Abstract
A Pasteurella haemolytica A1 gene was identified from a recombinant library clone that expressed hemolysis in host Escherichia coli cells. The gene, designated fnrP, had sequence identity to E. coli fnr, a global transcriptional regulator of genes required for conversion to anaerobic growth. FnrP complemented anaerobic deficiencies of a fnr-null mutant strain of E. coli and increased expression of the Fnr-dependent, anaerobic terminal reductase gene, frdA. FnrP was purified, identified by immunoblotting, and shown to be nonhemolytic. When FnrP was expressed in E. coli ΔsheA, a null mutant of the cryptic hemolysin SheA, the transformants were nonhemolytic, indicating that FnrP activates this silent hemolysin.
The most significant bacterial agent associated with the bovine respiratory disease complex of feedlot cattle is Pasteurella haemolytica A1 (29). Infection frequently results in severe fibrinopurulent bronchopneumonia characterized by abundant edema and fibrinocellular exudate (2). A variety of virulence factors have been described for P. haemolytica. Leukotoxin, the only described exotoxin, and lipopolysaccharide are considered to be the most important (5).
Leukotoxin is encoded on an operon composed of four genes, lktC, lktA, lktB, and lktD (12). The structural gene is lktA and requires the product of lktC for activation; the remaining genes, lktB and lktD, encode proteins that are involved in toxin secretion. Leukotoxin is hemolytic and cytotoxic to ruminant leukocytes and platelets (4, 22). A mutant strain of P. haemolytica, missing the 3′ half of lktC and all but the last 54 bp of lktA, was nonhemolytic, in contrast to the wild-type strain, which lysed bovine, ovine, and rabbit erythrocytes (17). Other strains of P. haemolytica that were unable to produce functional leukotoxin due to either insertional or deletional mutations generated within the lktC or lktA genes were also nonhemolytic (9, 26). These findings suggest that leukotoxin is responsible for the hemolytic property of P. haemolytica. Although leukotoxin appears to be the sole hemolysin of P. haemolytica, we discovered a gene, independent of the leukotoxin operon, that induces hemolysis when expressed in Escherichia coli.
Bacterial strains, growth conditions, recombinant DNA techniques, and PCR.
Strains used in this study are described in Table 1. P. haemolytica cells were grown in brain heart infusion broth (BHI; Difco Laboratories, Detroit, Mich.) or on BHI agar plates supplemented with 5% defibrinated ovine blood (Remel, Lenexa, Kans.). E. coli cells were routinely cultured in Luria-Bertani (LB) broth or on LB agar plates. Antibiotics were added as required at the following concentrations: tetracycline, 12.5 μg/ml; ampicillin, 100 μg/ml; chloramphenicol, 20 μg/ml; and kanamycin, 25 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or phenotype | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| CFP201 | MC4100 sheA::Tn5-2.1 | 7 |
| DH5α | F− φ80lacZΔM15 Δ(lacZYA-argF)U196 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ− gyrA96 relA1 | Bethesda Research Labs |
| JRG1728 | MC1000 Δ(lac)X74 galU galK rpsL Δ(ara leu) Δ(tyrR fnr rac trg) | 23 |
| JRG1787 | MC1000 Δ(lac)X74 galU galK rpsL Δ(ara leu) Δ(tyrR fnr rac trg) λ Φ(frdA′-lacZ) | 23 |
| M15 | Nals Strs Rifslac ara gal mtl recA+ uvr+ | Qiagen |
| XL1-Blue | thi-1 endA1 recA1 gyrA96 hsdR17 supE44 relA1 lac[F′ proAB lacIqZΔM15 Tn10(Tcr)] | Stratagene |
| P. haemolytica | ||
| OSU | Pathogenic isolate from an infected calf | 6 |
| Plasmids | ||
| pACYC184 | p15A replicon, ColE1 compatible, Cmr Tcr | 3 |
| pBluescript II KS(+) | ColE1 replicon, pUC19 origin, Apr | Stratagene |
| pBR322 | ColE1 replicon, Apr Tcr | 25 |
| pG4X4 | pBluescript II KS(+)::1,087-bp deletion clone of pGT3, fnrP+ | This work |
| pGS24 | pBR322::1.65-kb fnr+ fragment | 20 |
| pGT3 | pBluescript II KS(+)::1,852-bp Sau3A-XhoI P. haemolytica fragment, fnrP+ | This work |
| pGT3L | pACYC184::2,254-bp PvuII fragment of pGT3 | This work |
| pGU24 | pBluescript II KS(+)::1.65-kb HindIII-BamHI fnr+ fragment of pGS24 | This work |
| pH2 | pBluescript II KS(+)::15-kb Sau3A P. haemolytica fragment, fnrP+ | This work |
| pQE-30 | ColE1 replicon, Apr, E. coli phage T5 promoter with two lac operator sequences, pDS origin | Qiagen |
| pT7Blue | ColE1 replicon, pUC origin, t-tailed EcoRV site, Apr | Nolvagen |
P. haemolytica A1 and E. coli chromosomal DNAs were isolated as previously described (8, 30). Restriction endonucleases (Promega, Madison, Wis.), T4 DNA ligase (Promega), and calf intestine alkaline phosphatase (Boehringer Mannheim Corp., Indianapolis, Ind.) were used as recommended. Plasmids were introduced into E. coli strains and analyzed by standard methods (1). PCR was performed as described by Saiki et al. (18).
Identification of a hemolytic library clone and sequencing of fnrP.
A recombinant DNA library of P. haemolytica A1 chromosomal DNA was constructed by ligating partially digested, Sau3A fragments into the unique BamHI site of plasmid pBluescript II KS(+). The E. coli strain DH5α was transformed to ampicillin resistance with ligated DNA, and the resulting colonies were screened for hemolysis on LB agar supplemented with 5% blood. The phenotype of erythrolytic colonies was confirmed by incubating 20-μl samples of cell-free supernatants in 3-mm-diameter wells cut into LB agar containing 2.5% ovine blood. Clear zones surrounding the wells were evident after 16 h at 37°C. The plasmid pH2, containing a 15-kb insert, was isolated from a hemolytic colony. Plasmid pGT3 was constructed from pH2 by removing a 13.2-kb fragment by using XhoI sites within the vector and insert. Transfer of the 2.25-kb PvuII fragment of pGT3 to plasmid pACYC184 resulted in plasmid pGT3L. Subclones derived from plasmid pH2 all retained the ability to confer hemolysis to E. coli. Plasmid pGT3 was the smallest clone directly derived from pH2 that conferred hemolysis to E. coli.
The complete P. haemolytica insert from pGT3 was randomly labeled with digoxigenin–11-dUTP (Genius System; Boehringer Mannheim Corp.), detected by using disodium 3-(4-methoxyspiro{1,2-dioxetane-3,2′-(5′-chloro)tricyclo[3.3.1.13,7]decan}-4-yl)phenyl phosphate chemiluminescent substrate (Tropix, Bedford, Mass.), and found to hybridize to a 15-kb HindIII chromosomal fragment from P. haemolytica A1. The insert from plasmid pGT3 was sequenced in both directions by using 15 overlapping clones generated with exonuclease III (Ambion, Austin, Tex.) and mung bean nuclease (Promega) (11). Double-stranded DNA was sequenced by using Sequenase version 2.0 (United States Biochemical, Cleveland, Ohio) and 35S-labeled dATP (Dupont, Boston, Mass.). Sequence analysis used programs from the Genetics Computer Group Package version 7 and BLASTP from the Entrez Search System (National Center of Biotechnology Information).
The nucleotide sequence of the P. haemolytica chromosomal fragment from plasmid pGT3 had 1,852 bp and two open reading frames (ORF) (GenBank accession no. AF033119). A 774-bp ORF had strong sequence identity to E. coli fnr, Actinobacillus pleuropneumoniae hlyX, and other members of the fnr-like gene family. The ORF was named fnrP and has 66.1% homology with the E. coli fnr gene over a 763-bp overlap and 78.1% homology to A. pleuropneumoniae hlyX over 791 bp. A second, downstream ORF, of 455 bp, had 68.6% sequence identity over 605 bp to the E. coli envM gene that encodes short-chain alcohol dehydrogenase.
E. coli Fnr both positively and negatively regulates transcription of genes associated primarily with the transition from aerobic to anaerobic respiration (28). Fnr has a conserved C-terminal region coding for a helix-turn-helix motif involved in DNA binding and a cysteine-rich region near the N terminus that is believed to be involved in metal ion binding and capable of sensing changes in redox potential (21, 24, 27). Four cysteine residues (Cys20, 23, 29, and 122) in the Fnr protein are essential for activity (19). The comparison of the deduced amino acid sequence of FnrP with the sequences of Fnr and HlyX showed four conserved cysteine residues in FnrP (Cys19, 22, 28, and 121) and a strongly conserved region of 22 amino acids (positions 194 to 215) corresponding to the helix-turn-helix motif (19, 21). The sequence identity between FnrP and Fnr, the presence of conserved cysteine residues, and the putative DNA binding region suggest that FnrP is a member of the Fnr-like family of transcriptional regulators.
Determination of functional similarities between FnrP and Fnr.
The fnr-deficient E. coli strain JRG1728 was transformed with plasmid pG4X4, pGU24, or pBluescript II KS(+) for complementation studies (14). Plasmid pG4X4 is an exonuclease-generated sequencing clone that retains a hemolytic phenotype in JRG1728 but lacks the downstream ORF present in plasmid pGT3. Plasmid pGU24 consists of pBluescript II KS(+) with a 1.65-kb HindIII-BamHI fragment from plasmid pGS24 that encodes E. coli Fnr. Plasmid pBluescript II KS(+) was an Fnr-negative control. Bacterial growth was monitored by measuring optical density at 600 nm (OD600) and is reported as the average from three independent trials with three replicate samples per trial. Anaerobic growth of JRG1728 carrying E. coli fnr (OD600 = 0.444, standard deviation [SD] = 0.046) was significantly greater than that of the control (OD600 = 0.041, SD = 0.004) (P < 0.0001). There was no significant difference in growth between JRG1728 containing P. haemolytica fnrP (OD600 = 0.446, SD = 0.086) and JRG1728 containing E. coli fnr. Therefore, in E. coli, FnrP complemented the fnr mutation equally as well as the native gene.
The regulatory effect of FnrP on E. coli frdA was tested by using JRG1787 transformed with either plasmid pGT3L (fnrP) or plasmid pACYC184 (control). The fnr-deficient E. coli strain JRG1787 contains a lacZ chromosomal fusion with frdA, an Fnr-dependent, anaerobic terminal reductase gene. Bacteria were grown in M9 minimal media supplemented with 0.2% glycerol, 40 mM sodium fumarate, 0.1% Casamino Acids, and 0.00005% thiamine to an absorbance of 0.3 to 0.6, permeabilized with chloroform and sodium dodecyl sulfate (SDS), and tested for β-galactosidase activity (1, 13, 15). The frdA expression was measured as β-galactosidase specific activity (SA) and is reported as the average from three independent trials with three replicates per trial (15). Anaerobically grown JRG1787, carrying the recombinant fnrP, had significantly more β-galactosidase activity (SA = 332,561.1, SD = 51,390.0) than the fnr-negative control (SA = 39,678.0, SD = 5,858.4) (P < 0.0001). When cells were grown aerobically, there was no significant difference in frdA expression between JRG1787 carrying fnrP (SA = 19,139.5, SD = 3,144.6) and the JRG1787 control (SA = 16,479.8, SD = 3,455.9). These results indicate that like the Fnr protein, FnrP induces the expression of an anaerobic terminal reductase and can function as a regulatory protein that is activated during anaerobic growth.
FnrP is nonhemolytic.
Some members of the fnr-like family of genes, such as A. pleuropneumoniae hlyX, induce hemolysis when cloned into E. coli (14). Recently, HlyX was shown to activate the cryptic E. coli hemolysin HlyE (SheA) (GenBank accession no. U57430) (7, 10). To define the hemolytic properties of the pGT3 fragment, FnrP was expressed and purified by metal chelate affinity chromatography (QIAexpress System; Qiagen, Santa Clarita, Calif.). The structural gene for FnrP was amplified from plasmid pGT3 by PCR by using primers with BamHI sites on the 5′ ends (5′-GGATCCATGAAAATTGTATCAGAACCTAAAACAAGC and 5′-GGATCCGGCAAATGGAGTTAGCGGCTCATCG). The amplified products were cloned into the T-tailed site of pT7Blue (Novagen, Madison, Wis.) and transferred into the BamHI site of plasmid pQE-30 (Qiagen), thus adding six histidine residues to the amino terminus of FnrP.
Histidine-labeled FnrP was expressed in E. coli M15 carrying plasmid pQE-30::fnrP (Qiagen). This strain was hemolytic when grown on blood agar plates, indicating that additional histidine residues on the amino terminus of FnrP did not alter its hemolysin-inducing properties. Histidine-tagged FnrP was purified under both native and denaturing conditions by using Ni2+ charged Ni-nitrilotriacetic acid resin (QIAexpress kit; Qiagen). Denatured FnrP was isolated by eluting column-bound proteins with 15-ml aliquots of denaturing wash buffer (8 M urea, 0.1 M Na2HPO4, 0.01 M Tris-HCl) in a six-step gradient ranging from pH 6.5 to 4.0 (Qiagen). Denatured, purified FnrP eluted from the Ni-nitrilotriacetic acid column with a wash (pH 4.5). SDS-polyacrylamide gel electrophoresis (PAGE) analysis of this fraction revealed a solitary, 31-kDa protein. Native FnrP was isolated by the protocol for purification of native cytoplasmic proteins with the following modification: proteins were eluted in two fractions of wash buffer (50 mM Na2HPO4, 300 mM NaCl, 10% glycerol) at pH 6.0 and pH 4.0 (Qiagen). Native FnrP coeluted with contaminating proteins. Therefore, FnrP was detected immunologically by SDS-PAGE and immunoblotting.
Native FnrP was separated by SDS-PAGE and transferred onto nitrocellulose membranes (16). Membranes were sequentially reacted with a 1:2,400 dilution of FnrP polyclonal antiserum and a 1:2,000 dilution of affinity-purified horseradish peroxidase-labeled goat anti-rabbit immunoglobulin G (Kirkegaard and Perry Laboratories, Inc., Gaithersburg, Md.). 4-Chloro-1-naphthol (Bio-Rad Laboratories, Hercules, Calif.) and hydrogen peroxide (Sigma Chemical Co., St. Louis, Mo.) were added for color development. FnrP polyclonal antibodies used on the immunoblot were produced in New Zealand White rabbits by four injections, given at 10-day intervals, each containing 0.5 ml of a 1-mg/ml solution of filtered denatured protein with 0.5 ml of Freund’s incomplete adjuvant (Difco Laboratories).
On the immunoblots, anti-FnrP antibody reacted strongly with a 31-kDa protein that eluted at pH 4.0 (Fig. 1). No hemolysis was induced by the wash at pH 4.0 with solution containing FnrP when tested on blood agar plates. Proteins that eluted at pH 6.0 did not react with FnrP antibody, although eluent containing these proteins did produce a zone of hemolysis. These results show that during purification of recombinant FnrP, the fraction with hemolytic activity did not contain immunologically detectable amounts of FnrP whereas the fraction containing FnrP was not hemolytic. This finding suggests that the FnrP protein does not possess hemolytic activity but instead activates a cryptic hemolysin present in E. coli.
FIG. 1.
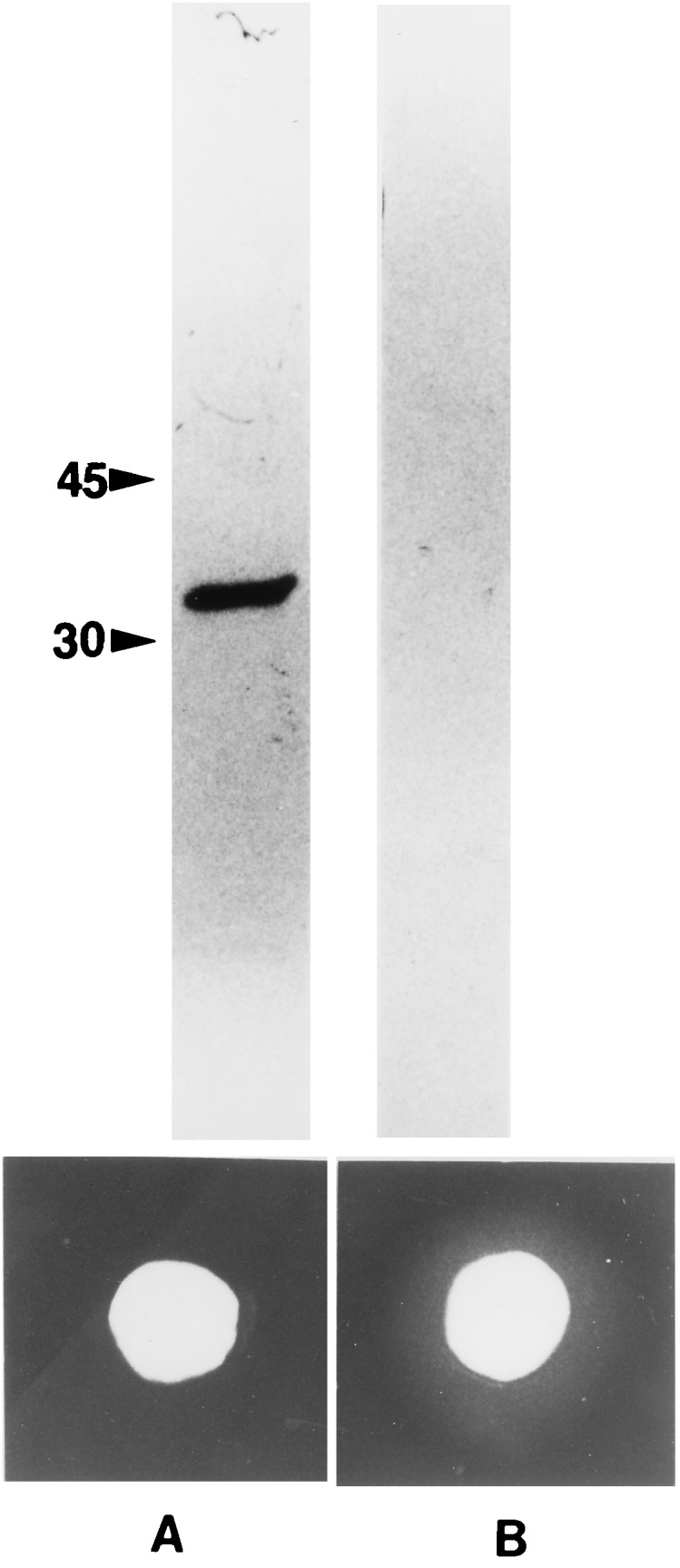
Identification of FnrP in the fractions eluted during affinity purification and their correlation with hemolysis. Top, immunoblot of the eluted column fractions collected during partial purification of FnrP. Proteins were separated by electrophoresis and blotted with anti-FnrP polyclonal antibody. Bottom, hemolytic activity of the eluted column fractions on 2.5% ovine blood agar. Column A, fraction eluted at pH 4.0; column B, fraction eluted at pH 6.0.
A sheA null mutant transformed with fnrP is nonhemolytic.
E. coli CFP201 contains a Tn5-2.1 insertion into sheA (hlyE), a gene that encodes a cryptic hemolysin in E. coli K-12 strains (7). When CFP201 was transformed with the hemolysis-inducing plasmid, pGT3 (FnrP), the resulting bacteria maintained a nonhemolytic phenotype on blood agar plates. This demonstrates activation of the silent hemolysin SheA by FnrP. In Southern hybridizations performed by using high-stringency washes at 65°C, the hlyE ORF failed to hybridize with DNA from P. haemolytica serotype 1, indicating that a SheA-like hemolysin may not be present in P. haemolytica.
FnrP appears to be a homolog of E. coli Fnr and may perform similar functions during anaerobic growth in P. haemolytica. However, like A. pleuropneumoniae hlyX, it also has the ability to induce a hemolytic phenotype in E. coli. This work indicates that E. coli SheA is the hemolytic protein activated by FnrP and suggests a possible role for FnrP in regulation of other virulence-associated genes.
Acknowledgments
We thank J. R. Guest (University of Sheffield, Sheffield, United Kingdom) for providing plasmid pGS24 and the E. coli strains JRG1728 and JRG1787. We also thank Francisco J. del Castillo (Unidad de Genetica Molecular, Hospital Ramón y Cajal, Madrid, Spain) for providing the E. coli strain CFP201. We thank Joan Rosch for assistance with manuscript preparation.
This work was supported in part by the Kansas Agricultural Experiment Station (KAES) Animal Health (Section 1433) Funds.
Footnotes
Contribution 99-409-J of the Kansas Agricultural Experiment Station.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Short protocols in molecular biology. New York, N.Y: Green Publishing Associates and John Wiley & Sons; 1992. [Google Scholar]
- 2.Breider M A, Walker R D, Hopkins F M, Schultz T W, Bowersock T L. Pulmonary lesions induced by Pasteurella haemolyticain neutrophil sufficient and neutrophil deficient calves. Can J Vet Res. 1988;52:205–209. [PMC free article] [PubMed] [Google Scholar]
- 3.Chang A C, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clinkenbeard K D, Upton M L. Lysis of bovine platelets by Pasteurella haemolyticaleukotoxin. Am J Vet Res. 1991;52:453–457. [PubMed] [Google Scholar]
- 5.Confer A W, Panciera R J, Clinkenbeard K D, Mosier D A. Molecular aspects of virulence of Pasteurella haemolytica. Can J Vet Res. 1990;54:S48–S52. [PubMed] [Google Scholar]
- 6.Corstvet R E, Panciera R J, Newman P. 21st Annual Proceedings of the American Association of Veterinary Laboratory Diagnosticians 1978. Buffalo, N.Y: The American Association of Veterinary Laboratory Diagnosticians; 1978. Vaccination of calves with Pasteurella multocida and Pasteurella haemolytica; pp. 67–90. [Google Scholar]
- 7.del Castillo F J, Leal S C, Moreno F, del Castillo I. The Escherichia coli K-12 sheAgene encodes a 34-kDa secreted haemolysin. Mol Microbiol. 1997;25:107–115. doi: 10.1046/j.1365-2958.1997.4391813.x. [DOI] [PubMed] [Google Scholar]
- 8.Dyer D W, Iandolo J J. Rapid isolation of DNA from Staphylococcus aureus. Appl Environ Microbiol. 1983;46:283–285. doi: 10.1128/aem.46.1.283-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fedorova N D, Highlander S K. Generation of targeted nonpolar gene insertions and operon fusions in Pasteurella haemolyticaand creation of a strain that produces and secretes inactive leukotoxin. Infect Immun. 1997;65:2593–2598. doi: 10.1128/iai.65.7.2593-2598.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Green J, Baldwin M L. The molecular basis for the differential regulation of the hlyE-encoded haemolysin of Escherichia coliby FNR and HlyX lies in the improved activating region 1 contact of HlyX. Microbiology. 1997;143:3785–3793. doi: 10.1099/00221287-143-12-3785. [DOI] [PubMed] [Google Scholar]
- 11.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 12.Highlander S K, Chidambaram M, Engler M J, Weinstock G M. DNA sequence of the Pasteurella haemolyticaleukotoxin gene cluster. DNA. 1989;8:15–28. doi: 10.1089/dna.1.1989.8.15. [DOI] [PubMed] [Google Scholar]
- 13.Lambden P R, Guest J R. Mutants of Escherichia coliK12 unable to use fumarate as an anaerobic electron acceptor. J Gen Microbiol. 1976;97:145–160. doi: 10.1099/00221287-97-2-145. [DOI] [PubMed] [Google Scholar]
- 14.MacInnes J I, Kim J E, Lian C-J, Soltes G A. Actinobacillus pleuropneumoniae hlyX gene homology with the fnr gene of Escherichia coli. J Bacteriol. 1990;172:4587–4592. doi: 10.1128/jb.172.8.4587-4592.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 16.Mosier D A, Simons K R, Confer A W, Panciera R J, Clinkenbeard K D. Pasteurella haemolyticaantigens associated with resistance to pneumonic pasteurellosis. Infect Immun. 1989;57:711–716. doi: 10.1128/iai.57.3.711-716.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy G L, Whitworth L C, Clinkenbeard K D, Clinkenbeard P A. Hemolytic activity of the Pasteurella haemolyticaleukotoxin. Infect Immun. 1995;63:3209–3212. doi: 10.1128/iai.63.8.3209-3212.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 19.Sharrocks A D, Green J, Guest J R. In vivo and in vitro mutants of FNR, the anaerobic transcriptional regulator of E. coli. FEBS Lett. 1990;270:119–122. doi: 10.1016/0014-5793(90)81248-m. [DOI] [PubMed] [Google Scholar]
- 20.Shaw D J, Guest J R. Amplification and product identification of the fnr gene of Escherichia coli. J Gen Microbiol. 1982;128:2221–2228. doi: 10.1099/00221287-128-10-2221. [DOI] [PubMed] [Google Scholar]
- 21.Shaw D J, Rice D W, Guest J R. Homology between CAP and Fnr, a regulator of anaerobic respiration in Escherichia coli. J Mol Biol. 1983;166:241–247. doi: 10.1016/s0022-2836(83)80011-4. [DOI] [PubMed] [Google Scholar]
- 22.Shewen P E, Wilkie B N. Cytotoxin of Pasteurella haemolyticaacting on bovine leukocytes. Infect Immun. 1982;35:91–94. doi: 10.1128/iai.35.1.91-94.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spiro S, Guest J R. Activation of the lac operon of Escherichia coliby a mutant FNR protein. Mol Microbiol. 1987;1:53–58. doi: 10.1111/j.1365-2958.1987.tb00526.x. [DOI] [PubMed] [Google Scholar]
- 24.Spiro S, Guest J R. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol Microbiol. 1988;2:701–707. doi: 10.1111/j.1365-2958.1988.tb00080.x. [DOI] [PubMed] [Google Scholar]
- 25.Sutcliffe J G. Complete nucleotide sequence of the Escherichia coliplasmid pBR322. Cold Spring Harbor Symp Quant Biol. 1979;43:77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Tatum F M, Briggs R E, Sreevatsan S S, Zehr E S, Hsuan S L, Whiteley L O, Ames T R, Maheswaran S K. Construction of an isogenic leukotoxin deletion mutant of Pasteurella haemolyticaserotype 1: characterization and virulence. Microb Pathog. 1998;24:37–46. doi: 10.1006/mpat.1997.0181. [DOI] [PubMed] [Google Scholar]
- 27.Unden G, Trageser M, Duchêne A. Effect of positive redox potentials (>+400mV) on the expression of anaerobic respiratory enzymes in Escherichia coli. Mol Microbiol. 1990;4:315–319. doi: 10.1111/j.1365-2958.1990.tb00598.x. [DOI] [PubMed] [Google Scholar]
- 28.Unden G, Becker S, Bongaerts J, Holighaus G, Schirawski J, Six S. O2-sensing and O2-dependent gene regulation in facultatively anaerobic bacteria. Arch Microbiol. 1995;164:81–90. [PubMed] [Google Scholar]
- 29.Wilkie B N, Shewen P. Defining the role that Pasteurella haemolytica plays in shipping fever. Vet. Med. 1988. pp. 1053–1058. [Google Scholar]
- 30.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F M, editor. Short protocols in molecular biology. New York, N.Y: Green Publishing Associates and John Wiley & Sons; 1992. [Google Scholar]


