Summary
Background
The annual mortality burden of antimicrobial resistant infections exceeds 1.27 million/year. With serious infections, every hour without effective antimicrobial therapy results in a 6.7% increased risk of death. New technology that delivers actionable pathology results in clinically-relevant timeframes is an urgent priority. We present the development and validation of an acoustic-enhanced flow cytometric (AFC) workflow that provides same-day confirmation of infection and antimicrobial susceptibility, using peritoneal dialysis (PD)-associated peritonitis as a demonstrative example.
Methods
In this cohort study, we analysed peritoneal dialysis effluent specimens using AFC to confirm the presence of infection and antimicrobial susceptibility of identified organisms. The primary outcome was the performance of the assay compared to conventional microbiology performed by the clinical laboratory. A secondary outcome was time to result.
Findings
AFC confirmed infection from primary specimens (n=116), with a sensitivity of 86% and specificity of 94% in ≤ one hour from arrival, including confirmation of infecting organisms in culture-negative cases. Combined with flow-cytometry-assisted antimicrobial susceptibility testing (FAST), we demonstrate same-day antimicrobial susceptibility profiles with an accuracy equivalent to conventional laboratory-based tests.
Interpretation
Application of AFC based assays to confirm infection and predict antimicrobial susceptibility can deliver actionable results with a performance that meets or exceeds currently utilised microbiological tests in clinically meaningful timeframes, as demonstrated for PD peritonitis. This technology shows potential for broad applicability to other time-critical serious infections.
Funding
Government of Western Australia (Department of Health), Government of Australia (National Health and Medical Research Council) and the Forrest Research Foundation.
Keywords: Peritoneal dialysis, PD, Peritonitis, Flow cytometry, Rapid diagnostics, Antimicrobial susceptibility
Research in context.
Evidence before this study
We searched PubMed and medRxiv for research articles between July 6th, 2020, and December 20th, 2021, with no language restrictions, using the search times “peritoneal dialysis”, “peritonitis”, “rapid diagnostic”, “confirmation of infection”, “detection”, “antimicrobial susceptibility”, “AST”, and “flow cytometry”. Current pathology practice for patients with peritoneal dialysis (PD) associated peritonitis is based on microscopy (to enumerate leucocytes present in fluids) and microbial culture, is too slow to provide actionable pathology evidence in clinically relevant timelines, and it is widely accepted that as many as 20% of peritonitis cases will be culture negative and provide no useful information for guiding clinicians. Effective antimicrobial therapy is crucial to patient and technique survival in PD peritonitis patients, however dysbiosis caused by over-use of broad-spectrum antimicrobials can be equally harmful. Rationalizing patients to targeted effective narrow-spectrum antimicrobials rapidly is a key goal of care. Attempts have been made to utilize 16S PCR for confirmation of infection have been made, but range from between 61% and 82% sensitivity, and are plagued by unacceptably high false positive rates. Cytokine based rapid detection methods (such as those measuring IL-6 and MMP8) have seen improvements for confirmation of infection, however such methods offer no prediction of antimicrobial susceptibility. Recent studies have shown the acceleration of the proliferation of antimicrobial resistance and its clinical consequences, with greater than 4 million deaths each year attributable to drug resistant infections. Expert consensus dictates that the development of rapid, phenotypic antimicrobial susceptibility tests is a key pillar in managing antimicrobial resistance. The International Society for Peritoneal Dialysis advocates for urgent development of rapid diagnostics for PD peritonitis in its peritonitis treatment guidelines.
Added value of this study
This cohort study demonstrated that flow cytometry-based diagnostics can be applied to the confirmation of peritoneal dialysis-associated peritonitis and antimicrobial susceptibility profiling of the organisms present in patient effluent. This methodological workflow is advantageous in that it provides rapid (<1 hour) confirmation of infection, and quantitative, phenotypic, same-day antimicrobial susceptibility profiles. No existing technique has demonstrated this combination of highly desirable features for PD peritonitis.
Implications of all the available evidence
Flow cytometry-assisted diagnostic tests for PD peritonitis display the required accuracy and rapidity to provide actionable diagnostic evidence for clinicians treating PD peritonitis. When employed alone, or as part of a diagnostic pipeline including cytokine-based detection methods, flow cytometry-assisted diagnostic methods have the potential to enable same-day rationalization of patients to targeted effective antimicrobial therapy. Future clinical studies must examine the realization of these potential benefits on rationalization of antimicrobial therapy, avoidance of unnecessary antibiotics, and improvements of patient outcomes.
Alt-text: Unlabelled box
Introduction
Infectious diseases are responsible for the second largest burden of ill health and 26% of all deaths worldwide.1 The O'Neill Report2 predicted that, by 2050, annual mortality associated with antimicrobial resistant infection would exceed 6 million deaths, however more recent data3 show that in 2019, that figure had already reached 4.95 million. Delays in diagnosis and commencement of appropriate treatment remain a major modifiable factor that has limited improvements in clinical outcomes. Traditional culture-based diagnostic microbiological methods are slow, with cultures usually taking 1-3 days to become positive, with many organisms either difficult or impossible to culture.4 This delay contributes to morbidity, mortality, increased health care costs and the emergence of drug-resistant microbes.5 While many new diagnostic methods are emerging,6 test performance remains variable, and these assays are often poorly suited to the analysis of more complex clinical specimens.7
End stage kidney disease affects more than 7 million people worldwide8 and in developed countries is responsible for a significant percentage of health expenditure.9 Of the two available dialysis therapies, haemodialysis, and peritoneal dialysis (PD), PD is associated with reduced costs in most countries, improved quality of life, and greater patient satisfaction.10 The major complication of PD is peritonitis, which incurs significant morbidity, mortality, and health care costs.11 PD peritonitis is also a leading cause of patient and clinician hesitancy to utilise this therapy.12 Most peritonitis episodes are caused by bacterial infections, with Gram positive organisms responsible for ∼ 65% of cases.13, 14, 15
Outcomes from peritonitis are improved with earlier administration of appropriate antibiotics,16 with empirical therapy advised by International Society of Peritoneal Dialysis (ISPD) guidelines to cover common Gram positive and Gram negative organisms.16 In up to 20% of cases no organisms can be cultured.17 Even in culture positive peritonitis, empirical therapy may be inadequate in 2% of Gram positive and 8% of Gram negative cases.15 As further decisions on therapy rely on microbiological culture and antibiotic susceptibility results being available, this information may take several days (or not be available at all in culture negative cases). This leaves clinicians reliant on extended empirical therapies, potentially contributing to antimicrobial resistance, and loss of normal commensal flora.18
Acoustic-enhanced flow cytometry (AFC) is a modification of the standard flow cytometry methodology that supports the detection of small particles (e.g., <2 µm), which in addition provides direct quantitation of particle numbers.19 We describe the validation of an AFC-based method for the rapid detection of serious infection, using PD peritonitis as a demonstrative example. Through further assessment of changes in particle size, and structure following exposure to antimicrobial agents we confirm that this method can provide a direct measure of antibiotic effectiveness. We demonstrate test performance equal to that obtained by conventional culture-based microbiological techniques with the additional benefit of unambiguous detection of culture negative peritonitis and results available within hours of sample arrival.
Methods
Ethics
Peritoneal dialysis effluent from patients in Western Australia was obtained under ethical approval (RGS0000003486 and HREC 2012-177) with written consent obtained from all patients, or for test development from spent clinical specimens in a manner exempt from ethical review. Patient demographics are presented, with comparison to ANZDATA 2019 patient cohort demographics, where possible (Table S1).
Study design
Specimens from patients with a clinical suspicion of peritonitis were submitted to the clinical pathology service for microbiological assessment. Upon specimen arrival in the laboratory, pathology staff decanted specimens, and aliquots were provided for current normal testing, and for AFC (Figure 1a). Specimens were collected in two groups: a “teaching set” to build the new diagnostic workflow, and a “testing set” to evaluate the performance of the method once validation was complete. Peritonitis was diagnosed according to ISPD guidelines. During the teaching phase (Figure 1b) samples were identified as culture positive (n=72) or culture negative peritonitis (n=16). Peritoneal dialysis effluent from in-patients with no clinical suspicion of peritonitis (n=16) was used as a negative control. In the testing phase (Figure 1c), 15 culture positive peritonitis specimens, and 5 culture negative specimens were analysed, in addition to 10 from patients without peritonitis. Causative organisms for culture positive peritonitis cases were recorded (Figure 1c) and used to select a panel of the most common organisms, and those most likely to cause severe disease for antimicrobial susceptibility testing (Figure 1d). The time to result for AFC was compared with existing pathology practice (Figure 1e).
Figure 1.
Flow Cytometry Diagnostics for Peritoneal Dialysis Study Design. a) Specimens with suspected peritonitis were submitted to the clinical pathology service. Once received, pathology staff decanted aliquots for normal processing (microscopy, culture, susceptibility testing) and for flow cytometric testing. b) 104 specimens were processed as part of our “training set”-of these 88 met the ISPD guidelines for peritonitis (72 with a causative organism cultured by routine pathology, 16 culture negative). c) 30 specimens were assayed as part of our “testing set” to demonstrate performance of the assay performed prospectively: 15 culture positive peritonitis, 5 culture negative peritonitis, and 10 from patients with clinical suspicion subsequently ruled to not have peritonitis. d) Identity of organisms cultured from peritonitis cases (CONS-Coagulase-negative Staphylococci).e) 35 PD cases were selected for rapid susceptibility testing from enrichment cultures of PDE. 25 culture positive peritonitis cases (representing the most common and most challenging organisms) were tested from the isolate, with 10 specimens tested direct from PD effluent. f) Flow cytometry-assisted Antimicrobial Susceptibility Test (FAST) returns results in 3-6 hours.
Specimen handling
Samples were aseptically decanted into 50 mL centrifuge tubes (Greiner, Kremsmünster, Austria) and specimens were allowed to settle to remove agglutinations of fibrin, then centrifuged at 200 xg for 5 minutes and supernatants retained. These supernatants were then centrifuged at 3,000 xg for 20 minutes. The pelleted material was resuspended in 0·1 µm filtered Hank's Balanced Salt Solution (HBSS - PathWest Media, PathWest Laboratory Medicine WA, Perth, Australia).
Culture results were obtained from the pathology service, having used incubation in aerobic and anerobic BD BACTEC™ (Becton, Dickinson, and Company, Franklin Lakes, New Jersey, USA) bottles per the manufacturer's instructions for use.
Antimicrobials
Antimicrobials were supplied as freeze-dried Sensititre™ 96-well well plates (Oxoid-ThermoFisher Scientific, Waltham, Massachusetts, United States), with custom formats (including 14 positive control wells) developed for FAST.
Antimicrobial exposure
Specimens were standardised to 0.5 McFarland using a nephelometer and processed as per Sensititre™ instructions. In the case where a sample, following clean-up protocol, contained too few cells, additional specimen was harvested (from the bulk fluid submitted for testing) and concentrated to allow for inoculation. 55 µL of this suspension was then added to Sensititre™ Mueller-Hinton broth and inoculated at 50 µL per well of the Sensititre™ plate using an AIM auto-inoculator (Oxoid-ThermoFisher Scientific, Waltham, Massachusetts, United States) in duplicate (i.e., one plate for FAST, one for broth microdilution-BMD). Both plates were then sealed and incubated at 37°C for 3 hours for FAST or 24 hours for BMD. BMD plates were imaged and recorded using a Sensititre™ Vizion (Oxoid- ThermoFisher Scientific, Waltham, Massachusetts, United States) to report the minimum inhibitory concentration (MICBMD). For Gram positive organisms, the following drugs and concentration ranges (inclusive) were tested: piperacillin-tazobactam (PT4: 0.25/ 4 mg/L-4 / 4 mg/L), benzyl-penicillin (PEN: 0.03 mg/L-1 mg/L), oxacillin (OXA: 0.12 mg/L-4 mg/L + 2% sodium chloride in all preparations), cefoxitin (FOX: 1 mg/L-16 mg/L), vancomycin (VAN: 0.12 mg/L-16 mg/L), teicoplanin (TEI: 0.5 mg/L-16 mg/L), gentamicin (GEN: 0.5 mg/L-8 mg/L), trimethoprim-sulfamethoxazole (SXT: 0.5 / 9.5 mg/L-32 / 608 mg/L), daptomycin (DAP: 0.25 mg/L-4 mg/L), erythromycin (ERY: 0.12 mg/L-8 mg/L), clindamycin (CLI: 0.12 mg/L-2 mg/L), amoxicillin (AMO: 0.25 mg/L-4 mg/L), linezolid (LZD: 0.5 mg/L-8 mg/L), and ceftriaxone (AXO: 0.12 mg/L-8 mg/L). For Gram negative organisms, the following drugs and concentration ranges (inclusive) were tested: amoxicillin (AMO: 4 mg/L-64 mg/L), gentamicin (GEN: 2 mg/L-32 mg/L), ciprofloxacin (CIP: 0.25 mg/L-4 mg/L), trimethoprim (TMP: 2 mg/L-32 mg/L), trimethoprim-sulfamethoxazole (SXT: 2 / 38 mg/L-32 / 608 mg/L), cefepime (FEP: 2 mg/L-32 mg/L), tigecycline (TGC: 0.25 mg/L-4 mg/L), ceftriaxone (AXO: 1 mg/L-16 mg/L), amikacin (AMI: 8 mg/L-128 mg/L), aztreonam (AZT: 2 mg/L-32 mg/L), amoxicillin-clavulanic acid (AUG: 1 / 2 mg/L-64 / 2 mg/L), piperacillin-tazobactam (PT4: 1 / 4 mg/L-128 / 4 mg/L), and meropenem (MER: 0.25 mg/L-64 mg/L).
Antimicrobial susceptibility testing quality control
Broth microdilution susceptibility results were quality controlled using the reference organisms ATCC 25922 Escherichia coli and ATCC 25923 Staphylococcus aureus were used to ensure that determinations fell within reference ranges for antimicrobials on the Gram negative and Gram positive antimicrobial exposure plates respectively.
Staining for flow cytometry
For detection and enumeration of microbial cells, enriched specimens of peritoneal dialysis effluent (PDE - as described above) were serially diluted to 10−4 in filtered HBSS. Two 1 mL aliquots were prepared: one unstained and one stained with 10µM SYTO® 9 (Life Technologies, ThermoFisher Scientific, Eugene, Oregon, USA), and 2 µL/mL Live/DEAD™ Fixable Violet viability stain (Life Technologies, ThermoFisher Scientific, Eugene, Oregon, USA). Samples were incubated in the presence of dye for ≥30 minutes in the dark before assessment by flow cytometry.
Flow cytometer setup
All data were acquired using a Attune™ NXT flow cytometer with an Attune™ plate autosampler. The instrument was configured using a 405 nm violet laser (VL1: 450/50 nm, VL2: 525/50 nm, VL3: 610/20 nm, VL4: 660/50 nm), a 488 nm blue laser (BL1: 530/30 nm, BL2: 590/40 nm, BL3: 695/40 nm), a 561 nm yellow laser (YL1: 585/16 nm, YL2: 620/15 nm, YL3: 695/40 nm, YL4: 780/60 nm), and a 637 nm red laser (RL1: 670/14 nm, RL2: 720/30 nm, RL3: 780/60 nm).
For enumeration experiments, the instrument settings were as follows: unstained - FSC 360 (Threshold 0.1×1000 AND), SSC 380, BL1 300, BL2 300, BL3 300, VL1 320, VL2 400, VL3 430, stained - FSC 360 (Threshold 0.1×1000 AND), SSC 380, BL1 300 (Threshold 0.1×1000 AND) BL2 300, BL3 300, VL1 320, VL2 400, VL3 430. The acquisition volume was set at 200 µL, measured at 25 µL per minute, and recorded for ≥25 µL.
Plate data for FAST were acquired with the following flow cytometer channel voltages: FSC 360 (Threshold 0.7×1000 AND), SSC 360 (Threshold 0.2×1000 AND), BL1 300 (Threshold 0.1×1000 AND), BL2 300, BL3 200). The acquisition volume was set at 125 µL/well, measured at 200 µL per minute, recorded for 36 µL, with one rinse between wells.
Flow cytometry gating for enumeration
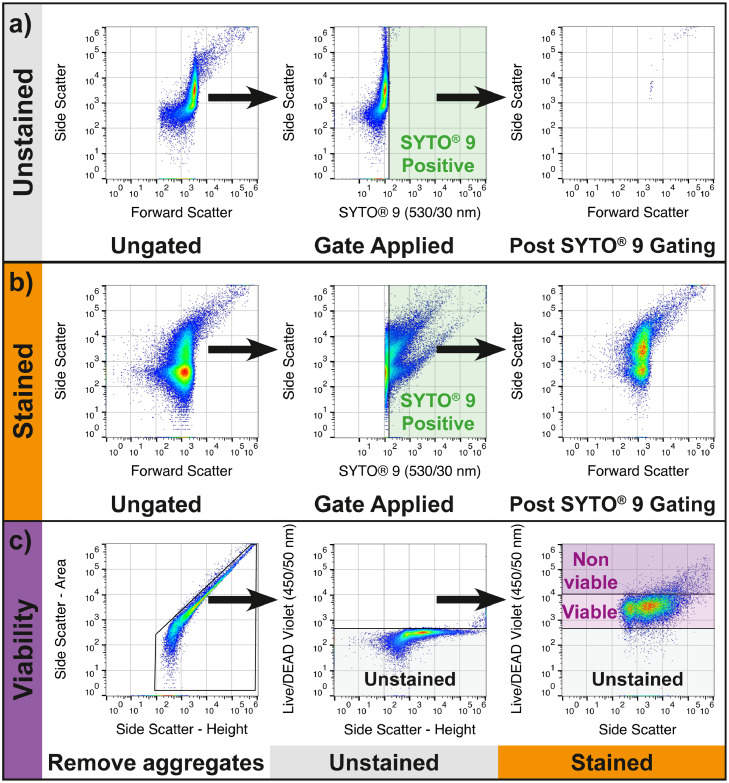
Data from stained and unstained aliquots of each PDE specimen were used to set gating in FlowJo v10.7.1 (FlowJo, Ashland, Oregon, USA). To determine positive SYTO® 9 staining, the unstained aliquot was used to define a positive staining gate (Figure 2a). This gate was applied to the stained aliquot and used to determine which events contained nucleic acid (Figure 2b). Data from the unstained specimen aliquot, and control aliquots of ATCC 25922 E. coli and ATCC 25923 S. aureus were used to set the unstained, live, and dead gates for the Live/DEAD™ Fixable Violet Viability Stain (Figure 2c). This strategy was validated using common organisms causing PD-associated peritonitis (Figure S1).
Figure 2.
Flow Cytometry Gating Workflow for Microbial Particle Enumeration. a) An unstained aliquot of the dialysis effluent specimen is measured, and a SYTO® 9 positive gate is set based on the upper limit of the unstained 488nm Ex 530/30 Em fluorescence. Sample is then backgated to ensure that no significant auto-fluorescent population remains. b) The SYTO® 9 positive gate is inherited to the data from a stained aliquot of the dialysis effluent. The specimen is backgated for demonstrative purposes (right panel). c) A bivariate plot (FSC-A vs FSC-H) is used to exclude doublets (left panel), unstained data is used to determine the threshold for Live/DEAD Violet staining (middle) and double positive gating applied to allow for determination of viability status (right).
Data analysis for flow cytometry-assisted antimicrobial susceptibility test
Flow cytometry data were analysed using FlowJo to provide susceptibility classifications as per the previously published method,20,21 with the addition of contemporaneous control wells (i.e., one well of antimicrobial unexposed bacteria per drug on the plate, measured immediately prior to the drug-exposed wells). In brief, control data were used to gate each sample in series: SYTO® 9 positive events were selected to exclude background, doublets were excluded, and distribution of the antimicrobial unexposed population was bounded using the FlowJo Autogate tool on a contour plot set at 10% (Figure S2). This gate was applied to all samples, and standardised comparisons of the events/µL falling in this gated region were used to determine the MICFAST. In brief, MICFAST was determined as first concentration in series in which there was less than a doubling of events observed in the gated region compared with the temporally nearest control well (Figure S3).
Antimicrobial susceptibility testing end-points
Susceptibility classification (i.e., is an isolate susceptible (S), susceptible with increased dose (I) or resistant (R)) was made using Clinical Laboratory Standards Institute (CLSI) M100 27th edition breakpoints. Values were deemed in essential agreement when numerical MIC values were within ± 1 dilution of each other. For categoric agreement, errors were categorised as follows: very major error-an organism classified as R by BMD was categorised as S by FAST, major error-an organism classified as S by BMD was classified as R by FAST, and minor error-an organism that is S or R by BMD is classified as I by FAST, or an organism that is classified as I by BMD is classified as S or R by FAST. Comparison of values for calculation of essential agreement and categoric agreement assumed that, in cases where both values were off scale in the same direction (i.e., both below or both above the tested range by BMD and FAST) were in treated as agreeing.
Statistics
GraphPad Prism v8·2·1 (GraphPad Software, San Diego, USA) was used to perform statistical tests. Normality and log-normality tests (Anderson-Darling, D'Agostino & Pearson, Shapiro-Wilk, Kolmogorov-Smirnov) were performed using a 0·05 significance level. Differences in cell-event enumerations between specimens (uninfected, culture negative peritonitis, culture positive peritonitis) were assessed using a Kruskal-Wallis non-parametric one-way ANOVA, with Dunn's multiple comparisons. Diagnostic sensitivity, specificity, positive predictive value, and negative predictive value were calculated defining the positive condition as “peritonitis” (i.e., agnostic to whether peritonitis was culture positive or negative) and negative as “not peritonitis”. These definitions were used to calculate a receiver-operator characteristic curve, with measures of area under the curve, standard error, and confidence interval.
Role of funders
The funders of this study had no role in study design, data collection, data analysis, interpretation, or preparation of this report.
Results
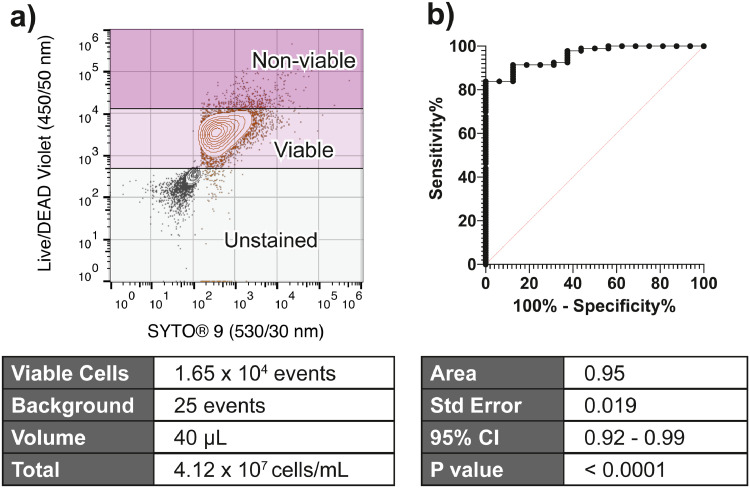
Using our AFC methodology, we quantified viable microbial “cell-like events” in PD effluent (the teaching data set - Figure 3a). We were able to classify samples as “not peritonitis”, or as “peritonitis” using the empirically determined cut-off of ≥ 9·5×104 events/mL determined by the ROC curve. The area under the ROC curve was 0·95, standard error 0·019, 95% confidence interval (0·92-0·99), P <0·001. This yielded an 84% sensitivity, 94% specificity, for the confirmation of peritonitis (95% confidence interval 0·72-1, Likelihood ratio 14 - Figure 3b).
Figure 3.
Enumerating cell-like events/mL predicts peritonitis. a) Using the gating strategy outlined in Figure 2, stained and unstained aliquots of all specimens were enumerated, and corrected for background. b) When a threshold of ≥9.5 × 104 microbial cell-like particles / mL was applied, acoustic-enhanced flow cytometry showed excellent prediction of peritonitis status (sens. 0.84, spec. 0.94, likelihood ratio 14).
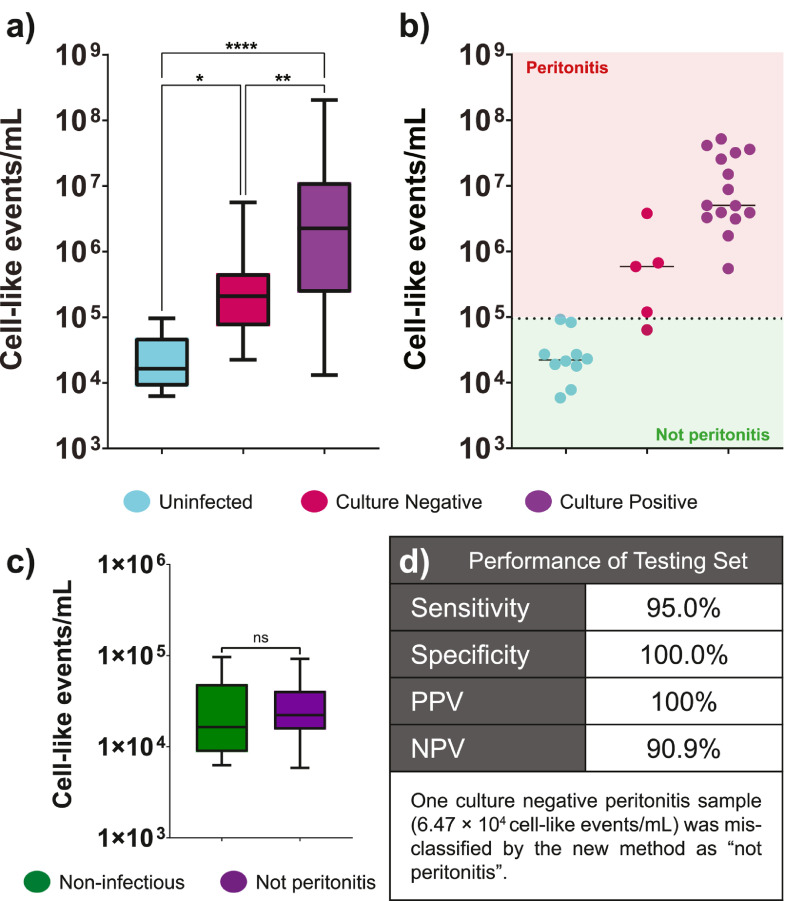
In the “training set”, there were significant differences in cell-like events/mL between specimens from culture positive, culture negative peritonitis, and uninfected specimens (Figure 4a-Kruskal–Wallis, P< 0.0001). When the threshold was applied to the “testing set” 29 out of 30 specimens were correctly classified for peritonitis status (Figure 4b). One specimen determined to be culture negative peritonitis by the clinical service was classified as “not peritonitis” by the AFC-based method. Across the 30 specimens in the “testing set” we determined sensitivity of 95·0%, specificity of 100%, positive predictive value of 100%, and negative predictive value of 90·9% for the confirmation of peritonitis (Figure 4d).
Figure 4.
Acoustic-enhanced flow cytometry can be used to confirm clinical suspicion of peritonitis. a) Using the method as outlined across a teaching set of data (n= 72 culture positive peritonitis specimens, n=16 culture negative peritonitis specimens, and n=16 non-infectious specimens taken from hospital in-patients with no clinical suspicion of peritonitis) there were significant differences in cell-like events/mL assayed for each group (Kruskal–Wallis P<0.001). b) Across a teaching set (n=15 culture positive peritonitis, n= 5 culture negative peritonitis, and n=10 specimens from patient submitted with suspicion of peritonitis but later found to be uninfected), using a cut-off value of 9.5×104 cell-like events/mL, 29 of 30 specimens were correctly classified as “peritonitis” or “not peritonitis”. c) Comparing non-infectious specimens from the teaching set with the “not peritonitis” specimens from the testing set, there were no statistically significant differences in numbers of cell-like events/mL (unpaired t-test, P= 0.94, ns). d) The testing set performance showed sensitivity and specificity in line with the teaching set optimisation (within appropriate confidence limits). Box plots, where presented, show mid-line as median, with whiskers as min-max.
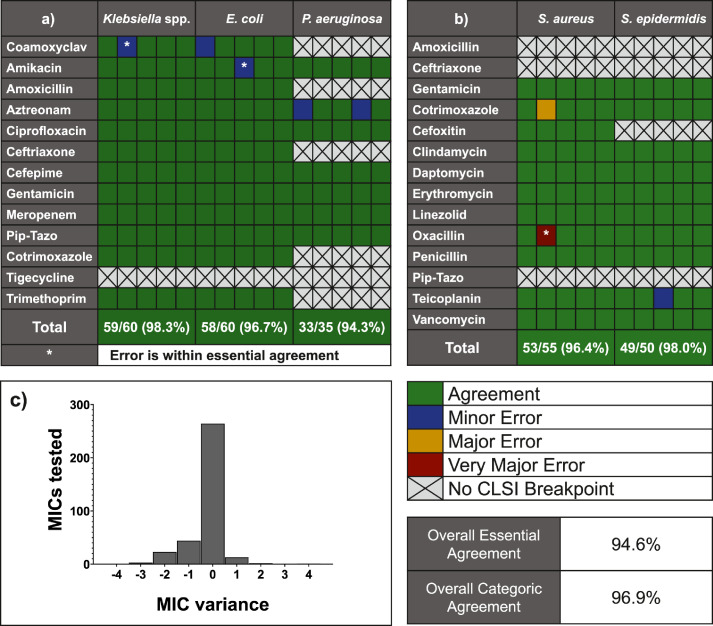
To test the application of FAST to PD-peritonitis, we selected 25 bacterial strains (5 each of Staphylococcus aureus, Staphylococcus epidermidis, Klebsiella pneumoniae, Escherichia coli, and Pseudomonas aeruginosa) isolated from PD peritonitis effluent. In brief, the underlying principle of FAST is that following a culture step in the presence of antimicrobials, organisms will display changes in biology that can be measured by AFC and converted into a prediction of MIC (Figures 5a and 5b). Each isolate was tested against 14 antimicrobials, with results available within 4 hours (Gram negative - Figure 6a, Gram positive Figure 6b) from inoculation. Results were compared with the Sensititre variant of the BMD test.
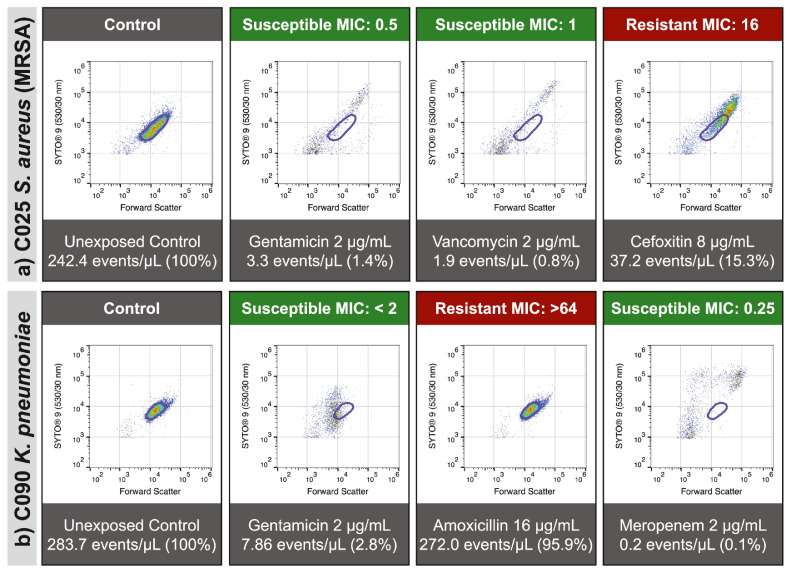
Figure 5.
Susceptibility-associated signatures are associated with antimicrobial effect. When assayed using our FAST protocol, population-level changes in multiparametric distribution can be observed as reproducible patterns of deviation from the antimicrobial unexposed population (bounded in purple). Absence of these patterns can be used to determine a lack of susceptibility (i.e., resistance) to a drug. These patterns are consistent, reproducible, and can be observed across a wide range of organisms and drugs: Staphylococcus aureus (a) and Klebsiella pneumoniae (b) are presented as demonstrative examples.
Figure 6.
Categorical agreement of Flow cytometry-assisted Antimicrobial Susceptibility Test assays performed on peritoneal dialysis-associated-peritonitis isolates with broth microdilution results. a) 5 Klebsiella spp., Escherichia coli, and P. aeruginosa were each tested against 14 antimicrobials (each column representing one isolate). For those organism-antimicrobial determinations where Clinical & Laboratory Standards Institute (CLSI) breakpoints exist, 150 of 155 results were in agreement with the reference method. b) 5 Staphylococcus aureus and Staphylococcus epidermidis species were tested against 14 antimicrobials. For those organism-antimicrobial determinations where CLSI breakpoints exist, 102 of 105 results were in agreement with the reference method. c) When comparing numerical minimum inhibitory concentration (MIC) values, 94.7% were within essential agreement (MICFAST is within ± 1 doubling dilution of the MICBMD result) between FAST and broth-microdilution methods.
Overall categoric agreement (i.e., whether both the new FAST method and the standard method classified the isolate as either sensitive, intermediate, or resistant) was 96·9% over 260 individual assay results. Numerical comparisons of minimum inhibitory concentration by FAST and by BMD showed that: 94·6% of all values were within essential agreement (±1 doubling dilution - Figure 6c). There was 1 false negative (a resistant isolate deemed sensitive by FAST (very major error), that could result in ineffective antimicrobial therapy being administered) for one S. epidermidis with oxacillin. This resulted from MIC values falling within one doubling dilution of the CLSI resistance breakpoint, noting that there was essential agreement between methods.
To further reduce the time until result availability, we also performed a series of targeted FAST assays to determine gentamicin and vancomycin susceptibility (i.e., to validate the appropriateness of the empirical antimicrobial therapy used in our institution) with direct analysis from PD effluent. We tested 10 specimens: 6 that were subsequently confirmed to be culture positive (S. epidermidis n=4, S. aureus n=1, E. coli n=1), and 4 culture negative. 100% of culture positive samples demonstrated essential and categoric agreement with broth-microdilution assays, with results available within 5 hours of sample receipt. For the four culture negative peritonitis samples no observable growth was recorded in positive control wells (rendering the control broth microdilution assay invalid) in 3, and the fourth assay returned results indicating an gentamicin resistant, vancomycin susceptible phenotype.
Discussion
We describe a methodology for the rapid confirmation of infection and prediction of antimicrobial susceptibility profiles direct from intact body site fluids, with verification of performance for patients with PD peritonitis. We chose to focus our technical demonstration on PD peritonitis for three reasons: patients can be routinely and non-invasively sampled, there are significant challenges in the microbiology of PD peritonitis (e.g., highly variable sample composition, culture negative specimens, etc.), and it is associated with high-healthcare costs. Our patient cohort was collected prospectively, with the laboratory scientists blind to the patient demographics and focused solely on the microbiology of the specimens, however data presented in Table S1 demonstrate trends that suggest our patient population samples are generally representative of the broader Australian and New Zealand peritoneal dialysis population.
As presented, our flow cytometric method has 84% sensitivity and 94% specificity for confirmation of PD peritonitis within 1 hour. While conventional teaching suggests that a much higher sensitivity is a requirement for new diagnostic tests to reduce false negatives and avoid patients failing to receive required treatment, this sensitivity represents a substantial improvement on current microbiology culture based methods (i.e., up to 20% culture negative). When combined with its high specificity, use of the workflow as presented gives confidence in a negative result. Future trials of the technology, potentially enhanced by simultaneous use of point-of care diagnostics for confirmation of the cytokine profiles of infection,22 may yield the level of confidence required to allow for commencement or cessation of empirical anti-microbial therapy within an hour of specimen receipt.
Our flow cytometric method to confirm infection gains value when paired with a FAST assay. We have demonstrated that for culture-positive peritonitis we can deliver accurate antimicrobial susceptibility profiles within 4 hours. These antimicrobial susceptibility profiles meet or exceed the required performance dictated by the most stringent international regulatory agencies.23 In addition, we have highlighted the potential that, in some cases, direct from specimen FAST assays are possible, reducing the time to result even further. Enumeration of cell-like event densities (as performed for the initial confirmation of infection) are essential in understanding the required technical steps to ensure success and should be developed further. The mixed success of the culture negative specimen testing highlights two key issues. Firstly, a lack of growth is likely to confound any rigorous phenotypic antimicrobial susceptibility test and underscores the necessity of further research into the biology of culture negative infections. Secondly, the success of the FAST assay for a culture negative specimen underscores a fundamental challenge in culture-independent diagnostic research: how to validate culture independent diagnostics for culture negative specimens when existing reference standards are all culture based?
Prevailing wisdom suggests that genotyping methods are the solution to this problem, however despite some recent advancements, nucleic acid amplification tests often fail, or have poor sensitivity/specificity in complex biological fluids. These limitations can bias results in either direction, false positives or false negatives for detection of organisms, without conclusive phenotypic (i.e., culture based) evidence to confirm. Microbial DNA is also ubiquitous in both patients and laboratory reagents,24,25 raising sampling and quality control issues that may further limit their application for diagnostic use. The power of acoustic-enhanced flow cytometry to enumerate microbial particles without culture, and quantify the viability status of those cells, avoids many of these shortcomings. Integrating flow cytometry with genotypic investigations, as is routine in many other areas of medicine, may represent a better framework for future development of new diagnostic tests.
Nucleic acid amplification tests are similarly problematic for prediction of antimicrobial susceptibility: while the presence of an antimicrobial resistance gene implies a drug resistant phenotype, it does little to provide a certain prediction of a susceptible phenotype.26 For a treating clinician, a rapid prediction of what drug will effectively treat their patient now is of greatest use: expert consensus is that quantitative phenotypic AST is necessary to support clinical decision making and currently genomic methods are not that.26
Our study has some acknowledged limitations. While sample preparation and operation of the flow cytometer can be performed by technicians of the skill level commonly found in clinical microbiology laboratories, interpretation of the cytometry data requires additional training. The need for automation of cytometry data analysis is a recognised challenge. Performance across the full spectrum of organisms that have been described to cause PD peritonitis is unknown and, as data has been generated from a pipeline that facilitates simultaneous processing by AFC/FAST in a tertiary centre, delays in sample arrival and variations in storage during transit might affect results. Historical inconsistencies observed with nucleic acid staining phenotypes across organism species and conditions, while not observed within the data presented within this study, further underscores the need for these expansive trials.27 As presented in this study, the range of organisms that cause PD peritonitis is quite limited, necessitating future studies focusing on addressing the microbiological challenges of each specific specimen type as part of a pathway to translation.
Similarly, our methods do not identify the organism/s contained within the specimen. Existing rapid solutions, such as MALDI-TOF analysis commonly used in tertiary centres, can be used to identify the organisms present in specimens. Organism identification is useful, but not strictly necessary, in selecting the format of the antimicrobial challenge: guided by the known epidemiology of the infection tested, in concert with guidelines from organisations such as CLSI28 and the European Committee on Antimicrobial Susceptibility Testing.29 Functionally, selection of an antimicrobial exposure plate in the current workflow requires only knowledge gleaned from a Gram stain (which can be performed as inocula are being prepared). AST from unknown microorganisms, or polymicrobial specimens, does not currently fit into the existing regulatory framework23 however the generation of single-cell resolution data by AFC offers exciting prospects for development of new frameworks that may be validated in future. Epidemiological concerns, while valid, should not influence the time critical challenges of commencing appropriate antimicrobial therapy and can still be tracked and quantified through processing of samples using conventional culture-based techniques.
Further validation of these technologies requires a significant broadening of scope for future studies: formal assessment of diagnostic and AST performance requires large studies, with prescriptive data acquisition strategies,30 that must be appropriately scoped and resourced. The breadth of these studies, combined with the high regulatory burden in many jurisdictions, remains a barrier to adoption to new and emerging technologies.
Our methods show potential for application to other infections and clinical sample types. While direct testing for bloodstream infections and of cerebrospinal fluids (for example) where organism numbers are low may necessitate a pre-enrichment step, quantitative cytometry protocols can still accelerate confirmation of infection and results of susceptibility tests.
In conclusion we demonstrate the potential for acoustic-enhanced flow cytometry to provide rapid confirmation of infection and AST results in PD peritonitis in time-frames that are relevant to clinical decision making. Future directions should examine the feasibility of performance by non-specialists, and the role of flow cytometry-based testing in comprehensive culture independent workflows.
Contributors
KM: Kieran Mulroney, MK: Margaret Kopczyk, CC: Christine Carson, TP: Teagan Paton, TI: Timothy Inglis, AC: Aron Chakera. Study Conceptualisation: KM, AC, TI, CC, Data Curation: KM, MK, TP, Formal Analysis: KM, MK, TP, Funding Acquisition: KM, AC, TI, CC, Investigation: KM, MK, CC, Methodology: KM, CC, AC, TI, Project Administration: KM, CC, AC, TI, Resources: KM, AC, TI, Software: KM, TP, Supervision: AC, CC, TI, Validation: KM, AC, CC, MK, TI, Visualisation: KM, Writing-Original Draft: KM, AC, Writing-Review and Editing: KM, AC, MK, TP CC, TI. KM, MK, and TP verified the underlying data used as part of this study. All authors read and approved the final version of the manuscript.
Data sharing statement
The datasets generated during and/or analysed during the current study are not publicly available due to metadata inclusion of potentially identifiable data, and for reasons of commercial sensitivity. However, redacted data are available from the corresponding author on reasonable request.
Declaration of interests
Kieran Mulroney (KM), Christine Carson (CC), Teagan Paton (TP), and Timothy Inglis (TI) declare financial interest arising from agreements pertaining to a provisional patent covering part of the work presented in this study. KM, CC, and TI declare loan of flow cytometry equipment and donation of reagents from ThermoFisher Scientific that partially supported the work presented in this study.
Acknowledgements
The authors gratefully acknowledge the provision of clinical specimens and microbiological strains by PathWest Laboratory Medicine WA. Hardware and consumable support were provided by Dr Michael Ward, of Life Technologies (ThermoFisher Scientific). The authors thank Professor Nicholas Topley for scientific and technical review of the project and manuscript. The work received funding from the Government of Western Australia, Department of Health via a Research Translation Project Grant (RTP2019R13), from the Government of Australia, National Health and Medical Research Council Ideas Grant (APP 2004298), and the Forrest Research Foundation via a Forrest Prospect Fellowship Awarded to Dr Kieran Mulroney.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104145.
Appendix. Supplementary materials
References
- 1.Pinheiro P, Mathers CD, Krämer A. In: Modern Infectious Disease Epidemiology Statistics for Biology and Health. Krämer A, Kretzschmar M, Krickeberg K, editors. Springer; New York: 2009. The global burden of infectious diseases; pp. 3–21. [Google Scholar]
- 2.O'Neill J. Tackling drug-resistant infections globally: final report and recommendations. Rev Antimicrob Resistance. 2016 https://amr-review.org/sites/default/files/160518_Final_paper_with_cover.pdf [Internet] [cited 2019 Jun 20]. Available from: [Google Scholar]
- 3.Murray CJ, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. http://www.thelancet.com/article/S01406736210272 40/fulltext [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan ZA, Siddiqui MF, Park S. Current and emerging methods of antibiotic susceptibility testing. Diagnostics. 2019;9(2):49. doi: 10.3390/diagnostics9020049. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6627445/ [Internet] [cited 2022 Jan 21] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolk DM, Dunne WM. New technologies in clinical microbiology. J Clin Microbiol [Internet]. Am Soc Microbiol J. 2011;49:62–67. http://jcm.asm.org/ [Internet] [cited 2021 May 4] Available from: [Google Scholar]
- 6.Benkova M, Soukup O, Marek J. Antimicrobial susceptibility testing: currently used methods and devices and the near future in clinical practice. J Appl Microbiol. 2020;129(4):806–822. doi: 10.1111/jam.14704. https://onlinelibrary.wiley.com/doi/full/10.1111/jam.14704 [Internet] [cited 2022 Jan 21] Available from: [DOI] [PubMed] [Google Scholar]
- 7.Anton-Vazquez V, Hine P, Krishna S, Chaplin M, Planche T. Rapid versus standard antimicrobial susceptibility testing to guide treatment of bloodstream infection. Cochrane Database Systemat Rev. 2021;5(5) doi: 10.1002/14651858.CD013235.pub2. https://pubmed.ncbi.nlm.nih.gov/34097767/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv JC, Zhang LX. Advances in Experimental Medicine and Biology. Springer; New York LLC: 2019. Prevalence and disease burden of chronic kidney disease.https://pubmed.ncbi.nlm.nih.gov/31399958/ p. 3–15. [Internet] [cited 2021 Jun 15] Available from: [DOI] [PubMed] [Google Scholar]
- 9.Cass A, White S, Snelling P, et al. The George Institute for Global Health; Melbourne: 2010. The Economic Impact of End-Stage Kidney Disease in Australia - Projections to 2020. [Google Scholar]
- 10.Li PKT, Chow KM, van de Luijtgaarden MWM, et al. Nature Reviews Nephrology. Nature Publishing Group; 2017. Changes in the worldwide epidemiology of peritoneal dialysis; pp. 90–103.www.nature.com/nrneph Vol. 13. [Internet] [cited 2021 Jun 15] Available from: [DOI] [PubMed] [Google Scholar]
- 11.Salzer WL. Peritoneal dialysis-related peritonitis: challenges and solutions. Int J Nephrol Renovasc Dis. 2018;11:173–186. doi: 10.2147/IJNRD.S123618. http://www.ncbi.nlm.nih.gov/pubmed/29928142 [Internet] [cited 2019 Jun 12] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morton RL, Snelling P, Webster AC, et al. Dialysis modality preference of patients with CKD and family caregivers: a discrete-choice study. Am J Kidney Dis. 2012;60(1):102–111. doi: 10.1053/j.ajkd.2011.12.030. https://linkinghub.elsevier.com/retrieve/pii/S0272638612001370 [Internet] [cited 2019 Jun 20] Available from: [DOI] [PubMed] [Google Scholar]
- 13.Santoianni JE, Predari SC, Verón D, Zucchini A. de Paulis a. N. A 15 year-review of peritoneal dialysis-related peritonitis: microbiological trends and patterns of infection in a teaching hospital in Argentina. Rev Argent Microbiol. 2008;40(1):17–23. [PubMed] [Google Scholar]
- 14.Vikrant S, Guleria RC, a Kanga, Verma BS, Singh D, Dheer SK. Microbiological aspects of peritonitis in patients on continuous ambulatory peritoneal dialysis. Indian J Nephrol. 2013;23(1):12–17. doi: 10.4103/0971-4065.107188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGuire A, Carson CF, Inglis T, Chakera A. Effects of a statewide protocol for the management of peritoneal dialysis-related peritonitis on microbial profiles and antimicrobial susceptibilities: a retrospective five-year review. Peritoneal Dialysis Int. 2015;35(7) doi: 10.3747/pdi.2014.00117. http://www.pdiconnect.com/content/early/2015/03/10/pdi.2014.00117.abstract Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muthucumarana K, Howson P, Crawford D, Burrows S, Swaminathan R, Irish A. The relationship between presentation and the time of initial administration of antibiotics with outcomes of peritonitis in peritoneal dialysis patients: the PROMPT study. Kidney Int Reports. 2016;1(2):65–72. doi: 10.1016/j.ekir.2016.05.003. https://www.sciencedirect.com/science/article/pii/S2468024916300092 [Internet] [cited 2019 Jun 19] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahim M, Hawley CM, McDonald SP, et al. Culture-negative peritonitis in peritoneal dialysis patients in Australia: predictors, treatment, and outcomes in 435 cases. Am J Kidney Dis. 2010;55(4):690–697. doi: 10.1053/j.ajkd.2009.11.015. https://pubmed.ncbi.nlm.nih.gov/20110144/ [Internet] [cited 2021 Jun 15] Available from: [DOI] [PubMed] [Google Scholar]
- 18.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiol. Microbiol (Reading) 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. https://pubmed.ncbi.nlm.nih.gov/20705661/ [Internet] [cited 2021 Jun 28] Available from: [DOI] [PubMed] [Google Scholar]
- 19.Ward MD, Kaduchak G. Fundamentals of acoustic cytometry. Curr Protocols Cytometry. 2018;84(1) doi: 10.1002/cpcy.36. https://pubmed.ncbi.nlm.nih.gov/30040220/ [Internet] [cited 2021 Jun 28] Available from: [DOI] [PubMed] [Google Scholar]
- 20.Inglis TJJ, Paton TF, Kopczyk MK, Mulroney KT, Carson CF. Same-day antimicrobial susceptibility test using acoustic-enhanced flow cytometry visualized with supervised machine learning. J Med Microbiol. 2020;69(5):657–669. doi: 10.1099/jmm.0.001092. https://pubmed.ncbi.nlm.nih.gov/31665100/ [Internet] [cited 2021 May 4] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mulroney KT, Hall JM, Huang X, et al. Rapid susceptibility profiling of carbapenem-resistant Klebsiella pneumoniae. Sci Reports. 2017;7(1):1903. doi: 10.1038/s41598-017-02009-3. http://www.nature.com/articles/s41598-017-02009-3 Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodlad C, George S, Sandoval S, et al. Measurement of innate immune response biomarkers in peritoneal dialysis effluent using a rapid diagnostic point-of-care device as a diagnostic indicator of peritonitis. Kidney Int. 2020;97(6):1253–1259. doi: 10.1016/j.kint.2020.01.044. [DOI] [PubMed] [Google Scholar]
- 23.The 510(k) Program: Evaluating Substantial Equivalence in Premarket Notifications [510(k)] Guidance for industry and food and drug administration staff [Internet]. [cited 2022 Jul 1]. Available from: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/510k-program-evaluating-substantial-equivalence-premarket-notifications-510k
- 24.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biology. 2014;12(1):1–12. doi: 10.1186/s12915-014-0087-z. https://link.springer.com/articles/10.1186/s12915-014-0087-z [Internet] [cited 2021 Jun 22] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwan BCH, Chow KM, Leung CB, et al. Circulating bacterial-derived DNA fragments as a marker of systemic inflammation in peritoneal dialysis. Nephrology Dialysis Transplant. 2013;28(8):2139–2145. doi: 10.1093/ndt/gft100. [DOI] [PubMed] [Google Scholar]
- 26.van Belkum A, Bachmann TT, Lüdke G, et al. Developmental roadmap for antimicrobial susceptibility testing systems. Nature Rev Microbiol. 2019;17(1):51–62. doi: 10.1038/s41579-018-0098-9. http://www.nature.com/articles/s41579-018-0098-9 [Internet] [cited 2018 Dec 24] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stiefel P, Schmidt-Emrich S, Maniura-Weber K, Ren Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO® 9 and propidium iodide. BMC Microbiol. 2015;15(1):36. doi: 10.1186/s12866-015-0376-x. http://www.biomedcentral.com/1471-2180/15/36 [Internet] [cited 2018 Sep 28] Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clinical and Laboratory Standards Institute . 29th Ed. The Clinical Laboratory Standard Institute; 2019. M100 - Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 29.The European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for the interpretation of MICs and zone diameters v 10.0. 2020. Available from: https://www.eucast.org/clinical_breakpoints/
- 30.ISO - ISO 20776-2:2021 - Clinical laboratory testing and in vitro diagnostic test systems — Susceptibility testing of infectious agents and evaluation of performance of antimicrobial susceptibility test devices — Part 2: Evaluation of performance of antimicrobial susceptibility test devices against reference broth micro-dilution [Internet]. [cited 2022 May 18]. Available from: https://www.iso.org/standard/79377.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.