.4. Molecular Docking Study of Compound 11b with MDM2 and HDAC1
In order to further investigate the inhibitory activity and the interaction manner of the most active compound 11b on both targets, molecular docking studies were performed. The proposed binding modes of 11b were analyzed using the CDOCKER method (embedded in the Accelrys Discovery Studio 3.5 package) and the MDM2 and HDAC1 protein structures were downloaded from the protein data bank (PDB) database (https://www.rcsb.org, PDB ID 4LWU).
Previous studies revealed that spirooxindole derivatives could project into the pockets usually occupied by the three key residues (Phe19, Trp23, and Leu26) of p53. As shown in Figure 7, we speculated that the oxindole framework projected into the Trp23 pocket, the 3’-benzoyl group and thiazolyl moiety of compound 11b projected into the Leu26 and Phe19 residues, respectively. Actually, compound 11b mimicked the interactions of p53 binding site on MDM2 via hydrogen-bonding effects, and the main hydrogen-bonding interaction between spirooxindole-based cap and MDM2 appears to involve residue Leu50. Moreover, the 3’-benzoyl moiety formed π−π stacking interaction with His92 residue.
Figure 7.
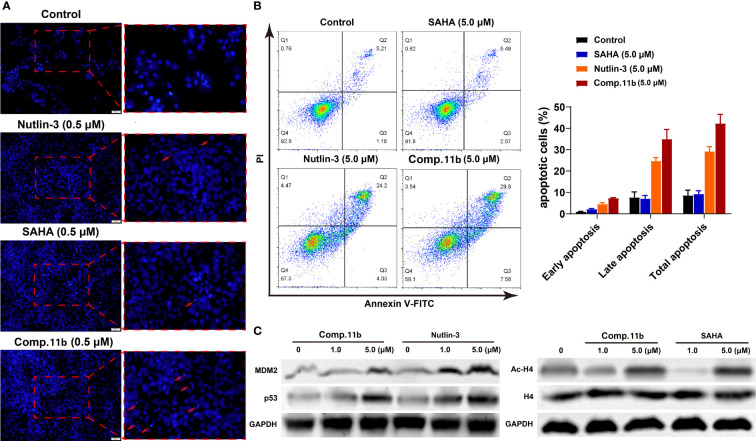
(A) TUNEL assay. MCF-7 cells were treated with 0.5 μM 11b. (B) MCF-7 cells were incubated with 5.0 μM 11b for 24 (h) Cell apoptosis was examined by Annexin V/PI double-stained assay. (C) Effects of 11b on expression levels of MDM2, p53, H4, Ac-H4 after 24 h incubation.
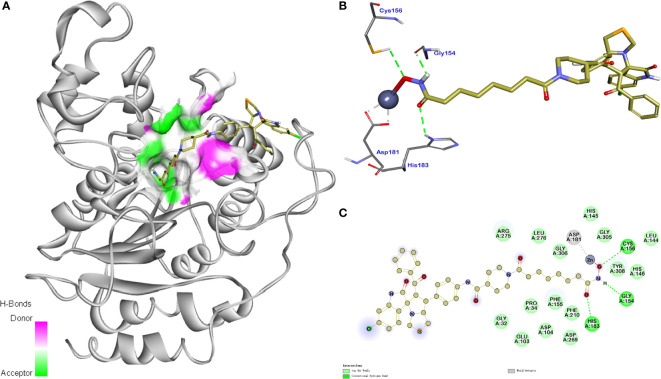
The molecular docking study of compound 11b with HDAC1 was shown in Figure 8, which further demonstrated their considerable HDAC inhibitory activities. We could see that the piperidine-based linker of compound 11b protrudes deeply into the hydrophobic cavity of HDAC1, the terminal hydroxamic acid group chelated with the Zn2+ in a bidentate manner, thereby forming hydrogen bonding interactions with Asp181. It’s also observed that the terminal group of compound 11b could form three important hydrogen bonds with Cys156, Gyl154, and His183 in the HDAC2 active site. The molecular docking studies provided a valid explanation for the interaction modes of compound 11b with MDM2 and HDAC1, which were consistent with enzyme inhibitory activities and in vitro antiproliferative assay.
Figure 8.
Synthesis of compounds 5a-5v. Reagents and conditions: (A) 1 (1.0 equiv), 2 (1.0 equiv), 3 (1.0 equiv), MeOH, 65°C, 4 h; (B) Fe (10.0 equiv), NH4Cl (5.0 equiv), MeOH/H2O = 2:1, 60°C, 6 h.
2.2.5. Effect of Compound 11b on Cell Apoptosis
It was reported that the inactivation of HDAC and/or MDM2 could increase the percentage of apoptosis cells (85–87). Therefore, three methods were applied to assess the effect of MDM2-HDAC dual inhibitor 11b on the apoptosis of MCF-7 cells. Firstly, as shown in Figure 9A, a significantly higher number of apoptotic positive cells were determined by TUNEL staining after treating MCF-7 cells with 0.5 μM 11b, compared with SAHA and Nutlin-3. This result was in accordance with previous studies. Subsequently, the annexin V-FITC/propidium iodide (PI) double staining method was carried out to quantitatively detect the effect of compound 11b on cell apoptosis (Figure 9B). According to the flow cytometry analysis, we found that the apoptotic cells reached 37.36% at a dose of 5.0 μM 11b, which was higher than that of reference compound SAHA (7.56%) and Nutlin-3 (28.23%), these results indicate that the simultaneously inhibit MDM2 and HDAC could increase the percentage of apoptosis cells.
Figure 9.
Synthesis of target compounds 7a-7y, 9a-9e, 11a-11b, 13a, 15a-15c. Reagents and conditions: (A) HATU (1.2 equiv), DIPEA (2.0 equiv), DCM, rt; (B) TFA/DCM=1:3, rt.
Western blot analysis was conducted to further confirm the dual-action targeting of MDM2 and HDAC compared to the MDM2 inhibitor Nutlin-3 and the HDAC inhibitor SAHA as control. As shown in Figure 9C (left), treatment with compound 11b in MCF-7 cells dose-dependently increased the protein level of MDM2 as well as p53 after 24 h incubation, which was much stronger than Nutlin-3 at the same concentration. And exposure MCF-7 cells to compound 11b for 24 h, the increased acetylation level of H4 in MCF-7 cells was also observed in a dose-dependent manner (Figure 9C, right), similar to SAHA on the acetylation-induced capacity at the same concentration. These results demonstrated that inhibition of intracellular MDM2 and HDAC were the main mechanisms of bioactivity of compound 11b.
3. Materials and methods
3.1. Chemistry
3.1.1. General information
Nuclear magnetic resonance (NMR) spectra were recorded in CDCl3 or DMSO-d6 on Bruker 400 MHz NMR instrument (at 400 MHz for 1H, and at 100 for 13C). Protonchemical shifts are reported in parts per million (δ scale). The 1H NMR chemical shifts are reported in ppm with the internal TMS signal at 0.0 ppm as standard. The 13C NMR chemical shifts were given using CDCl3 or DMSO-d6 as the internal standard (CDCl3: δ = 77.00 ppm; DMSO-d6 : δ = 39.51 ppm). Data are reported as follows: chemical shift [multiplicity (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad), coupling constant(s) (Hz), integration]. High-resolution mass spectra (HRMS) were obtained using Agilent P/N G1969-90010. High-resolution mass spectra were reported for the molecular ion [M+Na]+. X-ray diffraction experiment was carried out on Agilent Gemini or Agilent D8 QUEST and the data obtained were deposited at the Cambridge Crystallographic Data Centre. UV detection was performed at 254 nm. Column UV detection was performed at 254 nm. Column chromatography was performed on silica gel (200–300 mesh) using an eluent of ethyl acetate, petroleum ether, methanol, and dichloromethane. TLC was performed on glass-backed silica plates; products were visualized using UV light. All reagents and solvents were obtained from commercial sources and used without further purification. Oil baths were used as the heat source. Melting points were recorded on BUCHI Melting Point M-565 instrument.
3.1.2. General procedure for the preparation of 5a-5v
A mixture of compounds 1 (5.0 mmol), 4-nitrochalcone derivatives 2 (5.0 mmol), and amino acids 3 (5.0 mmol) were stirred in MeOH at 65°C. When the reaction was complete (based on TLC monitoring), the reaction mixture was poured into cold water and stirred for 20 min. The precipitate obtained was collected and washed with cold water. The solid was recrystal with MeOH dried at 50°C to constant weight affording the products 4a-4v. To a solution of compounds 4a-4v (1.0 mmol) in MeOH/H2O (2:1) was added iron powder (10.0 mmol), NH4Cl (5.0 mmol). The mixture was stirred at 60°C. When the reaction was complete (based on TLC monitoring), the reaction mixture was cooled to ambient and filtered through a pad of celite. The filtrate was washed with saturated saltwater and extracted with DCM (15 mL × 2). The combined organic layers were dried over anhydrous MgSO4, filtered, and concentrated under reduced pressure, the corresponding products 5a-5v were directly applied in the next step without further purification. The spectroscopic data are given in the supporting information.
3.1.3. General Procedure for the Preparation of Target Compounds 7a-7y, 9a-9e, 11a-11b, 13a, 15a-15c
To a solution of 16-20 (1.2 mmol) in DCM was added HATU (1.2 mmol),DIEA (2.0 mmol), the mixture was stirred at room temperature for 30 min. After that, compounds 5a-5v were added, and the mixture was stirred at room temperature until it was completed (based on TLC monitoring). Then the reaction mixture was concentrated and the residue was purified by flash chromatography on silica gel (DCM/MeOH = 50:1) to give intermediate 6a-6v, 8a-8e, 10a-10b, 12a, 14a-14c. The obtained intermediates (0.5 mmol) were then added TFA and stirred in DCM (TFA/DCM = 1:3) at room temperature. The reaction was monitored by TLC until it was completed, then saturated aqueous NaHCO3 solution was added to quench the reaction at room temperature. The mixture was extracted with EtOAc (15 mL × 2). The combined organic layers were dried over anhydrous MgSO4, filtered, and concentrated under reduced pressure. Then the residue was purified by flash chromatography on silica gel (DCM/MeOH = 20:1) to give the target products 7a-7y, 9a-9e, 11a-11b, 13a, 15a-15c.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7a)
Orange solid, 50% yield, dr > 99:1, m.p. 144.7-146.1 °C. 1H NMR (400 MHz, DMSO-d6 ): δ = 10.32 (s, 2H), 9.86 (s, 1H), 8.70 (s, 1H), 7.57–7.55 (m, 2H), 7.48 (t, J = 7.2 Hz, 1H), 7.44–7.38 (m, 3H), 7.35–7.26 (m, 4H), 7.11 (t, J = 7.6 Hz, 1H), 6.93 (t, J = 7.2 Hz, 1H), 6.49 (d, J = 8.0 Hz, 1H), 4.75 (d, J = 11.6 Hz, 1H), 4.12–4.08 (m, 1H), 3.82 (dd, J = 11.6, 9.6 Hz, 1H), 3.71 (d, J = 10.0 Hz, 1H), 3.33 (d, J = 10.4 Hz, 1H), 3.05–2.97 (m, 2H), 2.28–2.24 (m, 2H), 1.95–1.91 (m, 2H), 1.56–1.46 (m, 4H), 1.26–1.24 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d6 ): δ = 196.67, 178.79, 171.61, 169.53, 142.49, 138.79, 136.97, 134.13, 133.67, 130.18, 130.17, 128.78, 128.67, 128.39, 127.91, 123.44, 121.39, 119.87, 109.93, 74.82, 73.67, 61.78, 53.92, 50.94, 36.77, 36.22, 28.83, 25.52, 25.47 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C34H36N4NaO5S+ 635.2299, found 635.2305.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-5-methyl-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7b)
Orange solid, 41% yield, dr > 99:1, m.p. 144.2-145.6 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.29 (s, 2H), 10.03 (s, 1H), 8.79 (br, 1H), 7.59–7.57 (m, 2H), 7.47 (t, J = 7.6 Hz, 1H), 7.447–7.34 (m, 4H), 7.30–7.21 (m, 3H), 6.91 (d, J = 7.6 Hz, 1H), 6.40 (d, J = 8.0 Hz, 1H), 4.75 (d, J = 11.6 Hz, 1H), 4.12–4.07 (m, 1H), 3.81 (dd, J = 11.2 Hz, 9.2 Hz, 1H), 3.69 (d, J = 10.0 Hz, 1H), 3.32 (d, J = 10.0 Hz, 1H), 3.04–2.96 (m, 2H), 2.30–2.26 (m, 2H), 2.25 (s, 3H), 1.96–1.92 (m, 2H), 1.58–1.53 (m, 2H), 1.49–1.44 (m, 2H), 1.30–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.67, 178.78, 171.69, 169.62, 140.09, 138.84, 137.02, 134.09, 133.64, 130.41, 130.01, 128.89, 128.79, 128.60, 127.93, 123.59, 119.89, 109.66, 74.77, 73.60, 61.83, 53.69, 50.88, 36.75, 36.07, 32.68, 28.82, 25.55, 25.49, 21.30 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C35H38N4NaO5S+ 649.2455, found 649.2461.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-5-fluoro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7c)
Orange solid, 43% yield, dr > 99:1, m.p. 144.3-145.7 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.43 (s, 2H), 10.01 (s, 1H), 8.87 (br, 1H), 7.60–7.58 (m, 2H), 7.51 (t, J = 7.6 Hz, 1H), 7.46–7.40 (m, 4H), 7.34–7.26 (m, 3H), 7.00 (td, J = 8.8 Hz, 2.4 Hz, 1H), 6.53 (dd, J = 8.4 Hz, 4.4 Hz, 1H), 4.79 (d, J = 11.6 Hz, 1H), 4.13–4.09 (m, 1H), 3.81 (t, J = 11.2 Hz, 1H), 3.74 (d, J = 10.4 Hz, 1H), 3.36 (d, J = 10.4 Hz, 1H), 3.07–3.00 (m, 2H), 2.30–2.26 (m, 2H), 2.25 (s, 3H), 1.97–1.93 (m, 2H), 1.55–1.52 (m, 2H), 1.50–1.46 (m, 2H), 1.31–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.53, 178.63, 171.70, 169.63, 158.67, 156.32, 138.90, 138.79, 136.80, 133.95, 133.79, 128.93, 128.66, 128.01, 125.15, 125.07, 119.89, 116.87, 116.64, 116.10, 115.85, 110.78, 74.82, 74.24, 61.55, 54.15, 51.04, 36.76, 36.29, 32.69, 28.82, 25.55, 25.49 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C34H35FN4NaO5S+ 653.2204, found 653.2208.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-5-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7d)
Orange solid, 39% yield, dr > 99:1, m.p. 154.6-156.2 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 9.90 (s, 1H), 7.57–7.55 (m, 2H), 7.02 (t, J = 6.8 Hz, 1H), 7.46–7.44 (m, 3H), 7.40–7.39 (m, 2H), 7.34–7.30 (m, 2H), 7.19 (d, J = 7.6 Hz, 1H), 6.53 (d, J = 8.0 Hz, 1H), 4.79 (d, J = 11.6 Hz, 1H), 4.12–4.08 (m, 1H), 3.77 (t, J = 10.4 Hz, 1H), 3.73 (d, J = 10.4 Hz, 1H), 3.35 (d, J = 10.4 Hz, 1H), 3.06–2.99 (m, 2H), 2.28–2.25 (m, 2H), 1.96–1.92 (m, 2H), 1.57–1.46 (m, 4H), 1.29–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.58, 178.42, 171.63, 169.53, 141.51, 138.85, 136.79, 133.94, 133.71, 130.11, 128.92, 128.69,128.33, 127.98, 125.39, 119.86, 111.38, 74.78, 74.02, 61.51, 54.24, 51.10, 36.78, 36.28, 32.69, 28.81, 25.52, 25.48 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C34H35ClN4NaO5S+ 669.1909, found 669.1916.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-5-bromo-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7e)
Orange solid, 30% yield, dr > 99:1, m.p. 146.8-148.0 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 9.90 (s, 1H), 7.57–7.55 (m, 3H), 7.50 (t, J = 7.2 Hz, 1H), 7.47–7.45 (m, 2H), 7.41–7.39 (m, 2H), 7.34–7.30 (m, 3H), 6.49 (d, J = 8.0 Hz, 1H), 4.79 (d, J = 11.6 Hz, 1H), 4.12–4.08 (m, 1H), 3.79–3.77 (m, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.36 (d, J = 10.4 Hz, 1H), 3.07–2.99 (m, 2H), 2.29–2.25 (m, 2H), 1.96–1.92 (m, 2H), 1.57–1.46 (m, 4H), 1.30–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.61, 178.43, 171.64, 169.52, 142.19, 138.86, 136.82, 133.93, 133.73, 132.91, 131.01, 130.10, 128.93, 128.70, 127.99, 128.88, 119.87, 113.07, 111.93, 74.77, 73.96, 61.57, 54.24, 51.11, 36.79, 36.26, 32.71, 28.83, 25.53, 25.50 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C34H35BrN4NaO5S+ 713.1404, found 713.1404.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7f)
Orange solid, 39% yield, dr > 12:1, m.p. 150.2-151.2 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.48 (s, 2H), 10.04 (s, 1H), 8.77 (s, 1H), 7.60–7.58 (m, 2H), 7.52 (t, J = 7.2 Hz, 1H), 7.43–7.39 (m, 5H), 7.35–7.31 (m, 2H), 7.02 (dd, J = 8.0, 1.6 Hz, 1H), 6.56 (d, J = 2.0 Hz, 1H), 4.77 (d, J = 11.6 Hz, 1H), 4.12–4.08 (m, 1H), 3.82–3.77 (m, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.35 (d, J = 10.4 Hz, 1H), 3.06–2.98 (m, 2H), 2.30–2.26 (m, 2H), 1.97–1.93 (m, 2H), 1.59–1.51 (m, 2H), 1.49–1.44 (m, 2H), 1.30–1.24 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.55, 178.68, 171.71, 169.63, 144.16, 138.93, 136.82, 134.44, 133.96, 133.83, 129.94, 128.95, 128.64, 127.97, 122.32, 121.12, 119.91, 110.09, 74.82, 73.55, 61.54, 54.17, 51.17, 36.78, 36.36, 32.70, 28.85, 28.83, 25.56, 25.51 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C34H35ClN4NaO5S+ 669.1909, found 669.1908.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-bromo-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7g)
Orange solid, 34% yield, dr > 20:1, m.p. 149.8-152.1 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.39 (s, 1H), 9.96 (s, 1H), 8.76 (br, 1H), 7.58–7.56 (m, 2H), 7.52 (t, J = 7.2 Hz, 1H), 7.46–7.37 (m, 4H), 7.37–7.30 (m, 3H), 7.15 (dd, J = 8.0 Hz, 1.6 Hz, 1H), 6.67 (d, J = 2.0 Hz, 1H), 4.77 (d, J = 12.0 Hz, 1H), 4.11–4.07 (m, 1H), 3.78 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.71 (d, J = 10.4 Hz, 1H), 3.36 (s, 1H), 3.05–2.97 (m, 2H), 2.29–2.25 (m, 2H), 1.95–1.91 (t, J = 7.3 Hz, 2H), 1.56–1.45 (tm, 4H), 1.28–1.24 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.51, 178.68, 171.63, 169.56, 144.36, 138.82, 136.79, 133.91, 133.86, 130.23, 128.90, 128.66, 127.96, 123.98, 122.94, 122.77, 119.88, 112.82, 74.80, 73.64, 61.52, 54.17, 51.15, 36.78, 36.35, 32.69, 28.83, 28.81, 25.52, 25.48 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C34H35BrN4NaO5S+ 713.1404, found 713.1400.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-7-methyl-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7h)
Orange solid, 37% yield, dr > 20:1, m.p. 142.7-143.9 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.31 (s, 1H), 10.09 (s, 1H), 8.97 (br, 1H), 7.62–7.59 (m, 2H), 7.49–7.45 (m, 1H), 7.43–7.41 (m, 2H), 7.29–7.23 (m, 4H), 7.20 (d, J = 7.6 Hz, 1H), 6.93 (d, J = 7.6 Hz, 1H), 6.84 (t, J = 7.6 Hz, 1H), 4.73 (d, J = 11.6 Hz, 1H), 4.12–4.07 (m, 1H), 3.78 (dd, J = 11.2 Hz, 9.2 Hz, 1H), 3.71 (d, J = 10.0 Hz, 1H), 3.35 (d, J = 10.0 Hz, 1H), 3.05–2.96 (m, 2H), 2.31–2.27 (m, 2H), 1.97–1.93 (m, 2H), 1.86 (s, 3H), 1.59–1.52 (m, 2H), 1.50–1.44 (m, 2H), 1.29–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 197.09, 179.42, 171.72, 169.63, 141.13, 138.85, 137.23, 134.17, 133.34, 131.28, 128.62, 128.58, 127.71, 125.51, 123.22, 121.34, 119.90, 119.27, 74.93, 73.84, 62.46, 53.89, 50.71, 36.76, 36.19, 32.68, 28.82, 25.56, 25.50, 16.47 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C35H38N4NaO5S+ 649.2455, found 649.2462.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-7-bromo-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7i)
Orange solid, 40% yield, dr > 17:1, m.p. 137.5-138.9 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.10 (s, 1H), 7.62–7.60 (m, 2H), 7.53-7.48 (m, 1H), 7.44–7.42 (m, 2H), 7.39 (d, J = 7.6 Hz, 1H), 7.33–7.28 (m, 5H), 6.92 (t, J = 8.0 Hz, 1H), 4.77 (d, J = 11.6 Hz, 1H), 4.13–4.09 (m, 1H), 3.79 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.73 (d, J = 10.4 Hz, 1H), 3.38 (d, J = 10.4 Hz, 1H), 3.06–2.98 (m, 2H), 2.31–2.27 (m, 2H), 1.97–1.93 (m, 2H), 1.57–1.52 (m, 2H), 1.50–1.46 (m, 2H), 1.29–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.88, 178.87, 171.72, 169.61, 141.96, 138.93, 137.00, 133.82, 133.68, 133.03, 128.80, 128.63, 127.71, 127.27, 125.59, 123.00, 119.90, 102.74, 74.88, 74.53, 62.49, 53.91, 50.78, 36.75, 36.17, 32.68, 28.82, 25.55, 25.49 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C34H35BrN4NaO5S+ 713.1404, found 713.1403.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-5-methoxy-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7j)
Orange solid, 35% yield, dr > 20:1, m.p. 134.9-136.2 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.28 (s, 2H), 10.04 (s, 1H), 8.86 (br, 1H), 7.60–7.58 (m, 2H), 7.49 (t, J = 7.2 Hz, 1H), 7.44–7.39 (m, 4H), 7.32–7.29 (m, 2H), 7.04 (d, J = 2.4 Hz, 1H), 6.71 (dd, J = 8.4 Hz, 2.8 Hz, 1H), 6.44 (d, J = 8.4 Hz, 1H), 4.76 (d, J = 11.6 Hz, 1H), 4.14–4.09 (m, 1H), 3.79 (dd, J = 11.6 Hz, 9.6 Hz, 1H), 3.74–3.71 (m, 4H), 3.36 (d, J = 10.0 Hz, 1H), 3.05 –3.00 (m, 2H), 2.29 (t, J = 7.2 Hz, 2H), 1.95 (t, J = 7.2 Hz, 2H), 1.57–1.52 (m, 2H), 1.50–1.45 (m, 2H), 1.31–1.24 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.50, 178.67, 171.68, 169.62, 154.38, 138.86, 136.92, 135.76, 133.97, 133.73, 128.82, 128.63, 127.96, 124.66, 119.88, 115.60, 114.71, 110.18, 74.76, 74.13, 61.66, 55.98, 54.05, 50.98, 36.74, 36.17, 32.67, 28.81, 25.51, 25.48 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C35H38N4NaO6S+ 665.2404, found 665.2407.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-methoxy-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7k)
Orange solid, 29% yield, dr > 20:1, m.p. 139.4-136.2 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.03 (s, 1H), 7.60–7.58 (m, 2H), 7.50 (t, J = 7.6 Hz, 1H), 7.42–7.38 (m, 4H), 7.33–7.29 (m, 3H), 6.50 (dd, J = 8.4 Hz, 2.0 Hz, 1H), 6.07 (d, J = 2.0 Hz, 1H), 4.74 (d, J = 11.6 Hz, 1H), 4.10–4.05 (m, 1H), 3.81-3.75 (m, 1H), 3.70 (d, J = 10.0 Hz, 1H), 3.68 (s, 3H), 3.36 (d, J = 10.0 Hz, 1H), 3.05–2.96 (m, 2H), 2.28 (t, J = 7.2 Hz, 2H), 1.95 (t, J = 7.2 Hz, 2H), 1.59–1.52 (m, 2H), 1.50–1.44 (m, 2H), 1.31–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.68, 179.36, 171.67, 169.60, 160.91, 143.98, 138.83, 137.02, 134.15, 133.67, 129.47, 128.81, 128.60, 127.94, 119.88, 114.97, 106.42, 96.57, 74.80, 73.65, 61.35, 55.59, 54.15, 51.10, 36.75, 36.42, 32.68, 28.82, 25.54, 25.48 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C35H38N4NaO6S+ 665.2404, found 665.2411.
N1-hydroxy-N8-(4-((3R,6'S,7'R,7a'S)-6'-(3-methoxybenzoyl)-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)octanediamide (7l)
Orange solid, 31% yield, dr > 20:1, m.p. 121.6-122.8 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.38 (s, 1H), 9.97 (s, 1H), 8.81 (br, 1H), 7.59–7.57 (m, 2H), 7.44–7.42 (m, 2H), 7.38 (d, J = 7.2 Hz, 1H), 7.19 (t, J = 8.0 Hz, 1H), 7.12 (t, J = 7.6 Hz, 1H), 7.04 (dd, J = 8.4, 2.8 Hz, 1H), 6.95–6.92 (m, 2H), 6.85–6.84 (m, 1H), 6.53 (d, J = 7.6 Hz, 1H), 4.73 (d, J = 11.6 Hz, 1H), 4.12–4.07 (m, 1H), 3.81 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.70 (d, J = 10.0 Hz, 1H), 3.69 (s, 3H), 3.33 (d, J = 10.0 Hz, 1H), 3.05–2.97 (m, 2H), 2.28 (t, J = 7.2 Hz, 2H), 1.94 (t, J = 7.6 Hz, 2H), 1.57–1.51(m, 2H), 1.50–1.45 (m, 2H), 1.30–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.42, 178.87, 171.67, 169.59, 159.35, 142.59, 138.84, 138.32, 134.09, 130.17, 129.91, 128.68, 128.36, 123.50, 121.41, 120.26, 120.08, 119.90, 112.29, 109.98, 74.78, 73.71, 61.91, 55.57, 54.00, 51.04, 36.78, 36.27, 32.70, 28.85, 25.56, 25.50 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C35H38N4NaO6S+ 665.2404, found 665.2402.
N1-(4-((3R,6'S,7'R,7a'S)-6'-(4-(dimethylamino)benzoyl)-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7m)
Orange solid, 33% yield, dr > 16:1, m.p. 148.7-150.0 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.24 (s, 1H), 9.91 (s, 1H), 7.55–7.53 (m, 2H), 7.44 (d, J = 7.6 Hz, 1H), 7.39–7.34 (m, 4H), 7.10 (t, J = 7.6 Hz, 1H), 6.91 (t, J = 7.2 Hz, 1H), 6.58 (d, J = 7.2 Hz, 1H), 6.50 (d, J = 8.4 Hz, 2H), 4.64 (d, J = 11.6 Hz, 1H), 4.12–4.08 (m, 1H), 3.84 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.71 (d, J = 10.0 Hz, 1H), 3.32 (d, J = 10.0 Hz, 1H), 3.03–2.96 (m, 2H), 2.92 (s, 6H), 2.25 (t, J = 7.6 Hz, 2H), 1.93 (t, J = 7.2 Hz, 2H), 1.57–1.51 (m, 2H), 1.49–1.44 (m, 2H), 1.29–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 192.37, 179.12, 171.61, 169.50, 153.64, 142.44, 138.72, 130.32, 129.85, 128.86, 128.53, 124.51, 123.85, 121.18, 119.84, 110.82, 109.83, 74.77, 74.10, 60.36, 53.86, 51.46, 36.78, 36.26, 32.71, 28.83, 25.54, 25.50 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C36H41N5NaO5S+ 678.2721, found 678.2722.
N1-(4-((3R,6'S,7'R,7a'S)-6'-(3-bromobenzoyl)-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N8-hydroxyoctanediamide (7n)
Orange solid, 27% yield, dr > 10:1, m.p. 120.2-121.5 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.37 (s, 1H), 9.98 (s, 1H), 8.81 (br, 1H), 7.68 (d, J = 7.6 Hz, 1H), 7.60–7.58 (m, 2H), 7.47–7.45 (m, 2H), 7.38 (d, J = 7.6 Hz, 1H), 7.32–7.31 (m, 2H), 7.25 (t, J = 8.0 Hz, 1H), 7.15 (t, J = 7.6 Hz, 1H), 6.95 (t, J = 7.6 Hz, 1H), 6.52 (d, J = 7.6 Hz, 1H), 4.73 (d, J = 11.6 Hz, 1H), 4.11–4.07 (m, 1H), 3.78 (dd, J = 11.6 Hz, 9.6 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.34 (d, J = 10.4 Hz, 1H), 3.05–2.98 (m, 2H), 2.29 (t, J = 7.6 Hz, 2H), 1.95 (t, J = 7.2 Hz, 2H), 1.60–1.52 (m, 2H), 1.50–1.45 (m, 2H), 1.31–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.09, 178.61, 171.66, 169.58, 142.60, 138.97, 138.86, 136.15, 133.88, 130.91, 130.37, 130.30, 128.74, 128.24, 126.81, 123.27, 122.18, 121.48, 119.87, 110.03, 74.88, 73.64, 62.14, 54.06, 50.75, 36.79, 36.23, 32.70, 28.84, 25.55, 25.50 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C34H35BrN4NaO5S+ 713.1404, found 713.1404.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N4-hydroxysuccinamide (7o)
Orange solid, 20% yield, dr > 16:1, m.p. 145.1-146.3 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.38 (s, 1H), 10.10 (s, 1H), 8.79 (br, 1H), 7.58–7.56 (m, 2H), 7.48 (d, J = 7.2 Hz, 2H), 7.43–7.38 (m, 3H), 7.35–7.33 (m, 2H), 7.30–7.26 (m, 2H), 7.11 (d, J = 8.0 Hz, 1H), 6.93 (t, J = 7.2 Hz, 1H), 6.51 (d, J = 7.6 Hz, 1H), 4.76 (d, J = 11.6 Hz, 1H), 4.13–4.08 (m, 1H), 3.82 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.71 (d, J = 10.0 Hz, 1H), 3.33 (d, J = 10.0 Hz, 1H), 3.05–2.97 (m, 2H), 2.56–2.51 (m, 2H), 2.28 (t, J = 7.6 Hz, 2H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.67, 178.77, 170.62, 168.82, 142.53, 138.79, 136.98, 134.12, 133.69, 130.17, 128.79, 128.64, 128.38, 127.91, 123.45, 121.38, 119.80, 109.98, 74.82, 73.67, 61.79, 53.90, 50.96, 36.35, 36.23, 32.03, 28.05 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C30H28N4NaO5S+ 579.1673, found 579.1676.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N6-hydroxyadipamide (7p)
Orange solid, 26% yield, dr > 99:1, m.p. 140.3-141.7 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.37 (s, 2H), 10.00 (s, 1H), 8.75 (br, 1H), 7.60–7.58 (m, 2H), 7.50–7.39 (m, 3H), 7.36–7.34 (m, 2H), 7.31–7.27 (m, 2H), 7.11 (t, J = 7.6 Hz, 1H), 6.94 (t, J = 7.6 Hz, 1H), 6.51 (d, J = 7.6 Hz, 1H), 4.77 (d, J = 11.6 Hz, 1H), 4.13–4.09 (m, 1H), 3.83 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.71 (d, J = 10.0 Hz, 1H), 3.33 (d, J = 10.0 Hz, 1H), 3.06–2.98 (m, 2H), 2.29 (t, J = 6.8 Hz, 2H), 1.99 (t, J = 6.8 Hz, 2H), 1.59–1.49 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.67, 178.79, 171.51, 169.44, 142.51, 138.80, 136.97, 134.14, 133.67, 130.17, 128.79, 128.65, 128.39, 127.91, 123.45, 121.39, 119.89, 109.96, 74.83, 73.69, 61.78, 53.94, 50.97, 36.56, 36.24, 32.57, 25.31, 25.26 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C32H32N4NaO5S+ 607.1986, found 607.1986.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N10-hydroxydecanediamide (7q)
Orange solid, 35% yield, dr > 16:1, m.p. 131.9-132.1 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.39 (s, 2H), 10.01 (s, 1H), 8.80 (s, 1H), 7.60–7.58 (m, 2H), 7.48 (t, J = 7.2 Hz, 1H), 7.44–7.39 (m, 3H), 7.36–7.34 (m, 2H), 7.31–7.27 (m, 2H), 7.11 (t, J = 7.6 Hz, 1H), 6.93 (t, J = 7.6 Hz, 1H), 6.51 (d, J = 7.6 Hz, 1H), 4.76 (d, J = 11.6 Hz, 1H), 4.13–4.09 (m, 1H), 3.83 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.71 (d, J = 10.0 Hz, 1H), 3.33 (d, J = 10.0 Hz, 1H), 3.05–2.97 (m, 2H), 2.28 (t, J = 7.6 Hz, 2H), 1.94 (t, J = 7.2 Hz, 2H), 1.58–1.52 (m, 2H), 1.49–1.44 (m, 2H), 1.28–1.20 (m, 8H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.65, 17.8.78, 171.71, 169.61, 142.53, 138.86, 136.97, 134.08, 133.67, 130.16, 128.78, 128.62, 128.38, 127.91, 123.45, 121.37, 119.88, 109.97, 74.82, 73.68, 61.78, 53.92, 50.98, 36.81, 36.23, 32.70, 29.18, 29.10, 29.07, 29.01, 25.65, 25.57 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C36H40N4NaO5S+ 663.2612, found 663.2613.
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N6-hydroxyadipamide (7r)
Orange solid, 30% yield, dr > 13:1, m.p. 160.6-161.9 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.47 (s, 2H), 9.99 (s, 1H), 8.74 (br, 1H), 7.59–7.57 (m, 2H), 7.52 (t, J = 7.2 Hz, 1H), 7.44–7.39 (m, 5H), 7.35–7.31 (m, 2H), 7.02 (dd, J = 8.0 Hz, 2.0 Hz, 1H), 6.54 (d, J = 2.0 Hz, 1H), 4.78 (d, J = 11.6 Hz, 1H), 4.13–4.08 (m, 1H), 3.80 (dd, J = 11.6 Hz, 9.6 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.36 (d, J = 10.4 Hz, 1H), 3.06–2.98 (m, 2H), 2.29 (t, J = 6.4 Hz, 2H), 1.98 (t, J = 6.4 Hz, 2H), 1.59–1.49 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.56, 178.7, 171.53, 169.45, 144.12, 138.87, 136.84, 134.46, 133.94, 133.89, 129.94, 128.93, 128.67, 127.97, 122.33, 121.13, 119.90, 110.04, 74.83, 73.56, 61.57, 54.18, 51.14, 36.58, 36.36, 32.58, 25.32, 25.27 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C32H31ClN4NaO5S+ 641.1596, found 641.1596.
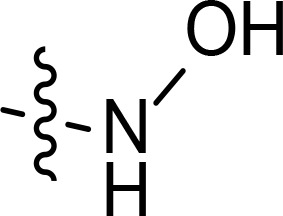
N1-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N10-hydroxydecanediamide (7s)
Orange solid, 37% yeld, dr > 99:1, m.p. 137.7-139.0 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.38 (s, 2H), 9.88 (s, 1H), 8.87 (br, 1H), 7.58–7.56 (m, 2H), 7.52 (t, J = 7.2 Hz, 1H), 7.44–7.39 (m, 5H), 7.35–7.31 (m, 3H), 7.02 (d, J = 8.0 Hz, 1H), 6.53 (s, 1H), 4.78 (d, J = 11.6 Hz, 1H), 4.13–4.08 (m, 1H), 3.80 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.36 (d, J = 10.4 Hz, 1H), 3.06–2.98 (m, 2H), 2.27 (t, J = 7.2 Hz, 2H), 1.93 (t, J = 7.2 Hz, 2H), 1.59–1.55 (m, 2H), 1.49–1.46 (m, 2H), 1.28–1.20 (m, 8H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.54, 178.72, 171.66, 169.57, 144.10, 138.85, 136.83, 134.46, 133.91, 133.87, 129.94, 128.91, 128.67, 127.96, 122.32, 121.13, 119.87, 110.01, 74.82, 73.54, 61.59, 54.16, 51.11, 36.83, 36.35, 32.71, 29.18, 29.08, 29.02, 25.63, 25.56 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C36H39ClN4NaO5S+ 697.2222, found 697.2227.
N1-(4-((1'R,2'S,3R,7a'R)-2'-benzoyl-6-chloro-2-oxo-1',2',5',6',7',7a'-hexahydrospiro[indoline-3,3'-pyrrolizin]-1'-yl)phenyl)-N8-hydroxyoctanediamide (7t)
Orange solid, 50% yield, dr > 25:1, m.p. 143.4-144.5 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.41 (s, 2H), 9.92 (s, 1H), 8.73 (br, 1H), 7.56–7.54 (m, 2H), 7.50 (d, J = 7.6 Hz, 1H), 7.43–7.31 (m, 6H), 7.26 (d, J = 8.4 Hz, 1H), 6.99 (d, J = 7.6 Hz, 1H), 6.57 (s, 1H), 4.85 (d, J = 11.6 Hz, 1H), 3.93-3.88 (m, 1H), 3.81 (t, J = 10.8 Hz, 1H), 2.55 (d, J = 8.0 Hz, 1H), 2.38-2.33 (m, 1H), 2.27 (t, J = 7.2 Hz, 2H), 1.95 (t, J = 7.6 Hz, 2H), 1.89-1.82 (m, 2H), 1.77–1.68 (m, 2H), 1.57–1.52 (m, 2H), 1.50–1.45 (m, 2H), 1.31–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 197.15, 179.76, 171.59, 169.58, 143.93, 138.52, 137.03, 134.55, 133.98, 133.77, 128.23, 128.90, 128.26, 127.92, 124.16, 121.23, 119.83, 110.08, 72.55, 71.83, 63.30, 52.27, 47.96, 36.76, 32.69, 29.95, 28.83, 27.22, 25.54, 25.48 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C35H37ClN4 651.2345, found 651.2347.
N-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-2-(2-(hydroxyamino)-2-oxoethoxy)acetamide (7u)
Orange solid, 36% yield, dr > 10:1, m.p. 167.1-168.5 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.55 (s, 2H), 9.95 (s, 1H), 9.07 (br, 1H), 7.61–7.58 (m, 2H), 7.53–7.46 (m, 3H), 7.43–7.36 (m, 3H), 7.33–7.29 (m, 2H), 7.01 (dd, J = 8.0, 2.0 Hz, 1H), 6.50 (d, J = 2.0 Hz, 1H), 4.78 (d, J = 11.6 Hz, 1H), 4.15–4.07 (m, 3H), 4.02 (s, 2H), 3.84–3.78 (m, 1H), 3.70 (d, J = 10.4 Hz, 1H), 3.33 (s, 1H), 3.05–2.99 (m, 2H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.59, 178.70, 168.05, 165.77, 144.10, 137.66, 136.84, 134.86, 133.93, 129.96, 128.92, 128.75, 127.98, 122.31, 121.15, 120.91, 110.02, 74.81, 73.57, 71.07, 69.81, 61.60, 54.18, 51.13, 36.36 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C30H27ClN4NaO6S+ 629.1232, found 629.1238.
N1-(4-((4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)carbamoyl)phenyl)-N6-hydroxyadipamide (7v)
Orange solid, 17% yield, dr > 16:1, m.p. 175.5-176.6 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.42 (s, 2H), 10.19 (s, 1H), 10.10 (s, 1H), 8.71 (br, 1H), 7.92–7.90 (m, 2H), 7.75–7.71 (m, 4H), 7.54–7.48 (m, 3H), 7.45–7.39 (m, 3H), 7.35–7.31 (m, 2H), 7.03 (dd, J = 8.0, 2.0 Hz, 1H), 6.52 (d, J = 2.0 Hz, 1H), 4.81 (d, J = 11.6 Hz, 1H), 4.15–4.11 (m, 1H), 3.83 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.73 (d, J = 10.4 Hz, 1H), 3.08–3.02 (m, 2H), 2.35 (t, J = 6.4 Hz, 2H), 1.99 (t, J = 6.4 Hz, 2H), 1.62–1.52 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.59, 178.71, 172.02, 169.39, 165.26, 144.08, 142.73, 138.80, 136.85, 134.47, 133.93, 129.95, 129.34, 129.02, 128.93, 128.61, 127.97, 122.33, 121.10, 118.62, 110.00, 74.83, 73.56, 61.62, 54.16, 51.15, 36.71, 36.38, 32.61, 25.32, 25.14 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C39H36ClN5NaO6S+ 760.1967, found 760.1968.
N1-(4-((3R,6'S,7'R,7a'S)-6-chloro-6'-(3-methoxybenzoyl)-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N6-hydroxyadipamide (7w)
Orange solid, 40% yield, dr > 99:1, m.p. 141.8-143.1 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 9.91 (s, 1H), 7.57–7.55 (m, 2H), 7.45–7.40 (m, 3H), 7.22 (t, J = 7.6 Hz, 1H), 7.06 (d, J = 8.0 Hz, 1H), 7.03–6.97 (m, 2H), 6.85 (m, 1H), 6.54 (m, 1H), 4.74 (d, J = 11.6 Hz, 1H), 4.11–4.07 (m, 1H), 3.80–3.75 (m, 2H), 3.71 (s, 3H), 3.37 (d, J = 10.4 Hz, 1H), 3.05–2.98 (m, 2H), 2.28 (t, J = 6.8 Hz, 2H), 1.98 (t, J = 6.8 Hz, 2H), 1.57–1.49 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.44, 178.96, 171.51, 169.43, 159.42, 144.40, 138.79, 138.21, 134.48, 133.95, 130.02, 129.84, 128.72, 122.42, 121.09, 120.36, 120.30, 119.89, 112.15, 110.06, 74.79, 73.58, 61.82, 55.55, 54.25, 51.14, 36.62, 36.39, 32.61, 25.33, 25.29 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C33H33ClN4NaO6S+ 671.1702, found 671.1694.
N1-(4-((3R,6'S,7'R,7a'S)-6-chloro-6'-(4-fluorobenzoyl)-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N6-hydroxyadipamide (7x)
Orange solid, 34% yield, dr > 13:1, m.p. 218.2-219.6°C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.43 (s, 2H), 9.90 (s, 1H), 8.73 (, 1H), 7.57–7.55 (m, 2H), 7.48–7.42 (m, 5H), 7.19–7.15 (m, 2H), 7.02 (d, J = 8.0 Hz, 1H), 6.55 (s, 1H), 4.76 (d, J = 11.6 Hz, 1H), 4.11–4.06 (m, 1H), 3.77 (t, J = 10.8 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.34 (d, J = 10.4 Hz, 1H), 3.05–2.99 (m, 2H), 2.28 (t, J = 6.8 Hz, 2H), 1.98 (t, J = 6.8 Hz, 2H), 1.57–1.49 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 195.24, 178.67, 171.48, 169.38, 164.22, 144.09, 138.81, 134.52, 133.79, 133.61, 131.05, 130.96, 129.92, 128.71, 122.22, 121.15, 119.86, 116.11, 115.89, 110.03, 74.85, 73.64, 61.62, 54.26, 51.02, 36.61, 36.35, 32.59, 25.31, 25.28 ppm; HRMS (ESI-TOF) m/z: [M + H]+ Calculated for C32H31ClFN4O5S+ 637.1682, found 637.1682.
N1-(4-((3R,6'S,7'R,7a'S)-6-chloro-6'-(4-(dimethylamino)benzoyl)-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N6-hydroxyadipamide (7y)
Orange solid, 37% yield, dr > 25:1, m.p. 178.2-179.9°C. 1H NMR (400 MHz, DMSO-d 6): δ = 9.87 (s, 1H), 7.53–7.47 (m, 3H), 7.41–7.34 (m, 4H), 6.99 (dd, J = 8.0 Hz, 2.0 Hz, 1H), 6.62 (d, J = 2.0 Hz, 1H), 6.54–6.52 (m, 2H), 4.66 (d, J = 12.0 Hz, 1H), 4.11–4.06 (m, 1H), 3.82 (dd, J = 12.0 Hz, 9.2 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.35 (d, J = 10.4 Hz, 1H), 3.05–2.99 (m, 2H), 2.94 (s, 6H), 2.26 (t, J = 7.2 Hz, 2H), 1.96 (t, J = 6.8 Hz, 2H), 1.58–1.48 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 192.16, 179.05, 171.46, 169.40, 153.78, 144.02, 138.71, 134.19, 134.10, 130.37, 128.55, 124.31, 122.71, 120.93, 119.83, 110.90, 109.89, 74.77, 73.99, 60.12, 54.15, 51.60, 36.60, 36.39, 32.60, 25.32, 25.28 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C34H36ClN5NaO5S+ 684.2018, found 684.2008.
N1-(2-aminophenyl)-N4-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)succinamide (9a)
White sold, 19% yield, dr > 99:1, m.p. 149.1-150.2 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.34 (s, 1H), 10.05 (s, 1H), 9.25 (s, 1H), 7.59–7.57 (m, 2H), 7.49–7.39 (m, 4H), 7.36–7.34 (m, 2H), 7.30–7.26 (m, 2H), 7.15-7.09 (m, 2H), 6.93 (t, J = 7.6 Hz, 1H), 6.88 (t, J = 7.6 Hz, 1H), 6.70 (d, J = 7.6 Hz, 1H), 6.53–6.49 (m, 2H), 4.90–4.88 (m, 2H), 4.77 (d, J = 11.6 Hz, 1H), 4.14–4.09 (m, 1H), 3.84 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.71 (d, J = 10.0 Hz, 1H), 3.33 (d, J = 10.0 Hz, 1H), 3.06–2.98 (m, 2H), 2.65 (s, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.69, 178.81, 170.97, 170.84, 142.70, 142.52, 138.76, 136.99, 134.18, 133.67, 130.18, 128.79, 128.68, 128.39, 127.92, 126.28, 126.02, 123.74, 123.47, 121.40, 119.87, 116.38, 116.04, 109.96, 74.84, 73.69, 61.82, 53.91, 50.95, 36.25, 32.16, 31.32 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C36H33N5NaO4S+ 654.2145, found 654.2150.
N1-(2-aminophenyl)-N8-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)octanediamide (9b)
White yield, 45% yield, dr > 99:1, m.p. 131.7-132.9 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.35 (s, 1H), 9.95 (s, 1H), 9.20 (s, 1H), 7.60–7.58 (m, 2H), 7.48 (t, J = 7.2 Hz, 1H), 7.46–7.39 (m, 3H), 7.36–7.33 (m, 2H), 7.30–7.26 (m, 2H), 7.17 (d, J = 7.6 Hz, 1H), 7.11 (t, J = 7.6 Hz, 1H), 6.93 (t, J = 7.6 Hz, 1H), 6.88 (t, J = 7.6 Hz, 1H), 6.71 (d, J = 8.0 Hz, 1H), 6.55–6.49 (m, 2H), 4.85 (s, 2H), 4.77 (d, J = 11.6 Hz, 1H), 4.13–4.09 (m, 1H), 3.83 (dd, J = 11.2 Hz, 9.2 Hz, 1H), 3.71 (d, J = 10.4 Hz, 1H), 3.33 (d, J = 10.0 Hz, 1H), 3.05–3.97 (m, 2H), 2.34–2.28 (m, 4H), 1.63–1.56 (m, 4H), 1.35–1.32 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.67, 178.80, 171.67, 171.64, 142.52, 142.28, 138.84, 136.99, 134.11, 133.67, 130.17, 128.79, 128.65, 128.40, 127.91, 126.04, 125.69, 124.10, 123.45, 121.39, 119.91, 116.58, 116.33, 109.96, 74.84, 73.70, 61.80, 53.94, 50.98, 36.82, 36.25, 36.22, 28.95, 28.91, 25.71, 25.59 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C40H41N5NaO4S+ 710.2771, found 710.2772.
N1-(2-aminophenyl)-N6-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)adipamide (9c)
White yield, 24% yield, dr > 20:1, m.p. 148.9-150.0 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.53 (s, 1H), 9.99 (s, 1H), 9.21 (s, 1H), 7.60–7.58 (m, 2H), 7.52 (t, J = 7.2 Hz, 1H), 7.45–7.39 (m, 5H), 7.34–7.31 (m, 2H), 7.17 (d, J = 8.0 Hz, 1H), 7.02 (dd, J = 8.0, 2.0 Hz, 1H), 6.88 (t, J = 7.6 Hz, 1H), 6.72 (d, J = 8.0 Hz, 1H), 6.55–6.51 (m, 2H), 4.86 (s, 2H), 4.78 (d, J = 11.6 Hz, 1H), 4.13–4.09 (m, 1H), 3.80 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.36 (d, J = 10.4 Hz, 1H), 3.06–2.98 (m, 2H), 2.38–2.33 (m, 4H), 1.66-1.60 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.56, 178.72, 171.57, 171.49, 144.12, 142.31, 138.86, 136.84, 134.46, 133.91, 129.94, 128.92, 128.67, 127.97, 126.09, 125.73, 124.02, 122.33, 121.13, 119.92, 116.57, 116.32, 110.03, 74.82, 73.56, 61.59, 54.17, 51.13, 36.66, 36.36, 36.03, 25.46, 25.35 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C38H36ClN5NaO4S+ 716.2069, found 716.2070.
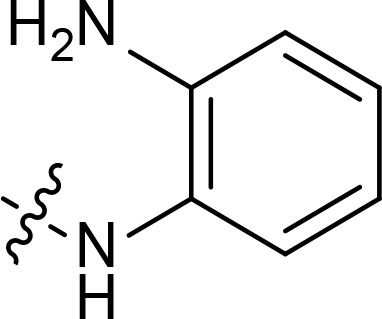
N1-(2-aminophenyl)-N8-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)octanediamide (9d)
White yield, 30% yield, dr > 99:1, m.p. 145.2-146.4 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.53 (s, 1H), 9.92 (s, 1H), 9.16 (s, 1H), 7.59–7.57 (m, 2H), 7.52 (t, J = 7.6 Hz, 1H), 7.44–7.39 (m, 5H), 7.34–7.31 (m, 2H), 7.17 (d, J = 7.6 Hz, 1H), 7.02 (dd, J = 8.0 Hz, 2.0 Hz, 1H), 6.88 (t, J = 7.6 Hz, 1H), 6.72 (d, J = 7.6 Hz, 1H), 6.55-6.51 (m, 2H), 4.84 (s, 2H), 4.78 (d, J = 11.6 Hz, 1H), 4.13–4.08 (m, 1H), 3.80 (dd, J = 11.2 Hz, 9.2 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.36 (d, J = 10.4 Hz, 1H), 3.06–2.98 (m, 2H), 2.34–2.27 (m, 4H), 1.63–1.56 (m, 4H), 1.35–1.32 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.54, 178.70, 171.65, 171.62, 144.08, 142.29, 138.87, 136.83, 134.46, 133.91, 133.85, 129.93, 128.91, 128.66, 127.95, 126.07, 125.70, 124.07, 122.31, 121.13, 119.89, 116.59, 116.33, 110.01, 74.81, 73.54, 61.58, 54.17, 51.12, 36.82, 36.35, 36.21, 28.94, 28.91, 25.70, 25.58 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C40H40ClN5NaO4S+ 744.2382, found 744.2386.
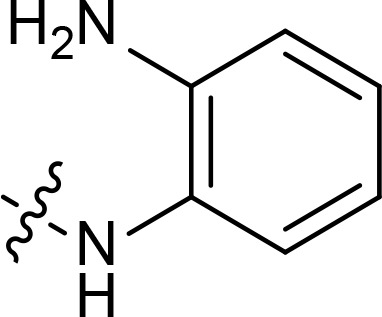
N1-(2-aminophenyl)-N10-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)decanediamide (9e)
White solid, 36% yield, dr > 99:1, m.p. 132.4-133.9 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.52 (s, 1H), 9.91 (s, 1H), 9.16 (s, 1H), 7.58–7.56 (m, 2H), 7.52 (t, J = 7.6 Hz, 1H), 7.44–7.39 (m, 5H), 7.34–7.30 (m, 2H), 7.16 (d, J = 7.2 Hz, 1H), 7.02 (dd, J = 8.0 Hz, 2.0 Hz, 1H), 6.88 (t, J = 7.2 Hz, 1H), 6.71 (d, J = 7.6 Hz, 1H), 6.55–6.51 (m, 2H), 4.84 (s, 2H), 4.77 (d, J = 11.6 Hz, 1H), 4.13–4.08 (m, 1H), 3.79 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.36 (d, J = 10.8 Hz, 1H), 3.06–2.98 (m, 2H), 2.33–2.26 (m, 4H), 1.60-1.56 (m, 4H), 1.29 (s, 8H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.54, 178.71, 171.68, 171.66, 144.09, 142.29, 138.87, 136.83, 134.46, 133.91, 133.85, 129.94, 128.91, 128.66, 127.96, 126.07, 125.68, 122.32, 121.14, 119.88, 116.61, 116.35, 110.01, 74.82, 73.55, 61.59, 54.16, 51.12, 36.84, 36.36, 36.24, 29.20, 29.18, 29.12, 29.09, 25.78, 25.65 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C42H44ClN5NaO4S+ 772.2695, found 772.2700.
N-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-1-(6-(hydroxyamino)-6-oxohexanoyl)piperidine-4-carboxamide (11a)
Orange solid, 24% yield, dr > 16:1, m.p. 170.3-171.5 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.43 (s, 2H), 9.92 (s, 1H), 8.71 (s, 1H), 7.58–7.56 (m, 2H), 7.52 (t, J = 7.2 Hz, 1H), 7.45–7.38 (m, 5H), 7.34–7.31 (m, 2H), 7.02 (d, J = 8.0 Hz, 1H), 6.52 (s, 1H), 4.78 (d, J = 11.6 Hz, 1H), 4.41 (d, J = 12.4 Hz, 1H), 4.12–4.08 (m, 1H), 3.91 (d, J = 13.2 Hz, 1H), 3.80 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.35 (s, 1H), 3.05-2.98 (m, 3H), 2.33–2.54 (m, 2H), 2.33–2.26 (m, 2H), 1.99–1.95 (m, 2H), 1.81–1.75 (m, 2H), 1.58–1.42 (m, 6H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.53, 178.71, 173.35, 170.74, 169.46, 144.07, 138.78, 136.83, 134.47, 134.05, 133.93, 129.95, 128.92, 128.71, 127.96, 122.31, 121.15, 119.98, 110.00, 74.81, 73.55, 61.61, 54.17, 51.12, 44.90, 43.14, 40.97, 36.34, 32.59, 32.50, 29.34, 28.65, 25.37, 24.91 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C38H40ClN5NaO6S+ 752.2280, found 752.2284.
N-(4-((3R,6'S,7'R,7a'S)-6'-benzoyl-6-chloro-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-1-(8-(hydroxyamino)-8-oxooctanoyl)piperidine-4-carboxamide (11b)
Orange solid, 31% yield, dr > 17:1, m.p. 137.3-138.6 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.47 (s, 1H), 10.32 (s, 1H), 9.91 (s, 1H), 8.64 (s, 1H), 7.56–7.54 (m, 2H), 7.50 (t, J = 7.2 Hz, 1H), 7.43–7.36 (m, 5H), 7.33–7.29 (m, 2H), 7.00 (dd, J = 8.0, 1.6 Hz, 1H), 6.50 (d, J = 2.0 Hz, 1H), 4.75 (d, J = 12.0 Hz, 1H), 4.39 (d, J = 12.8 Hz, 1H), 4.10–4.05 (m, 1H), 3.89 (d, J = 13.2 Hz, 1H), 3.78 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.70 (d, J = 10.4 Hz, 1H), 3.04–3.99 (m, 2H), 2.55 (t, J = 12.8 Hz, 2H), 2.29–2.25 (m, 2H), 1.94–1.90 (m, 2H), 1.79–1.73 (m, 2H), 1.50–1.42 (m, 4H), 1.29–1.20 (m, 8H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 196.51, 178.68, 173.33, 170.85, 169.58, 144.05, 138.76, 136.81, 134.44, 134.01, 133.91, 129.92, 128.90, 128.67, 127.93, 122.29, 121.12, 119.95, 109.98, 74.79, 73.52, 61.58, 54.13, 51.09, 44.89, 43.10, 40.92, 36.30, 32.75, 32.69, 29.32, 28.95, 28.89, 28.64, 25.49, 25.23 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C40H44ClN5NaO6S+ 780.2593, found 780.2596.
N1-(4-((3R,3'S,4'R)-3'-benzoyl-6-chloro-1'-methyl-2-oxospiro[indoline-3,2'-pyrrolidin]-4'-yl)phenyl)-N8-hydroxyoctanediamide (13a)
Orange solid, 50% yield, dr > 20:1, m.p. 126.7-127.9 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.47 (s, 2H), 9.96 (s, 1H), 8.76 (s, 1H), 7.57–7.55 (m, 2H), 7.47 (t, J = 7.6 Hz, 1H), 7.40–7.38 (m, 2H), 7.34–7.28 (m, 4H), 6.95–6.88 (m, 2H), 6.51 (s, 1H), 4.42 (d, J = 9.2 Hz, 1H), 4.29 (dd, J = 16.8 Hz, 9.2 Hz, 1H), 3.42 (d, J = 8.8 Hz, 1H), 3.35 (d, J = 8.0 Hz, 1H), 2.28 (t, J = 7.2 Hz, 2H), 2.07 (s, 3H), 1.95 (t, J = 7.2 Hz, 2H), 1.59–1.54 (m, 2H), 1.50–1.45 (m, 2H), 1.31–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 197.54, 179.33, 171.61, 169.59, 144.05, 138.56, 136.97, 135.81, 133.72, 128.93, 128.25, 127.91, 127.65, 126.23, 121.91, 119.79, 109.71, 72.75, 62.07, 60.18, 44.07, 36.75, 34.91, 32.69, 28.84, 25.50 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C33H35ClN4 625.2188, found 625.2188.
N1-(4-((3R,3'S,4'R,5'S)-3'-benzoyl-6-chloro-2-oxo-5'-phenylspiro[indoline-3,2'-pyrrolidin]-4'-yl)phenyl)-N8-hydroxyoctanediamide (15a)
Orange solid, 15% yield, dr > 10:1, m.p. 146.5-147.6 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.38 (s, 2H), 9.81 (s, 1H), 8.86 (br, 1H), 7.47–7.45 (m, 2H), 7.40–7.38 (m, 2H), 7.34–7.16 (m, 11H), 6.94 (d, J = 8.4 Hz, 1H), 6.45 (s, 1H), 4.94 (dd, J = 10.4 Hz, 4.4 Hz, 1H), 4.57 (d, J = 10.4 Hz, 1H), 4.08 (d, J = 4.8 Hz, 1H), 3.94 (t, J = 10.4 Hz, 1H), 2.23 (t, J = 7.2 Hz, 2H), 1.92 (t, J = 7.2 Hz, 2H), 1.53 (t, J = 6.8 Hz, 2H), 1.47 (t, J = 6.8 Hz, 2H), 1.26–1.23 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 197.47, 182.21, 171.55, 169.52, 143.46, 141.87, 138.50, 137.05, 133.95, 133.72, 133.32, 129.70, 128.91, 128.80, 128.50, 127.92, 127.79, 127.63, 127.59, 121.73, 119.63, 109.47, 67.56, 67.27, 62.31, 55.25, 36.76, 32.69, 28.84, 25.49 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C38H37ClN4 687.2345, found 687.2342.
N1-(4-((3R,6'S,7'R,7a'S)-6-chloro-6'-(furan-2-carbonyl)-2-oxo-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N6-hydroxyadipamide (15b)
Orange solid, 34% yield, dr > 25:1, m.p. 151.3-152.4 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.38 (s, 1H), 9.88 (s, 1H), 8.86 (br, 1H), 7.83 (s, 1H), 7.55–7.53 (m, 2H), 7.46 (d, J = 8.0 Hz, 1H), 7.39–7.37 (m, 2H), 7.16 (d, J = 3.2 Hz, 1H), 7.01 (d, J = 8.0 Hz, 1H), 6.70 (s, 1H), 6.59 (s, 1H), 4.45 (d, J = 11.6 Hz, 1H), 4.11–4.06 (m, 1H), 3.78 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.72 (d, J = 10.4 Hz, 1H), 3.38 (d, J = 10.4 Hz, 1H), 3.05–2.96 (m, 2H), 2.29–2.26 (m, 2H), 1.99–1.96 (m, 2H), 1.57–1.50 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 182.78, 178.86, 171.49, 169.43, 151.78, 149.24, 144.06, 138.89, 134.51, 133.54, 130.12, 128.65, 122.29, 121.20, 120.19, 119.90, 113.12, 110.17, 74.64, 73.83, 61.91, 53.90, 50.60, 36.63, 36.16, 32.61, 25.31, 25.30 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C30H29ClN4NaO6S+ 631.1389, found 631.1383.
N1-(4-((3R,6'S,7'R,7a'S)-6-chloro-2-oxo-6'-(thiophene-2-carbonyl)-1',6',7',7a'-tetrahydro-3'H-spiro[indoline-3,5'-pyrrolo[1,2-c]thiazol]-7'-yl)phenyl)-N6-hydroxyadipamide (15c)
Orange solid, 36% yield, dr > 20:1, m.p. 151.6-153.8 °C. 1H NMR (400 MHz, DMSO-d 6): δ = 10.49 (s, 2H), 9.86 (s, 1H), 8.67 (s, 1H), 7.89 (d, J = 4.8 Hz, 1H), 7.63 (d, J = 3.6 Hz, 1H), 7.54–7.52 (m, 2H), 7.47 (d, J = 8.4 Hz, 1H), 7.40–7.38 (m, 2H), 7.12 (t, J = 4.4 Hz, 1H), 7.03 (dd, J = 8.0 Hz, 1H), 6.63 (d, J = 2.0 Hz, 1H), 4.60 (d, J = 12.0 Hz, 1H), 4.11–4.06 (m, 1H), 3.77 (dd, J = 11.6 Hz, 9.2 Hz, 1H), 3.72 (d, J = 10.8 Hz, 1H), 3.38 (d, J = 10.8 Hz, 1H), 3.05–2.97 (m, 2H), 2.26 (t, J = 6.4 Hz, 2H), 1.95 (t, J = 6.4 Hz, 2H), 1.52–1.49 (m, 4H) ppm; 13C NMR (100 MHz, DMSO-d 6): δ = 188.07, 179.08, 171.55, 169.49, 144.25, 143.85, 138.89, 136.78, 133.53, 130.19, 129.10, 128.67, 122.33, 121.18, 119.93, 110.22, 74.72, 74.20, 62.36, 54.25, 51.13, 36.61, 36.32, 32.61, 25.32, 25.30 ppm; HRMS (ESI-TOF) m/z: [M + Na]+ Calculated for C30H29ClN4NaO5 647.1160, found 647.1161.
3.2. Biological evaluation
3.2.1. Enzyme inhibition assay
The enzymatic assay of test compounds is using the HTRF-based method provided by Cisbio Co. Ltd. In brief, the target compounds to be tested were prepared into samples with a concentration of 1 μM, which was added to GST-tagged MDM2 and HDAC1 proteins, respectively, then biotinylated substrate p53 peptide and HTRF detection reagents were added. The data obtained were processed by the ratio method, and the inhibition rates of MDM2 and HDAC1 were calculated. The detailed experimental procedures are according to the manufacturer’s protocols.
3.2.2. Cell culture and cell viability assay
The human epidermal cancer cells A431, human breast cancer cells MCF-7, and human colorectal cancer cells HCT116 were all derived from Chinese Center for Type Culture Collection (Wuhan, China) and cultured in our laboratory, cells were incubated with Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) in a humidified atmosphere including 5% CO2 under a sterile condition at 37°C (v/v) (GIBCO, Waltham, MA, USA). The cytotoxicity assay was performed with MTT method as previously described (73, 88, 89).
3.2.3. Cell apoptosis analysis using TUNEL staining assay
TUNEL assay was performed using in situ apoptosis detection kit (Promega, USA), according to the manufacturer’s protocols. Briefly, MCF-7 cells were seeded in 6-well plates for 12 h, and then treated with compound SAHA, Nutlin-3, 11b (0.5 μM, respectively) or vehicle (DMSO). After 24 h incubation, cells were plated onto the slides, fixed in 4% paraformaldehyde in PBS for 25 min, and then washed with PBS twice. After that, the cells were incubated in TUNEL reaction mixture at 37°C for 60 min. Then nuclei were counterstained with DAPI, and cells were checked under an immunofluorescent confocal microscopy (Olympus FV1000).
3.2.3. Cell apoptosis analysis using annexin V-FITC/PI double staining
MCF-7 cells were seeded in 6-well plates for 12 h, and then treated with compound SAHA, Nutlin-3, 11b (5 μM, respectively) or vehicle (DMSO) for 24 h. Cells were collected after incubation, then fixed with 75% ice-cold ethanol and stored at -20°C for 1 h. The cells were then harvested with 0.25% trypsin (without EDTA) and centrifuged at 2000 rpm for 5min. Then, the harvested cells were washed with ice-cold PBS twice and resuspended in 500 μL of 1× binding buffer, which was then added to 5 μL of annexin V-FITC and incubated at room temperature in the dark for 15 min. After adding 10 μL of propidium iodide (PI), the cells were incubated at room temperature in the dark. After another 15 min incubation, the cells were collected and analyzed by flow cytometry.
3.2.5. Molecular docking study
The crystal structures of MDM2 and HDAC1 were obtained from PDB (Protein Data Bank, http://www.pdb.org) database. Water molecules and cocrystallized ligands were removed from the proteins and the polar hydrogens were added to the protein structure. The molecular docking study of 11b was carried out via Discovery Studio 3.5 according to the previously reported protocol (90, 91).
3.2.6. Western blot assay
MCF-7 cells were seeded in 6-well plates for 24 h and treated with compounds SAHA, Nutlin-3, 11b (5 μM, respectively) or DMSO for 24 h. The cells were collected and washed with cold PBS twice, then added the protease and phosphatase inhibitors contained lysis buffer. Total cell lysates were prepared in lysis RIPA buffer (Invitrogen, CA, USA) on ice for 30 min, followed by centrifugation at 12000 rpm for 10 min at 4°C. After collecting supernatant, protein concentration was determined by a BCA protein assay. The total proteins were loaded into the 10%-15% SDS-PAGE gel and separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (PAGE). After electrophoresis, the protein was transferred to the PVDF membranes and rinsed three times in PBST buffer for 10 min each time, then sealed with 5% BSA Buffer for 1h. The membranes were incubated at 4°C for 24 h with primary antibodies and washed with TBST three times. Then incubated at room temperature for 2 h with secondary antibodies (1:10,000). The incubated PVDF membranes were taken out and rinsed with TBST three times. The protein bands were visualized through enhanced chemiluminescence (ECL) WB substrate (Millipore, MA, USA) and analyzed by ChemiScope. The antibodies including p53, MDM2, H4, and GAPDH were purchased from Abcam (Cambridge, MA, USA).
4. Conclusions
In summary, a series of MDM2-HDAC dual small-molecule inhibitors derivatives were designed and synthesized on the basis of the pharmacophore hybrid strategy. Among the target compounds, one spirooxindole-based hydroxamic acid 11b exhibited comparable inhibitory activities among MDM2 and HDAC, it also possessed good in vitro activity toward the tested three cancer cells, especially MCF-7, reaching the IC50 value of 1.37 ± 0.45 μM. Moreover, compound 11b exhibited remarkable selectivity for HDAC1 and HDAC2, with nanomolar IC50 of 58 nM and 64 nM, respectively. Further antitumor mechanism studies indicated that compound 11b could significantly induce the apoptosis of MCF-7 cells, and exerted potent anticancer effects. According to the western blot analysis, we demonstrated the most active compound 11b effectively and simultaneously inhibited MDM2 and HDAC through up-regulating the expression of p53 and Ac-H4 in MCF-7 cancer cells in a dose-dependent manner. These results highlighted that the simultaneous blockade of MDM2 and HDAC proteins with a single small-molecule compound represents a promising method for the discovery of new anticancer drugs.
Data availability statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.
Author contributions
GH and BH conceived and designed the experiments, QZ, S-SX, and CC performed the experiments, S-SX, H-PZ, QZ, and XX analyzed the data, CP, QZ, and BH wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (82073998, 22107015, 22177084 and 82104376), Sichuan Science and Technology Program (2022JDRC0045 and 2022JDRC0125), and the China Postdoctoral Science Foundation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.972372/full#supplementary-material
References
- 1. Medina-Franco JL, Giulianotti MA, Welmaker GS, Houghten RA. Shifting from the single to the multitarget paradigm in drug discovery. Drug Discov Today (2013) 18(9-10):495–501. doi: 10.1016/j.drudis.2013.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Peters JU. Polypharmacology - foe or friend? J Med Chem (2013) 56(22):8955–71. doi: 10.1021/jm400856t [DOI] [PubMed] [Google Scholar]
- 3. Anighoro A, Bajorath J, Rastelli G. Polypharmacology: Challenges and opportunities in drug discovery. J Med Chem (2014) 57(19):7874–87. doi: 10.1021/jm5006463 [DOI] [PubMed] [Google Scholar]
- 4. Zhou J, Jiang X, He S, Jiang H, Feng F, Liu W, et al. Rational design of multitarget-directed ligands: Strategies and emerging paradigms. J Med Chem (2019) 62(20):8881–914. doi: 10.1021/acs.jmedchem.9b00017 [DOI] [PubMed] [Google Scholar]
- 5. Kamb A, Wee S, Lengauer C. Why is cancer drug discovery so difficult? Nat Rev Drug Discov (2007) 6(2):115–20. doi: 10.1038/nrd2155 [DOI] [PubMed] [Google Scholar]
- 6. Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 7. Roth BL, Sheffler DJ, Kroeze WK. Magic shotguns versus magic bullets: Selectively non-selective drugs for mood disorders and schizophrenia. Nat Rev Drug Discov (2004) 3(4):353–9. doi: 10.1038/nrd1346 [DOI] [PubMed] [Google Scholar]
- 8. Brötz-Oesterhelt H, Brunner NA. How many modes of action should an antibiotic have? Curr Opin Pharmacol (2008) 8(5):564–73. doi: 10.1016/j.coph.2008.06.008 [DOI] [PubMed] [Google Scholar]
- 9. Knight ZA, Lin H, Shokat KM. Targeting the cancer kinome through polypharmacology. Nat Rev Cancer (2010) 10(2):130–7. doi: 10.1038/nrc2787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haber DA, Gray NS, Baselga J. The evolving war on cancer. Cell (2011) 145(1):19–24. doi: 10.1016/j.cell.2011.03.026 [DOI] [PubMed] [Google Scholar]
- 11. Trusolino L, Bertotti A. Compensatory pathways in oncogenic kinase signaling and resistance to targeted therapies: Six degrees of separation. Cancer Discov (2012) 2(10):876–80. doi: 10.1158/2159-8290.Cd-12-0400 [DOI] [PubMed] [Google Scholar]
- 12. Ciceri P, Müller S, O'Mahony A, Fedorov O, Filippakopoulos P, Hunt JP, et al. Dual kinase-bromodomain inhibitors for rationally designed polypharmacology. Nat Chem Biol (2014) 10(4):305–12. doi: 10.1038/nchembio.1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shang E, Yuan Y, Chen X, Liu Y, Pei J, Lai L. De novo design of multitarget ligands with an iterative fragment-growing strategy. J Chem Inf Model (2014) 54(4):1235–41. doi: 10.1021/ci500021v [DOI] [PubMed] [Google Scholar]
- 14. Shaveta, Mishra S, Singh P. Hybrid molecules: The privileged scaffolds for various pharmaceuticals. Eur J Med Chem (2016) 124:500–36. doi: 10.1016/j.ejmech.2016.08.039 [DOI] [PubMed] [Google Scholar]
- 15. Francisco AP, Mendes E, Santos AR, Perry MJ. Anticancer triazenes: From bioprecursors to hybrid molecules. Curr Pharm Design (2019) 25(14):1623–42. doi: 10.2174/1381612825666190617155749 [DOI] [PubMed] [Google Scholar]
- 16. Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol (2010) 28(10):1069–78. doi: 10.1038/nbt.1678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet (2016) 17(10):630–41. doi: 10.1038/nrg.2016.93 [DOI] [PubMed] [Google Scholar]
- 18. Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: Molecular mechanisms of action. Oncogene (2007) 26(37):5541–52. doi: 10.1038/sj.onc.1210620 [DOI] [PubMed] [Google Scholar]
- 19. Hesham HM, Lasheen DS, Abouzid KAM. Chimeric hdac inhibitors: Comprehensive review on the hdac-based strategies developed to combat cancer. Med Res Rev (2018) 38(6):2058–109. doi: 10.1002/med.21505 [DOI] [PubMed] [Google Scholar]
- 20. Ho TCS, Chan AHY, Ganesan A. Thirty years of hdac inhibitors: 2020 insight and hindsight. J Med Chem (2020) 63(21):12460–84. doi: 10.1021/acs.jmedchem.0c00830 [DOI] [PubMed] [Google Scholar]
- 21. Bass AKA, El-Zoghbi MS, Nageeb EM, Mohamed MFA, Badr M, Abuo-Rahma GEA. Comprehensive review for anticancer hybridized multitargeting hdac inhibitors. Eur J Med Chem (2021) 209:112904. doi: 10.1016/j.ejmech.2020.112904 [DOI] [PubMed] [Google Scholar]
- 22. Qiu X, Zhu L, Wang H, Tan Y, Yang Z, Yang L, et al. From natural products to hdac inhibitors: An overview of drug discovery and design strategy. Bioorgan Med Chem (2021) 52:116510. doi: 10.1016/j.bmc.2021.116510 [DOI] [PubMed] [Google Scholar]
- 23. Qiu L, Meng Y, Wang L, Gunewardena S, Liu S, Han J, et al. Histone lysine demethylase 4b regulates general and unique gene expression signatures in hypoxic cancer cells. MedComm (2021) 2(3):414–29. doi: 10.1002/mco2.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin a. J Biol Chem (1990) 265(28):17174–9. doi: 10.1016/S0021-9258(17)44885-X [DOI] [PubMed] [Google Scholar]
- 25. Zagni C, Floresta G, Monciino G, Rescifina A. The search for potent, small-molecule hdacis in cancer treatment: A decade after vorinostat. Med Res Rev (2017) 37(6):1373–428. doi: 10.1002/med.21437 [DOI] [PubMed] [Google Scholar]
- 26. Mandl-Weber S, Meinel FG, Jankowsky R, Oduncu F, Schmidmaier R, Baumann P. The novel inhibitor of histone deacetylase resminostat (Ras2410) inhibits proliferation and induces apoptosis in multiple myeloma (Mm) cells. Briti J Haematol (2010) 149(4):518–28. doi: 10.1111/j.1365-2141.2010.08124.x [DOI] [PubMed] [Google Scholar]
- 27. Wang H, Yu N, Chen D, Lee KC, Lye PL, Chang JW, et al. Discovery of (2e)-3-{2-Butyl-1-[2-(Diethylamino)Ethyl]-1h-Benzimidazol-5-Yl}-N-Hydroxyacrylamide (Sb939), an orally active histone deacetylase inhibitor with a superior preclinical profile. J Med Chem (2011) 54(13):4694–720. doi: 10.1021/jm2003552 [DOI] [PubMed] [Google Scholar]
- 28. Lu X, Ning Z, Li Z, Cao H, Wang X. Development of chidamide for peripheral T-cell lymphoma, the first orphan drug approved in China. Intractable Rare Dis Res (2016) 5(3):185–91. doi: 10.5582/irdr.2016.01024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aggarwal R, Thomas S, Pawlowska N, Bartelink I, Grabowsky J, Jahan T, et al. Inhibiting histone deacetylase as a means to reverse resistance to angiogenesis inhibitors: Phase I study of abexinostat plus pazopanib in advanced solid tumor malignancies. J Clin Oncol (2017) 35(11):1231–9. doi: 10.1200/jco.2016.70.5350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bretz AC, Parnitzke U, Kronthaler K, Dreker T, Bartz R, Hermann F, et al. Domatinostat favors the immunotherapy response by modulating the tumor immune microenvironment (Time). J Immunother Cancer (2019) 7(1):294. doi: 10.1186/s40425-019-0745-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liang X, Zang J, Li X, Tang S, Huang M, Geng M, et al. Discovery of novel janus kinase (Jak) and histone deacetylase (Hdac) dual inhibitors for the treatment of hematological malignancies. J Med Chem (2019) 62(8):3898–923. doi: 10.1021/acs.jmedchem.8b01597 [DOI] [PubMed] [Google Scholar]
- 32. Cheng C, Yun F, Ullah S, Yuan Q. Discovery of novel cyclin-dependent kinase (Cdk) and histone deacetylase (Hdac) dual inhibitors with potent in vitro and in vivo anticancer activity. Eur J Med Chem (2020) 189:112073. doi: 10.1016/j.ejmech.2020.112073 [DOI] [PubMed] [Google Scholar]
- 33. Mehndiratta S, Lin MH, Wu YW, Chen CH, Wu TY, Chuang KH, et al. N-Alkyl-Hydroxybenzoyl anilide hydroxamates as dual inhibitors of hdac and Hsp90, downregulating ifn-Γ induced pd-L1 expression. Eur J Med Chem (2020) 185:111725. doi: 10.1016/j.ejmech.2019.111725 [DOI] [PubMed] [Google Scholar]
- 34. Peng X, Sun Z, Kuang P, Chen J. Recent progress on hdac inhibitors with dual targeting capabilities for cancer treatment. Eur J Med Chem (2020) 208:112831. doi: 10.1016/j.ejmech.2020.112831 [DOI] [PubMed] [Google Scholar]
- 35. Thakur A, Tawa GJ, Henderson MJ, Danchik C, Liu S, Shah P, et al. Design, synthesis, and biological evaluation of quinazolin-4-One-Based hydroxamic acids as dual Pi3k/Hdac inhibitors. J Med Chem (2020) 63(8):4256–92. doi: 10.1021/acs.jmedchem.0c00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pan T, Dan Y, Guo D, Jiang J, Ran D, Zhang L, et al. Discovery of 2,4-pyrimidinediamine derivatives as potent dual inhibitors of alk and hdac. Eur J Med Chem (2021) 224:113672. doi: 10.1016/j.ejmech.2021.113672 [DOI] [PubMed] [Google Scholar]
- 37. Ren Y, Li S, Zhu R, Wan C, Song D, Zhu J, et al. Discovery of Stat3 and histone deacetylase (Hdac) dual-pathway inhibitors for the treatment of solid cancer. J Med Chem (2021) 64(11):7468–82. doi: 10.1021/acs.jmedchem.1c00136 [DOI] [PubMed] [Google Scholar]
- 38. Romanelli MN, Borgonetti V, Galeotti N. Dual Bet/Hdac inhibition to relieve neuropathic pain: Recent advances, perspectives, and future opportunities. Pharmacol Res (2021) 173:105901. doi: 10.1016/j.phrs.2021.105901 [DOI] [PubMed] [Google Scholar]
- 39. Schäker-Hübner L, Warstat R, Ahlert H, Mishra P, Kraft FB, Schliehe-Diecks J, et al. 4-acyl pyrrole capped hdac inhibitors: A new scaffold for hybrid inhibitors of bet proteins and histone deacetylases as antileukemia drug leads. J Med Chem (2021) 64(19):14620–46. doi: 10.1021/acs.jmedchem.1c01119 [DOI] [PubMed] [Google Scholar]
- 40. Liang X, Tang S, Liu X, Liu Y, Xu Q, Wang X, et al. Discovery of novel Pyrrolo[2,3-D]Pyrimidine-Based derivatives as potent Jak/Hdac dual inhibitors for the treatment of refractory solid tumors. J Med Chem (2022) 65(2):1243–64. doi: 10.1021/acs.jmedchem.0c02111 [DOI] [PubMed] [Google Scholar]
- 41. Hainaut P, Hollstein M. P53 and human cancer: The first ten thousand mutations. Adv Cancer Res (2000) 77:81–137. doi: 10.1016/s0065-230x(08)60785-x [DOI] [PubMed] [Google Scholar]
- 42. Chène P. Inhibiting the P53-Mdm2 interaction: An important target for cancer therapy. Nat Rev Cancer (2003) 3(2):102–9. doi: 10.1038/nrc991 [DOI] [PubMed] [Google Scholar]
- 43. Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. Oncoprotein Mdm2 conceals the activation domain of tumour suppressor P53. Nature (1993) 362(6423):857–60. doi: 10.1038/362857a0 [DOI] [PubMed] [Google Scholar]
- 44. Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of P53. Nature (1997) 387(6630):296–9. doi: 10.1038/387296a0 [DOI] [PubMed] [Google Scholar]
- 45. Kubbutat MH, Jones SN, Vousden KH. Regulation of P53 stability by Mdm2. Nature (1997) 387(6630):299–303. doi: 10.1038/387299a0 [DOI] [PubMed] [Google Scholar]
- 46. Midgley CA, Lane DP. P53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene (1997) 15(10):1179–89. doi: 10.1038/sj.onc.1201459 [DOI] [PubMed] [Google Scholar]
- 47. Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, et al. Discovery and cocrystal structure of benzodiazepinedione Hdm2 antagonists that activate P53 in cells. J Med Chem (2005) 48(4):909–12. doi: 10.1021/jm049137g [DOI] [PubMed] [Google Scholar]
- 48. Ding Q, Zhang Z, Liu JJ, Jiang N, Zhang J, Ross TM, et al. Discovery of Rg7388, a potent and selective P53-Mdm2 inhibitor in clinical development. J Med Chem (2013) 56(14):5979–83. doi: 10.1021/jm400487c [DOI] [PubMed] [Google Scholar]
- 49. Sun D, Li Z, Rew Y, Gribble M, Bartberger MD, Beck HP, et al. Discovery of amg 232, a potent, selective, and orally bioavailable Mdm2-P53 inhibitor in clinical development. J Med Chem (2014) 57(4):1454–72. doi: 10.1021/jm401753e [DOI] [PubMed] [Google Scholar]
- 50. Wang S, Sun W, Zhao Y, McEachern D, Meaux I, Barrière C, et al. Sar405838: An optimized inhibitor of Mdm2–P53 interaction that induces complete and durable tumor regression. Cancer Res (2014) 74(20):5855–65. doi: 10.1158/0008-5472.CAN-14-0799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aguilar A, Lu J, Liu L, Du D, Bernard D, McEachern D, et al. Discovery of 4-((3′R,4′S,5′R)-6″-Chloro-4′-(3-Chloro-2-Fluorophenyl)-1′-Ethyl-2″-Oxodispiro[Cyclohexane-1,2′-Pyrrolidine-3′,3″-Indoline]-5′-Carboxamido)Bicyclo[2.2.2]Octane-1-Carboxylic acid (Aa-115/Apg-115): A potent and orally active murine double minute 2 (Mdm2) inhibitor in clinical development. J Med Chem (2017) 60(7):2819–39. doi: 10.1021/acs.jmedchem.6b01665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Miller JJ, Gaiddon C, Storr T. A balancing act: Using small molecules for therapeutic intervention of the P53 pathway in cancer. Chem Soc Rev (2020) 49(19):6995–7014. doi: 10.1039/d0cs00163e [DOI] [PubMed] [Google Scholar]
- 53. Iancu-Rubin C, Mosoyan G, Glenn K, Gordon RE, Nichols GL, Hoffman R. Activation of P53 by the Mdm2 inhibitor Rg7112 impairs thrombopoiesis. Exp Hematol (2014) 42(2):137–45.e5. doi: 10.1016/j.exphem.2013.11.012 [DOI] [PubMed] [Google Scholar]
- 54. Siu LL, Italiano A, Miller WH, Blay J-Y, Gietema JA, Bang Y-J, et al. Phase 1 dose escalation, food effect, and biomarker study of Rg7388, a more potent second-generation Mdm2 antagonist, in patients (Pts) with solid tumors. J Clin Oncol (2014) 32(15_suppl):2535–. doi: 10.1200/jco.2014.32.15_suppl.2535 [DOI] [Google Scholar]
- 55. Wagner AJ, Banerji U, Mahipal A, Somaiah N, Hirsch H, Fancourt C, et al. Phase I trial of the human double minute 2 inhibitor mk-8242 in patients with advanced solid tumors. J Clin Oncol (2017) 35(12):1304–11. doi: 10.1200/jco.2016.70.7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jiang L, Malik N, Acedo P, Zawacka-Pankau J. Protoporphyrin ix is a dual inhibitor of P53/Mdm2 and P53/Mdm4 interactions and induces apoptosis in b-cell chronic lymphocytic leukemia cells. Cell Death Discov (2019) 5:77. doi: 10.1038/s41420-019-0157-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lee B, Min JA, Nashed A, Lee SO, Yoo JC, Chi SW, et al. A novel mechanism of irinotecan targeting Mdm2 and bcl-xl. Biochem Bioph Res Commun (2019) 514(2):518–23. doi: 10.1016/j.bbrc.2019.04.009 [DOI] [PubMed] [Google Scholar]
- 58. Wang W, Cheng J-W, Qin J-J, Hu B, Li X, Nijampatnam B, et al. Mdm2-Nfat1 dual inhibitor, Ma242: Effective against hepatocellular carcinoma, independent of P53. Cancer Lett (2019) 459:156–67. doi: 10.1016/j.canlet.2019.114429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Conlon IL, Drennen B, Lanning ME, Hughes S, Rothhaas R, Wilder PT, et al. Rationally designed polypharmacology: α-helix mimetics as dual inhibitors of the oncoproteins mcl-1 and Hdm2. ChemMedChem (2020) 15(18):1691–8. doi: 10.1002/cmdc.202000278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wang B, Peng F, Huang W, Zhou J, Zhang N, Sheng J, et al. Rational drug design, synthesis, and biological evaluation of novel chiral tetrahydronaphthalene-fused spirooxindole as Mdm2-Cdk4 dual inhibitor against glioblastoma. Acta Pharm Sin B (2020) 10(8):1492–510. doi: 10.1016/j.apsb.2019.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liu SJ, Zhao Q, Peng C, Mao Q, Wu F, Zhang FH, et al. Design, synthesis, and biological evaluation of nitroisoxazole-containing Spiro[Pyrrolidin-oxindole] derivatives as novel glutathione peroxidase 4/Mouse double minute 2 dual inhibitors that inhibit breast adenocarcinoma cell proliferation. Eur J Med Chem (2021) 217:113359. doi: 10.1016/j.ejmech.2021.113359 [DOI] [PubMed] [Google Scholar]
- 62. Wu Z, Gu L, Zhang S, Liu T, Lukka PB, Meibohm B, et al. Discovery of n-(3,4-Dimethylphenyl)-4-(4-Isobutyrylphenyl)-2,3,3a,4,5,9b-Hexahydrofuro[3,2-C]Quinoline-8-Sulfonamide as a potent dual Mdm2/Xiap inhibitor. J Med Chem (2021) 64(4):1930–50. doi: 10.1021/acs.jmedchem.0c00932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang S, Yan Z, Li Y, Gong Y, Lyu X, Lou J, et al. Structure-based discovery of Mdm2/4 dual inhibitors that exert antitumor activities against Mdm4-overexpressing cancer cells. J Med Chem (2022) 65(8):6207–30. doi: 10.1021/acs.jmedchem.2c00095 [DOI] [PubMed] [Google Scholar]
- 64. Condorelli F, Gnemmi I, Vallario A, Genazzani AA, Canonico PL. Inhibitors of histone deacetylase (Hdac) restore the P53 pathway in neuroblastoma cells. Brit J Pharmacol (2008) 153(4):657–68. doi: 10.1038/sj.bjp.0707608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. McCormack E, Haaland I, Venås G, Forthun RB, Huseby S, Gausdal G, et al. Synergistic induction of P53 mediated apoptosis by valproic acid and nutlin-3 in acute myeloid leukemia. Leukemia (2012) 26(5):910–7. doi: 10.1038/leu.2011.315 [DOI] [PubMed] [Google Scholar]
- 66. Micelli C, Rastelli G. Histone deacetylases: Structural determinants of inhibitor selectivity. Drug Discov Today (2015) 20(6):718–35. doi: 10.1016/j.drudis.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 67. Traoré MDM, Zwick V, Simões-Pires CA, Nurisso A, Issa M, Cuendet M, et al. Hydroxyl ketone-based histone deacetylase inhibitors to gain insight into class I hdac selectivity versus that of Hdac6. ACS Omega (2017) 2(4):1550–62. doi: 10.1021/acsomega.6b00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhang L, Zhang J, Jiang Q, Zhang L, Song W. Zinc binding groups for histone deacetylase inhibitors. J Enzyme Inhib Med Chem (2018) 33(1):714–21. doi: 10.1080/14756366.2017.1417274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. He S, Dong G, Wu S, Fang K, Miao Z, Wang W, et al. Small molecules simultaneously inhibiting P53-murine double minute 2 (Mdm2) interaction and histone deacetylases (Hdacs): Discovery of novel multitargeting antitumor agents. J Med Ch (2018) 61(16):7245–60. doi: 10.1021/acs.jmedchem.8b00664 [DOI] [PubMed] [Google Scholar]
- 70. Yu B, Yu DQ, Liu HM. Spirooxindoles: Promising scaffolds for anticancer agents. Eur J Med Chem (2015) 97:673–98. doi: 10.1016/j.ejmech.2014.06.056 [DOI] [PubMed] [Google Scholar]
- 71. Yu B, Zheng YC, Shi XJ, Qi PP, Liu HM. Natural product-derived spirooxindole fragments serve as privileged substructures for discovery of new anticancer agents. Anti-Cancer Agents Me (2016) 16(10):1315–24. doi: 10.2174/1871520615666151102093825 [DOI] [PubMed] [Google Scholar]
- 72. Gupta AK, Bharadwaj M, Kumar A, Mehrotra R. Spiro-Oxindoles as a promising class of small molecule inhibitors of P53-Mdm2 interaction useful in targeted cancer therapy. Topics Curr Chem (2017) 375(1):3. doi: 10.1007/s41061-016-0089-0 [DOI] [PubMed] [Google Scholar]
- 73. Zhao Q, Peng C, Huang H, Liu SJ, Zhong YJ, Huang W, et al. Asymmetric synthesis of tetrahydroisoquinoline-fused spirooxindoles as ras-gtp inhibitors that inhibit colon adenocarcinoma cell proliferation and invasion. Chem Commun (2018) 54(60):8359–62. doi: 10.1039/c8cc04732d [DOI] [PubMed] [Google Scholar]
- 74. Peng F, Zhao Q, Huang W, Liu S-J, Zhong Y-J, Mao Q, et al. Amine-catalyzed and functional group-controlled chemo- and regioselective synthesis of multi-functionalized Cf3-benzene Via a metal-free process. Green Chem (2019) 21(22):6179–86. doi: 10.1039/C9GC02694K [DOI] [Google Scholar]
- 75. Li J-L, Liu Y-Q, Zou W-L, Zeng R, Zhang X, Liu Y, et al. Radical acylfluoroalkylation of olefins through n-heterocyclic carbene organocatalysis. Angew Chem Int Ed (2020) 59(5):1863–70. doi: 10.1002/anie.201912450 [DOI] [PubMed] [Google Scholar]
- 76. Zhang X, Li X, Li J-L, Wang Q-W, Zou W-L, Liu Y-Q, et al. Regiodivergent construction of medium-sized heterocycles from vinylethylene carbonates and allylidenemalononitriles. Chem Sci (2020) 11(11):2888–94. doi: 10.1039/C9SC06377C [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Han B, He X-H, Liu Y-Q, He G, Peng C, Li J-L. Asymmetric organocatalysis: An enabling technology for medicinal chemistry. Chem Soc Rev (2021) 50(3):1522–86. doi: 10.1039/D0CS00196A [DOI] [PubMed] [Google Scholar]
- 78. Luo M-L, Huang W, Zhu H-P, Peng C, Zhao Q, Han B. Advances in indole-containing alkaloids as potential anticancer agents by regulating autophagy. BioMed Pharmacother (2022) 149:112827. doi: 10.1016/j.biopha.2022.112827 [DOI] [PubMed] [Google Scholar]
- 79. Mahboobi S, Dove S, Sellmer A, Winkler M, Eichhorn E, Pongratz H, et al. Design of chimeric histone deacetylase- and tyrosine kinase-inhibitors: A series of imatinib hybrides as potent inhibitors of wild-type and mutant bcr-abl, pdgf-rβ, and histone deacetylases. J Med Chem (2009) 52(8):2265–79. doi: 10.1021/jm800988r [DOI] [PubMed] [Google Scholar]
- 80. Cai X, Zhai H-X, Wang J, Forrester J, Qu H, Yin L, et al. Discovery of 7-(4-(3-Ethynylphenylamino)-7-Methoxyquinazolin-6-Yloxy)-N-Hydroxyheptanamide (Cudc-101) as a potent multi-acting hdac, egfr, and Her2 inhibitor for the treatment of cancer. J Med Chem (2010) 53(5):2000–9. doi: 10.1021/jm901453q [DOI] [PubMed] [Google Scholar]
- 81. Qian C, Lai C-J, Bao R, Wang D-G, Wang J, Xu G-X, et al. Cancer network disruption by a single molecule inhibitor targeting both histone deacetylase activity and phosphatidylinositol 3-kinase signaling. Clin Cancer Res (2012) 18(15):4104–13. doi: 10.1158/1078-0432.CCR-12-0055 [DOI] [PubMed] [Google Scholar]
- 82. Ko KS, Steffey ME, Brandvold KR, Soellner MB. Development of a chimeric c-src kinase and hdac inhibitor. ACS Med Chem Lett (2013) 4(8):779–83. doi: 10.1021/ml400175d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Giacomini E, Nebbioso A, Ciotta A, Ianni C, Falchi F, Roberti M, et al. Novel antiproliferative chimeric compounds with marked histone deacetylase inhibitory activity. ACS Med Chem Lett (2014) 5(9):973–8. doi: 10.1021/ml5000959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yang EG, Mustafa N, Tan EC, Poulsen A, Ramanujulu PM, Chng WJ, et al. Design and synthesis of janus kinase 2 (Jak2) and histone deacetlyase (Hdac) bispecific inhibitors based on pacritinib and evidence of dual pathway inhibition in hematological cell lines. J Med Chem (2016) 59(18):8233–62. doi: 10.1021/acs.jmedchem.6b00157 [DOI] [PubMed] [Google Scholar]
- 85. Henderson C, Mizzau M, Paroni G, Maestro R, Schneider C, Brancolini C. Role of caspases, bid, and P53 in the apoptotic response triggered by histone deacetylase inhibitors trichostatin-a (Tsa) and suberoylanilide hydroxamic acid (Saha)*. J Biol Chem (2003) 278(14):12579–89. doi: 10.1074/jbc.M213093200 [DOI] [PubMed] [Google Scholar]
- 86. Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, et al. In vivo activation of the P53 pathway by small-molecule antagonists of Mdm2. Science (2004) 303(5659):844–8. doi: 10.1126/science.1092472 [DOI] [PubMed] [Google Scholar]
- 87. Berghauser Pont LME, Naipal K, Kloezeman JJ, Venkatesan S, van den Bent M, van Gent DC, et al. DNA Damage response and anti-apoptotic proteins predict radiosensitization efficacy of hdac inhibitors saha and Lbh589 in patient-derived glioblastoma cells. Cancer Lett (2015) 356(2, Part B):525–35. doi: 10.1016/j.canlet.2014.09.049 [DOI] [PubMed] [Google Scholar]
- 88. Zhu HP, Xie K, He XH, Huang W, Zeng R, Fan Y, et al. Organocatalytic diastereoselective [3+2] cyclization of mbh carbonates with dinucleophiles: Synthesis of bicyclic imidazoline derivatives that inhibit Mdm2-P53 interaction. Chem Commun (2019) 55(76):11374–7. doi: 10.1039/c9cc05916d [DOI] [PubMed] [Google Scholar]
- 89. Yu M, Zeng M, Pan Z, Wu F, Guo L, He G. Discovery of novel Akt1 inhibitor induces autophagy associated death in hepatocellular carcinoma cells. Eur J Med Chem (2020) 189:112076. doi: 10.1016/j.ejmech.2020.112076 [DOI] [PubMed] [Google Scholar]
- 90. Ouyang L, Zhang L, Liu J, Fu L, Yao D, Zhao Y, et al. Discovery of a small-molecule bromodomain-containing protein 4 (Brd4) inhibitor that induces amp-activated protein kinase-modulated autophagy-associated cell death in breast cancer. J Med Chem (2017) 60(24):9990–10012. doi: 10.1021/acs.jmedchem.7b00275 [DOI] [PubMed] [Google Scholar]
- 91. Pan Z, Li X, Wang Y, Jiang Q, Jiang L, Zhang M, et al. Discovery of Thieno[2,3-D]Pyrimidine-Based hydroxamic acid derivatives as bromodomain-containing protein 4/Histone deacetylase dual inhibitors induce autophagic cell death in colorectal carcinoma cells. J Med Chem (2020) 63(7):3678–700. doi: 10.1021/acs.jmedchem.9b02178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/ Supplementary Material . Further inquiries can be directed to the corresponding authors.