Highlights
-
•
Silencing GoPGF using a seed-specific promoter led to a glandless phenotype with an ultra-low gossypol content in the cottonseeds.
-
•
The phenotype of a low gossypol content (lower than 450 ppm) in cottonseeds is stable and heritable.
-
•
The engineered “glanded plant and glandless seed” cottonseeds could be used directly for human nutrition and feed for animals.
Keywords: Cottonseed, Gene silencing, Glandless seed, Low gossypol, Human nutritious food
Abstract
After fiber, cottonseed is the second most important by-product of cotton production. However, high concentrations of toxic free gossypol deposited in the glands of the cottonseed greatly hamper its effective usage as food or feed. Here, we developed a cotton line with edible cottonseed by specifically silencing the endogenous expression of GoPGF in the seeds, which led to a glandless phenotype with an ultra-low gossypol content in the seeds and nearly normal gossypol in other parts of the plants. This engineered cotton maintains normal resistance to insect pests, but the gossypol content in the seeds dropped by 98%, and thus, it can be consumed directly as food. The trait of a low gossypol content in the cottonseeds was stable and heritable, while the protein, oil content, and fiber yield or quality were nearly unchanged compared to the transgenic receptor W0. In addition, comparative transcriptome analysis showed that down-regulated genes in the ovules of the glandless cotton were enriched in terpenoid biosynthesis, indicating the underlying relationship between gland formation and gossypol biosynthesis. These results pave the way for the comprehensive utilization of cotton as a fiber, oil, and feed crop in the future.
1. Introduction
Cotton is one of the most important economic crops in the world, which provides natural fiber used in the textile industry and cottonseed used in food, livestock feed etc. For every kilogram of fiber collected, about 1.65 kg of cottonseeds is concomitantly produced (Cai et al., 2010). Cottonseed contains plenty of protein, oil, carbohydrates, cellulose, and mineral elements with high utilization value. After seed delinting and hull removal, the kernels contain around 38 % protein (dry wt.) and 35 % oil (dry wt.). Cottonseed protein has been widely used in various food processing and is regarded as a nutritious food additive (Zhuge et al., 1988) and high-quality high protein feed (Rogers et al., 2002). Based on a total annual global production of 44 million tonnes of cottonseed, 10 million tonnes of protein will be produced, enough to satisfy the daily protein needs of around 600 million people (Kumar et al., 2021). In addition to its high-quality protein, cottonseed oil is the most valuable source of relatively high-quality, which consists of approximately 70 % unsaturated and 30 % saturated fatty acids and sees extensive use for edible purposes (Dowd et al., 2010). Linoleic acid is the main component of cottonseed oil, which is an essential unsaturated fatty acid for the human body that can reduce cholesterol and blood lipids in the blood. Cottonseed oil is also a natural shortening without trans fatty acids. The finished oil made of low phenol cottonseed has a light golden color and flavor and is a healthy oil with high nutritional value. Worldwide, the annual production of cottonseed oil stands at about 5.7 M tons and is valued at $7.4B. The top three countries with the highest cottonseed oil consumption are India (1.6 M tons), China (1.4 M tons), and Pakistan (470 K tons), together comprising 63 % of global consumption (Khan et al., 2020).
Despite the high protein content of cottonseeds, their direct usage as food and feed is seriously hindered because cottonseeds contain a large amount of gossypol, accounting for 0.4 ∼ 1.7 % of the whole kernel, which can cause poisoning in humans and monogastric animals (Zhou et al., 2013). According to reports, pigs, chickens, and dogs are all affected by gossypol toxicity, and gossypol can impact the male reproductive system as well (Randel et al., 1992, Coutinho, 2002). But since ruminants have a high tolerance to gossypol, cottonseeds are mainly used as feed for sheep, cattle, and other ruminants (Zhang et al., 2007). All told, gossypol is the single biggest factor hampering the direct utilization and thus the value of cottonseed. On the other hand, gossypol and related terpenoids are regarded as natural insecticides, being produced by the cotton plant to protect itself against insects, field mice, and other rodents. A feeding study using glandular and non-glandular leaves demonstrated that beet armyworms have a strong preference for glandless leaves, and the larvae prefer the top leaves of undamaged plants (McAuslane and Alborn, 1998). In another study, beet armyworm larvae feeding on old leaves induced increased levels of hemigossypolone and the heliocides 1 and 4 in the glands of newly-grown leaves (Bezemer et al., 2004). The production of gossypol and its related terpenoids can also be induced upon infection of plants by nematodes (Agriotes lineatus) or microorganisms, such as Verticillium (Townsend et al., 2005). Notably, the knockdown of CAD, which encodes δ-cadinene synthase, an enzyme involved in terpenoid biosynthesis, reduces gossypol content, and impairs plant resistance to Verticillium dahliae (Gao et al., 2013). These results indicate that gossypol and associated terpenoids deposited in cotton glands serve as a natural defence mechanism against various pests and fungal and bacterial pathogens.
Gossypol and its related terpenoids are phenolic compounds (Kenar, 2006) that mainly stored in a specialized structure called pigment gland, which is one of the unique traits of the tribe Gossypieae. The glands derive from the subepidermal cells of the leaf, stem, petal, and other parts of the plant. Based on its gland trait, cotton was classified into three categories as follows: glandless in the whole plant; glandless in parts of the plant; and delayed gland (the dormant seeds are glandless but the germinated cotyledons are glanded). Because of their positive relationship to gossypol content, glands are often regarded as an indicator of gossypol level (Mohan et al., 1995). Uncovering the gene network that regulates the differentiation and forming of glands is of considerable research interest; such knowledge will facilitate understanding the close relationship between trichomes or glands and their secretions or deposits. Genetic analyses reveal that the complete absence of a gland is controlled by two pairs of duplicate recessive genes (gl2 and gl3) (McMichael, 1954). At least six independent loci (gl1-gl6) for gland traits have been identified (McMichael, 1954, McMichael, 1960, Lee, 1962, Murray, 1965), indicating the complexity of gland formation. Among the six loci, gl2 and gl3 play a major role, while the other genes only play a minor role. Gl2e is a dominant allele at the Gl2 locus that is produced by 32P irradiation (Afifi et al., 1966). In a previous study, we first isolated the key gland gene PIGMENT GLAND FORMATION (GoPGF) underlying the Gl2e locus using map-based cloning (Ma et al., 2016). After silencing GoPGF employing virus-induced gene silencing (VIGS), the glands nearly completely disappeared, and the gossypol content was significantly decreased to a safe level.
Several feasible strategies have been developed to eliminate the gossypol content in cottonseeds, such as the development of gossypol-free cotton varieties by transferring the glandless trait from mutants into commercial varieties. An early report by McMichael (1954) reported the first discovered glandless cotton mutant that was selected from the “Hopi Moencopi” variety. Then, this glandless line was crossed with commercial cotton to develop glandless cotton seeds. This was the real beginning of glandless cotton breeding. Continuing McMichael’s work, Cooper developed several glandless strains in the Acala-type cotton (Lusas & Jividen, 1987). Subsequently, breeders in many cotton-producing countries in the world have carried out the breeding of low-gossypol cotton. Many low-gossypol cotton varieties were developed in the 1990s. However, these commercial glandless cotton varieties did not last long, because they were extraordinarily susceptible to insect and rodent damage on account of having lost the protection provided by gossypol (Sunilkumar et al., 2006). Antisense and RNAi methods have been successfully used to eliminate gossypol content in cottonseeds by silencing the gene that encodes (+)-δ-cadinene synthase, which catalyzes the first reaction involving the cyclization of farnesyl diphosphate to (+)-δ-cadinene (Sunilkumar et al., 2006). A transgenic RNAi line with ultra-low gossypol cottonseeds was determined to be genetically stable and did not have adverse effects on cotton fiber quality and yield (Palle et al., 2013, Rathore et al., 2012). Rathore's group (2019) released a new low-gossypol variety of cotton TAM66274 that produces ultra-low gossypol via silencing cadinene synthase using a seed-specific promoter (the gossypol levels in seed reduced by 97 %). TAM66274 was approved by the FDA in the US for human and animal consumption. Such lines have value for a wide range of applications (Rathore et al., 2020). As the presence or absence of glands affected gossypol content, an ideal low-gossypol cotton variety should be a glanded plant (high gossypol content) but have glandless seeds that of broad application prospects (Cai et al., 2010). In this study, considering the positive relationship between glands and gossypol, we report the development of a novel “glanded plant and glandless seed” cotton by interfering the transcription of a key gland forming gene GoPGF, which can not only be planted for conventional purposes, such as a fiber and oil crops, but also grown directly as a feed crop without the need for physical or chemical detoxification.
2. Materials and methods
2.1. Plant materials
Transgenic plants and the recipient Gossypium hirsutum accession W0 were cultivated in the field with normal practices at the Dangtu Breeding Station, Zhejiang University (Dangtu, Anhui, China), and in the greenhouse facility of Zhejiang University (Hangzhou Zhejiang, China) at 28/18 °C (day/night) with a 16-h photoperiod. The day of anthesis was defined as 0 DPA (days post anthesis). Developing seeds at 10, 20, and 30 DPA and three tissues (roots, stems, and leaves) from 14-day-old seedlings were harvested for RT-PCR. Mature seeds in the field were harvested to determine the gossypol, oil, and protein content.
2.2. Design and construction of RNAi gene silencing vectors
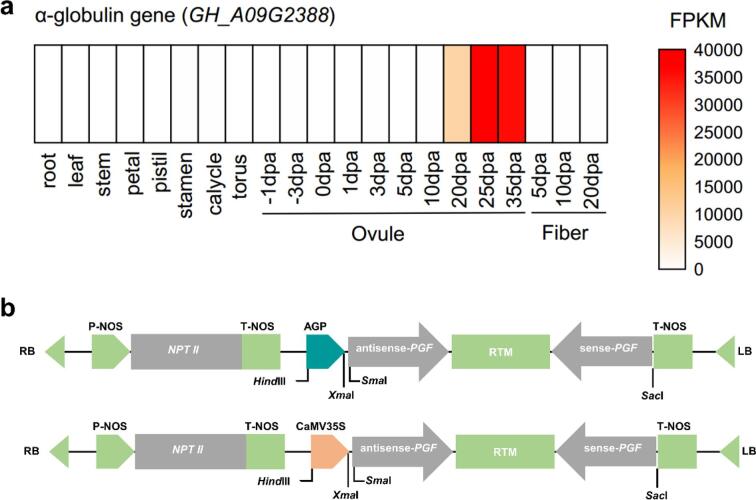
We designed RNAi vectors consisting of an inverted repeat structure of partial GoPGF DNA sequences (GH_A12G2598) driven by a seed-specific promoter derived from the α-globulin gene (GH_A09G2388) or by a CaMV35S promoter. We first obtained the trigger DNA fragment corresponding to nucleotides 1,037 to 1,396 of GoPGF by PCR and added Sma I and Xba I restriction sites to the ends via primers (Table S1). The amplified PCR fragment was then inserted into the pBSK vector, after which the 360-bp inverse repeat sequence plus 750 bp of an adjacent DNA sequence acting as a spacer was amplified by PCR. The resulting amplicon was inserted into the pBI121 vector to generate the 35S::GoPGF RNAi vector. Primers K4360F and K4360R for the α-globulin promoter (Sunilkumar et al., 2002) were designed with Primer premier 5 (Table S1), appended with the Hind III and Sma I restriction sites, and used to clone the α-globulin promoter (AGP) using DNA from the TM-1 ovule as the template. The AGP fragment and 35S::GoPGF RNAi vector were double digested with Hind III and Sma I, after which the target fragments were separated by electrophoresis, extracted, and connected with T4 ligase to generate the AGP::GoPGF RNAi vector, which was verified by sequencing. Finally, the constructed vectors were transformed into the Agrobacterium tumefaciens strain LB4404 for the transformation of cotton plants.
2.3. Cotton transformation and transgenic plant selection
The cotton transformation was performed by generating an embryogenic callus from cotton hypocotyl as previously described (Li et al., 2009), using G. hirsutum accession W0 as the recipient variety. After about eight months of tissue culture, putative transgenic seedlings were transferred to the greenhouse. Plants positive for the NPTII gene by PCR analysis and the glandless trait were identified. Finally, six independent and stable transformants were obtained for each construct. Homozygous transgenic lines were developed by pedigree selection and used for further analysis.
2.4. Southern blot analysis
The copy number of the T1 transgenic lines with positive results was identified by Southern blot. First, genomic DNA was extracted and digested with the restriction enzyme EcoR Ι. Then, the digested DNA was fractionated on a 0.8 % agarose gel and transferred to nylon membranes. The PCR product of NPTII was used as the probe. Standard procedures for Southern blot analysis, probe labeling, and detection were used following the DIG DNA Labeling and Detection Kit (Roche).
2.5. Real-time RT-PCR expression analysis
Seven different tissues (root, stem, leaf, and ovule at 10 DPA, 15 DPA, 20 DPA, and 30 DPA) and germinated seeds from the T3 transgenic plants with the same growth trend at the flowering stage were ground into a powder with liquid nitrogen for RNA extraction. Total RNA was isolated from the powdered tissues and purified using the Biospin Plant Total RNA Extraction Kit (BioFlux, cat: BSC65S1) according to the manufacturer’s instructions. Gene expression profiling was carried out by real-time reverse transcription polymerase chain reaction (qRT-PCR) using cotton Histone 3 (GenBank accession no. AF024716) as the internal control according to a previously described method (Schmittgen & Livak, 2008). The mean and standard deviation were calculated from three biological replicates.
2.6. Content determination for gossypol and related terpenoids
The leaf, stem, and root were collected at the seedling stage and six different tissues (leave, stem, bract, sepal, petal, and 3-dpa boll shell) at the flowering stage. These were ground into a powder with liquid nitrogen, to which 1 ml of leaf extraction liquid was added per 100 mg of powder. Ultrasonic extraction was performed for 30 min, and the resulting solution was centrifuged for 5 min at 12,500 rpm. The supernatant was carefully extracted and filtered with a 0.22-μm filter for HLPC detection. For seed determination, about 8–10 kernels were ground into a fine powder, to which 1 ml of cottonseed extraction liquid per 200 mg of sample was added. After soaking for 1 h at room temperature and centrifugation for 5 min at 12,500 rpm, the supernatant was carefully extracted and filtered with a 0.22-μm filter for HLPC detection.
The compositions of the solutions used are as follows: cottonseed extraction liquid, ethanol: ether: water: acetic acid = 59: 17: 24: 0.2; leaf extraction liquid, acetonitrile: water: phosphoric acid = 80: 20: 0.1; and HPLC mobile phase, ethanol: methanol: isopropanol: acetonitrile: ethyl acetate: dimethylformamide: phosphoric acid = 16.7: 4.6: 12.1: 20.2: 37.4: 3.8: 5.1: 0.1. The HPLC analysis was performed on an Agilent Technologies 1200 liquid chromatograph, and the gossypol and related terpenoids content was calculated based on a standard curve (Benson et al., 2001, Stipanovic et al., 1988).
2.7. Determination of fatty acid component in seeds
Mashed hulled cotton seeds (0.1 g dry weight) were combined with 1 ml of n-hexane (analytical pure), vigorously shaken for 0.5 min, and incubated for 5 h at room temperature. Afterward, the supernatant was transferred into another 2-ml centrifuge tube and mixed with 0.5 ml of the methyl esterification reagent to carry out the methyl esterification reaction. The reaction mixture was shaken for 2 min and incubated for 1 h at room temperature, after which the supernatant was removed and centrifuged for 6 min at a relative centrifugal force of 6000 r/min. The fatty acid composition was determined by a gas chromatography-mass spectrometry (gas chromatograph, TRACE 1310; mass spectrometer, TSQ 9000) (Liu et al., 2017).
2.8. Determination of seed oil content
Cottonseed oil content was measured with a nuclear magnetic resonance oil content meter, which uses nuclear magnetic resonance technology to directly quantify liquid hydrogen nuclei in the sample, from which the oil content is then calculated. Briefly, the hulled seeds were dried under forced air at 35 °C, equilibrated to room temperature in a glass dryer, and then analyzed. The sample size was about 17 g, and the average of two readings from each sample was used as the final measurement of oil content, with three replicates for each material. A commercial cottonseed oil sample was used as a standard control (Acros, CAS: 8001-29-4).
2.9. Determination of cottonseed protein content
Finely ground and dried cotton kernels (0.2 g) were combined with 5 ml of HCl (6 mol/L), dried by blowing nitrogen gas, and reacted in an oven at 110 °C for 24 h. Afterward, the sample was diluted to 10 ml with water. A 1-ml aliquot was dried by blowing nitrogen, then diluted using 10 ml of HCl (0.02 mol/L) and filtered with a 0.22-μm filter. The filtered liquid (1–1.5 ml) was then placed in a sample bottle for protein content determination. The analysis conditions were as follows: ion exchange column specification 4.6 × 60 mm; Hitachi’s special 3 μm ion exchange resin; column temperature 135 °C; pump flow rate 0.00∼0. 999 ml/min; and pump pressure 0∼20 kpa. The analysis time was about 30 min, and the injection volume was 20 μL. Protein content was measured using a Hitachi L-8900 automatic amino acid analyzer and EZChrom Elite for HITACHI analysis systems.
2.10. Yield-related and fiber quality traits measurement
Plants of the transgenic lines T182-36 and T183-88 and the non-transgenic W0 were cultivated for fiber samples in DBS/ZJ in 2018, 2019, and 2020. For each line, 25 bolls were hand-harvested from the internal middle parts of the plants in the middle of each row. The evaluated yield-related traits were seed index, lint index, boll weight, and lint percentage. Fiber quality traits were measured on an AFIS single-fiber analyzer at the Supervision, Inspection, and Test Center of Cotton Quality, Ministry of Agriculture in China and consisted of micronaire (MIC, a measurement of fiber fineness and maturity), the average length of the upper half (UHM, average length of the longer half fiber), uniformity index (UI, the ratio of average length to UHM), strength (STR; the force needed to break a 1-tex-unit bundle of fibers), and elongation (ELO, the elongation of the fiber before breaking when measuring its strength).
2.11. RNA-seq
Ovules were collected at 20 and 30 DPA from three biological replicates of the glandless (GoPGF-RNAi plant) and glanded cotton (W0) were quickly frozen in liquid nitrogen and stored at −80 °C. RNA extraction and evaluation of purity and integrity were performed as previously described (Hu et al., 2019). The Illumina TruseqTM RNA Sample Prep Kit was used to construct the sequencing library, and an Illumina NovaSeq 6000 was used for transcriptome sequencing. Illumina reads were aligned to the reference TM-1 genome (v2.1) (Hu et al., 2019) by hisat2 (2.1.1) (Kim et al., 2019) to obtain the mapped reads, and quantification of gene expression was performed with subread (2.0.1) (Liao et al., 2014). The analysis of differentially expressed genes (DEGs) was performed using the R package DESeq2 (1.30.1) (Love et al., 2014) with an absolute value of log2 (fold change) ≥ 1 and a P value ≤ 0.05. Gene Ontology (GO) enrichment analysis was performed using the R package clusterProfiler (3.18.1) (Yu et al., 2012). Heatmaps were generated using the R package pheatmap (1.0.12).
3. Results
3.1. Development of GoPGF-silenced cotton plants
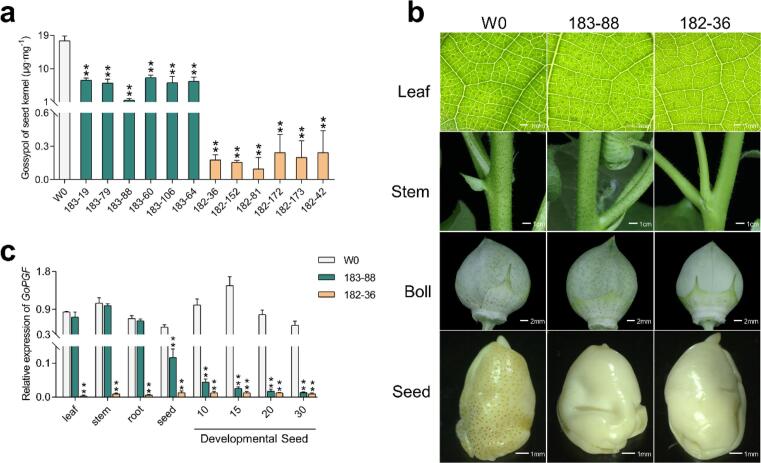
To construct the RNAi vectors, the specific 3′-end fragment (from 1037 to 1396 nt) of GoPGF was amplified from the G. hirsutum genetic standard line TM-1 and bidirectionally inserted into binary transformation vector pBI121 under the control of two different promoters, the highly cotton ovule-specific α-globulin gene promoter (AGP) (Fig. 1a) and the constitutive CaMV35S promoter, respectively, generating the AGP::GoPGF and 35S::GoPGF vectors (Fig. 1b). The constructs were introduced into the receptor line W0 via Agrobacterium tumefaciens-mediated transformation. All the resulting independent transformants were phenotypically screened first for the reduced presence of the small, darkly-pigmented lysigenous glands and then for reduced seed gossypol levels. After screening, we successfully obtained 12 independently-transgenic lines having substantially reduced gossypol content in the T1 generation seed kernels (Fig. 2a). Regarding gossypol content variation among these transgenic cotton plants, two representative transgenic lines were chosen for further analysis, the “glanded plant and almost glandless seed” line T183-88 (AGP::GoPGF) and the “glandless whole plant” line T182-36 (CaMV35S::GoPGF). Compared with the receptor W0, gossypol levels in the cottonseed were approximately decreased by 95 % in T183-88 and 99 % in T182-36 (Fig. 2a), respectively. Further, hybridization assays revealed that the T-DNA had integrated as two copies in T183-88 and a single copy in T182-36 (Fig. S1).
Fig. 1.
Construction of the vectors for transformation. (a) The expression profile of α-globulin gene (GH_A09G2388) in distinct tissues of cotton, including root, stem and leaf, in ovule and fiber development stages based on transcriptome datasets of G. hirsutum acc. TM-1 (Hu et al., 2019). (b) The structure of AGP::GoPGF RNAi vector and CaMV35S::GoPGF RNAi vector.
Fig. 2.
Decreased expression of GoPGF gene leads to glandless phenotype (a) Glandular phenotype of the T1 transgenic lines and W0 (the recipient variety). (b) Glandular phenotype of leaf, stem, boll, and seeds of transgenic lines 183–88 and 182–36. (c) Relative expression level of GoPGF gene in different tissues of the transgenic lines. W0 was the recipient variety used for transformation, here served as a non-transgenic control. Transgenic line 183–88 transformed by AGP::GoPGF RNAi vector maintains the glanded leaf, stem, boll but glandless kernel. Transgenic line 182–36 transformed by 35S::GoPGF RNAi vector is glandless of the whole plant. Black dot represents gland in the figure. **P < 0.01; Student’s t-test, n = 3. Error bars are S.D. of three biological repeats.
3.2. Gland status and distribution in the transgenic plants
Homozygous plants of the T183-88 line exhibited glands in the plant tissues, including the leaf, stem, and boll shell, but nearly no glands in the mature seed (Fig. 2b). Correspondingly, consistent with the gland distribution, GoPGF expression was dramatically decreased in the developing ovules and mature seeds, but no significant differences relative to wild-type counterparts were observed in the leaf, stem, or root (Fig. 2c). Meanwhile, the T182-36 line exhibited almost no visible black glands throughout the whole plant, including in seeds (Fig. 2b), and the GoPGF transcripts were greatly suppressed in all the tissues tested (Fig. 2c).
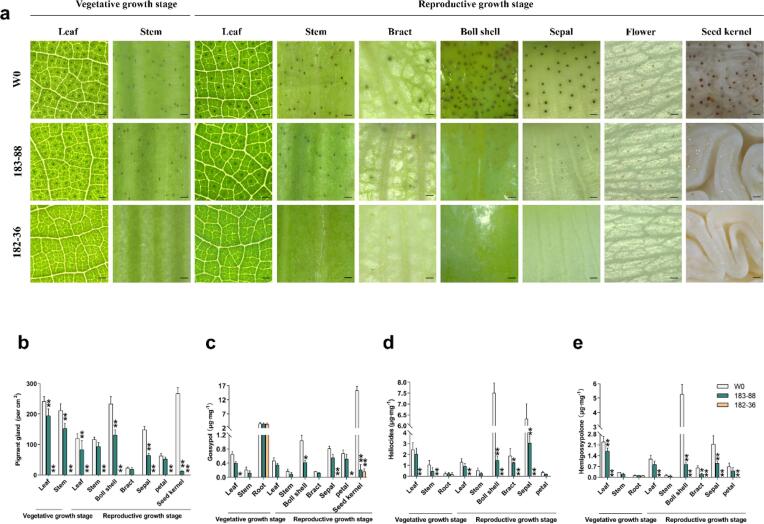
We further examined gland density and the levels of gossypol and related terpenoids in various tissues and organs throughout the plants’ growth and development. We observed distinctly different gland densities between the transgenic homozygous lines and the non-transgenic control W0 in different tissues and organs or at different development stages (Fig. 3a). In wild-type seedlings, gland density on the leaf and stem remained comparatively high during the vegetative stage (leaf > stem) (Fig. 3b). Then, in the reproductive growth stage, gland density greatly decreased in the leaf and stem, while the seed kernel contained the greatest number of glands (seed kernel > boll shell > sepal > leaf > stem > petal > bract) (Fig. 3b). These findings indicated that gland presence was variable and changed dynamically along with plant growth and development. In contrast, for the T182-36 plants, the glands were almost undetectable in all the examined tissues and organs (Fig. 3a), and for the homozygous T183-88 progeny, the gland density in the seed kernel was significantly decreased (Fig. 3b).
Fig. 3.
The gland density and level of gossypol, other terpenoids on the different parts of transgenic line 182–36, 183–88 and non-transgenic plants W0. (a) Gland distribution on the different parts leaf, stem, boll shell, bract, sepal and seed kernel of transgenic line 182–36, 183–88 and non-transgenic WT plants grown under field conditions. Bar is 0.5 mm. (b) Gland density. (c) Gossypol levels. (d) Hemigossypolone levels. (e) Heliocides levels.
3.3. Transgenic cottonseeds exhibit a significant reduction in gossypol level
We used HPLC to measure gossypol content in cottonseeds and other plant parts. As expected, based on gland density, the gossypol content of T183-88 was dramatically and specifically reduced in the seeds, but a certain level of gossypol, hemigossypolone, and other terpenoids was retained in other parts of the plant, which preserves its terpenoid-based defense capabilities (Fig. 3c-e). For T182-36, the content of gossypol and other terpenoids was strikingly reduced in all the examined tissues (Fig. 3c-e). Interestingly, no significant difference was observed in the gossypol content of the roots of the transgenic plants (T182-36 and T183-88) compared to the wild-type control (Fig. 3c), suggesting that gossypol synthesis in the root was not affected by GoPGF silencing despite the obvious impact on gland formation in the aerial parts. This finding clearly demonstrated that gossypol synthesis and gland formation were controlled by different molecular mechanisms. The cottonseeds exclusively stored gossypol (Fig. 3c), while other terpenoids derived from the same synthesis pathway, such as hemigossypolone and heliocides (H1, H2, H3, and H4), were mainly deposited in other plant parts, such as the boll shell (Fig. 3d-e).
To test the stability of the low-gossypol traits in cotton, we measured gossypol content in seeds from sequential T3, T4, and T5 progeny of the transgenic lines T183-88 and T182-36 grown under field conditions. The results demonstrated that the glandless or low-seed-gossypol trait was successfully inherited and stably maintained across multiple generations when grown in various field conditions. The free gossypol content of the transgenic cottonseeds from the T182-36 was 90–120 ppm, and 190–210 ppm for that of T183-88 lines, which is within the established limits for safe consumption of below 450 ppm (FDA/USA) and 600 ppm (FAO/UN), and conforms to the Chinese cottonseed food safety standard (gossypol concentrations below 0.02 %). Gossypol content decreased respectively by 98.6 % in T183-88 and 99.3 % in T182-36 line compared with W0 (14,100–15,800 ppm) (Table 1). In addition, the gossypol contents of the transgenic lines T182-36 (90–120 ppm) and T183-88 (190–210 ppm) were slightly lower than that of the three ULGCS lines 66-49B, 66–81 and 66–250 (140, 380, and 450 ppm) (Palle et al., 2013).
Table 1.
The content (g/kg or 1000 ppm) of gossypol, oil, and protein in seed kernel obtained from mature seeds grown in the field.
| 2018 |
2019 |
2020 |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Gossypol** | Oil | Protein | Gossypol** | Oil | Protein | Gossypol** | Oil | Protein | |
| W0 | 14.1 ± 1.93 | 282 ± 11 | 432 ± 9 | 15.8 ± 0.93 | 272 ± 5 | 421 ± 4 | 15.4 ± 1.78 | 293 ± 3 | 417 ± 13 |
| 183–88 | 0.19 ± 0.05 | 279 ± 13 | 420 ± 9 | 0.23 ± 0.04 | 265 ± 7 | 415 ± 9 | 0.21 ± 0.06 | 286 ± 8 | 419 ± 14 |
| 182–36 | 0.12 ± 0.04 | 276 ± 3 | 429 ± 14 | 0.09 ± 0.03 | 286 ± 4 | 418 ± 12 | 0.09 ± 0.08 | 318 ± 2 | 402 ± 8 |
Data represent mean ± SE,*P < 0.05; **P < 0.01; Student’s t-test n = 3 rows (10 plants/row). Error bars are s.d. of three biological repeats. Plants grown in the field in 2018, 2019, and 2020.
3.4. Nutritional oil and protein content in the transgenic cottonseeds
Since the main utilization value of cottonseed is in its protein and oil, we also investigated the protein and oil contents and fatty acid composition of cottonseeds from sequential T3, T4, and T5 progeny of the transgenic lines T183-88 and T182-36 grown under field conditions. In the T183-88 seeds, the fatty acid composition and oil content were similar to the non-transgenic plant, while seeds from the T4 and T5 progeny of T182-36 had a slightly higher oil content, 7 %∼12 % more linolenic acid, and 9 %∼16 % less palmitic acid (Table 1 and Table S2). The seed protein level was stable, and the seed protein content in the transgenic lines was like the non-transgenic control (Table 1). Though, He et al. (2022) observed that storage protein was affected by gossypol because that gossypol interacts with the free epsilon-amino groups from lysine and arginine likely promoting the formation of protein aggregates, which make not only the protein hard to extract. So, the distribution pattern and abundance of the storage proteins in the transgenic cottonseeds need to be investigated in future research.
3.5. Fiber quality and yield-related traits of the transgenic plants
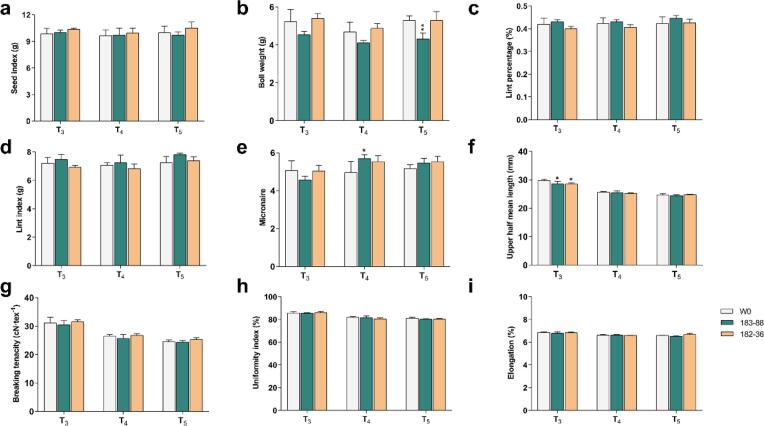
As an economic crop, the main value of cotton is its fiber. To evaluate fiber quality and yield, we harvested open bolls by hand from the transgenic homozygous lines and W0, then dried and weighed them before detaching the fibers from the seeds. We measured the various fiber quality parameters using an HFT9000 high-capacity instrument. Yield-related traits were stable, with no significant difference in seed index, lint percentage, or lint index between the transgenic lines and W0, except that the single boll weight of T183-88 was a little lower (Fig. 4a-d). These results indicated that gossypol content had no direct effect on the traits valued in cotton fiber production. In addition, we evaluated the five most important quality traits: micronaire, upper half mean length, uniformity index, strength, and elongation. Micronaire was higher for T183-88 versus W0 in the T4 generation. The upper half mean length was lower for T183-88 and T182-36 in the T3 generation. In general, the fiber quality of the transgenic lines was almost similar in the different generations (Fig. 4e-i).
Fig. 4.
Main agricultural traits, each from non-transgenic W0 plants and the transgenic lines (183–88 and 182–36) grown under field conditions. Yield traits include Seed index (a), lint index (b), boll weight (c), lint percentage (d). fiber traits include micronaire (e), upper half mean length (f), breaking tenacity (g), uniformity (h), and fiber elongation determined (i) by HFT 9000 High Volume Instrumentation for cotton fiber. Data represent mean ± SE,*P < 0.05; **P < 0.01; Student’s t-test n = 3 rows (10 plants/row). Error bars are s.d. of three biological repeats. T3, T4 and T5 represents plants grown in the field in 2018, 2019, and 2020.
3.6. Comparative transcriptome analysis of developing ovules from transgenic glandless and non-transgenic cotton
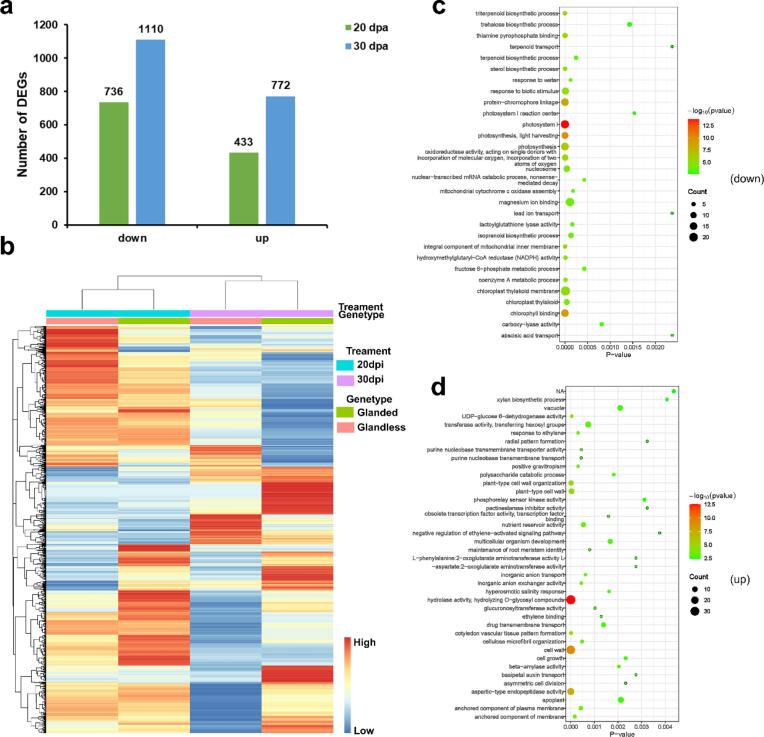
To identify genes associated with GoPGF that are also involved in gland formation, we conducted a comparative transcriptome analysis of cotton ovules from the glandless transgenic lines T182-36 and W0 at 20 and 30 DPA, which represent the stages of gland development and gossypol filling, respectively. A total of 512 million quality filtered paired reads were obtained, and more than 97 % of the data was mapped to the reference genome G. hirsutum acc. TM-1 v2.1 (Table S3). In total, 2,850 differentially expressed genes (DEGs) were identified, including 433 up-regulated and 736 down-regulated genes at 20 DPA and 772 up-regulated and 1,110 down-regulated genes at 30 DPA (Fig. 5a, b). Gene ontology (GO) enrichment analysis revealed that the down-regulated genes were significantly enriched in the terms terpenoid biosynthesis, photosystem, magnesium ion binding, and chloroplast (Fig. 5c). As expected, many important genes involved in the gossypol biosynthetic pathway and gland formation were remarkably downregulated, such as the terpenoid synthase genes (Huang et al., 2020, Tian et al., 2018, Tian et al., 2018), CGF2 (Janga et al., 2019), CGP1 (Gao et al., 2020) and ERF105 (Wu et al, 2021) (Fig. S2). Notably, the up-regulated genes were related to hydrolyzing O-glycosyl compounds, cell wall, and aspartic-type endopeptidase activity (Fig. 5d). These results suggest that the disruption of glands reduces the biosynthesis of gossypol and increases cell wall formation.
Fig. 5.
Comparative transcriptome analysis of ovules at 20 DPA and 30 DPA from CaMV35S::GoPGF RNAi (glandless) and W0 (glanded). (a) Number of differentially expressed genes (DEGs) (P < 0.05, |log2(FC)| > 2) in comparison of ovules at 20 DPA and 30 DPA between CaMV35S::GoPGF RNAi (glandless) and W0 (glanded). (b) Heat map of DEGs from 20 DPA and 30 DPA comparison sets. (c,d) Gene ontology (GO) enrichment analysis of all down-regulated (c) or all up-regulated genes (d). TOP 30 significantly enriched biological process GO terms are shown. Three biological replicates were included for each treatment.
4. Discussion
With the continuous growth of the global population, the output of grain, fiber, and feed has gradually increased. Cottonseed includes many valuable by-products, such as its shell, linter, cottonseed oil, cottonseed protein, cottonseed meal, and these co-products account for between 15 and 25 % of the value of the cotton crop (Dowd, 2015). Like other excellent cooking oils such as olive oil, sesame oil, coconut oil, cottonseed oil has abundant natural antioxidant compounds and poly-unsaturated linoleic acid. It was considered as a health oil and American Heart Association (AHA) has included it in the “OK FOOD” products and approved it as a nutritious and “Heart Oil” food (Riaz et al., 2021). Cottonseed oil is generally used in food cooking i.e. potato frying and processed food industry due to its special high-temperature frying characteristics, which helps extend the shelf life (Sekhar and Rao, 2011). Besides being utilized as cooking oil, cottonseed oil also has applications in many fields such as biofuel, livestock, cosmetics, agriculture, and chemicals (Riaz et al., 2021). Cottonseed protein and meal is the other important by-product of cotton production. Cottonseed protein contains a balanced proportion of necessary amino acids and have been widely applied as food additives rich in nutrients (Zhuge et al., 1988) and other added-value products such as coating, adhensives, bioplastics and films, antioxidant fraction etc. (He et al., 2022). As we all know, the presence of toxic gossypol greatly limited cottonseed’s utilization especially for cottonseed protein and meal. Several physical and chemical methods have been applied to detoxify cottonseed such as solvent extraction, water extraction, enzyme assisted, and ethanol extraction (Delgado et al., 2019, Pelitire et al., 2014, Qian et al., 2008). With the development of methods for oil extraction and refining, large amount of seeds have been used as the source for oil. However, gossypol removing from the cotton meal is more difficult. As reported by Dowd, isolate yield of cottonseed protein was 2.5-fold greater from the glandless defatted meal (23.6 %) than from the galnded defatted meal (10.6 %), because gossypol cross-link protein, which could promote protein aggregates and make it more difficult to extract (Dowd & Hojilla-Evangelista, 2013). Additionally, these treatments result in varying degrees of nutritional loss, and the complicated nature of the processing contributes to high cost, low food security and environment problem. As such, the cultivation of glandless cotton (having low gossypol) is of considerable interest. Research on the commercializing glandless cotton began in the 1960s (Lusas & Jividen, 1987). By 1980, many glandless cotton varieties were successively developed and cultivated in the USA, China, Egypt, and elsewhere. Nonetheless, the production of glandless cotton has remained limited since the resulting plants are very sensitive to insects, pests, rats, and pathogens because they lack the protection provided by the toxic gossypol. Therefore, it is essential to apply different breeding approaches to develop new cotton varieties. To aim it, as a result of the long-term effort, several low gossypol/ glandless Upland cultivars were released. For example, long-staple Acala 1517-18 GLS, medium staple NuMex COT 15 GLS, and NuMex COT 17 GLS with Fusarium wilt race 4 resistance with improved yield and disease resistance were developed via introgression of the dominant glandless allele Gl2e from G.barbadense into Upland cotton (Zhang & Wedegaertner, 2021). Another example is to employ engineering technology to genetically modify the critical genes that involved in gossypol biosynthesis like cadinene synthase to develop gossypol-free seeds (Palle et al., 2013). Besides these methods, as an alternative approach, impeding gland formation in the seeds could fundamentally prevent the production and storage of gossypol. Our present study developed a glanded plant/glandless seed cotton variety by interfering with the gland-forming gene GoPGF and demonstrated its stable inheritance over several generations. Field experiments showed that the transgenic lines were not impacted in terms of their main characteristics, except for the glands and sesquiterpenoids. A transcriptome analysis revealed that the decreased GoPGF expression directly affected the terpenoid synthesis pathway. By interfering with expression of GoPGF specifically in the seeds of the transgenic plants they still had abundant glands and terpenoids in their non-seed parts, which helps with resistance against pests and diseases. All told, cotton plants with specific silencing of gland formation in the seeds have considerable potential as an important source of oil, protein and other value-added nutrition-rich food (Fig. 6). In the future, the production and application prospect of glandless cottonseed will significantly increase its economic benefits, making cotton a fiber, feed, and grain crop.
Fig. 6.
Schematic chart shows the cultivation of GoPGF- RNAi line with glanded plant and glandless kernel, showing its potential as fiber, oil and feed crop in the future.
Data availability
The transcriptomic data sets were deposited under PRJNA729575 for the detection of differently expressed genes.
6. Contributions
Y.H. and T.Z.Z. conceived the study and designed the experiments. W.H.G., X.F.Z., L.Y.D., C.Y., Z.F.S., F.D. performed the most experiments. Y.G. performed the test of transgenic plants. B.L.L performed the construction of RNAi vector. L.Y.D. performed the cotton transformation. L.F. and X.Y.G. performed the expression analysis. S.J.Z contributed to the writing. Y.H., W.H.G and T.Z.Z. wrote the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported in part by grants National Natural Science Foundation of China (31970320), the Xinjiang Production and Construction Corps (2021AB008), Hainan Yazhou Bay Seed Lab (B21HJ0223), the Fundamental Research Funds for the Central Universities (226-2022-00100) and Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang (2019R01002).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochms.2022.100130.
Contributor Information
Tianzhen Zhang, Email: cotton@zju.edu.cn.
Yan Hu, Email: 0016211@zju.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Table S1 Primers used in this study. Table S2 Fatty acid composition of transgenic lines 183-88 and 182-36 in three successive progeny generations. Table S3 RNA-seq reads for glanded (W0) and glandless T182-36 ovules at 20 and 30 DPA and their mapping rate to the reference genome TM-1. Fig. S1 Molecular detection of the transgenic lines. Fig. S2 qRT-PCR validation of the expression level of gossypol pathway genes and gland genes in the ovules at 20 and 30DPA of the glanded cotton W0 and glandless transgenic line T182-36.
References
- Afifi A., Bary A., Kamel S.A. Bahtim 110, a new strain of Egyptian cotton free from gossypol. Empire Cotton Growing Review. 1966;43:112–120. [Google Scholar]
- Benson C.G., Wyllie S.G., Leach D.N., Mares C.L., Fitt G.P. Improved method for the rapid determination of terpenoid aldehydes in cotton. Journal of Agricultural and Food Chemistry. 2001;49:2181–2184. doi: 10.1021/jf0010836. [DOI] [PubMed] [Google Scholar]
- Bezemer T.M., Wagenaar R., Van Dam N.M., Van Der Putten W.H., Wackers F.L. Above- and below-ground terpenoid aldehyde induction in cotton, Gossypium herbaceum, following root and leaf injury. Journal of Chemical Ecology. 2004;30:53–67. doi: 10.1023/b:joec.0000013182.50662.2a. [DOI] [PubMed] [Google Scholar]
- Cai Y.F., Xie Y.F., Liu J.G. Glandless seed and glanded plant research in cotton. A review. Agronomy for Sustainable Development. 2010;30:181–190. [Google Scholar]
- Coutinho E.M. Gossypol: A contraceptive for men. Contraception. 2002;65:259–263. doi: 10.1016/s0010-7824(02)00294-9. [DOI] [PubMed] [Google Scholar]
- Delgado E., Valverde-Quiroz L., Lopez D., Cooke P., Valles-Rosales D., Flores N. Characterization of Soluble glandless cottonseed meal proteins based on electrophoresis, functional properties, and microscopic structure. J Food Sci. 2019;84:2820–2830. doi: 10.1111/1750-3841.14770. [DOI] [PubMed] [Google Scholar]
- Dowd M.K., Boykin D.L., Meredith W.R., Campbell B.T., Zhang J. Fatty acid profiles of cottonseed genotypes from the National Cotton Variety Trials. Journal of Cotton Science. 2010;14:64–73. [Google Scholar]
- Dowd, M. K., & Hojilla-Evangelista, M. P. (2013). Preparation and characterization of protein isolate from glandless and glanded cottonseed.
- Dowd M.K. In: Cotton, agronomy monograph #57. 2nd ed. Fang D.D., Percy R.G., editors. American Society of Agronomy, Crop Science of America, and Soil Society of America; Madison, WI: 2015. Cottonseed; pp. 745–781. [Google Scholar]
- Gao W., Long L., Zhu L.F., Xu L., Gao W.H., Sun L.Q.…Zhang X.L. Proteomic and Virus-induced Gene Silencing (VIGS) Analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Molecular & Cellular Proteomics. 2013;12:3690–3703. doi: 10.1074/mcp.M113.031013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Xu F.C., Long L., Li Y., Zhang J.L., Chong L.…Song C.P. The gland localized CGP1 controls gland pigmentation and gossypol accumulation in cotton. Plant Biotechnology Journal. 2020;18:1573–1584. doi: 10.1111/pbi.13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Zhang D., Mattison C.P. Quantitative comparison of the storage protein distribution in glandless and glanded cottonseeds. Agricultural & Environmental Letters. 2022;7:e20076. [Google Scholar]
- Hu Y., Chen J.D., Fang L., Zhang Z.Y., Ma W., Niu Y.C.…Zhang T.Z. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nature Genetics. 2019;51:739–748. doi: 10.1038/s41588-019-0371-5. [DOI] [PubMed] [Google Scholar]
- Huang J.Q., Fang X., Tian X., Chen P., Lin J.L., Guo X.X.…Chen X.Y. Aromatization of natural products by a specialized detoxification enzyme. Nature Chemical Biology. 2020;16:250–256. doi: 10.1038/s41589-019-0446-8. [DOI] [PubMed] [Google Scholar]
- Janga M.R., Pandeya D., Campbell L.M., Konganti K., Villafuerte S.T., Puckhaber L.…Rathore K.S. Genes regulating gland development in the cotton plant. Plant Biotechnology Journal. 2019;17:1142–1153. doi: 10.1111/pbi.13044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenar J.A. Reaction chemistry of gossypol and its derivatives. Journal of the American Oil Chemists' Society. 2006;83:269–302. [Google Scholar]
- Khan M.A., Wahid A., Ahmad M., Tahir M.T., Ahmed M., Ahmad S., Hasanuzzaman M. In: Cotton production and uses: Agronomy, crop protection, and postharvest technologies. Ahmad S., Hasanuzzaman M., editors. Springer; Singapore: 2020. World cotton production and consumption: An overview; pp. 1–7. [DOI] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Tomar M., Punia S., Grasso S., Amarowicz R. Cottonseed: A sustainable contributor to global protein requirements. Trends in Food Science & Technology. 2021;111:100–113. [Google Scholar]
- Lee J.A. Genetical studies concerning the distribution of pigment glands in the cotvledens and leaves of upland cotton. Genetics. 1962;47:134–142. doi: 10.1093/genetics/47.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F.F., Wu S.J., Chen T.Z., Zhang J., Wang H.H., Guo W.Z., Zhang T.Z. Agrobacterium-mediated co-transformation of multiple genes in upland cotton. Plant Cell Tissue Organ Culture. 2009;97:225–235. [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu Q., Wu M., Zhang B.L., Shrestha P., Petrie J., Green A.G., Singh S.P. Genetic enhancement of palmitic acid accumulation in cotton seed oil through RNAi down-regulation of ghKAS2 encoding beta-ketoacyl-ACP synthase II (KASII) Plant Biotechnology Journal. 2017;15:132–143. doi: 10.1111/pbi.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:38. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusas E.W., Jividen G.M. Glandless cottonseed: A review of the first 25 years of processing and utilization research. Journal of the American Oil Chemists Society. 1987;64:839–854. [Google Scholar]
- Ma D., Hu Y., Yang C.Q., Liu B.L., Fang L., Wan Q.…Zhang T.Z. Genetic basis for glandular trichome formation in cotton. Nature Communications. 2016;7:10456. doi: 10.1038/ncomms10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAuslane H.J., Alborn H.T. Systemic induction of allelochemicals in glanded and glandless isogenic cotton by Spodoptera exigua feeding. Journal of Chemical Ecology. 1998;24:399–416. [Google Scholar]
- McMichael S.C. Glandless boll in upland cotton and its use on the study of natural crossing. Agronomy Journal. 1954;46:527–528. [Google Scholar]
- McMichael S.C. Combined effect of glandless genes gl2 and gl3 on pigment glands in the cotton plant. Agronomy Journal. 1960;52:385–386. [Google Scholar]
- Mohan P., Singh P., Dongre A.B., Narayanan S.S. Gossypol-gland density and free gossypol content in seed and cotyledonary leaf of upland cotton (Gossypium hirsutum) Indian Journal of Agricultural Sciences. 1995;65:66–68. [Google Scholar]
- Murray J.C. A new locus for glanded stem in tetraploid cotton. Journal of Heredity. 1965;56:42–44. [Google Scholar]
- Palle S.R., Campbell L.M., Pandeya D., Puckhaber L., Tollack L.K., Marcel S.…Rathore K.S. RNAi-mediated ultra-low gossypol cottonseed trait: Performance of transgenic lines under field conditions. Plant Biotechnology Journal. 2013;11:296–304. doi: 10.1111/pbi.12013. [DOI] [PubMed] [Google Scholar]
- Pelitire S.M., Dowd M.K., Cheng H.N. Acidic solvent extraction of gossypol from cottonseed meal. Animal Feed Science and Technology. 2014;195:120–128. [Google Scholar]
- Qian J., Wang F., Liu S., Yun Z. In situ alkaline transesterification of cottonseed oil for production of biodiesel and nontoxic cottonseed meal. Bioresour Technol. 2008;99:9009–9012. doi: 10.1016/j.biortech.2008.04.059. [DOI] [PubMed] [Google Scholar]
- Randel R.D., Chase C.C., Wyse S.J. Effects of gossypol and cottonseed products on reproduction of mammals. Journal of Animal Science. 1992;70:1628–1638. doi: 10.2527/1992.7051628x. [DOI] [PubMed] [Google Scholar]
- Rathore, K. S., Pandeya, D., Campbell, L. M., & Palle, S.R. Cotton transgenic event TAM66274. US patent Application 2019/0008113 A1.
- Rathore K.S., Pandeya D., Campbell L.M., Wedegaertner T.C., Puckhaber L., Stipanovic R.D.…Hake K. Ultra-low gossypol cottonseed: Selective gene silencing opens up a vast resource of plant-based protein to improve human nutrition. Critical Reviews in Plant Sciences. 2020;39:1–29. [Google Scholar]
- Rathore K.S., Sundaram S., Sunilkumar G., Campbell L.M., Puckhaber L., Marcel S.…Wedegaertner T.C. Ultra-low gossypol cottonseed: Generational stability of the seed-specific, RNAi-mediated phenotype and resumption of terpenoid profile following seed germination. Plant Biotechnology Journal. 2012;10:174–183. doi: 10.1111/j.1467-7652.2011.00652.x. [DOI] [PubMed] [Google Scholar]
- Riaz T., Iqbal M.W., Mahmood S., Yasmin I., Leghari A.A., Rehman A.…Bilal M. Cottonseed oil: A review of extraction techniques, physicochemical, functional, and nutritional properties. Critical Reviews in Food Science and Nutrition. 2021:1–19. doi: 10.1080/10408398.2021.1963206. [DOI] [PubMed] [Google Scholar]
- Rogers G.M., Poore M.H., Paschal J.C. Feeding cotton products to cattle. Veterinary Clinics of North America Food Animal Practice. 2002;18:267–294. doi: 10.1016/s0749-0720(02)00020-8. [DOI] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C-T method. Nature Protocol. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Sekhar S.C., Rao B. Cottonseed oil as health oil. Pertanika Journal of Tropical Agricultural Science. 2011;34:17–24. [Google Scholar]
- Stipanovic R.D., Altman D.W., Begin D.L., Greenblatt G.A., Benedict J.H. Terpenoid aldehydes in upland cottons: Analysis by aniline and HPLC methods. Journal of Agricultural and Food Chemistry. 1988;36:509–515. [Google Scholar]
- Sunilkumar G., Campbell L.M., Puckhaber L., Stipanovic R.D., Rathore K.S. Engineering cottonseed for use in human nutrition by tissue-specific reduction of toxic gossypol. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18054–18059. doi: 10.1073/pnas.0605389103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunilkumar G., Connell J.P., Smith C.W., Reddy A.S., Rathore K.S. Cotton alpha-globulin promoter: Isolation and functional characterization in transgenic cotton, Arabidopsis, and tobacco. Transgenic Research. 2002;11:347–359. doi: 10.1023/a:1016322428517. [DOI] [PubMed] [Google Scholar]
- Tian X., Ruan J.X., Huang J.Q., Yang C.Q., Fang X., Chen Z.W., Hong H., Wang L.J., Mao Y.B., Lu S., Zhang T.Z., Chen X.Y. Characterization of gossypol biosynthetic pathway. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:E5410–E5418. doi: 10.1073/pnas.1805085115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend B.J., Poole A., Blake C.J., Llewellyn D.J. Antisense suppression of a (+)-delta-cadinene synthase gene in cotton prevents the induction of this defense response gene during bacterial blight infection but not its constitutive expression. Plant Physiology. 2005;138:516–528. doi: 10.1104/pp.104.056010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Cheng H., Li S., Zuo D., Lin Z., Zhang Y.…Song G. Molecular cloning and characterization of GhERF105, a gene contributing to the regulation of gland formation in upland cotton (Gossypium hirsutum L.) BMC Plant Biology. 2021;21(102) doi: 10.1186/s12870-021-02846-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G.C., Wang L.G., Han Y.Y., He Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wedegaertner T. Genetics and breeding for glandless upland cotton with improved yield potential and disease resistance: A review. Frontiers in Plant Science. 2021;12:753426. doi: 10.3389/fpls.2021.753426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.J., Xu Z.R., Pan X.L., Yan X.H., Wang Y.B. Advances in gossypol toxicity and processing effects of whole cottonseed in dairy cows feeding. Livestock Science. 2007;111:1–9. [Google Scholar]
- Zhou M., Zhang C., Wu Y., Tang Y. Metabolic engineering of gossypol in cotton. Applied Microbiology and Biotechnology. 2013;97:6159–6165. doi: 10.1007/s00253-013-5032-5. [DOI] [PubMed] [Google Scholar]
- Zhuge Q., Posner E.S., Deyoe C.W. Production study of a low-gossypol protein product from cottonseed meal. Journal of Agricultural & Food Chemistry. 1988;36:153–155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Primers used in this study. Table S2 Fatty acid composition of transgenic lines 183-88 and 182-36 in three successive progeny generations. Table S3 RNA-seq reads for glanded (W0) and glandless T182-36 ovules at 20 and 30 DPA and their mapping rate to the reference genome TM-1. Fig. S1 Molecular detection of the transgenic lines. Fig. S2 qRT-PCR validation of the expression level of gossypol pathway genes and gland genes in the ovules at 20 and 30DPA of the glanded cotton W0 and glandless transgenic line T182-36.
Data Availability Statement
The transcriptomic data sets were deposited under PRJNA729575 for the detection of differently expressed genes.