Benjamin D. Greenbaum and David Hoyos discuss the challenges of antigen modeling to design vaccines for infectious diseases and cancer.
Abstract
Advances in genomics and precision measurement have continued to demonstrate the importance of the immune system across many disease types. At the heart of many emerging approaches to leverage these insights for precision immunotherapies is the computational antigen prediction problem. We propose a threefold approach to improving antigen predictions: further defining the geometry of the antigen landscape, incorporating the coupling of antigen recognition to other cellular processes, and diversifying the training sets used for models that predict immunogenicity.
Immune-based therapies are finding successful application across many diseases, most famously cancer and COVID-19 (Wolchok et al., 2009; Oliver et al., 2020). The goal of such therapies is to induce an immune response against specific antigens and thereby prime the immune system to recognize those antigens in the near or distant future. At the heart of this goal lies a mathematical prediction problem: can we predict which antigens, when introduced today, will best prepare us for tomorrow? Historically, at a time when vaccines contained entire live or inactivated pathogens, this task was more straightforward and focused on an epidemiological problem, namely predicting which strains were likely to be circulating when a vaccine went into production. Once a potential new strain was identified, the computational challenge was to determine whether the antigens derived from this new strain were sufficiently different from the original strain to warrant updating the vaccine. The practical question then became whether the new and original strains diverged enough to warrant the cost, time, and labor involved in changing the vaccine, which typically required many months (Lambert and Fauci, 2010).
Our adaptive immune system has sophisticated mechanisms for detecting proteins that are “non-self,” resulting from viral proteins or mutated peptides in tumor genomes (Goldberg and Rock, 1992). Emerging platforms, such as mRNA vaccines and engineered T cells, target specific antigen subsets rather than an entire strain or genome (Plotkin, 2014). As has become painfully clear during the COVID-19 pandemic, new pathogen strains can rapidly evolve in response to selective pressures and escape acquired immunity. However, the speed and specificity of emerging technologies creates a potential to update precise targets almost in real time. Due to mRNA vaccine technology and rapid viral genome sequencing, we went from the identification of the SARS-CoV-2 spike protein to an effective vaccine in less than a year. Real-time global surveillance, advances in computational models, and high-throughput immune monitoring and genome sequencing have moved computational antigen prediction from epidemiology into the field of evolutionary modeling, requiring, among others, tools from machine learning, evolutionary dynamics, and biophysics (Morris et al., 2018).
The data and mathematical toolkit for defining and powering these models, while beginning to emerge, are still largely lacking. As a result, investigators often repurpose existing datasets and black-box modeling approaches that were not designed for this specific problem. We outline three “gaps” where better datasets and appropriate mathematical methods are needed.
Quantifying the geometry of the antigen landscape
Technological advances in the last few decades have brought an explosion of genomic data and, for the first time in human history, we are able to quantify the molecular etiology of diseases such as cancer at large scale. Accurate mutation calling, a prerequisite for precise vaccine development, remains difficult to resolve from heterogeneous tumor samples from bulk DNA and RNA sequences. Evidence suggests subclonal mutations may be a source of targetable neoantigens (Roudko et al., 2020); however, it is often difficult to estimate which percentage of the tumor is targetable, due to sequencing bias and varying purity (Aran et al., 2015). State-of-the-art algorithms such as NetMHC (Andreatta and Nielsen, 2016) infer the affinity of an antigen to an MHC molecule, a critical step required for productive immune recognition. Still, there remain unknowns around the degree to which the T cell receptor interaction provides additional predictive information (Łuksza et al., 2017), in part due to the low throughput of T cell validation assays. Moreover, recent work has proposed biases in the “distance from self” of various antigens, suggesting that the antigen landscape, in part resulting from T cell cross-reactivity, is highly non-uniform and likely poorly quantified (Bradley and Thomas, 2019). We therefore need to further specify the “geometry of the antigen landscape,” which defines which antigenic distances determine immunogenicity.
Understanding an antigen’s evolutionary and ecological context
There has been a tendency to look at the immunogenicity of antigens as separate from other cellular processes. Although most efforts to predict antigenicity focus on the peptide-MHC binding process, factors such as the concentration of the antigen in question, and the immune microenvironment in which it appears, are equally important. Protein expression is non-uniform for viruses and cancer cells. In cancer, mutations and copy number changes in particular genes, such as TP53 and KRAS, result in alterations in protein concentration, and recent work has suggested intrinsic trade-offs in the pro-proliferative activity and immune vulnerability of driver genes (Hoyos et al., 2022). This suggests tumors that contain aggressive pro-tumor driver mutations may have immunogenic clonal mutations that result in stronger responses than otherwise expected by affinity inferences. Likewise, the ecological niche in which an antigen occurs may be immunosuppressive, dampening the ability of a potentially immunogenic peptide to ultimately lead to clearance of an infected cell or cancer cell. Defining the context in which antigens exist will help us better understand how to manipulate the trade-offs between their immunogenicity and other cellular processes to boost responses.
Creating equitable training datasets
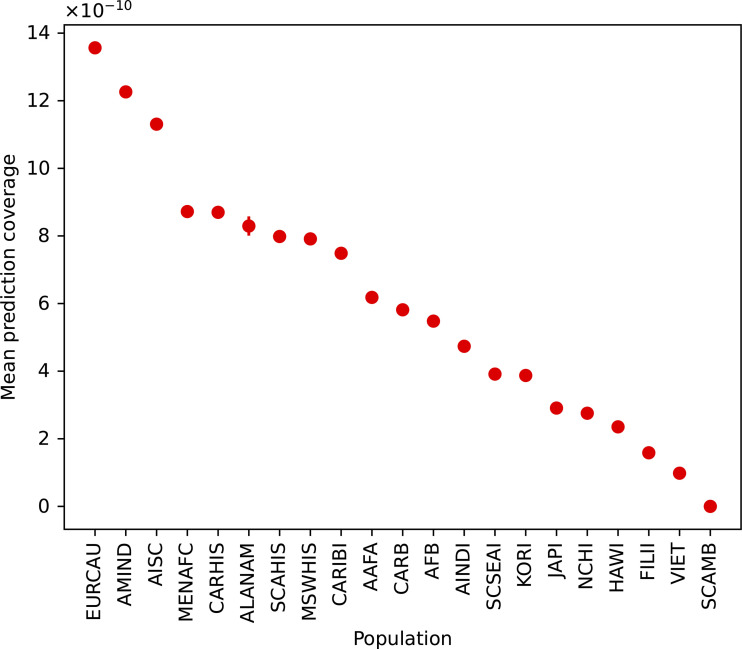
Despite cancer cases rising in certain population groups, there is a marked underrepresentation of tumors from diverse demographics in the databases most often studied and used for computational models and machine learning (Clegg et al., 2002). These methods are constrained by available training datasets (Fig. 1), which inevitably introduces bias in the accuracy of predictive models that can only be ameliorated with large, carefully assembled training data. For example, MHC prediction algorithms are most accurate for A*02:01, the most common MHC-I molecule in the American population of European ancestry, which reflects the abundance of immunological assays restricted to this particular HLA type (Liu et al., 2021). Not only could this bias the accuracy of therapeutics, which may then be potentially less effective in different populations, scientifically it will limit our understanding for how therapeutic targets in viruses and cancer cells evolve. Our ability to predict emerging pathogens and the evolution of global pandemics requires accurately measuring the selective pressures the immune system exerts on them. If such predictions are poorer for substantial portions of the global population, our ability to predict the future evolution and emergence of pandemics and how to respond to them effectively will suffer.
Figure 1.
Disparities in the accurate coverage of computational antigen predictions for diverse HLA-I populations. The mean fraction of a population that is covered by the NetMHC 4.0 software is plotted for diverse population groups. The mean and 95% confidence interval are denoted. The population groups are derived from the National Marrow Donor Program (Gragert et al., 2013). All HLA-I haplotypes constructed from the HLA-I molecules covered by the NetMHC 4.0 software were simulated and population frequencies inferred (Hoyos et al., 2022).
Discussion
We are currently amid a scientific and clinical revolution of historic proportions. As the molecular etiologies of infectious diseases and cancer are being discerned, we are experiencing the efficacy of mRNA vaccine technology to halt a global pandemic. Encouraged by these results, many are eager to expand precision immunotherapies. However, we remain limited by gaps in quantifying the antigen landscape, the evolutionary and ecological context of antigenicity, and the generation of large, equitable training sets. In addressing these gaps, we can move towards perfected predictions and increase the reach of precision immunotherapies to underserved populations in the process.
Acknowledgments
The authors would like to thank Nicole Rusk and Jedd Wolchok for their helpful comments and feedback.
Disclosures: The authors declare no competing interests exist.
References
- Andreatta, M., and Nielsen M.. 2016. Bioinformatics. 10.1093/bioinformatics/btv639 [DOI] [Google Scholar]
- Aran, D., et al. 2015. Nat. Commun. 10.1038/ncomms9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, P., and Thomas P.G.. 2019. Annu. Rev. Immunol. 10.1146/annurev-immunol-042718-041757 [DOI] [PubMed] [Google Scholar]
- Clegg, L.X., et al. 2002. Arch. Intern. Med. 10.1001/archinte.162.17.1985 [DOI] [Google Scholar]
- Goldberg, A.L., and Rock K.L.. 1992. Nature. 10.1038/357375a0 [DOI] [Google Scholar]
- Gragert, L., et al. 2013. Hum. Immunol. 10.1016/j.humimm.2013.06.025 [DOI] [PubMed] [Google Scholar]
- Hoyos, D., et al. 2022. Nature. 10.1038/s41586-022-04696-z [DOI] [Google Scholar]
- Lambert, L.C., and Fauci A.S.. 2010. N. Engl. J. Med. 10.1056/NEJMra1002842 [DOI] [PubMed] [Google Scholar]
- Liu, B., et al. 2021. Immun. Inflamm. Dis. 10.1002/iid3.416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łuksza, M., et al. 2017. Nature. 10.1038/nature24473 [DOI] [Google Scholar]
- Morris, D.H., et al. 2018. Trends Microbiol. 10.1016/j.tim.2017.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, S.E., et al. 2020. Morb. Mortal Wkly. Rep. 10.15585/mmwr.mm6950e2 [DOI] [Google Scholar]
- Plotkin, S. 2014. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1400472111 [DOI] [Google Scholar]
- Roudko, V., et al. 2020. Cell. 10.1016/j.cell.2020.11.004 [DOI] [Google Scholar]
- Wolchok, J.D., et al. 2009. Clin. Cancer Res. 10.1158/1078-0432.CCR-09-1624 [DOI] [Google Scholar]