This cohort study assesses the self-reported awareness of Omicron variant infection in employees and patients of an academic medical center in Los Angeles County, California.
Key Points
Question
What proportion of individuals who recently contracted the SARS-CoV-2 Omicron variant were aware of their infectious status?
Findings
In this cohort study of 210 adults with evidence of seroconversion during a regional Omicron variant surge, 56% reported being unaware of any recent Omicron variant infection.
Meaning
Findings of this study suggest that low rates of Omicron variant infection awareness may be a key contributor to rapid transmission of the virus within communities.
Abstract
Importance
Some individuals who were infected by the SARS-CoV-2 Omicron variant may have been completely unaware of their infectious status while the virus was actively transmissible.
Objective
To examine awareness of infectious status among individuals during the recent Omicron variant surge in a diverse and populous urban region of Los Angeles County.
Design, Setting, and Participants
This cohort study analyzed the records of adult employees and patients of an academic medical center who were enrolled in a longitudinal COVID-19 serological study in Los Angeles County, California. These participants had 2 or more serial anti-nucleocapsid IgG (IgG-N) antibody measurements at least 1 month apart, with the first occurring after the end of a regional Delta variant surge (September 15, 2021) and a subsequent one occurring after the start of a regional Omicron variant surge (December 15, 2021). Adults with evidence of new SARS-CoV-2 infection occurring during the Omicron variant surge period through May 4, 2022, were included in the present study sample.
Exposures
Recent Omicron variant infection as evidenced by SARS-CoV-2 seroconversion.
Main Outcomes and Measures
Awareness of recent SARS-CoV-2 infection was ascertained from review of self-reported health updates, medical records, and COVID-19 testing data.
Results
Of the 210 participants (median [range] age, 51 (23-84) years; 136 women [65%]) with serological evidence of recent Omicron variant infection, 44% (92) demonstrated awareness of any recent Omicron variant infection and 56% (118) reported being unaware of their infectious status. Among those who were unaware, 10% (12 of 118) reported having had any symptoms, which they attributed to a common cold or other non–SARS-CoV-2 infection. In multivariable analyses that accounted for demographic and clinical characteristics, participants who were health care employees of the medical center were more likely than nonemployees to be aware of their recent Omicron variant infection (adjusted odds ratio, 2.46; 95% CI, 1.30-4.65).
Conclusions and Relevance
Results of this study suggest that more than half of adults with recent Omicron variant infection were unaware of their infectious status and that awareness was higher among health care employees than nonemployees, yet still low overall. Unawareness may be a highly prevalent factor associated with rapid person-to-person transmission within communities.
Introduction
The rapid spread of the SARS-CoV-2 Omicron variant across communities worldwide has been attributed to several factors, including transmissibility features of the viral variant, limited coverage by available vaccines, and changes in face mask use and socializing behaviors.1,2 In particular, alterations of the Omicron variant have been implicated because of its greater transmissibility compared with previous SARS-CoV-2 variants, as demonstrated by higher infection rates within households.3,4 Furthermore, available vaccines have had variable efficacy against the Omicron variant, and waning levels of immunity provided by vaccinations administered up to 1 year earlier likely also compounded the magnitude of the Omicron variant surge.5 Beyond the specific factors associated with the variant and vaccines, public health guidance on masking and social distancing has been changing along with adherence to public health policies in general.6
Despite all of the known factors in the Omicron variant surge, there remain unknown variables. Prominent among these unknown variables is the extent to which infected individuals may be unaware of their infectious status. Multiple previous studies have indicated that asymptomatic or minimally symptomatic SARS-CoV-2 infections are likely triggers of outbreaks as well as ongoing rapid person-to-person transmission across communities.7,8,9 Although rapid antigen testing kits, which were distributed to facilitate self-diagnosis, are now widely available throughout the US, the extent to which some individuals with Omicron variant infection are unaware of their infection while it is actively transmissible is unknown.
We aimed to examine awareness of infectious status among individuals during the recent Omicron variant surge in a diverse and populous urban region of Los Angeles County in California. To this end, we assessed participants in a large observational cohort for whom serial SARS-CoV-2 serological assays had been performed before or after the onset of the Omicron variant surge in this region.
Methods
All participants in the source study provided written informed consent for all protocols, which were approved by the Cedars-Sinai Medical Center Institutional Review Board. The approval and informed consent also applied to the present cohort study. We followed the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
Study Cohort
Individuals who were registered as adult employees or patients at Cedars-Sinai Medical Center, an academic medical center in Southern California, were enrolled in the longitudinal serological study of COVID-19 risks and outcomes. Participants completed surveys on exposures and symptoms at baseline and at serial time points over the course of the study according to protocols described in previous publications.10 All participants were invited to complete monthly health update surveys (eAppendix in the Supplement) and to contact study coordinators by telephone or email at any time regarding health status updates, including interval SARS-CoV-2 infections contracted or doses of COVID-19 vaccines received. All participants were also invited to contribute to monthly blood draws for COVID-19 serological assays.11
The sampling strategy for the current study is outlined in the eFigure in the Supplement. Of the 6385 participants enrolled in the source study, 2479 provided plasma samples for serological assays; these samples were collected from at least 2 serial time points, including at least 1 time point after the end of the Delta variant surge (September 15, 2021) and at least 1 subsequent time point after the start of the Omicron variant surge (December 15, 2021) in the region.12 Of this subset, 264 participants met the criterion for seroconversion during the Omicron variant surge period (from December 15, 2021, through May 4, 2022).
We excluded 52 participants who had not completed at least 1 health update survey within 5 weeks before or any time after collection of the second time point blood draw.13 We also excluded 2 participants who received pre-exposure monoclonal antibody treatment (given its potential to markedly increase antibody levels in the absence of infection). We verified that all 210 participants who made up the final sample for the present analysis had completed an in-person brief health history update during the intake registration for the second time point (ie, postseroconversion) blood draw visit.
Clinical and Serological Assessment
Participants completed surveys on medical history, exposures, and symptoms at baseline and at serial time points over the course of the source study. To verify self-reported absence or presence of comorbidities for participants, we reviewed medical records using the electronic health record (EHR) system at Cedars-Sinai Medical Center.14 Serological measures were performed using the Abbott Architect immunoassay15 (Abbott Laboratories) to quantify circulating levels of SARS-CoV-2 anti-nucleocapsid IgG antibody (IgG-N), with an index value of 1.4 or higher considered to be the threshold indicating previous SARS-CoV-2 infection.16 All primary data were extracted from the EHR using algorithm-based scripts and from the REDCap electronic survey database (Vanderbilt University). For SARS-CoV-2 infection or COVID-19 vaccine events that were not captured in the EHR because they occurred outside of Cedars-Sinai Medical Center, manual verification was performed by study staff with training in medical record review and data abstraction procedures relevant to COVID-19 research study protocols. Self-reported SARS-CoV-2 infection events were verified from external records of polymerase chain reaction (PCR) or other testing. Self-reported type, dose, and date of vaccinations were ascertained from vaccination cards or external documents. All staff involved in manual verification protocols used standardized data capture instruments with explicitly defined variable fields, and all verification procedures were performed in batch fashion for the source study as a whole; thus, all staff were blinded to the study hypothesis for the present cohort study.
Classification of Recent Omicron Variant Infection
We classified participants as having had recent Omicron variant infection if their IgG-N index level changed from lower than 1.2 to 1.4 or higher after the surge onset (n = 210) in accordance with the high sensitivity and specificity reported for the 1.4 or higher index threshold for the Abbott Architect assay used.16 In sensitivity analyses, given the possibility that values close to (albeit lower than) the 1.4 index threshold could represent diminishing IgG-N levels from previous SARS-CoV-2 infection or (less likely) recent-onset SARS-CoV-2 infection and could be an indicator of newly increasing levels, we used more conservative criteria to define seroconversion events on the basis of a lower preseroconversion IgG-N level. Accordingly, for sensitivity analyses, we classified 195 participants as having had recent Omicron variant infection if their pre- to post-IgG-N value change was from lower than 1.0 to 1.4 or higher (ie, a lower threshold for defining previous negative status). We also classified 159 participants as having had recent Omicron variant infection if their IgG-N value change was from lower than 0.4 to 1.4 or higher (ie, the lowest threshold for defining previous negative status).
Assessment of Infection Awareness
Participants were invited to complete an electronic health update survey using a standardized REDCap instrument.17,18 The survey included questions regarding recent vaccinations, infections, and symptoms as well as other changes in health status. Participants were specifically asked whether they had developed any known new infections since completing the previous health update survey or study visit (eAppendix in the Supplement). In the source study of 2479 participants with 2 or more serological assays, which were appropriately timed to detect possible seroconversion during the Omicron variant surge, 1937 participants completed a correspondingly timed health update survey and 542 did not. The frequency of seroconversion was 16% (316 of 1937) in the former group and 18% (96 of 542) in the latter group.
Participants who met the criteria for seroconversion and inclusion in the final sample of the present study (eFigure in the Supplement) were categorized as aware or unaware of recent Omicron variant infection on the basis of their responses to the health update survey, which was completed after the preconversion blood draw. For participants who completed more than 1 monthly health update survey (eTable 1 in the Supplement), we selected the survey closest in timing to the postseroconversion blood draw. Given that the timing and up to monthly frequency of electronic surveys might not completely capture the information on Omicron variant infection awareness, for participants who initially appeared unaware according to their survey responses alone, we also reviewed other records that could have captured such data, including study-related telephone and email logs and the EHR for any diagnostic testing or codes relevant to possible recent Omicron variant infection. In addition, we reviewed the in-person health history update (which included screening for interval COVID-19 vaccination or SARS-CoV-2 infection) that was conducted as part of the intake registration for each blood draw visit, including the postseroconversion blood draw visit attended by all participants in the current study sample.
Statistical Analysis
To compare preexisting demographic and clinical characteristics between participants who were aware vs unaware of recent Omicron variant infection, we used the unpaired, 2-tailed t test to analyze continuous variables and χ2 tests for categorical variables. We then used logistic regression analyses to examine the association of demographic and clinical characteristics with infection awareness. After initially examining the associations in unadjusted models, we used a multivariable regression model with stepwise selection to identify demographic and clinical variables associated with awareness while accounting for the effects of other covariates. Race and ethnicity were self-reported by participants and were included as covariates in analyses given their potential to represent factors associated with variation in access to COVID-19 testing resources, including at home testing kits. The race categories were identified as Asian, Black or African American, Native Hawaiian or Other Pacific Islander, White, multiple, other, or unknown. The ethnicity categories were identified as Hispanic, non-Hispanic, or other.
Given the small sample size, we used P = .10 as the threshold for model entry or exit for all of the variables considered in the stepwise selection. We conducted all statistical analyses using Stata, version 15.1 (StataCorp LLC), and we considered a 2-tailed P < .05 to be statistically significant.
Results
Among the 210 adult participants in the sample with serological evidence of recent Omicron variant infection, the median (range) age was 51 (23-84) years and there were 136 women (65%) and 74 men (35%). Compared with participants in the source cohort, the individuals included in the present study were similar in demographic and most clinical characteristics but were less likely to have autoimmune disease (eTable 2 in the Supplement). Before seroconversion, 94% of participants (197 of 210) in this sample had received any COVID-19 vaccine, and 93% of previously vaccinated individuals (183 of 197) received a vaccination series that included at least 1 mRNA vaccine (eTable 3 in the Supplement). Compared with individuals with Omicron variant infection that was identified using standard seroconversion criteria, those whose infection was identified with more stringent criteria (using the lowest preseroconversion IgG-N levels) were similar in demographic and clinical characteristics (eTable 4 in the Supplement).
Overall, of all participants with seroconversion during the Omicron variant surge, only 44% (92 of 210) demonstrated awareness of any recent Omicron variant infection, whereas 56% (118 of 210) were unaware (Table; Figure 1). Of participants who were aware of such an infection, 73% (67 of 92) had an interval positive PCR result that was recorded in the EHR, self-reported, or both.
Table. Characteristics of the Study Sample.
| Characteristic | Participants, No. (%) | P value | ||
|---|---|---|---|---|
| Overall | Unaware of Omicron variant infection | Aware of Omicron variant infection | ||
| No. of participants | 210 | 118 (56) | 92 (44) | |
| Age, median (range), y | 51 (23-84) | 53 (24-84) | 47 (23-77) | .02 |
| Sex | ||||
| Female | 136 (65) | 81 (69) | 55 (60) | .20 |
| Male | 74 (35) | 37 (31) | 37(40) | |
| Ethnicitya | ||||
| Hispanic | 19 (9) | 8 (7) | 11 (12) | .42 |
| Non-Hispanic | 189 (90) | 109 (92) | 80 (87) | |
| Unknown | 2 (1) | 1 (1) | 1 (1) | |
| Racea | ||||
| Asian | 24 (11) | 14 (12) | 10 (11) | .18 |
| Black or African American | 6 (3) | 4 (3) | 2 (2) | |
| Native Hawaiian or Other Pacific Islander | 1 (1) | 1 (1) | 0 | |
| White | 164 (78) | 95 (81) | 69 (75) | |
| Multiple | 8 (4) | 3 (3) | 5 (5) | |
| Otherb | 5 (3) | 0 | 5 (5) | |
| Unknown | 2 (1) | 1 (1) | 1 (1) | |
| Health care employee | 78 (37) | 33 (28) | 45 (49) | .002 |
| Medical history | ||||
| Obesity | 31 (15) | 17 (14) | 14 (15) | .87 |
| Hypertension | 49 (23) | 28 (24) | 21 (23) | .88 |
| Diabetes | 11 (5) | 5 (4) | 6 (7) | .46 |
| Coronary disease or heart failure | 28 (13) | 14 (12) | 14 (15) | .48 |
| Asthma or COPD | 37 (18) | 21 (18) | 16 (17) | .94 |
| Cancer | 31 (15) | 19 (16) | 12 (13) | .54 |
| Autoimmune disease | 13 (6) | 9 (8) | 4 (4) | .33 |
| Organ transplant recipient | 4 (2) | 3 (3) | 1 (1) | .44 |
| No. of vaccination doses | ||||
| 0 | 13 (6) | 8 (7) | 5 (5) | .16 |
| 1 | 6 (3) | 5 (4) | 1 (1) | |
| 2 | 18 (9) | 10 (9) | 8 (9) | |
| 3 | 141 (67) | 72 (61) | 69 (75) | |
| 4 | 32 (15) | 23 (20) | 9 (10) | |
| Received ≥3 vaccination doses | 173 (82) | 95 (81) | 78 (85) | .42 |
Abbreviation: COPD, chronic obstructive pulmonary disease.
Race and ethnicity were self-reported by participants.
No subcategories were specified in Other.
Figure 1. Awareness of SARS-CoV-2 Omicron Variant Infection.
Of the 210 adults with contemporaneously available self-reported, medical, and testing data on SARS-CoV-2 infection, only 92 (44%) demonstrated awareness of any recent Omicron variant infection (Table) and 118 (56%) were unaware of their infectious status. Rates of awareness were generally similar across demographic and clinical characteristics.
In sensitivity analyses, rates of unawareness of Omicron variant infection were similar at 54% (105 of 195) for individuals whose seroconversion was identified using the more conservative lower IgG-N index values and 51% (81 of 159) for individuals whose seroconversion was identified with the lowest preinfection IgG-N thresholds. Among the 92 individuals who were aware of recent Omicron variant infection, 67% (62) reported awareness on their health update survey; of these 62 respondents, 90% (56) reported attributable symptoms and 10% (6) reported being asymptomatic. Among the 118 participants with seroconversion and who were unaware of recent Omicron variant infection, 10% (12) reported experiencing interval symptoms during the seroconversion period that they attributed to a common cold or other non–SARS-CoV-2 infection.
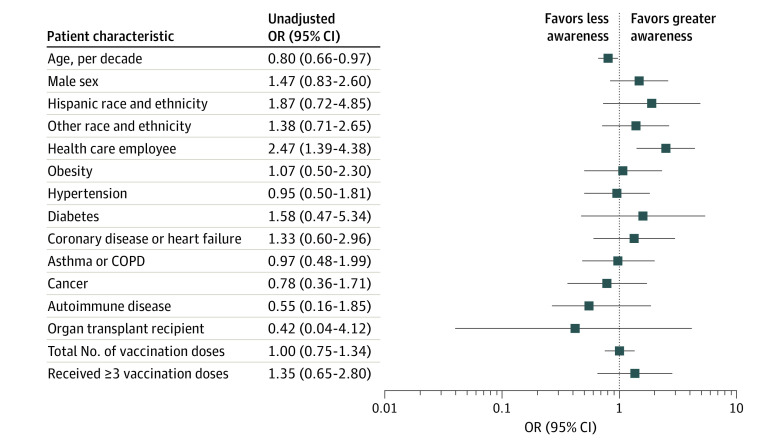
In unadjusted analyses, individuals aware of recent Omicron variant infection compared with those who were unaware were more likely to be younger (median [range] age, 47 [23-77] years vs 53 [24-84] years; P = .02) and more likely to be health care employees of Cedars-Sinai Medical Center (45 [49%] vs 33 [28%]; P = .002) (Table; Figure 2). Both groups of individuals were otherwise similar in demographic and clinical characteristics (Table) as well as the number of vaccination doses or types of vaccines received (eTable 3 in the Supplement). In stepwise multivariable-adjusted analyses, health care employee status (odds ratio [OR], 2.46; 95% CI, 1.30-4.65; P = .006) and male sex (OR, 1.87; 95% CI, 1.01-3.45; P = .05) were associated with greater awareness, whereas older age was associated with less awareness (OR, 0.78; 95% CI, 0.64-1.00; P = .05) (Figure 3). These overall results were similar in sensitivity analyses conducted for individuals whose seroconversion was defined using more conservative IgG-N thresholds (eTables 5 and 6 in the Supplement). Across all analyses in this study, comorbidities were not consistently significantly associated with infection awareness.
Figure 2. Unadjusted Logistic Regression Analysis of Association Between Preexisting Characteristics and Awareness of SARS-CoV-2 Omicron Variant Infection .
COPD indicates chronic obstructive pulmonary disease; OR, odds ratio.
Figure 3. Multivariable-Adjusted Logistic Regression Analysis of Association Between Preexisting Characteristics and Awareness of SARS-CoV-2 Omicron Variant Infection .
All variables were considered as candidate variables, and P = .10 was the criterion for model entry or exit in the stepwise selection. OR indicates odds ratio.
Discussion
In analyzing a large, longitudinal cohort study of ambulatory adults for whom SARS-CoV-2 serological assays were performed, we found that, among individuals with Omicron variant infection indicated by recent seroconversion, most were unaware of their COVID-19 status. Of those who were unaware, most were asymptomatic, with only 10% reporting symptoms that were attributed to a common cold or other non–SARS-CoV-2 infection. Lack of awareness of Omicron variant infection, either owing to the relative absence of symptoms or lack of timely testing, likely had a role in rapid transmission within the communities in Los Angeles County.
These findings extend the results from previous investigations of asymptomatic and undiagnosed SARS-CoV-2 infection and are recognized as critical factors in SARS-CoV-2 transmission.7 Studies relying predominantly on COVID-19 testing data have estimated that 25% to 48% of community cases are asymptomatic, with proportions of up to 73% and 81% reported in highly exposed populations.19,20,21 Studies of seroprevalence data have estimated even higher proportions, with undiagnosed infections, with or without symptoms, making up at least 50% and up to 83% of all SARS-CoV-2 infections occurring in the general population within the first 6 months of the COVID-19 pandemic.22,23
Because most previous studies were conducted earlier in the pandemic, considerably less is known about the Omicron variant specifically. One study of adults in South Africa found high rates of asymptomatic carriage of the Omicron variant, with an approximate 23% prevalence; furthermore, in the first week after a positive PCR result, 60% of participants were asymptomatic.9 In a similar population, only 2% of those infected with pre-Omicron variants were found to be asymptomatic, suggesting a higher rate of asymptomatic infection for the Omicron variant in this population.9
The current study is a detailed assessment of Omicron variant infection awareness, which, to our knowledge, has not previously been characterized in a diverse and populous urban region in the US. By examining timing-based evidence of seroconversion, answers to a comprehensive symptoms survey, and active surveillance data, we found a relatively high level of unawareness of Omicron variant infection, which we recognize depends on both symptoms and variations in the use of diagnostic testing. The overall concordance of the results with reported rates of undiagnosed COVID-19 from pre-Omicron variants may be attributed to similarly high proportions of asymptomatic cases; although general access to testing resources have improved over time, the Omicron variant has also been associated with less severe illness compared with previous variants.24,25,26 In the present study, a 10th of the individuals who recently contracted Omicron variant infection but were unaware of this status reported interval symptoms but attributed them to a non–COVID-19 source. We found that participants who worked for Cedars-Sinai Medical Center were more likely to be aware of their recent Omicron variant infection than nonemployees. This finding may be explained not only by the differences in general health awareness and literacy between employees and nonemployees but also by the mandatory daily screening protocols for employees that are supported by COVID-19 sick pay policies, which were broadened at the institution during the Omicron variant surge. We also observed that younger and male individuals were more likely to be more aware of recent Omicron variant infection.
If these results are found to be generalizable, they could be associated with the tendency of certain individuals to develop a greater symptomatic response or more severe forms of Omicron variant infection.27,28 The extent to which self-reported awareness of Omicron variant or symptoms of infection appeared lower in older participants could be associated with worse recall in survey assessments.
Limitations
This study has several limitations. The ability to detect seroconversion may have been limited by the technical sensitivity of the assay used and by the potential delay or attenuation of IgG-N antibody response in individuals who were immunocompromised or received mRNA vaccination.29,30 Thus, the established criterion for seroconversion used in this analysis may have underestimated the true prevalence of seroconversion during the study period. The modest sample size had limited statistical power to detect multiple factors associated with Omicron variant infection awareness; larger studies could identify additional potentially important factors. We attributed seroconversion during the regional Omicron variant surge to the Omicron variant or its subvariants, although we were unable to confirm the actual strain of the virus through sequencing. In addition, seroconversion events could not be uniformly verified by concurrent or contemporary PCR testing in every case. Seventy-three percent of participants with awareness of their Omicron variant infection had positive PCR results that were contemporaneously captured in the EHR, self-reported, or both. The serology assays we used have demonstrated overall high specificity,16 and our analyses of awareness of infection, defined using different IgG-N thresholds, yielded consistent results.
A limitation encountered in similar previous studies22,31 is that the specific timing of SARS-CoV-2 infection compared with reported symptoms could not be directly verified. Because the data collection for repeated serological assays and infection surveillance was more complete than the data collection for repeated health update surveys (eTable 1 in the Supplement), we focused the current study on awareness of infection rather than on presence or absence of symptoms. Future studies with more complete serial collection of symptoms data are needed to elucidate the extent to which seroconversion events are asymptomatic or symptomatic. Given that most participants received mRNA vaccines, additional studies are needed to examine whether findings would be similar in populations who received predominantly non-mRNA vaccines. The study was conducted among employees and patients at a single health system in Los Angeles County; thus, additional studies are needed to assess the generalizability of the study results.
Conclusions
The study findings demonstrate how serological assessments can complement household4 and wastewater32 surveillance programs to understand patterns of susceptibility to emerging SARS-CoV-2 variants. This cohort study revealed that, of the individuals with evidence of seroconversion during the Omicron variant surge in Los Angeles County, most were unaware of their infectious status, a factor that has likely played a role in the surge.33 Given that unawareness of active infection precludes self-initiated interventions, such as testing and self-isolation, even modest levels of undiagnosed infection can contribute to substantial population-level transmission.34 The greater availability and use of rapid antigen testing could be associated with augmented awareness and, in turn, reduced rates of person-to-person transmission of the Omicron variant. Further studies are needed to more completely understand the factors associated with unawareness of infectious status, particularly factors that could be mitigated by available interventions.
eTable 1. Number of Monthly Electronic Health Update Surveys Completed by Participants During the Study Period
eTable 2. Characteristics of the Source Study Cohort Participants, by Inclusion/Exclusion From the Main Analyses
eTable 3. Types of Vaccination Series Received by Study Participants Prior to Omicron Infection
eTable 4. Characteristics of the Study Samples With Seroconversion From Omicron Defined Using More Conservative Pre-Seroconversion IgG-N Antibody Level Thresholds
eTable 5. Pre-existing Characteristics Associated With Awareness of COVID Infection, for the Sample With Omicron Seroconversion Defined Using a Pre-Seroconversion IgG-N Antibody Index <1.0 (N=195)
eTable 6. Pre-existing Characteristics Associated With Awareness of COVID Infection, for the Sample With Omicron Seroconversion Defined Using a Pre-Seroconversion IgG-N Antibody Index <0.4 (N=159)
eFigure. Sampling Strategy Flow Diagram
eAppendix. EMBARC Health Survey
References
- 1.Saxena SK, Kumar S, Ansari S, et al. Characterization of the novel SARS-CoV-2 Omicron (B.1.1.529) variant of concern and its global perspective. J Med Virol. 2022;94(4):1738-1744. doi: 10.1002/jmv.27524 [DOI] [PubMed] [Google Scholar]
- 2.Garcia-Beltran WF, St Denis KJ, Hoelzemer A, et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185(3):457-466.e4. doi: 10.1016/j.cell.2021.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Madewell ZJ, Yang Y, Longini IM Jr, Halloran ME, Dean NE. Household secondary attack rates of SARS-CoV-2 by variant and vaccination status: an updated systematic review and meta-analysis. JAMA Netw Open. 2022;5(4):e229317. doi: 10.1001/jamanetworkopen.2022.9317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker JM, Nakayama JY, O’Hegarty M, et al. SARS-CoV-2 B.1.1.529 (Omicron) variant transmission within households—four U.S. jurisdictions, November 2021-February 2022. MMWR Morb Mortal Wkly Rep. 2022;71(9):341-346. doi: 10.15585/mmwr.mm7109e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collie S, Champion J, Moultrie H, Bekker LG, Gray G. Effectiveness of BNT162b2 vaccine against Omicron variant in South Africa. N Engl J Med. 2022;386(5):494-496. doi: 10.1056/NEJMc2119270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petherick A, Goldszmidt R, Andrade EB, et al. A worldwide assessment of changes in adherence to COVID-19 protective behaviours and hypothesized pandemic fatigue. Nat Hum Behav. 2021;5(9):1145-1160. doi: 10.1038/s41562-021-01181-x [DOI] [PubMed] [Google Scholar]
- 7.Sayampanathan AA, Heng CS, Pin PH, Pang J, Leong TY, Lee VJ. Infectivity of asymptomatic versus symptomatic COVID-19. Lancet. 2021;397(10269):93-94. doi: 10.1016/S0140-6736(20)32651-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim EY, Choe YJ, Park H, et al. Community transmission of SARS-CoV-2 Omicron variant, South Korea, 2021. Emerg Infect Dis. 2022;28(4):898-900. doi: 10.3201/eid2804.220006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garrett N, Tapley A, Andriesen J, et al. ; Ubuntu Study Team . High asymptomatic carriage with the Omicron variant in South Africa. Clin Infect Dis. 2022;ciac237. doi: 10.1093/cid/ciac237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebinger JE, Joung S, Liu Y, et al. Demographic and clinical characteristics associated with variations in antibody response to BNT162b2 COVID-19 vaccination among healthcare workers at an academic medical centre: a longitudinal cohort analysis. BMJ Open. 2022;12(5):e059994. doi: 10.1136/bmjopen-2021-059994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ebinger JE, Fert-Bober J, Printsev I, et al. Antibody responses to the BNT162b2 mRNA vaccine in individuals previously infected with SARS-CoV-2. Nat Med. 2021;27(6):981-984. doi: 10.1038/s41591-021-01325-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Los Angeles County Department of Public Health. Delta and Omicron variants as a percentage of all specimens sequenced for baseline variant surveillance [slide]. February 24, 2022. Accessed May 15, 2022. http://www.publichealth.lacounty.gov/media/Coronavirus/docs/media/02_24_22_Slides.pdf
- 13.Falzone L, Gattuso G, Tsatsakis A, Spandidos DA, Libra M. Current and innovative methods for the diagnosis of COVID-19 infection (Review). Int J Mol Med. 2021;47(6):100. doi: 10.3892/ijmm.2021.4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebinger JE, Botwin GJ, Albert CM, et al. Seroprevalence of antibodies to SARS-CoV-2 in healthcare workers: a cross-sectional study. BMJ Open. 2021;11(2):e043584. doi: 10.1136/bmjopen-2020-043584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abbott Laboratories, Diagnostics Division. SARS-CoV-2 IgG II Quant Assay—instructions for use—FDA. December 2020. Accessed June 15, 2022. https://www.fda.gov/media/137383/download
- 16.Chew KL, Tan SS, Saw S, et al. Clinical evaluation of serological IgG antibody response on the Abbott Architect for established SARS-CoV-2 infection. Clin Microbiol Infect. 2020;26(9):1256.e9-1256.e11. doi: 10.1016/j.cmi.2020.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris PA, Taylor R, Minor BL, et al. ; REDCap Consortium . The REDCap consortium: building an international community of software platform partners. J Biomed Inform. 2019;95:103208. doi: 10.1016/j.jbi.2019.103208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap): a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377-381. doi: 10.1016/j.jbi.2008.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alene M, Yismaw L, Assemie MA, et al. Magnitude of asymptomatic COVID-19 cases throughout the course of infection: a systematic review and meta-analysis. PLoS One. 2021;16(3):e0249090. doi: 10.1371/journal.pone.0249090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Q, Liu J, Liu Q, et al. Global percentage of asymptomatic SARS-CoV-2 infections among the tested population and individuals with confirmed COVID-19 diagnosis: a systematic review and meta-analysis. JAMA Netw Open. 2021;4(12):e2137257-e2137257. doi: 10.1001/jamanetworkopen.2021.37257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Syangtan G, Bista S, Dawadi P, et al. Asymptomatic SARS-CoV-2 carriers: a systematic review and meta-analysis. Front Public Health. 2021;8:587374. doi: 10.3389/fpubh.2020.587374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalish H, Klumpp-Thomas C, Hunsberger S, et al. Undiagnosed SARS-CoV-2 seropositivity during the first 6 months of the COVID-19 pandemic in the United States. Sci Transl Med. 2021;13(601):eabh3826. doi: 10.1126/scitranslmed.abh3826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stout RL, Rigatti SJ. Seroprevalence of SARS-CoV-2 antibodies in the US adult asymptomatic population as of September 30, 2020. JAMA Netw Open. 2021;4(3):e211552-e211552. doi: 10.1001/jamanetworkopen.2021.1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wolter N, Jassat W, Walaza S, et al. Early assessment of the clinical severity of the SARS-CoV-2 Omicron variant in South Africa: a data linkage study. Lancet. 2022;399(10323):437-446. doi: 10.1016/S0140-6736(22)00017-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maisa A, Spaccaferri G, Fournier L, et al. ; regional COVID-19 investigation team; EMERGEN consortium . First cases of Omicron in France are exhibiting mild symptoms, November 2021-January 2022. Infect Dis Now. 2022;52(3):160-164. doi: 10.1016/j.idnow.2022.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menni C, Valdes AM, Polidori L, et al. Symptom prevalence, duration, and risk of hospital admission in individuals infected with SARS-CoV-2 during periods of Omicron and Delta variant dominance: a prospective observational study from the ZOE COVID Study. Lancet. 2022;399(10335):1618-1624. doi: 10.1016/S0140-6736(22)00327-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brosh-Nissimov T, Hussein K, Wiener-Well Y, et al. Hospitalized patients with severe COVID-19 during the Omicron wave in Israel - benefits of a fourth vaccine dose. Clin Infect Dis. 2022;ciac501. doi: 10.1093/cid/ciac501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auvigne V, Vaux S, Strat YL, et al. Severe hospital events following symptomatic infection with Sars-CoV-2 Omicron and Delta variants in France, December 2021-January 2022: a retrospective, population-based, matched cohort study. EClinicalMedicine. 2022;48:101455. doi: 10.1016/j.eclinm.2022.101455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Follmann D, Janes HE, Buhule OD, et al. Anti-nucleocapsid antibodies following SARS-CoV-2 infection in the blinded phase of the mRNA-1273 Covid-19 vaccine efficacy clinical trial. medRxiv. 2022:2022.2004.2018.22271936. doi: 10.1101/2022.04.18.22271936 [DOI] [PMC free article] [PubMed]
- 30.Burbelo PD, Riedo FX, Morishima C, et al. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of Severe Acute Respiratory Syndrome Coronavirus 2 in patients with Coronavirus Disease 2019. J Infect Dis. 2020;222(2):206-213. doi: 10.1093/infdis/jiaa273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogawski McQuade ET, Guertin KA, Becker L, et al. Assessment of seroprevalence of SARS-CoV-2 and risk factors associated with COVID-19 infection among outpatients in Virginia. JAMA Netw Open. 2021;4(2):e2035234-e2035234. doi: 10.1001/jamanetworkopen.2020.35234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaplan EH, Zulli A, Sanchez M, Peccia J. Scaling SARS-CoV-2 wastewater concentrations to population estimates of infection. Sci Rep. 2022;12(1):3487. doi: 10.1038/s41598-022-07523-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oran DP, Topol EJ. Prevalence of asymptomatic SARS-CoV-2 infection: a narrative review. Ann Intern Med. 2020;173(5):362-367. doi: 10.7326/M20-3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson MA, Quandelacy TM, Kada S, et al. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw Open. 2021;4(1):e2035057-e2035057. doi: 10.1001/jamanetworkopen.2020.35057 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Number of Monthly Electronic Health Update Surveys Completed by Participants During the Study Period
eTable 2. Characteristics of the Source Study Cohort Participants, by Inclusion/Exclusion From the Main Analyses
eTable 3. Types of Vaccination Series Received by Study Participants Prior to Omicron Infection
eTable 4. Characteristics of the Study Samples With Seroconversion From Omicron Defined Using More Conservative Pre-Seroconversion IgG-N Antibody Level Thresholds
eTable 5. Pre-existing Characteristics Associated With Awareness of COVID Infection, for the Sample With Omicron Seroconversion Defined Using a Pre-Seroconversion IgG-N Antibody Index <1.0 (N=195)
eTable 6. Pre-existing Characteristics Associated With Awareness of COVID Infection, for the Sample With Omicron Seroconversion Defined Using a Pre-Seroconversion IgG-N Antibody Index <0.4 (N=159)
eFigure. Sampling Strategy Flow Diagram
eAppendix. EMBARC Health Survey