Abstract
Objective:
Osteoclasts are bone-resorbing cells that play a critical role in bone homeostasis including bone remodeling and repair. Osteoclast differentiation or osteoclastogenesis occurs when receptor activator of nuclear factor B ligand (RANKL) stimulates osteoclast precursor cells such as monocyte/macrophage and induces expression of osteoclast-related genes including Nfatc1. Although transcriptional regulation of these osteoclast-related genes are well studied, the involvement of epigenetic regulation in this process is not fully understood. The objective of this study is to identify and verify the histone modifier during osteoclastogenesis.
Method:
Murine macrophage-like cell line, RAW 264.7 cells, or murine bone marrow macrophages (BMMs) were treated RANKL alone or RANKL with M-CSF, respectively, to induce osteoclast differentiation. qRT-PCR was used to screen different arrays of histone demethylases. Chromatin immunoprecipitation (ChIP) assay was used to examine occupancy of Jmjd7 in the promoter regions of different osteoclast-related genes. Jmjd7 was knocked down using siRNA. Dentin slice assay was used to evaluate bone-resorptive functions.
Result:
Among the screened histone demethylases, Jmjd7 was significantly down-regulated during osteoclast differentiation. The occupancy of Jmjd7 at the promoter regions of osteoclast-related genes was also decreased. Knockdown of Jmjd7 in RAW 264.7 cells and BMMs enhanced osteoclast differentiation and to increased the expression of osteoclast-related genes, such as c-fos, Dc-stamp, CtsK, Acp5, and Nfatc1. Bone resorptive functions of the cells were also increased.
Conclusion:
Our study shows that Jmjd7, a histone demethylase, functions as a negative regulator of osteoclastogenesis, and may be a therapeutic target of bone-related diseases.
Keywords: Jmjd7, osteoclast differentiation, RANKL, histone demethylases
INTRODUCTION
Osteoclasts are the only bone resorbing cells that play a critical role in bone homeostasis including bone remodeling and repair 1. Osteoclast formation occurs through terminal differentiation of myelogenic precursor cells, monocyte/macrophage, by two key cytokines, macrophage colony-stimulating factor (M-CSF) and the receptor activator of nuclear factor B ligand (RANKL).
M-CSF binds to its receptor c-Fms and activates receptor tyrosine kinase pathway, and it is known to mediate survival and proliferation of monocyte/macrophage. On the other hand, RANKL binds to its receptor RANK and induces the mitogen-activating kinase and Nf-kB pathways via recruitment of a series of linker proteins 2.
RANKL stimulation to osteoclast precursor cells leads to activation of transcriptional factors including nuclear factor–activated T cells (NFATc1) which is indispensable to osteoclast formation and to regulation of osteoclast-specific genes such as Jdp2 (Jun dimerization protein 2), Acp5 (acid phosphatase 5, tartrate resistant), DC-STAMP (dendritic cell specific transmembrane protein), and Ctsk (capthesin K). 3. Increasing lines of evidence suggest that osteoclast-related genes epigenetics play important regulatory roles in osteoclast formation. In particular, histone demethylase Jmjd5 has been reported to attenuate osteoclastogenesis through post-translational hydroxylation of Nfatc1, and targeting Nfatc1 for proteasomal degradation 4. Jmjd3 has been shown to bind to the TSS of Nfatc1, and its repression reduced demethylation of histone H3K27me3 at the transcription start site of Nfatc1, inhibiting osteoclast formation 5. Nonetheless, a detail mechanism of epigenetic regulation in osteoclast differentiation is not fully understood.
In searching for additional epigenetic regulators in osteoclast differentiation, we screened for the expression of different histone demethylases during the RANKL-stimulated osteoclastogenesis and report that jumonji domain containing 7 (Jmjd7) histone demethylase plays an important role in osteoclast differentiation.
MATERIALS AND METHODS
Reagents
Receptor activator of nuclear factor κB ligand (RANKL) and macrophage-colony stimulating factor (M-CSF) was purchased from R&D Systems (Minneapolis, MN, USA). -Minimum essential medium (a-MEM), gentamycin, and fetal bovine serum (FBS) were purchased from Life Technologies (Carlsbad, CA, USA). Jmjd7 antibody (sc-134351), Nfatc1 antibody, and Jmjd7 siRNA (sc-148668) were purchased from Santa Cruz Biotech (Santa Cruz, CA, USA).
Cell Culture and Osteoclast Differentiation
RAW 264.7 cell line was purchased from ATCC. Murine bone marrow macrophages (BMM) were collected from the tibia of normal mice. BMMs and RAW cells were cultured in a -MEM/10% FBS with 1% gentamycin. RAW cells were grown on 60.8 cm2 plates, and passed when cells reached 75% confluence. RAW cells were kept under 30 passages for the experiment. BMMs were cultured and used once. For osteoclast differentiation, cells were seeded at a density of 1 x 10^4 cells/ well in 6-well plates. RAW cells were differentiated into osteoclasts in the presence of RANKL (100 ng/ml) for 4-5 days to generate osteoclasts. Osteoclast formation with BMMs were accomplished with RANKL (100 ng/ml) and M-CSF (50 ng/ml) addition to the culture medium for 4-5 days. Cells were fixed with acetone and stained for tartrate resistant acid phosphatase TRAP per manufacturer’s instructions. TRAP positive cells containing more than three or more nuclei were counted as osteoclasts.
Virus generation and transduction
For the gene silencing of Jmjd7, RAW 264 cells and BMM cells were transfected with Jmjd7 siRNA by Lipofectamine RNAimax (Invitrogen) according the manufacturer's protocol. After 12 h, culture media were changed to fresh media. Cells were allowed to recover for another 12 hours at which point pellets were obtained at various time points post- transfection. Knockdown was confirmed via qPCR. For experiments that looked at osteoclastogenesis of Jmjd7 knockdowns, cells were seeded to 48-well plates at an initial density of 3.3 x 10^3 cells. Induction medium containing RANKL (100ng/ml) was added immediately after transfection medium was removed. TRAP staining was conducted on Day 4-5 post RANKL addition.
Real-time quantitative PCR
RNA was extracted from collected cell pellets via RNAeasy mini kit. (Qiagen) cDNA synthesis was completed using oligo(dTs) and random primer via SuperScript II Reverse Transcriptase (Invitrogen) to produce approximately 5 micrograms of cDNA per RNA sample. Subsequent qPCR was performed using Sybrgreen (Roche) per manufacturer’s instructions on a ABI Prism Sequence Detection System (Life Technologies). Subsequent data analysis was conducted using second derivative quantification method. All results were averaged from triplicates. Gapdh (glyceraldehyde 3-phosphate dehydrogenase) was used to control for differences in the quantity of cDNA between samples.
Western blot
Cells were washed with PBS and protein lysate was extracted using lysis buffer and centrifugation. Protein concentration was determined using the Bradford assay using the Bio-Rad protein assay dye reagent concentrate.. Protein quantification follows manufacturer’s recommendation including measurement at 595 nm and performing spectrophotometric analysis using a standard curve. Equal amounts of protein (~50ug) were loaded for SDS-Page polyacrylamide gel. The contents of the gel were transferred to a polyvinylidene fluoride (PVDF) membrane after methanol activation. The membranes were blocked with 5% skim milk in TBST, and 1:100 concentration of primary antibody was bound overnight at 4 °C.
After washing with TBST, horseradish peroxidase (HRP)-conjugated secondary antibody (Santa Cruz Biotechnology) was incubated at room temperature for 1 hour. Immuno-positive signals were detected with enhanced chemiluminescence (ELC) substrate (Promega). Gapdh served as an internal control.
Chromatin immunoprecipitation (ChIP) assay
Cells were fixed at room temperature for 10 min in the culture medium containing 1% formaldehyde, and ChIP assay was performed using the MAGnify ChIP System (Invitrogen) according to the manufacturer’s instructions. Jmjd7 antibody was coupled to Dynabeads, and the antibody-coupled dynabeads were incubated with the sheared chromatin. Chromatin-antibody-Dynabeads complexes were then washed with washing buffers, and the crosslinks were reversed in the presence of Proteinase K. The un-crosslinked DNA was purified using the DNA Purification Magnetic Beads. The purified DNA fragments were amplified using primer sets that target the promoter regions of indicated genes.
Statistical analysis.
The results are expressed as means ± standard deviation. For the comparison, the outcome measurements were compared to the control group using the Student t-test. p-values that are less than 0.05 are considered significant. * p < 0.05; ** p < 0.01; and *** p < 0.001.
RESULTS
Expression of histone demethylases during osteoclast differentiation
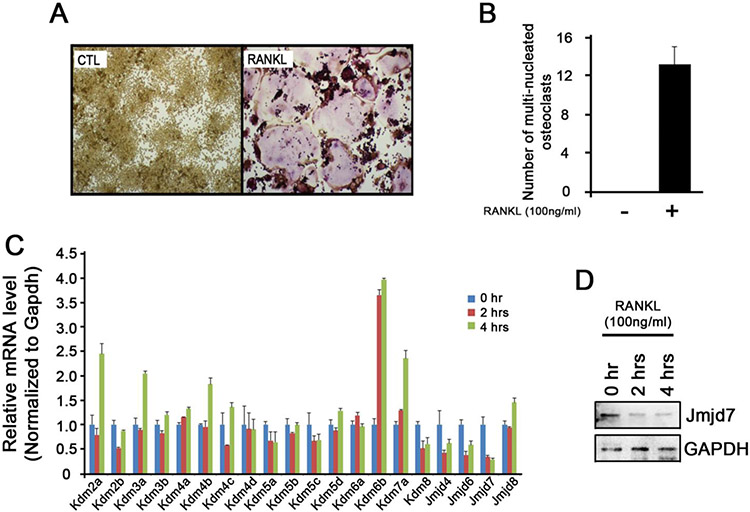
To examine the expression patterns of epigenetic regulators during osteoclast differentiation, we first confirmed that RANKL stimulation alone can induce osteoclast differentiation in RAW cells (Fig. 1A and 1B). When we treated RAW cells with RANKL for 2 and 4 hours and analyzed for a panel of 20 histone demethylases using real-time RT-PCR, we found an increased expression of Kdm6b (Jmjd3) and a decreased expression of Kdm8 (Jmjd5) expression, two of which were previously reported to play a role during osteoclast differentiation 4, 5. Interestingly, we noted that Jmjd7 expression dropped dramatically as early as 2 hours post- RANKL stimulation (Fig. 1C). Such reduction was also confirmed at the protein level (Fig. 1D), indicating that the loss of Jmjd7 expression occur during osteoclast differentiation.
Figure 1. Expression of histone demethylases during osteoclast differentiation.
(A) RAW 264.7 cells were treated with or without RANKL (100 ng/ml) for 5 days and stained for TRAP. (B) TRAP positive cells containing more than three or more nuclei were counted as osteoclasts. (C) RAW 264.7 cells were treated with RANKL (100 ng/ml) for 0, 2, and 4 hours. Cells were harvested and subjected to RNA isolation, cDNA synthesis, and qRT-PCR for the expression of different histone demethylases. (D) The same cells were subjected to Western blotting for Jmjd7 expression.
Jmjd7 selectively binds to the promoter of the genes associated with osteoclast differentiation
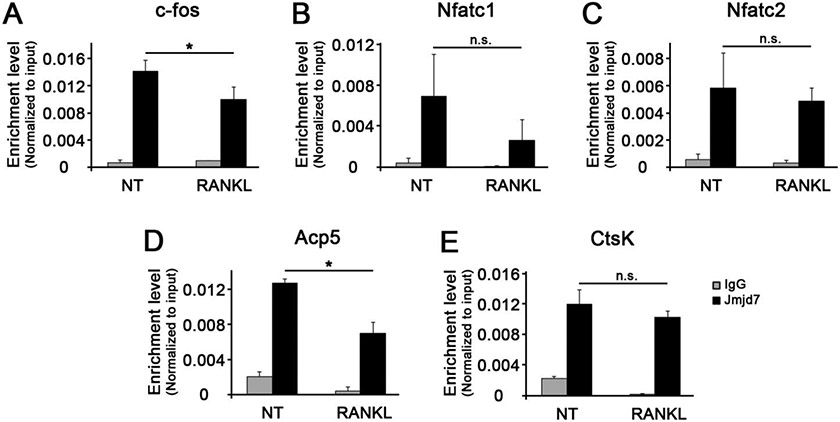
Jmjd7 is one of the members of the histone demethylase that has a highly conserved protein with JmjC domain 6. To examine the dynamics of Jmjd7 occupancy on osteoclastic gene promoters during osteoclast differentiation, we performed ChIP assay after stimulating RAW cells with RANKL for 4 hours. The occupancy of Jmjd7 on the promoters of osteoclastic genes was all significantly high in the absence of RANKL treatment, suggesting that Jmjd7 reside in the promoters of these genes in the resting state (Fig. 2A-2E, left panels). When RANKL was challenged, the occupancy of Jmjd7 was diminished only in the promoters of the selective genes (e.g., c-fos and Acp5) when others were insignificant (e.g., Nfatc1, Nfatc2, and CtsK). These data suggest that the loss of Jmjd7 occupancy during osteoclast differentiation occurs in a highly specific manner.
Figure 2. The occupancy of Jmjd7 at the osteoclast-associated gene promoters.
ChIP assay was performed for Jmjd7 occupancy at the promoters of c-fos (A), Nfatc1 (B), Nfatc2 (C), Acp5 (D), and CtsK (E).
Jmjd7 knockdown enhanced osteoclast differentiation and functions in RAW cells
To examine the functional role of Jmjd7 during osteoclast differentiation, we knocked down Jmjd7 in RAW cells using siRNA and confirmed its suppressed expression (Fig. 3A). When RAW cells were stimulated with RANKL for 4 days, the cells with Jmjd7 knockdown undergo osteoclast differentiation more readily as demonstrated by TRAP staining (Fig. 3B and 3C). Consistent with this observation, expressions of osteoclastic genes were induced more in cells with Jmjd7si when compared to those with CTLsi (Fig. 3D). Furthermore, cells with Jmjd7si induced more bone resorption as demonstrated by dentin slice assay when compared to the cells with CTLsi (Fig. 3E and 3F). These results suggest that knockdown of Jmjd7 enhances osteoclast differentiation and function.
Figure 3. Jmdj7 knockdown enhances osteoclastogenesis in RAW 264.7 cells.
(A) Control siRNA (CTLsi) and Jmjd7 siRNA (Jmjd7si) were transfected into the RAW 264.7 cells, and Jmjd7 expression was examined using qRT-PCR. (B) Transfected RAW 264.7 cells were treated with or without RANKL (100 ng/ml) for 5 days and stained for TRAP. (C) TRAP positive cells containing more than three or more nuclei were counted as osteoclasts. (D) Transfected RAW 264.7 cells were treated with or without RANKL (100 ng/ml) for 3 days. Cells were harvested and subjected to RNA isolation, cDNA synthesis, and qRT-PCR for the expression of genes associated with osteoclast differentiation. (E) Transfected RAW 264.7 cells were treated with RANKL (100 ng/ml) on the dentin slices. After 10 days, the cells were scraped off, and the dentin slices were stained with Hematoxylin. (F) The numbers of stained lacunae were quantified.
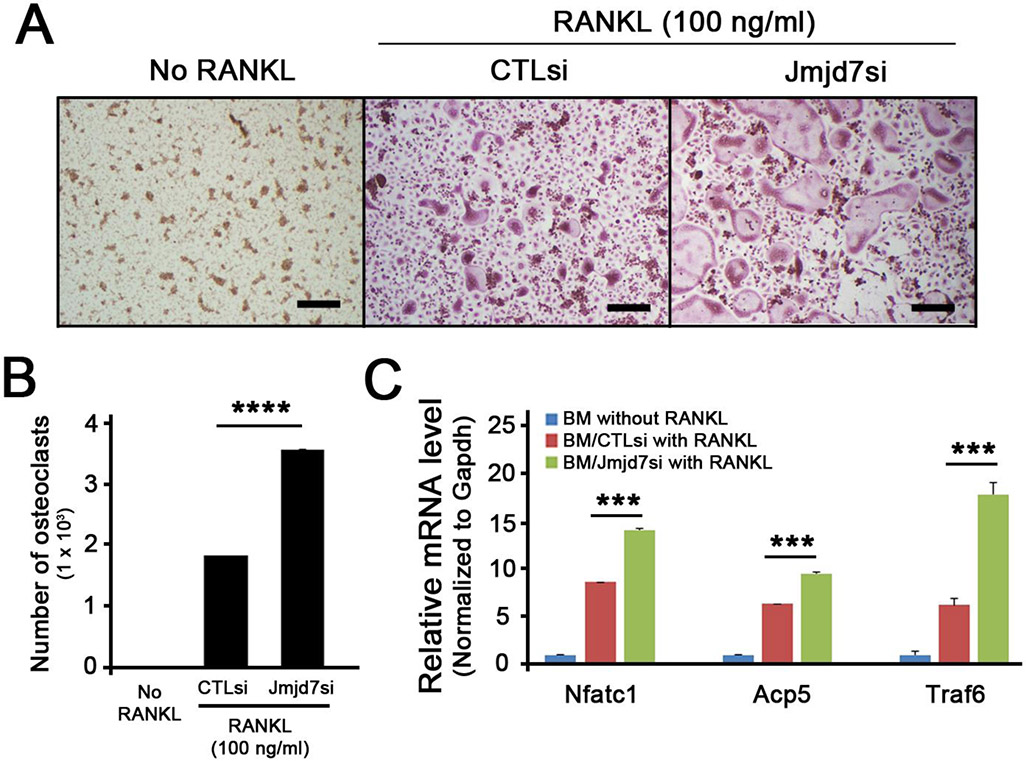
Jmjd7 knockdown enhanced osteoclast differentiation in BMM cells
To further show the effect of Jmjd7 in normal cells, we conducted similar experiments using murine bone marrow macrophages (BMMs) isolated from mice. BMMs were transfected with CTLsi or Jmjd7si, and the cells were stimulated with RANKL and M-CSF to undergo osteoclast differentiation. Similar to our previous observation, BMMs with Jmjd7si undergo osteoclast differentiation more readily than those with CTLsi (Fig. 4A and 4B). Furthermore, genes associated with osteoclast differentiation such as Nfatc1, Acp5, and Traf6 were all significantly induced in BMMs transfected with Jmjd7si when compared to those with CTLsi. These data indicates that Jmjd7 plays a functional role during osteoclast differentiation.
Figure 4. Jmdj7 knockdown enhances osteoclastogenesis in BMMs.
(A) Control siRNA (CTLsi) and Jmjd7 siRNA (Jmjd7si) were transfected into the primary BMMs. Transfected BMMs were treated with or without RANKL (100 ng/ml) in the presence of M-CSF (50 ng/ml) for 5 days and stained for TRAP. (B) TRAP positive cells containing more than three or more nuclei were counted as osteoclasts. (C) CTLsi- or Jmjd7si-transfected primary BMMs were treated with or without RANKL (100 ng/ml) in the presence of M-CSF (50 ng/ml) for 3 days. Cells were harvested and subjected to RNA isolation, cDNA synthesis, and qRT-PCR for the expression of Nfac1, Acp5, and Traf6.
DISCUSSION
The role of epigenetic regulation in osteoclasts has long been proposed based on the fact that NFATc1, the master regulator of osteoclast differentiation, is auto-amplified via its promoter binding specific to the transcription start site (TSS) of NFATc1 gene but not to the NFATc2 gene, despite the identical base sequences of both TSSs 7. Indeed, the terminal ends of histones, a group of proteins that physically binds to DNA to regulate accessibility of transcriptional machinery, are subject to transient modifications, such as acetylation, methylation, phosphorylation, and ubiquitination. These distinct modification states of histones influence the accessibility of transcription factors to the genome site, and therefore can control gene activation or repression. However, little is known about detail mechanisms of epigenetic regulation during osteoclast differentiation.
In this study, we screened for the expression of histone demethylases during osteoclast differentiation and identified Jmjd7 as one of the genes that are significantly downregulated upon RANKL stimulation. Knock down of Jmjd7 enhanced osteoclast differentiation and bone resorptive function in both RAW cells and BMMs, suggesting that Jmjd7 plays a functional role in osteoclast differentiation. To the best of our knowledge, this is the first report demonstrating that Jmjd7 plays a functional role in modulating osteoclast differentiation.
Our study showed that Jmjd7 expression decreased as early 2 hours and 4 hours post RANKL treatment at both mRNA and protein levels (Fig. 1). Such observation indicates that Jmjd7 may be an early player in the epigenetics regulation of osteoclastogenesis. Indeed, other histone demethylases such as Jmjd5 is also known to play essential roles in early event in osteoclast differentiation 4, 5, suggesting that epigenetic regulation is involved in osteoclast differentiation at the initial stages following RANKL stimulation.
When Jmjd7 was knockdown, we found that transcript levels of osteoclast differentiation genes, including Nfatc1, Jdp2, Acp5, DC-STAMP, and Ctsk, were further increased. Nfatc1 is known to regulate expression of these genes at the transcriptional level 8. Jdp2, an AP-1 family of transcription factor, promotes transcription of secretory protein such as TRAP and Ctsk 9, 10. Acp5 and DC-STAMP induce osteoclastogenesis by upregulating osteoclast specific marker TRAP 11. Interestingly, Jmjd7 occupancy on the promoters of these genes was decreased upon RANKL treatment in a selective manner; Jmjd7 occupancy was diminished only in c-fos and Acp5 but not Nfatc1, Nfatc2, and CtsK (Fig. 2). Although detail mechanisms are yet to be demonstrated, a possibility exist wherein Jmjd7, the JmjC-domain-containing histone demethylation protein 6, may regulate these gene expression at the epigenetic level in a gene-specific manner. Indeed, Jmjd7 is recently shown to have divalent cation-dependent protease functions to cleave histone tails containing methylated arginines and increase an accessibility of transcription factors 12. Further investigation is needed to clarify the differential involvement of Jmjd7 on transcriptional regulation of these genes.
Our study also showed that effect of Jmjd7 is not cell-type specific; knockdown of Jmjd7 in both RAW cells and BMMs showed enhanced osteoclast differentiation (Fig. 3 and 4). This finding underscores the involvement of Jmjd7 in osteoclast differentiation in general.
Recent studies showed that Jmjd7 is involved in cells other than osteoclasts. In prostate cancer cells, Jmjd7 plays a role in the evasion of apoptosis in prostatic cancer cells 13 where as in head and neck squamous cell carcinoma, Jmjd7 plays a role in cell proliferation and survival 14. It remains to be elucidated whether Jmjd7 also plays similar roles in osteoclasts.
REFERENCES
- 1.Boyce BF, Rosenberg E, de Papp AE, Duong le T. The osteoclast, bone remodelling and treatment of metabolic bone disease. Eur J Clin Invest 2012;42(12):1332–41. [DOI] [PubMed] [Google Scholar]
- 2.Kobayashi K, Takahashi N, Jimi E, et al. Tumor necrosis factor alpha stimulates osteoclast differentiation by a mechanism independent of the ODF/RANKL-RANK interaction. J Exp Med 2000;191(2):275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyce BF. Advances in the regulation of osteoclasts and osteoclast functions. J Dent Res 2013;92(10):860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Youn MY, Yokoyama A, Fujiyama-Nakamura S, et al. JMJD5, a Jumonji C (JmjC) domain-containing protein, negatively regulates osteoclastogenesis by facilitating NFATc1 protein degradation. J Biol Chem 2012;287(16):12994–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yasui T, Hirose J, Tsutsumi S, et al. Epigenetic regulation of osteoclast differentiation: possible involvement of Jmjd3 in the histone demethylation of Nfatc1. J Bone Miner Res 2011;26(11):2665–71. [DOI] [PubMed] [Google Scholar]
- 6.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 2006;7(9):715–27. [DOI] [PubMed] [Google Scholar]
- 7.Yasui T, Hirose J, Aburatani H, Tanaka S. Epigenetic regulation of osteoclast differentiation. Ann N Y Acad Sci 2011;1240:7–13. [DOI] [PubMed] [Google Scholar]
- 8.Neal JW, Clipstone NA. A constitutively active NFATc1 mutant induces a transformed phenotype in 3T3-L1 fibroblasts. J Biol Chem 2003;278(19):17246–54. [DOI] [PubMed] [Google Scholar]
- 9.Kawaida R, Ohtsuka T, Okutsu J, et al. Jun dimerization protein 2 (JDP2), a member of the AP-1 family of transcription factor, mediates osteoclast differentiation induced by RANKL. J Exp Med 2003;197(8):1029–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lotinun S, Kiviranta R, Matsubara T, et al. Osteoclast-specific cathepsin K deletion stimulates S1P-dependent bone formation. J Clin Invest 2013;123(2):666–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kukita T, Wada N, Kukita A, et al. RANKL-induced DC-STAMP is essential for osteoclastogenesis. J Exp Med 2004;200(7):941–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Wang C, Lee S, et al. Clipping of arginine-methylated histone tails by JMJD5 and JMJD7. Proc Natl Acad Sci U S A 2017;114(37):E7717–E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu S, Xu Y, Song M, et al. PRDM16 is associated with evasion of apoptosis by prostatic cancer cells according to RNA interference screening. Mol Med Rep 2016;14(4):3357–61. [DOI] [PubMed] [Google Scholar]
- 14.Cheng Y, Wang Y, Li J, Chang I, Wang CY. A novel read-through transcript JMJD7-PLA2G4B regulates head and neck squamous cell carcinoma cell proliferation and survival. Oncotarget 2017;8(2):1972–82. [DOI] [PMC free article] [PubMed] [Google Scholar]