SUMMARY
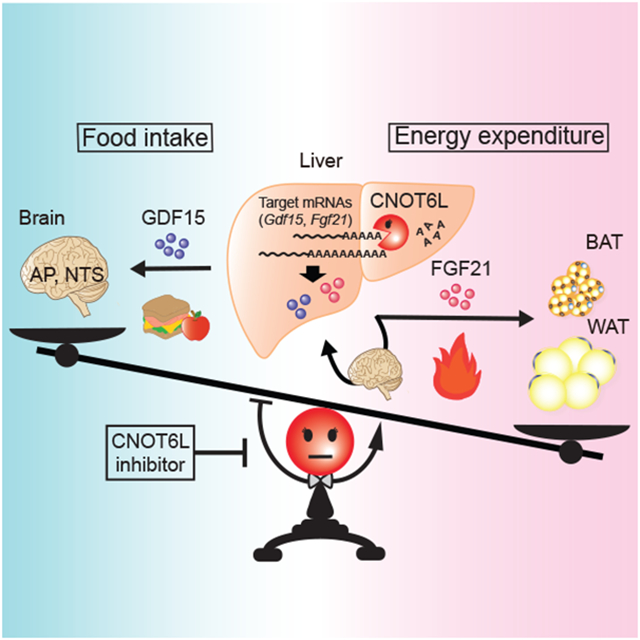
Hepatokines, secretory proteins from the liver, mediate inter-organ communication to maintain a metabolic balance between food intake and energy expenditure. However, molecular mechanisms by which hepatokine levels are rapidly adjusted following stimuli are largely unknown. Here, we unravel CNOT6L deadenylase switches off hepatokine expression after responding to the stimuli (e.g., exercise and food) to orchestrate energy intake and expenditure. Mechanistically, CNOT6L inhibition stabilizes hepatic Gdf15 and Fgf21 mRNAs, increasing corresponding serum protein levels. The resulting up-regulation of GDF15 stimulates the hindbrain to suppress appetite, while increased FGF21 affects the liver and adipose tissues to induce energy expenditure and lipid consumption. Despite the potential of hepatokines to treat metabolic disorders, their administration therapies have been challenging. Using small-molecule screening, we identified a CNOT6L inhibitor enhancing GDF15 and FGF21 hepatokine levels, which dramatically improves diet-induced metabolic syndrome. Our discovery, therefore, lays the foundation for an unprecedented strategy to treat metabolic syndrome.
Graphical Abstract
eTOC Blurb
Katsumura et al. demonstrate that expression of GDF15 and FGF21 hepatokines that mediate inter-organ communication is switched off by hepatic CNOT6L deadenylase following multiple stimuli. Hepatic CNOT6L inhibition in mice modulates food intake and energy expenditure through the brain and peripheral tissues, thereby ameliorating diet-induced obesity and metabolic disorders.
INTRODUCTION
Organokines, such as hepatokines from the liver and adipokines from adipose tissues, have been coined to indicate a group of hormones that mediate inter-organ cross-talk through the autocrine, paracrine, and/or endocrine systems (Karsenty and Olson, 2016; Meex and Watt, 2017; Priest and Tontonoz, 2019; Scheja and Heeren, 2019; Stefan and Haring, 2013; Villarroya et al., 2017). The liver is one of the most metabolically active tissues, and a large number of transcripts in the liver encode secretory proteins, including hepatokines (Lai et al., 2008; Uhlen et al., 2015). Hepatokines are abundant among organokines and maintain metabolic balance through communication from the liver to the brain and peripheral tissues (e.g., adipose tissues and muscles) (Meex and Watt, 2017; Priest and Tontonoz, 2019; Stefan and Haring, 2013). Although the transcriptional control for switching “on” expression of organokines in response to external cues (e.g., food and exercise) has been well-documented (Meex and Watt, 2017; Priest and Tontonoz, 2019; Stefan and Haring, 2013), mechanisms that switch “off” their expression are still elusive.
Growth differentiation factor 15 (GDF15) and fibroblast growth factor 21 (FGF21) are emerging as key secretory proteins stimulating the brain and peripheral tissues to decrease food intake and increase energy expenditure and lipid consumption (Flippo and Potthoff, 2021; Kliewer and Mangelsdorf, 2019; Mullican and Rangwala, 2018; Patel et al., 2019; Tsai et al., 2018). Under physiological stimuli (e.g., exercise and inflammation), GDF15 is released mainly from the liver and kidney to decrease appetite and body weight through hindbrain activation (Coll et al., 2020; Mullican and Rangwala, 2018; Tsai et al., 2018). FGF21 stimulates the brain and peripheral tissues, such as the liver and adipose tissues, in response to a wide variety of stresses (e.g., fasting and exercise) to enhance energy expenditure and lipid consumption (Flippo and Potthoff, 2021; Kliewer and Mangelsdorf, 2019; Sekiyama et al., 2019).
The CCR4-NOT deadenylase complex degrades the 3’ poly(A) tail and is regarded as the off-switch for gene expression, whereby it is thought that the 3’ poly(A) tail protects mRNAs from decay and enhances translational efficiency (Collart, 2016; Mugridge et al., 2018; Nicholson and Pasquinelli, 2019; Passmore and Coller, 2022). The CCR4-NOT complex consists of 8 subunits, including two deadenylase catalytic subunits CNOT6/CNOT6L and CNOT7/CNOT8, a scaffold protein CNOT1, and regulatory subunits CNOT2, CNOT3, CNOT9, CNOT10, and CNOT11 (Basquin et al., 2012; Bhaskar et al., 2013; Boland et al., 2013; Raisch et al., 2019; Ukleja et al., 2016). Of the two deadenylase subunits, CNOT6/CNOT6L is defined as the predominant factor for mRNA deadenylation (Webster et al., 2018; Yi et al., 2018). Deadenylation catalyzed by CNOT6/CNOT6L is thought to be the rate-limiting step of mRNA decay (Collart, 2016; Mugridge et al., 2018). Deadenylase activity of the CCR4-NOT complex is regulated by various stimuli, such as nutrients, suggesting its potential involvement in metabolic regulation and the development of metabolic disorders (Li et al., 2017; Morita et al., 2011; Morita et al., 2019; Mostafa et al., 2020; Takahashi et al., 2015; Takahashi et al., 2019; Watanabe et al., 2014). In particular, CNOT3 is increased in response to food intake and stimulates the CCR4-NOT deadenylase activity (Boland et al., 2013; Raisch et al., 2019). Recent transcriptomics studies of genetically engineered mice suggested that the CCR4-NOT deadenylase complex in the liver controls whole-body metabolic homeostasis through inter-organ communication (Morita et al., 2019; Takahashi et al., 2020).
Here, we reveal that hepatokines are post-transcriptionally turned “off” by CNOT6L following stimuli, including fasting and exercise. CNOT6L inhibition prevents hepatic Gdf15 and Fgf21 mRNA degradation, increases their serum protein levels, and thereby stimulates the brain and peripheral tissues. This leads to reduced food intake and body weight, and enhanced energy expenditure and lipid consumption. Furthermore, our findings open a hitherto unappreciated therapeutic avenue to alleviate metabolic syndrome, a cluster of conditions increasing the risk of heart disease, stroke, and other health problems. Organokines have become attractive targets for anti-obesity therapies; however, their use has been limited due to low protein stabilities, route of administration, emerging resistance, and side-effects (Kliewer and Mangelsdorf, 2019; Mullican and Rangwala, 2018; Tsai et al., 2018). As a possible solution to these limitations, we present the first-in-class CNOT6L inhibitor that increases circulating levels of hepatokines and improves metabolic syndrome. In summary, we provide mechanistic insight into hepatokine-mediated inter-organ communication and identify a new lead compound for the potential treatment of metabolic syndrome.
RESULTS
Small molecule screening with FRET-based deadenylase assay identifies the CNOT6L inhibitor
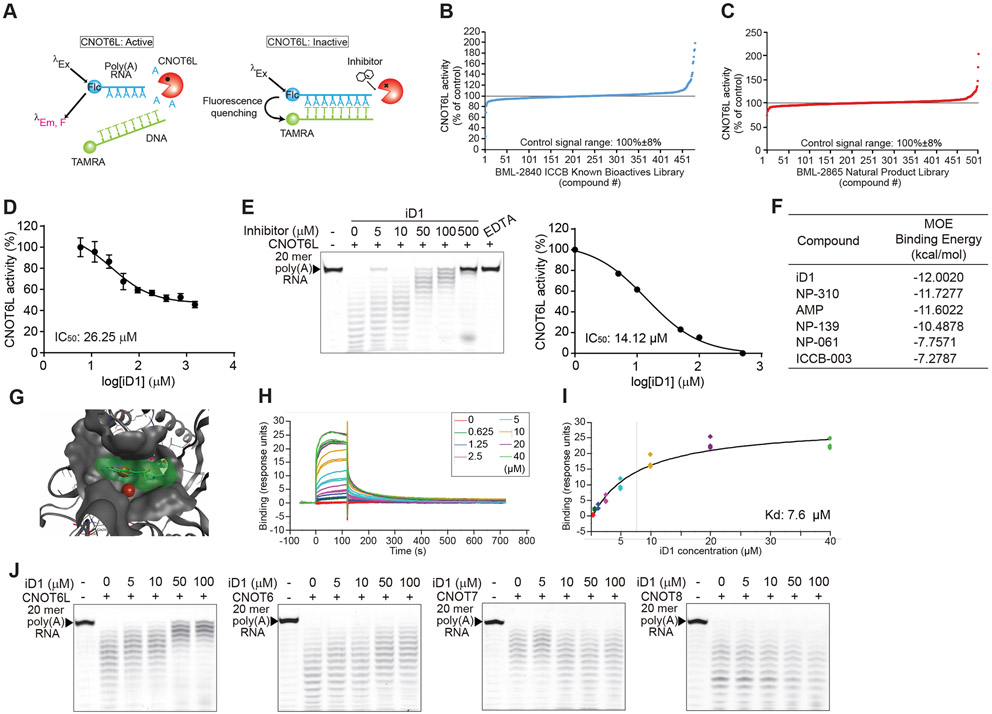
To explore potential pharmacological means to modulate CNOT6L deadenylase activity, we screened small molecule inhibitors using a high-throughput fluorescence resonance energy transfer (FRET)-based deadenylase assay (Maryati et al., 2014) (Figure 1A). To prioritize compounds evaluated in the assay, in-silico screening with the Molecular Operating Environment (MOE) software was performed on the CNOT6L crystal structure (PDB: 3NGQ and 3NGN) (Wang et al., 2010) using chemical libraries of bioactive compounds (see materials and methods). The two libraries, BML-2840 ICCB Known Bioactives Library and BML-2865 Natural Product Library, contained most compounds with low binding energies and were therefore selected for the high-throughput screening (HTS) (Figures 1B and 1C). Out of the 982 compounds screened, 8 with varying inhibitory effects on CNOT6L were selected for further analysis (Figure S1A). AMP was used as a positive control as it was previously shown to inhibit the activity of CNOT6L (Figure S1A) (Zhang et al., 2016). A dose-dependent FRET-based deadenylase assay showed five compounds possessing inhibitory activity (IC50 ≤ 1000 μM) (one compound in Figure 1D and the remaining four in Figure S1B). To validate the identified compounds as bona fide CNOT6L inhibitors, we performed a gel-based deadenylase assay. In this assay, compound ICCB-475 was identified as a more potent inhibitor of CNOT6L (IC50: 14.12 μM for 2.5 μM of CNOT6L protein) than AMP (IC50: 76.32 μM) (Figures 1E and S1C), and was termed iD1 (inhibitor of CNOT6L Deadenylase 1). Consistently, the in-silico docking model showed the lowest binding energy of iD1 to CNOT6L (Figure 1F) and mapped its binding to a cleft of the CNOT6L deadenylase domain (Figure 1G). To confirm the direct interaction of iD1 to the CNOT6L catalytic domain and measure their binding kinetics, we performed surface plasmon resonance (SPR) using recombinant CNOT6L protein. SPR demonstrated a stable interaction of iD1 with CNOT6L (Kd = 7.6 μM) (Figures 1H and 1I). To further examine the specificity of iD1 to CNOT6L, we compared its inhibitory activity toward other deadenylases in the CCR4-NOT complex, including CNOT6, CNOT7, and CNOT8, using a gel-based deadenylase assay. CNOT6, a homolog of CNOT6L, was found to be modestly inhibited by iD1 compared to CNOT6L, while iD1 did not inhibit CNOT7 or CNOT8 (Figure 1J).
Figure 1. Identification of the CNOT6L inhibitor.
(A) Schematic diagram of FRET-based deadenylase assay. When CNOT6L activity was suppressed by a compound, 6-FAM fluorescence of the intact substrate was quenched upon probe hybridization because of the proximity of the TAMRA fluorophore.
(B and C) FRET-based HTS for CNOT6L inhibitor candidates from two compound libraries, BML-2840 ICCB Known Bioactives Library (B) and BML-2865 Natural Product Library (C).
(D) FRET-based deadenylase assay showing the dose-dependent inhibition curve of CNOT6L activity with iD1. CNOT6L protein and iD1 at the indicated concentration were incubated with 6-FAM-poly(A)12 RNA and TAMRA-oligo(dT) DNA. Fluorescence intensity was measured to calculate IC50 of iD1. n = 3 per group.
(E) Gel-based deadenylase assay of CNOT6L with iD1. 2.5 μM of CNOT6L protein and iD1 at the indicated concentration were incubated with 5’-FITC-poly(A)20 RNA. Labeled RNAs were visualized on a denaturing sequencing gel (left). The dose-dependent inhibition curve of CNOT6L activity with iD1 was calculated from the left panel (right). EDTA was used as a positive control.
(F) List of the binding energy of the indicated compounds with CNOT6L. Binding energy was calculated in the MOE software.
(G) The docking model of iD1 to a cleft of the CNOT6L deadenylase domain. The docking model of the active-site pocket of CNOT6L (PDB: 3NGO) (grey) with iD1 (green) and Mg2+ (red) was built in the MOE software.
(H and I) The normalized steady-state affinity curve showing the binding of iD1 to CNOT6L protein on the sensor chip surface. Serial dilutions of iD1 were used to calculate Kd = 7.6 μM.
(J) Gel-based deadenylase assay of the indicated deadenylase with iD1. CNOT6L, CNOT6, CNOT7, or CNOT8 protein and iD1 at the indicated concentration were incubated with 5’-FITC-poly(A)20 RNA.
See also Figures S1.
The first-in-class CNOT6L inhibitor stabilizes hepatic GDF15 and FGF21 mRNAs, increasing levels of corresponding proteins in serum
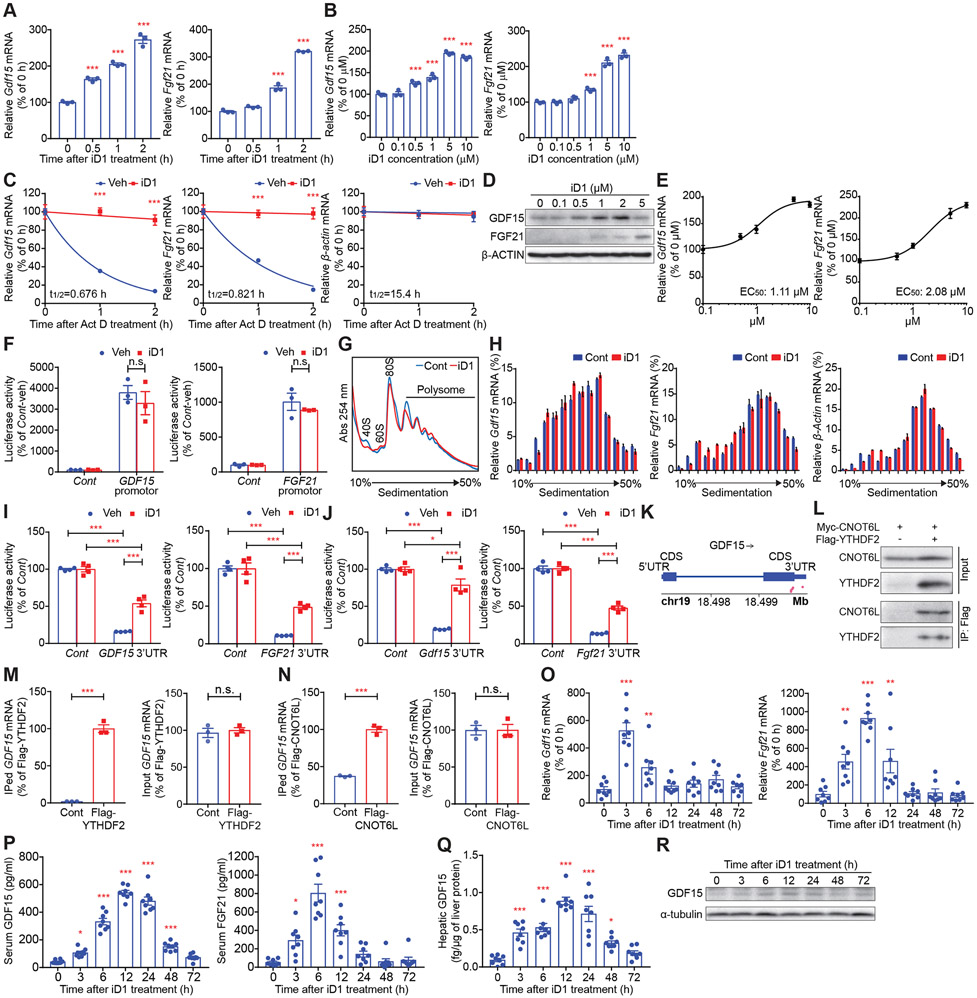
To identify hepatic secretory proteins potentially switched “off” by CNOT6L, we searched for hepatokine mRNAs bound to CNOT6L by re-analyzing our data from the RNA immunoprecipitation (RIP)-transcriptomics in mouse primary hepatocytes (Morita et al., 2019). We found 16 CNOT6L-associated mRNAs encoding proteins secreted from hepatocytes, such as GDF15 and FGF21 (Table S1). Next, we investigated the dose- and time-dependent effect of the CNOT6L inhibitor on hepatokine-encoding mRNAs in mouse primary hepatocytes and Huh7 human liver cancer cells. iD1 increased levels of hepatokine mRNAs, including GDF15, FGF21, INHBE, ADM, ADAMTS1, and HBEGF mRNAs, in a dose- and time-dependent manner (Figures 2A, 2B, and S1D-S1G). We then examined the effects of iD1 on the stability of these mRNAs following treatment with actinomycin D (Act D), an inhibitor of transcription. iD1 potently suppressed degradation of GDF15 and FGF21 mRNAs in primary hepatocytes and Huh7 cells (Figures 2C and S2A-S2C). This was in contrast to Inhbe mRNA that was partially stabilized by iD1 and other mRNAs which appeared to be unaffected (Figure S2D). These results suggest that GDF15 and FGF21 mRNAs are most likely to be direct targets downstream of CNOT6L. Consistent with the induction of GDF15 and FGF21 mRNA levels, iD1 increased GDF15 and FGF21 protein abundance (Figure 2D). The EC50 of iD1 for Gdf15 and Fgf21 mRNAs in primary hepatocytes as well as GDF15 and FGF21 mRNAs in Huh7 cells are 1.11 μM, 2.08 μM, 0.530 μM, and 1.61 μM, respectively (Figures 2E, S2E, and S2F). To exclude the possibility that iD1 also affects the promoter activity of GDF15 and FGF21, we inserted the promoter region of these genes into a luciferase plasmid and performed a luciferase reporter assay. iD1 did not alter the promoter activity of GDF15 and FGF21 genes (Figure 2F). Poly(A) tail is thought to be critical for mRNA translation as well as stability (Nicholson and Pasquinelli, 2019; Passmore and Coller, 2022). We therefore investigated the effect of iD1 on translational efficacy of hepatokine mRNAs using the polysome profiling technique (Gandin et al., 2014). iD1 did not affect global translation, as illustrated by the comparable polysomal engagement of ribosomes in control and iD1-treated samples (Figure 2G). Translational efficacy of Gdf15 and Fgf21 mRNAs was also not affected by iD1, as illustrated by the similar distribution of these mRNAs across the sucrose gradient in the presence or absence of iD1 (Figure 2H). This suggests that deadenylation is not correlated to alterations in translational efficacy in Gdf15 and Fgf21 mRNAs. To further explore the molecular mechanism of CNOT6L-dependent degradation of hepatokine mRNAs, we performed a luciferase assay using 3’ untranslated regions (UTRs) of human GDF15 and FGF21 and mouse Gdf15 and Fgf21 mRNAs (Figures 2I and 2J). The 3’ UTRs of these mRNAs decreased luciferase activities, which was reversed by the iD1 treatment (Figures 2I and 2J). This corroborates that iD1 protects Gdf15 and Fgf21 mRNAs from CNOT6L-dependent degradation. Recent epitranscriptomics studies revealed that GDF15 mRNA is N6-methylated at adenosines (m6A) (Ke et al., 2017; Wang et al., 2014b; Zaccara and Jaffrey, 2020). Analysis of an available dataset of m6A sites (Ke et al., 2017) showed five m6A sites in GDF15 mRNA (Figure 2K). An m6A RNA-binding protein, YTHDF2, was reported to interact with the CCR4-NOT complex to destabilize mRNAs (Du et al., 2016; Park et al., 2019; Zaccara and Jaffrey, 2020). We confirmed the interaction between CNOT6L and YTHDF2 (Figure 2L). Moreover, RNA-immunoprecipitation followed by quantitative PCR (RIP-qPCR) demonstrated that both YTHDF2 and CNOT6L associate with GDF15 mRNA (Figures 2M and 2N), suggesting that CNOT6L is likely to interact with YTHDF2 to recognize the 3’UTR of GDF15 mRNA for decay.
Figure 2. The CNOT6L/CNOT6 inhibitor iD1 increases levels of GDF15 and FGF21 mRNAs through their stabilization.
(A and B) Levels of the indicated mRNAs in mouse primary hepatocytes treated with iD1 at 5 μM for the indicated time (A) or at the indicated concentration for 1 h (B). mRNA levels were normalized to Hprt mRNA. n = 3 per group.
(C) Stability of the indicated mRNAs in primary hepatocytes incubated with 5 μg/mL of Act D and Veh or 5 μM of iD1 for the indicated time. mRNA levels were normalized to Hprt mRNA. n = 3 per group.
(D) Levels of GDF15 and FGF21 proteins in Huh7 cells treated with iD1 at the indicated concentration. β-ACTIN protein was used as a loading control.
(E) EC50 of iD1 for Gdf15 and Fgf21 mRNA levels in primary hepatocytes. Levels of the indicated mRNAs in primary hepatocytes treated with iD1 at the indicated concentration were normalized to Hprt mRNA. n = 3 per group.
(F) Relative luciferase activity of the Firefly luciferase construct without (Cont) or with a promotor of the human GDF15 or FGF21 gene in Huh7 cells treated with Veh or iD1. Values represent percent changes in normalized Firefly/Renilla luciferase activities. n = 3 per group.
(G and H) Polysome profiles of livers of mice treated without (Cont) or with iD1 (iD1) (G). 40S, 60S, 80S, and polysome denote the positions of corresponding ribosomal subunits, monosome, and polysome. Distribution of the indicated mRNAs in polysome profiles from (G) was determined by RT-qPCR (H). n = 3 per group.
(I and J) Relative luciferase activities of the Firefly luciferase construct without (Cont) or with the 3’UTR of the indicated human (I) or mouse (J) mRNAs in Huh7 cells treated with Veh or iD1. Values represent percent changes in normalized Firefly/Renilla luciferase activities. n = 4 per group.
(K) m6A site distribution on GDF15 mRNA was analyzed from the public data of methylation individual-nucleotide resolution using crosslinking and immunoprecipitation (miCLIP). Pink dots represent m6A sites.
(L) Interaction of Myc-CNOT6L with Flag-YTHDF2 examined by co-immunoprecipitation with anti-Flag antibody, followed by immunoblotting with anti-Myc and anti-Flag antibody.
(M and N) Association of YTHDF2 or CNOT6L with the indicated mRNA in cells transfected with Flag-YTHDF2 (M) or Flag-CNOT6L (N). Immunoprecipitated (IPed) and input mRNAs were isolated from anti-Flag immunoprecipitates and total cell lysates, respectively. Levels of IPed (left) and input (right) mRNAs were determined by qPCR. IPed mRNA levels were normalized to those of input. n = 3 per group.
(O-R) Levels of Gdf15 and Fgf21 mRNAs in livers (O), serum GDF15 and FGF21 proteins (P), and hepatic GDF15 protein (Q and R) of WT mice injected with iD1 for the indicated time. mRNA levels were normalized to Hprt mRNA. α-tubulin was used as a loading control. n = 8 per group.
Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; p values by one-way ANOVA with Dunnett's multiple comparison test for (A, B, and O-Q), two-way ANOVA with Bonferroni's multiple comparison test for (C, F, and H-J), or Student’s t-test for (M and N).
See also Figures S1 and S2.
To examine the in vivo effect of iD1 on hepatokine levels, we performed one-time intraperitoneal (IP) administration of iD1 into wild-type (WT) C57BL/6J mice (Figure S2G). iD1 significantly increased levels of hepatic Gdf15 and Fgf21 mRNAs and serum protein levels of GDF15 and FGF21 3 h after iD1 injection (Figures 2O and 2P). Consistently, levels of GDF15 protein in livers were up-regulated in response to iD1 administration (Figures 2Q and 2R). Gdf15 and Fgf21 mRNAs were reported to be induced by high-intensity exercise through the activating transcription factor 4 (ATF4), followed by degradation via unknown mechanisms within 3 h after exercise in mice and humans (Kim et al., 2013; Klein et al., 2021; Kleinert et al., 2018; Patel et al., 2019). To interrogate the effects of iD1 on preventing hepatokine mRNA decay under physiological conditions, we exposed WT mice to intense treadmill running and recapitulated degradation of Gdf15 and Fgf21 mRNAs during the post-exercise (Figure S2H). High-intensity endurance exercise increased hepatic Gdf15 and Fgf21 as well as Atf4 mRNAs (Figure S2I). Importantly, iD1 treatment attenuated degradation of Gdf15 and Fgf21 mRNAs during the 3 h post-exercise recovery period, while not altering Atf4 mRNA levels (Figure S2I). This is consistent with the above findings that iD1 prevents Gdf15 and Fgf21 mRNA degradation while not affecting transcription of corresponding genes. To investigate whether high-intensity exercise also affects the CCR4-NOT deadenylase complex, we examined levels of its components. In contrast to the induction of ATF4 transcription factor, high-intensity exercise decreased hepatic CNOT3, an activator of CCR4-NOT deadenylase (Raisch et al., 2019), whereas CNOT3 was up-regulated during the recovery period (Figure S2J). Exercise did not alter levels of CNOT1 scaffold protein and CNOT6L deadenylase (Figure S2J). These data suggest that Gdf15 and Fgf21 mRNAs are increased during exercise through transcriptional activation and suppression of their degradation. High-intensity endurance exercise provokes a series of perturbations, such as inflammation and energetic stress (Hawley et al., 2014; Sanford et al., 2020). To this end, we next assessed other stimuli reported to affect GDF15 and FGF21 levels. Fgf21 mRNA in the liver is known to be induced by fasting and rapidly suppressed after refeeding (Figure S2K) (Fisher et al., 2017; Inagaki et al., 2007; Kliewer and Mangelsdorf, 2019). Importantly, iD1 attenuated Fgf21 mRNA suppression induced by refeeding (Figure S2K). In contrast, hepatic CNOT3 protein was decreased by fasting and induced by refeeding, while CNOT1 and CNOT6L were not affected by fasting and refeeding (Figure S2L). Gdf15 mRNA was reported to be induced by inflammatory stimuli, such as poly(I:C) (Luan et al., 2019). Administration of poly(I:C) increased the hepatic Gdf15 mRNA, which was cleared 3 h after the induction (Figure S2M). iD1 slowed the clearance of Gdf15 mRNA, but did not alter Atf4 mRNA levels in the liver (Figure S2M). To further examine the relationship between transcription and degradation of Gdf15 and Fgf21 mRNAs, we treated Huh7 cells with a combination of iD1 with compounds that induce GDF15 or FGF21 transcription, including an endoplasmic reticulum (ER) stress inducer, thapsigargin (Tg), and a PPARα agonist, WY-14643 (WY) (Inagaki et al., 2008; Patel et al., 2019). As reported previously, thapsigargin and WY-14643 increased GDF15 and FGF21 mRNAs, respectively (Figures S2N and S2O). Significantly, iD1 potentiated the effects of thapsigargin and WY-14643 on bolstering GDF15 and FGF21 mRNA levels without affecting ATF4 expression (Figures S2N and S2O). Collectively, these data support a model whereby GDF15 and FGF21 hepatokines are turned on by transcription factors while being post-transcriptionally switched off by CNOT6L deadenylase in response to a variety of external stimuli.
CNOT6L inhibition decreases body weight by reducing food intake and enhancing energy expenditure and fat utilization
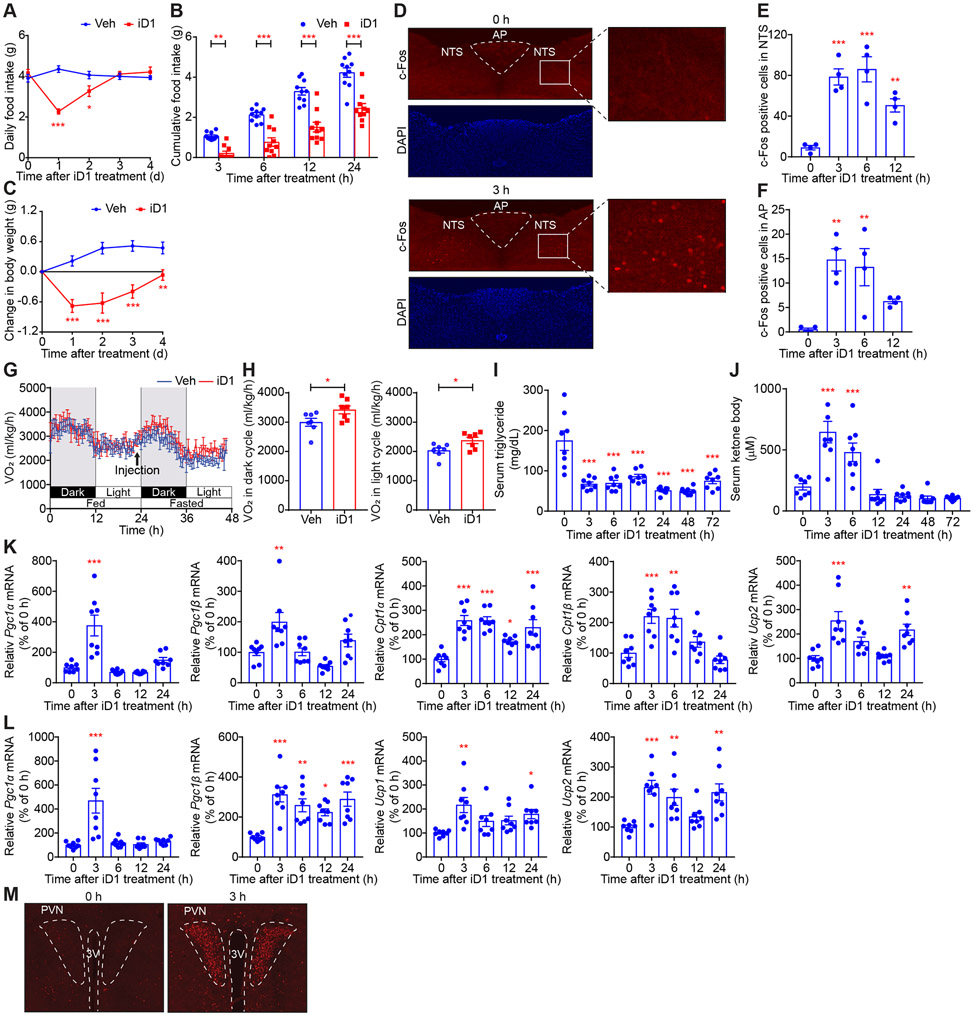
GDF15 suppresses food intake and induces body weight loss (Mullican and Rangwala, 2018; Tsai et al., 2018). Consistent with the induced expression of GDF15 in the iD1-treated mice, a single administration of iD1 resulted in a 55% decrease in food intake after 1 day compared to vehicle (Veh)-treated mice (Figures 3A and 3B). Notably, body weight was significantly reduced after administration of iD1 (Figure 3C). The EC50 of iD1 for food intake and body weight were 1.43 and 1.23 mg/kg, respectively (Figures S3A-S3D). The effects of 2 mg/kg of iD1 on food intake and body weight lasted for 2 and 3 days, respectively (Figures S3A-S3D). GDF15 has been reported to activate neurons in the nucleus tractus solitaries (NTS) and the area postrema (AP) of the hindbrain to decrease appetite, as evidenced by an increase in c-Fos positive cells (Emmerson et al., 2017; Hsu et al., 2017; Mullican et al., 2017; Yang et al., 2017). Consistent with this, robust c-Fos staining within NTS and AP was detected 3 h after iD1 administration (Figures 3D-3F).
Figure 3. CNOT6L controls food intake, energy expenditure, and lipid metabolism through regulation of hepatokines.
(A-C) Daily food intake (A), cumulative food intake for the indicated time (B), and change in body weight (C) after a single administration of Veh or iD1 in WT mice. n = 10 per group.
(D-F) Immunofluorescence (IF) staining of c-Fos in NTS and AP of the hindbrain of WT mice treated with iD1 for the indicated time (D). Quantification of c-Fos positive cells in NTS (E) and AP (F) of mice treated with iD1 for the indicated time. n = 4 per group.
(G and H) Oxygen consumption (VO2) for 48 h (G) and average VO2 of dark- or light-cycle during fasting after a single administration of Veh or iD1 into WT mice (H). VO2 was normalized to body weight. n = 7 per group.
(I-L) Levels of serum triglycerides (I), serum ketone bodies (J), and the indicated mRNAs in livers (K) and BAT (L) in WT mice treated with iD1 for the indicated time. mRNA levels were normalized to Hprt mRNA. n = 8 per group.
(M) IF staining of c-Fos in PVN of the hypothalamus of WT mice treated with iD1 for the indicated time. Representative images of three independent experiments are presented. 3rd ventricle (3V).
Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; p values by one-way ANOVA with Dunnett's multiple comparison test for (E, F, and I-L), two-way ANOVA with Bonferroni's multiple comparison test for (A-C), or Student’s t-test for (H).
See also Figure S3.
FGF21 stimulates the brain and peripheral tissues, such as the liver and adipose tissue, to enhance lipid catabolism and oxygen consumption (Flippo and Potthoff, 2021; Hashimoto et al., 2018; Kliewer and Mangelsdorf, 2019; Sekiyama et al., 2019). Mice treated with iD1 showed a trend of consuming more oxygen than Veh-treated mice despite their lower food intake (Figures S3E-S3F). Food intake stimulates energy expenditure, referred to as diet-induced thermogenesis, which is a critical component of daily oxygen consumption (Feldmann et al., 2009; Kazak et al., 2017). To exclude the iD1 effect on food intake, we removed food from mice treated with Veh or iD1 and measured the oxygen consumption rate. iD1-treated mice consumed more oxygen relative to Veh-treated counterparts under a fasted condition (Figures 3G and 3H). Fasting decreased the respiratory exchange rate (RER) nearly to 0.7, as reported previously, while iD1 significantly reduced RER under a fed condition (Figure S3G and S3H), suggesting that CNOT6L inhibition enhances fat utilization. Moreover, iD1 treatment reduced serum triglycerides while increasing serum ketone body levels (Figures 3I and 3J). Hepatic triglyceride increased transiently following iD1 injection and then decreased to 10% of the basal level (Figure S3I). The effects of iD1 on hepatic triglycerides were similar to those observed with an FGF21 analog (LY2405319) (Figures S3J and S3K). iD1 treatment also resulted in a decrease in serum insulin levels (Figures S3L and S3M), which appeared comparable to the effect of FGF21 (Flippo and Potthoff, 2021; Kliewer and Mangelsdorf, 2019; Lan et al., 2017). Furthermore, iD1 injection increased levels of hepatic mRNAs relevant to energy expenditure and fatty acid oxidation, including Pgc1α/β, Cpt1α/β, and Ucp2 mRNAs, while decreasing mRNAs related to lipid synthesis, such as Srebf1 and Fasn mRNAs (Figures 3K and S3N). In contrast, iD1 did not change mRNA levels of transcription factors, ATF4 and PPARα (Figure S3N), that promote Gdf15 and Fgf21 transcription (Day et al., 2019; Fisher et al., 2017; Inagaki et al., 2007; Kliewer and Mangelsdorf, 2019; Patel et al., 2019). In addition to the effects in the liver, iD1 led to alterations in expression of genes relevant to energy expenditure and lipid metabolism in brown adipose tissues (BAT) and inguinal white adipose tissues (ingWAT) (Figures 3L, S3O, and S3P). The effect of FGF21 on lipid-lowering and energy expenditure is thought to be mediated through activation of the hypothalamus, but not through its direct stimulation on the liver and adipose tissues (Flippo and Potthoff, 2021; Geng et al., 2020). Robust c-Fos staining within the paraventricular nucleus (PVN) in the hypothalamus was detected after iD1 administration (Figure 3M). These results suggest that CNOT6L inhibition by iD1 reduces food intake and increases lipid consumption and energy expenditure through the induction of hepatic GDF15 and FGF21, leading to loss of body weight.
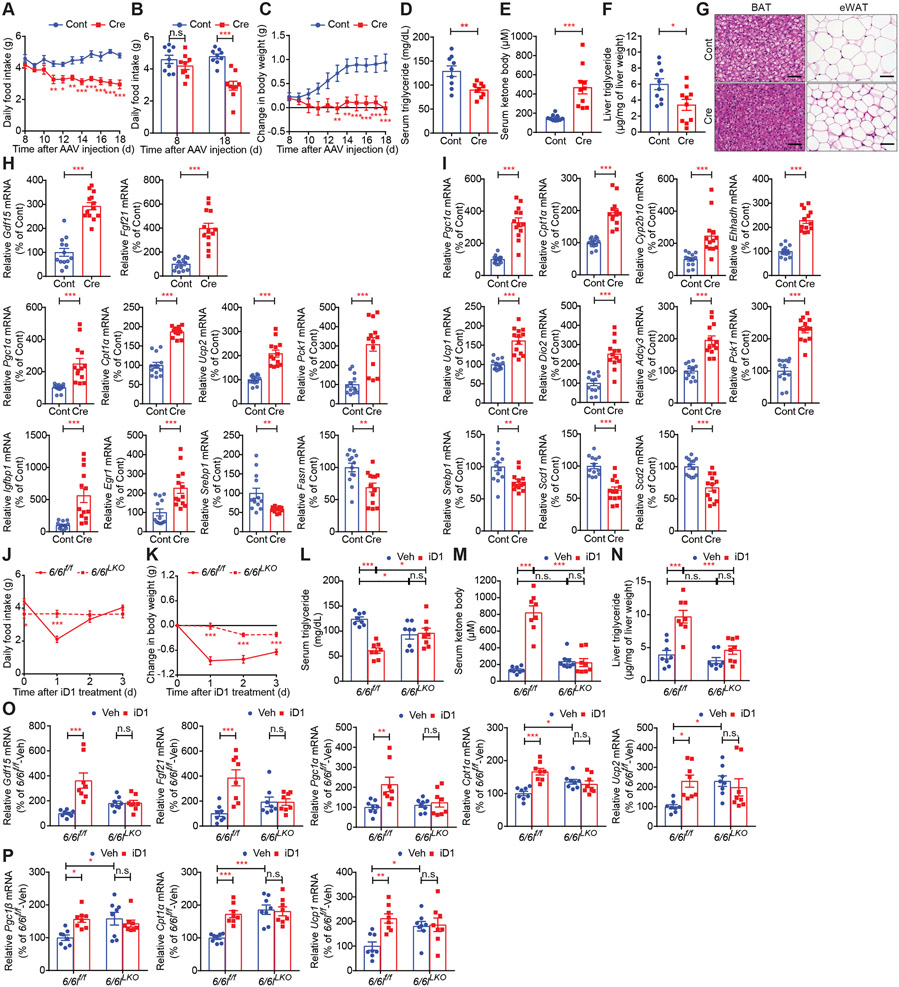
To further corroborate results obtained with iD1, we employed genetic approaches whereby we took advantage of knockdown (KD)/knockout (KO) of CNOT6 and CNOT6L. We first targeted hepatic CNOT6 and CNOT6L deadenylases using adenovirus-delivered shRNAs. This resulted in depletion of Cnot6 and Cnot6l mRNAs in the liver (58.33% and 73.14% decrease, respectively) (Figure S4A), but not in other tissues, including BAT, ingWAT, kidney, muscle, and pancreas (Figure S4B). Consistent with the effects of iD1, food intake and weights of body, livers, and WAT significantly decreased upon depletion of hepatic CNOT6/CNOT6L (Figures S4C-S4G). Hepatic CNOT6/CNOT6L depletion reduced serum triglycerides and increased serum ketone bodies (Figures S4H and S4I). KD of CNOT6/CNOT6L enhanced levels of serum GDF15 and FGF21 (Figures S4J and S4K) and induced expression of hepatic Pgc1α/β, Cpt1α, Ucp2, Pck1, and Igfbp1 mRNAs (Figure S4L). Significantly, in BAT and ingWAT, levels of mRNAs encoding factors implicated in energy expenditure and fat utilization were also altered following hepatic CNOT6/CNOT6L KD (Figures S4M and S4N). To further confirm these results, we generated conditional Cnot6/Cnot6l KO (6/6lf/f) mice and injected adeno-associated virus 8 (AAV8) expressing Cre recombinase under the liver-specific TBG promoter (AAV8-TBG-Cre) (Reizel et al., 2020). Tail vein administration of AAV8-TBG-Cre resulted in a dramatic reduction in hepatic Cnot6 and Cnot6l mRNAs levels (90.87% and 91.51% decrease, respectively) (Figure S4O). Liver-specific ablation of CNOT6/CNOT6L decreased food intake, body weight, and serum triglycerides, increased serum ketone bodies, and reduced hepatic triglycerides and lipid contents in BAT and epididymal WAT (eWAT) (Figures 4A-4G). Levels of hepatic Gdf15 and Fgf21 mRNAs were significantly up-regulated by CNOT6/CNOT6L deletion (Figure 4H). Liver-specific CNOT6/CNOT6L deletion induced hepatic expression of genes relevant to energy expenditure and fatty acid oxidation while reducing mRNAs encoding proteins involved in lipid synthesis (Figures 4H and S4P). This was accompanied by altered expression of genes pertinent to energy expenditure and lipid metabolism, such as Pgc1α/β, Cpt1α/β, Ucp1/2, Dio2, and Srebf1 mRNAs in BAT (Figures 4I, S4Q, and S4R). Thus, our data show that hepatic CNOT6/CNOT6L orchestrates inter-organ communication from the liver to other tissues, such as the brain, BAT, and WAT, to regulate food intake and energy expenditure.
Figure 4. Hepatic Cnot6 and Cnot6l double knockout induces loss of food intake and body weight and stimulates lipid consumption and ketogenesis through an increase in hepatokines.
(A-I) Daily food intake (A), daily food intake 0 and 18 days after AAV injection (B), change in body weight (C), levels of serum triglycerides (D), serum ketone bodies (E), and liver triglycerides (F), representative H&E staining images of the indicated tissues (G), and levels of the indicated mRNAs in livers (H) and BAT (I) of Cnot6/Cnot6l conditional KO mice injected with AAV8 harboring mock (Cont) or Cre recombinase (Cre) under the liver-specific TBG promoter. mRNA levels were normalized to Hprt mRNA. n = 9-13 per group.
(J-P) Daily food intake (J), change in body weight (K), and levels of serum triglycerides (L), serum ketone bodies (M), liver triglycerides (N), and the indicated mRNAs in livers (O) and BAT (P) of control (6/6lf/f) and Cnot6/Cnot6l liver-specific double KO (6/6lLKO) mice after a single administration of Veh or iD1. mRNA levels were normalized to Hprt mRNA. n = 8 per group
Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; p value by two-way ANOVA with Bonferroni's multiple comparison test for (A-C and J-P) or Student’s t-tests for (D-F, H, and I).
See also Figures S4 and S5.
To examine whether the iD1 effects are mediated by CNOT6L, we treated wild-type and Cnot6l whole-body KO (Cnot6l−/−) mice with iD1 (Figure S5A). This revealed that in Cnot6l−/− mice, iD1 did not induce significant effects on food intake, body weight, serum triglycerides, serum ketone bodies, levels of hepatic Gdf15 and Fgf21 mRNAs, and expression of genes relevant to energy expenditure and lipid metabolism in livers and BAT (Figures S5B-S5G). To further investigate whether hepatic CNOT6/CNOT6L are required for the actions of iD1, we generated Cnot6/Cnot6l liver-specific double KO (6/6lLKO) mice by crossing conditional Cnot6/Cnot6l KO (6/6lf/f) mice with Alb-cre transgenic mice (Figures S5H and S5I) and treated control (6/6lf/f) and 6/6lLKO mice with iD1. Hepatic deletion of Cnot6/Cnot6l diminished the impact of iD1 on food intake, body weight, serum triglycerides, serum ketone bodies, liver triglycerides, levels of hepatic Gdf15 and Fgf21 mRNAs, and expression of genes relevant to energy expenditure and lipid metabolism in livers and BAT (Figures 4J-4P, S5J, and S5K).
GDF15 and FGF21 mediate the effects of hepatic CNOT6L on food intake and energy expenditure, respectively
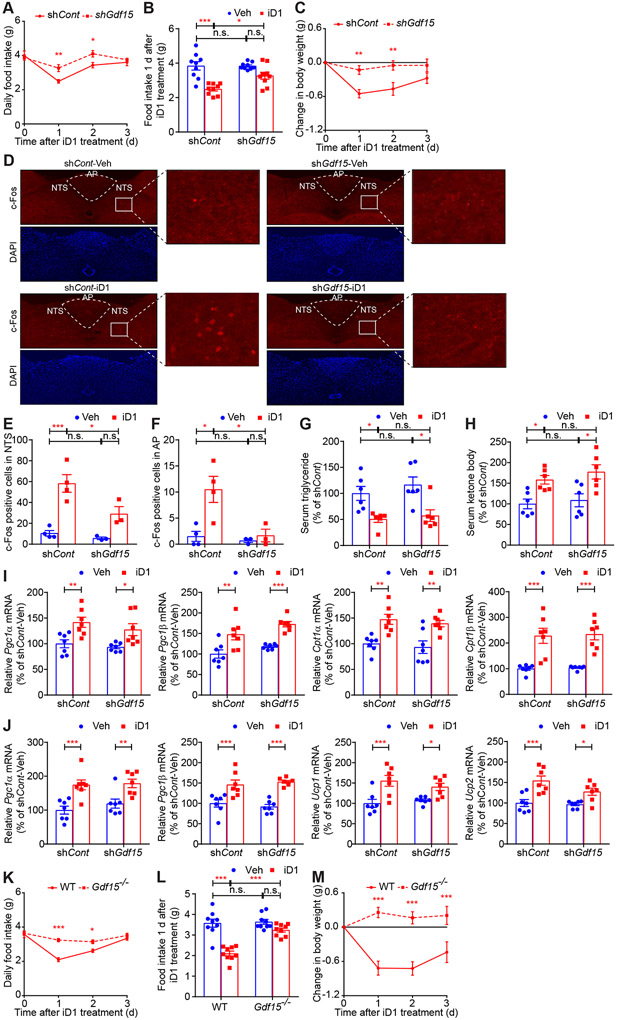
To determine whether GDF15 mediates the effect of hepatic CNOT6L on food intake, we depleted Gdf15 mRNA in the liver (74.17% decrease) using adenovirus expressing shRNA against Gdf15 mRNA and injected iD1 into control and GDF15 KD mice (Figures S6A-S6C). The ability of iD1 to decrease food intake was partially impaired by the depletion of hepatic GDF15 (Figures 5A and 5B). Significantly, GDF15 KD attenuated the iD1 efficacy in lowering body weight (Figure 5C), suggesting that, in this context, food intake is likely to represent a significant determinant of body weight. c-Fos activation in NTS and AP neurons after iD1 administration was decreased by GDF15 KD (Figures 5D-5F). In contrast, GDF15 KD did not change the effect of iD1 on lipid metabolism, as demonstrated by decreased serum triglycerides and increased serum ketone bodies following iD1 treatment (Figures 5G and 5H). Furthermore, GDF15 depletion failed to reverse the effect of iD1 on levels of mRNAs encoding factors implicated in energy expenditure and fat metabolism in livers, BAT, and ingWAT (Figures 5I, 5J, and S6D-S6F). Consistent with the hepatic KD of GDF15, whole-body KO of Gdf15 in mice completely rescinded the effects of iD1 on decreased food intake and body weight (Figures 5K-5M). These findings support the tenet that GDF15 is a major mediator of the effect of hepatic CNOT6L deadenylase on food intake and body weight.
Figure 5. GDF15 mediates the effect of CNOT6L inhibition on loss of food intake and body weight.
(A-J) Daily food intake (A), food intake 1 day after the treatment (B), change in body weight (C), IF staining of c-Fos in NTS and AP of the hindbrain (D), quantification of c-Fos positive cells in NTS (E) and AP (F), and levels of serum triglycerides (G), serum ketone bodies (H), and the indicated mRNAs in livers (I) and BAT (J) of WT mice injected with adenovirus expressing shRNA against Gfp (shCont) or Gdf15 mRNA (shGdf15) after a single administration of Veh or iD1. mRNA levels were normalized to Hprt mRNA. n = 3-9 per group.
(K-M) Daily food intake (K), food intake 1 d after the treatment (L), and change in body weight (M) of WT and Gdf15 KO (Gdf15−/−) mice after a single administration of Veh or iD1. n = 9 per group.
Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; p values by two-way ANOVA with Bonferroni's multiple comparison test.
See also Figure S6.
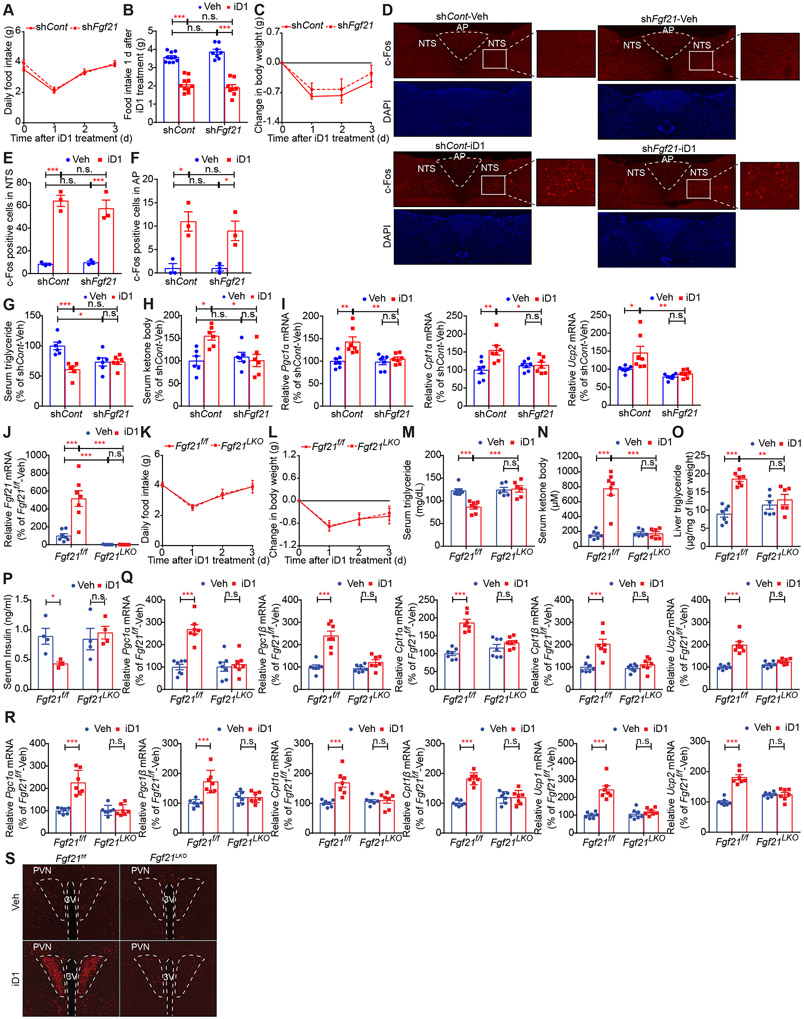
To identify the mediators of the effects of hepatic CNOT6L on energy expenditure and lipid metabolism, we depleted Fgf21 mRNA in the liver (74.17% decrease) but not in other tissues using adenovirus-delivered shRNA (Figures S6G-S6I). FGF21 KD did not change the effects of a single administration of iD1 on food intake and body weight (Figures 6A-6C). iD1-induced activation of neurons in NTS and AP was also not affected by FGF21 KD (Figures 6D-6F). In contrast, FGF21 depletion diminished the iD1 effects on serum triglyceride and ketone body levels (Figures 6G and 6H). Similarly, hepatic Pgc1α/β, Cpt1α/β, and Ucp2 mRNA expression were not induced by iD1 in FGF21-depleted mice (Figures 6I and S6J). Finally, the impact of iD1 on expression of genes relevant to energy expenditure and fat metabolism in BAT and ingWAT were dampened by FGF21 KD (Figures S6K and S6L), suggesting that hepatic FGF21 mediates the iD1 effect on adipose tissues. To further investigate the role of FGF21 on the effects of iD1 in the liver and adipose tissues, we treated control (Fgf21f/f) and Fgf21 liver-specific KO (Fgf21LKO) mice (Markan et al., 2014; Potthoff et al., 2009) with iD1 (Figure 6J). Although iD1 decreased food intake and body weight in both Fgf21f/f and Fgf21LKO mice (Figures 6K and 6L), hepatic FGF21 deletion attenuated the iD1 effects on serum triglycerides, serum ketone bodies, hepatic triglycerides, and serum insulin (Figures 6M-6P). iD1 increased levels of Pgc1α/β, Cpt1α/β, and Ucp1/2 mRNAs in livers and BAT of Fgf21f/f, but not in Fgf21LKO mice (Figures 6Q and 6R). Moreover, Fgf21LKO decreased iD1-induced activation of neurons in PVN in the hypothalamus (Figure 6S), suggesting that hepatic FGF21 mainly mediates the effect of iD1 on fat utilization, not on food intake. Altogether, these data demonstrate that iD1 decreases appetite via hepatic GDF15 while enhancing energy expenditure and lipid consumption through hepatic FGF21, which in conjunction leads to body weight loss.
Figure 6. FGF21 mediates the iD1 effect on lipid metabolism and energy expenditure but not on food intake.
(A-I) Daily food intake (A), food intake 1 d after the treatment (B), change in body weight (C), IF staining of c-Fos in NTS and AP of the hindbrain (D), quantification of c-Fos positive cells in NTS (E) and AP (F), and levels of serum triglycerides (G), ketone bodies (H), and the indicated mRNAs in livers (I) of WT mice injected with adenovirus expressing shRNA against Gfp (shCont) or Fgf21 (shFgf21) mRNA after a single administration of Veh or iD1. n = 3-10 per group.
(J-S) Levels of Fgf21 mRNA in livers (J), daily food intake (K), change in body weight (L), levels of serum triglycerides (M), serum ketone bodies (N), liver triglycerides (O), serum insulin (P), and the indicated mRNAs in livers (Q) and BAT (R), and IF staining of c-Fos in PVN of the hypothalamus (S) of control (Fgf21f/f) and Fgf21 liver-specific KO (Fgf21LKO) mice after a single administration of Veh or iD1. mRNA levels were normalized to Hprt mRNA. n = 6-9 per group.
Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; p values by two-way ANOVA with Bonferroni's multiple comparison test.
See also Figure S6.
Targeting CNOT6L deadenylase has a therapeutic potential to treat diet-induced metabolic disorders
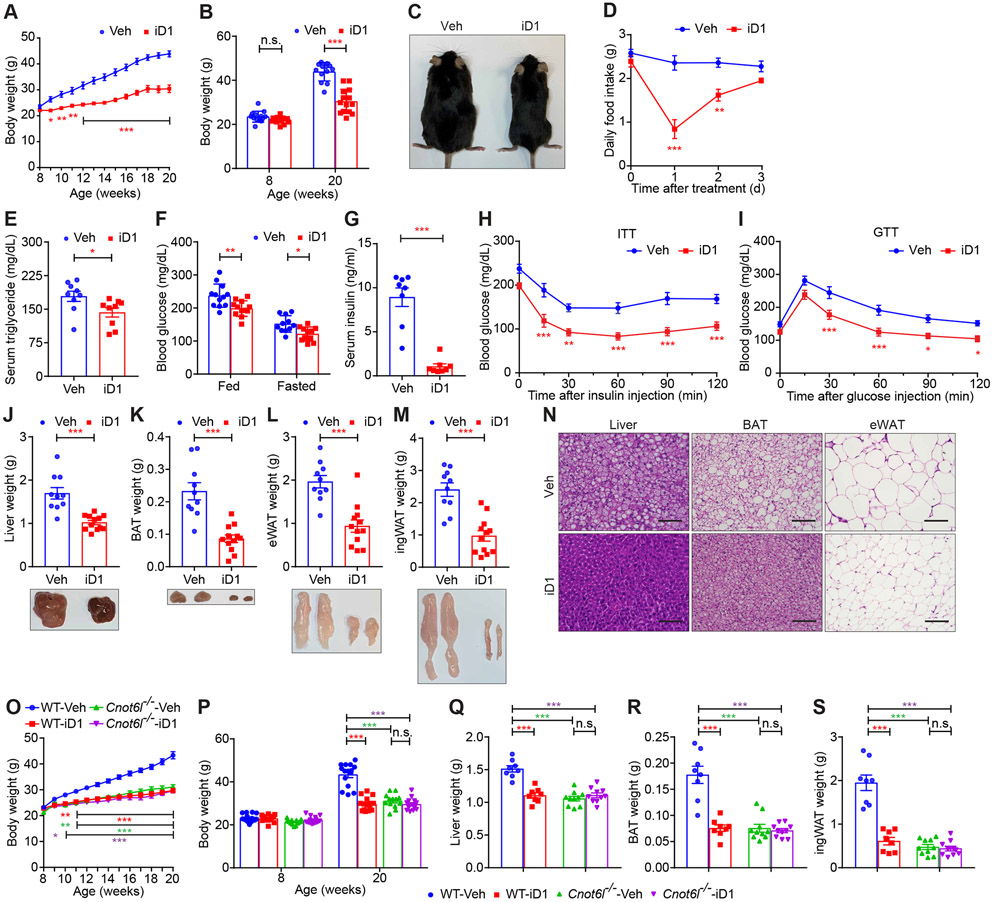
Administration of recombinant secretory proteins, such as GDF15 and FGF21, can ameliorate metabolic disorders in rodents, monkeys, and humans (Charles et al., 2019; Flippo and Potthoff, 2021; Kliewer and Mangelsdorf, 2019; Patel et al., 2019; Sanyal et al., 2019; Tsai et al., 2018). However, it has been challenging due to low protein stabilities and emerging resistance. Therefore, there is heightened interest in developing alternative and clinically applicable approaches to enhance levels of organokines. To test whether this may be achievable via CNOT6L repression, we intraperitoneally administrated iD1 twice a week into male and female C57BL/6J WT mice kept on a high-fat diet (HFD) (Figures 7 and S7). In both male and female mice, iD1 treatment dramatically decreased HFD-induced gain of body weight and suppressed food intake (Figures 7A-7D, S7A, and S7B). iD1 treatment ameliorated obesity-induced hyperlipidemia, hyperglycemia, and hyperinsulinemia (Figures 7E-7G). Insulin sensitivity and glucose tolerance were also improved by long-term iD1 treatment in male and female mice (Figures 7H, 7I, S7C, and S7D). Hepatic steatosis and lipid accumulation in adipose tissues induced in obese mice were reversed by iD1 administration (Figures 7J-7N and S7E). Interestingly, iD1 treatment did not affect weights of other tissues except for livers, BAT, and WAT or body length irrespectively of the sex of the animal (Figures S7F and S7G). To support our concept that targeting CNOT6L improves metabolic syndrome, we depleted hepatic CNOT6L (76.7 % decrease) using adenovirus-delivered shRNA (Figure S7H). Consistent with the iD1 effects on metabolic syndrome shown above, KD of hepatic Cnot6l improved diet-induced obesity and hepatic steatosis (Figures S7I and S7J), decreased weights of tissues, including livers, BAT, and WAT (Figures S7K and S7L), and induced Gdf15 and Fgf21 mRNA expression in the liver (Figure S7M).
Figure 7. CNOT6L inhibition has a therapeutic potential to ameliorate diet-induced metabolic disorders.
(A-I) Body weight curves for 12 weeks (A), starting (8-week-old) and ending (20-week-old) body weight (B), a representative 20-week-old mouse image (C), daily food intake (D), levels of serum triglycerides (E), fed/fasted blood glucose (F), and serum insulin (G), insulin tolerance tests (ITTs) (H), glucose tolerance tests (GTTs) (I), tissue weights and representative images of livers (J), BAT (K), eWAT (L), and ingWAT (M), and representative H&E staining image of the indicated tissues (N) of WT male mice fed with HFD and treated with Veh or iD1 for 12 weeks. HFD feeding and iD1 treatment started at 8 weeks of age. n = 4-14 per group. Scale bars represent 50 μm.
(O-S) Body weight curve (O), starting and ending body weight (P), and weights of livers (Q), BAT (R), and ingWAT (S) of WT and Cnot6l−/− male mice fed with HFD and treated with Veh or iD1 for 12 weeks. HFD feeding started at 8-week-old. n = 8-15 per group.
Data represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001; p value by Student’s t-tests for (E, G, and J-M) or two-way ANOVA with Bonferroni's multiple comparison test for (A, B, D, F, H, I, and O-S).
See also Figure S7.
To confirm that CNOT6L is required for the iD1 action on diet-induced obesity, we treated HFD-fed Cnot6l whole-body KO mice with iD1. In support of this postulate, iD1 decreased a diet-induced gain of body weight in WT but not in Cnot6l−/− mice (Figures 7O and 7P). Similarly, Cnot6l deletion abolished the effect of iD1 on hepatic steatosis and lipid accumulation in BAT and WAT (Figures 7Q-7S). These findings indicate that iD1 may be effective in ameliorating diet-induced metabolic syndrome through inhibition of CNOT6L.
DISCUSSION
Herein, we provide evidence that the hepatic CNOT6L deadenylase governs expression of hepatic secretory proteins, including GDF15 and FGF21, that stimulate the brain and peripheral tissues to modulate food intake and energy expenditure. Previous studies showed that expression of Gdf15 mRNA is induced by ATF4 and CHOP, which are activated by exercise and inflammation (Day et al., 2019; Mullican and Rangwala, 2018; Patel et al., 2019). Also, several transcription factors, such as PPARα, ATF4, ChREBP, were identified to stimulate Fgf21 mRNA expression during fasting and exercise (Fisher et al., 2017; Inagaki et al., 2007; Kliewer and Mangelsdorf, 2019; Patel et al., 2019). However, the “switch-off” mechanism for hepatokine mRNAs was unknown. Herein, we report such a “switch-off” mechanism for down-regulating GDF15 hepatokine mRNA via recognition of its 3’UTR by the CNOT6L and YTHDF2 complex (Figure 2). YTHDF2 recognizes m6A, the most abundant modification in RNAs, and has been reported to contribute to metabolic regulation, cell proliferation, and differentiation, highlighting its role in obesity and other human diseases (Zaccara et al., 2019; Zhao et al., 2017). The best-established molecular function of m6A is thought to be the destabilization of target mRNAs (Zaccara et al., 2019). Our results suggest that YTHDF2, together with the CCR4-NOT deadenylase complex, may play a critical role in destabilizing mRNA encoding hepatokine and modulating whole-body metabolism.
mRNA levels are considered to be maintained by the balance between transcription and degradation (Rabani et al., 2011; Schwanhausser et al., 2011). To recapitulate the synthesis-degradation program of Gdf15 and Fgf21 mRNAs under physiological conditions, we exposed mice to a variety of stresses, such as exercise, fasting, and inflammation (Figure S2). In response to these stimuli, mRNA levels of Gdf15 and Fgf21 were up-regulated and rapidly decreased to baseline expression levels, which was attenuated by iD1 (Figure S2). In contrast, CCR4-NOT deadenylase is likely to be suppressed during these stimuli (e.g., exercise and fasting) through a decrease in its activator, CNOT3 (Figures S2J and S2L) (Raisch et al., 2019). Thus, the data suggest that transcription and mRNA degradation act in an inversely correlated manner to rapidly up- and down-regulate expression of hepatokine mRNAs in response to acute stimuli, such as exercise, food, and inflammation. In turn, iD1 did not affect translational efficiency of Gdf15 and Fgf21 mRNAs (Figure 2H), nor did it affect levels of transcription factors (e.g., ATF4) that promote their transcription (Figures S2I, S2M, S2N, and S3N). Of note, although the poly(A) tail is thought to be a central player for translational regulation, the lack of the iD1 effects on translation of Gdf15 and Fgf21 mRNAs is consistent with the recent model that deadenylation is not necessarily coupled with translation efficiency (Chang et al., 2014; Park et al., 2016; Subtelny et al., 2014). Collectively, our data show that iD1 drives an increase in GDF15 and FGF21 protein levels principally at the level of mRNA stability.
Intriguingly, we observed a delay between an iD1-induced increase in hepatic Gdf15 mRNA levels (peak: 3 h) and levels of GDF15 protein in serum (peak: 12-24 h) (Figures 2O and 2P). A half-life of circulating GDF15 protein was reported to be 3 h (Xiong et al., 2017), which suggests that GDF15 protein may accumulate in the liver after mRNA translation. Indeed, GDF15 protein levels in the liver peaked 12h after CNOT6L inhibition (Figures 2Q and 2R), thus indicating that GDF15 protein is stable and stored in the liver until it is released to the bloodstream. Therefore, these findings support our model that the strategy to increase hepatic GDF15 protein levels by inhibiting its mRNA decay may have a longer-lasting effect compared to direct administration of GDF15 protein.
Our findings show that hepatic CNOT6/CNOT6L contributes to whole-body metabolic homeostasis through hepatokine-dependent regulation of inter-organ communication. Hepatic Cnot6/Cnot6l knockout mimicked the effects of iD1 largely but not completely (Figures 4A-4I and S4O-S4R), suggesting that iD1 may also influence CNOT6/CNOT6L in other organs or other proteins. Consistent with these observations, adipose tissue- and pancreas-specific deletion of the components of the CCR4-NOT deadenylase complex has been shown to affect whole-body metabolism (Li et al., 2017; Mostafa et al., 2020). These data suggest that CNOT6/CNOT6L deadenylase has a critical role in regulating metabolic homeostasis through inter-organ cross-talk mediated by other secretory proteins, such as adipokines and myokines. Further experiments, including RNA-immunoprecipitation-sequencing (RIP-seq) in WAT, BAT, and muscles, are anticipated to reveal additional CCR4-NOT deadenylase-mediated inter-organ communication.
Obesity and its associated metabolic disorders are thought to be a result of excess food intake and/or insufficient activity. The therapeutic potentials of GDF15 and FGF21 have been explored in the clinic (Charles et al., 2019; Kliewer and Mangelsdorf, 2019; Patel et al., 2019; Sanyal et al., 2019; Tsai et al., 2018). Administration of GDF15 causes loss of appetite and body weight without altering energy expenditure (Mullican and Rangwala, 2018; Tsai et al., 2018). In contrast, FGF21 treatment induces energy expenditure and lipid consumption, enhances insulin sensitivity, and modulates macronutrient preference, leading to weight loss despite no effect on appetite (Fisher and Maratos-Flier, 2016; Flippo and Potthoff, 2021; Kharitonenkov and Adams, 2014; Kliewer and Mangelsdorf, 2019). However, clinical trials using organokine-based therapy have struggled in improving metabolic disorders due to limited protein stability, route of administration, and emerging resistance (Kliewer and Mangelsdorf, 2019; Mullican and Rangwala, 2018; Tsai et al., 2018; Xiong et al., 2017). As indicated above, it is anticipated that iD1 may exert longer-lasting effects compared to the administration of GDF15 protein. In addition, iD1 may thus be more effective than organokine-based therapies inasmuch as it simultaneously targets multiple hepatokines, which is expected to reduce the risk of emerging resistance. In summary, the inhibitor of CNOT6L, iD1, can ameliorate diet-induced metabolic disorders through modulating food intake and energy and lipid consumption via GDF15 and FGF21. Overall, our findings may provide the molecular basis for a previously unexplored therapeutic strategy to treat metabolic syndrome.
Limitations of study
This study identified the inhibitor of CNOT6L deadenylase, iD1, whose specificity was explored by employing 6/6lLKO mice (Figures 4 and S5) and in vitro deadenylase assay with another family of deadenylases, CNOT7 and CNOT8 (Figure 1). Our data suggest that iD1 suppresses CNOT6/CNOT6L but not CNOT7/CNOT8 in vitro and requires CNOT6/CNOT6L for its impact on food intake, body weight, lipid metabolism, and expression of hepatokine mRNAs. Furthermore, we demonstrated that iD1 up-regulated Gdf15 and Fgf21 mRNA stability without affecting other mRNA degradation pathways (Table S2). Nonetheless, we acknowledge that the off-target effects of iD1 have not been investigated completely. To this end, our future plan is to carry out the structure-guided optimization of iD1 to develop more specific and potent CNOT6/CNOT6L inhibitors.
We observed that GDF15 and FGF21 are major mediators of the effect of iD1 on metabolic regulation (Figures 5 and 6); however, our RIP-transcriptomics identified other mRNAs as CNOT6L-targets. Our results suggest that CNOT6L preferentially targets mRNAs with a shorter half-life (1-2 h of half-life), such as Gdf15 and Fgf21 transcripts (Figures 2 and S2), compared to other mRNAs (9 h of the median half-life) (Schwanhausser et al., 2011). Future research on other CNOT6L-target mRNAs in several tissues (e.g., livers, adipose tissues, and muscles) is required to comprehensively define CNOT6L-mediated inter-organ metabolic communication.
STAR★METHOD
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, M.M. (moritam@uthscsa.edu).
Materials availability
Mice and plasmids generated in this study are available on reasonable request.
Data and code availability
The bulk RNA-seq data are publicly available at GEO under accession number (GSE185851). The data generated in this study will be shared by the lead contact upon reasonable request.
This study did not generate new code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAIL
Mice
Mice were maintained on a 12-hr light-dark cycle with access to food and water ad libitum and fed on a standard diet (SD) (D12450B, Research Diets, Inc.) or a high-fat diet (HFD) (D12492, Research Diets, Inc.). We used WT mice with a C57BL/6J background (Stock No: 000664, Jackson laboratory). Cnot6l whole-body knockout (Cnot6l−/−) mice (RIKEN, accession number CDB0648K) were described as previously (Morita et al., 2019). To generate Fgf21 liver-specific KO (Fgf21LKO) mice, Fgf21f/f (Stock No: 022361) mice were purchased from the Jackson Laboratory and crossed with Alb-cre (Stock No: 003574, Jackson Laboratory). To generate Cnot6l conditional knockout (Cnot6lf/f) mice, Cnot6l potential conditional KO (Cnot6ltm1a(EUCOMM)Wtsi) mice were obtained from the European Mouse Mutant Archive (EMMA) and crossed with ACTB-FLPe mice (Stock No: 005703, Jackson Laboratory) (Coleman et al., 2015). To generate Cnot6 conditional knockout (Cnot6lf/f) mice, Cnot6 potential conditional KO (Cnot6tm1a(KOMP)Wtsi) was obtained from the Mutant Mouse Resource & Research Centers (MMRRC) supported by NIH with a C57BL/6N background, back-crossed to C57BL/6J more than 10 times, and crossed with ACTB-FLPe mice. To generate Cnot6/Cnot6l conditional double knockout (6/6lf/f) mice, Cnot6f/f and Cnot6f/f mice were crossed. To generate Cnot6/Cnot6l liver-specific double knockout (6/6lLKO) mice, 6/6lf/f mice were crossed with Alb-cre mice. Gdf15 KO mouse line (C57BL/6N-Gdf15tm1a(KOMP)Wtsi) mice were obtained from MMRRC supported by NIH with a C57BL/6N background. All experiments were conducted according to the guidelines for animal use issued by the Institutional Animal Care and Use Committee (IACUC), University of Texas Health Science Center at San Antonio.
METHOD DETAILS
In silico screening of small chemical compounds
To select compound libraries for our FRET-based HTS, we first performed a virtual screening of libraries of small molecule compounds that were known to be bioactive (Rabinowitz et al., 2008). For each compound library, an SDF (MDLMOL) file was supplied from the vendor and was imported into a MDB format (Molecular Operating Environment [MOE], Chemical Computing Group, Montreal Canada). Counterions and neutral state protonation were derived using the wash protocol in MOE, followed by a 3D geometry minimization using the default Amber 10/EHT force-field (Rabinowitz et al., 2008). The crystal structures of the human CNOT6L were obtained from the Brookhaven Protein Repository (www.RCSB.org) (Wang et al., 2010). The protein structure was prepared using LigX (Pinzi et al., 2018) with default settings for tethers, protonation, and refinement. The structure was optimized using the default Amber10/EHT force-field(Chaaban et al., 2018). MOE's SiteFinder algorithm (Hu et al., 2018) was used to identify the dominant binding site, which was subsequently used for docking site placement. All docking was performed in MOE using the generalized docking workflow with rigid receptor constraints. The Triangle placement and London-dG scoring function were used for placement, followed by force-field refinement and rescoring using the GVBI/WSA scoring function. All ligands sets were combined and docked with their source database annotations and subsequently rank-ordered by ascending docking scores (kCal/mol). The docking output was summarized by deck source using MS-Excel's PivotTable functionality to identify compound collections with the highest number of putative hits. The decks containing compounds with the lowest energy were selected for our FRET-based HTS. The binding model of the active-site pocket in CNOT6L (PDB: 3NGO) with iD1 was built in the MOE software.
FRET-based HTS of in vitro deadenylase assay
The catalytic domain of CNOT6L was purified as previously described (Wang et al., 2010). For the FRET-based screening, we adapted an in vitro deadenylase assay as previously described (Maryati et al., 2014). A single point assay was performed using 10 μl of CNOT6L WT (2 μM) protein in the deadenylase buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10% glycerol, 1 mM DTT, 5 mM MgCl2), mixed with 100 nl of a compound, and incubated for 60 min at 37°C. 10 μl of 5’-6-carboxyfluorescein (6-FAM)-labeled poly(A) RNA substrate (2 μM) (Sigma-Aldrich) in the deadenylase buffer was added into the protein and compound mixture and incubated for 60 min at 37°C. 5 μl of 20 μM 3’-tetramethylrhodamine (TAMRA)-labeled oligo (dT) DNA probe (diluted in 1% SDS, 20 mM Tris-HCl [pH 8.0], 0.5 mM EDTA) (Sigma-Aldrich) was then added for a final of >5-fold molar excess vs RNA concentration and incubated for 30 min at room temperature. The final concentrations for each component in the deadenylase buffer were 0.8 μM protein, 0.8 μM RNA substrate, 4 μM DNA probe, 1% DMSO, and (on average) 40 μM compound. Fluorescence intensity was measured at room temperature with an M1000 Infinite Pro plate reader (Tecan, CA) (Excitation 485nm; Emission 520nm, Gain 165, BW 5). A total of 982 compounds from two libraries, BML-2840 ICCB Known Bioactives Library (480 compounds) and BML-2865 Natural Product Library (502 compounds), were tested. All samples were plated into 384-well non-treated plates (Corning) using a BioTek EL406 plate washer (BioTek). All samples were run in duplicates. Compounds were pin-transferred (V&P Scientific, CA, pin tool mounted onto Tecan Freedom Evo 150 MCA96 head, Tecan, CA) into duplicate assay plates. Per plate, there are 32 wells of a DMSO control and 32 wells of a positive control compound (AMP, Sigma-Aldrich). For dose-dependent inhibition curves, the FRET-based HTS selected 8 compounds, such as endocannabinoid derivatives, naphtoquinones derivatives, and xanthones derivatives (Fowler, 2021; Klein-Junior et al., 2020; Tu et al., 2021; Widhalm and Rhodes, 2016). For dose-dependent inhibition curves, compound stocks from the libraries were plated in 384-well format in at least 100-fold concentration ranges. Dose-dependent inhibition curves of CNOT6L activity were generated by GraphPad Prism 7.
Gel-based in vitro deadenylation assay
The catalytic domain of human CNOT6L (158–555 a.a.) and CNOT6 (153-557 a.a.) (Wang et al., 2010) and the full length of human CNOT7 and CNOT8 (Yang et al., 2008) were purified from Escherichia coli strain BL21. Cells were grown in Lysogeny Broth (LB) medium containing 50 mg/ml ampicillin at 37°C. Protein expression was induced by 0.1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 16 h at 16°C. The glutathione-S-transferase (GST)-tagged CNOT6L, CNOT6, CNOT7, and CNOT8 were purified using 50% glutathione-Sepharose 4B (GenScript) for 3 h at 4°C. The GST tag was cleaved using HRV GST-3C protease (Acrobiosystem), and the proteins were dialyzed with the deadenylase buffer using Amicon Ultra-4 10K centrifuge filter (Millipore).
Gel-Based deadenylation assay of 20 mer poly (A) RNA was previously described (Morita et al., 2019; Wang et al., 2010). CNOT6L, CNOT6, CNOT7, or CNOT8 at a final concentration of 2.5 μM and a CNOT6L deadenylase inhibitor at the indicated concentration were incubated for 1 h at 37°C, and after this binding reaction, 5’ fluorescein-5-isothiocyanate (FITC)-labeled poly(A)20 RNA at a final concentration of 50 nM (Sigma-Aldrich) were incubated for 1.5 h at 37°C. The labeled RNAs were analyzed on a 7 M urea-25% sequence polyacrylamide denaturing gel in 1 x TBE buffer. After electrophoresis, gels were scanned with iBright FL1000 Imaging System (Thermo Fisher Scientific Inc.). Dose-dependent inhibition curves of CNOT6L activity were generated by GraphPad Prism 7.
SPR and LC-MS/MS
A Biacore system (GE Healthcare) was used for SPR experiments. The CNOT6L protein purified from E. coli was immobilized on the sensor chip CM5 using the amine coupling method. To immobilize the protein, 20 μM of CNOT6L in 10 mM sodium acetate [pH 5.5] was injected and immobilized until about 17,000 RU. Prior to determining binding affinities, serial dilutions of 8 concentrations of the iD1 compound (0.3125, 0.625, 1.25, 2.5, 5, 10, 20, 40 μM) were prepared in the SPR running buffer (10 mM HEPES [pH 7.5], 150 mM NaCl, 0.05% Surfactant P20, 5% DMSO). The compound dilutions were injected onto the surface of the chip. Seonsorgrams were analyzed using the Biacore evaluation software. The binding affinities were determined based on a steady-state model. The equilibrium response units were plotted against the iD1 concentrations. The concentration at 50% saturation represents Kd.
For measurement of serum and haptic levels of iD1, the iD1 concentrations were determined by LC-MS/MS using LCMS8050 (Shimadzu, Kyoto, Japan) equipped with an LC40-XR series HPLC system (Shimadzu) and a COSMOSIL 3C18-MS-II (2.0 mm × 50 mm; Nacalai Tesque, Kyoto, Japan). The mobile phase consisted of 0.1% (v/v) formic acid in distilled water (A) and 0.1% (v/v) formic acid in acetonitrile (B). The flow rate of the mobile phase was 0.4 mL/min with a gradient elution starting from 20% of B and increased to 98% of B in 0.6-2.8 min and then kept at 98% of B until 4.6 min and finally returned to the initial conditions in 4.6-4.61 min and kept at 2.4 min. The wash eluent consisted of 0.1% of formic acid in isopropanol/methanol/acetonitrile/water = 1/1/1/1 (v/v/v/v, %). The injection volume was 5 μL. The column temperature was maintained at 40°C, and the autosampler was maintained at 10°C. The retention times of iD1 and juglone were 2.71, and 2.14 min, respectively. Multiple reaction monitoring (MRM) transitions were measured at negative ion mode at m/z 287.1→218.2 for iD1 and 172.8→145.0 for juglone. Data were processed using LabSolutions software (version 5.97 SP1, Shimadzu).
RIP-transcriptomics and data analysis
RNA-immunoprecipitation-transcriptomics (RIP-transcriptomics) was described previously (Morita et al., 2019). In brief, primary hepatocytes were isolated from 8-week-old C57BL/6J male mice by non-recirculating collagenase liver perfusion. 5 mg of hepatocyte lysate was pre-cleared and incubated with the complex of 5 μg of anti-CNOT6L antibody (#694-3) and 50 μl of Dynabeads Protein G (Invitrogen) for 4 h at 4°C. Beads were washed 5 times with the lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40) using a Magnetic Separator (Invitrogen). The lysate from Cnot6l−/− hepatocytes was used as a negative control. Input and immunoprecipitated RNA were extracted with TRIzol Reagent and GlycoBlue Coprecipitant (Thermo Fisher Scientific Inc.) and purified with RNeasy Kit (Qiagen) according to the manufacturer’s instructions. The quality of RNA was determined using the Bioanalyzer nanochip (Agilent Technologies, Inc.). cDNA and biotin-labeled cRNA were synthesized according to the manufacturer’s instruction (Affymetrix). After cRNA fragmentation, biotin-labeled cRNA was hybridized to the GeneChip Mouse Genome 430 2.0 Array (Affymetrix). All experiments were done in triplicates with immunoprecipitated and input RNA from WT or Cnot6l−/− hepatocytes (a total of 12 microarrays).
For analysis of transcriptomics data, we extracted and normalized data using robust multi-array averaging implemented in the R package “affy” (www.r-project.org). We used updated probe set definitions because they improved precision and accuracy (Sandberg and Larsson, 2007). To identify immunoprecipitated mRNAs, we used the analysis of partial variance (APV) and applied the random variance model (RVM) as implemented in the ANalysis Of Translational Activity (anota) R package (Larsson et al., 2010, 2011). The resulting genes were further filtered to identify those enriched in immunoprecipitated mRNAs from WT cells. The complete dataset reported herein was deposited to the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE62365. To identify hepatic secretory proteins among the 195 CNOT6L-associated mRNAs, we manually curated the mouse secretory gene list from several available datasets (Wang et al., 2014a; Xiong et al., 2019) and listed 16 CNOT6L-associated mRNAs encoding hepatic secretory proteins (Table S1).
RNA isolation, RT-qPCR, mRNA stability assay, RIP-qPCR, RNA-seq
Total RNAs were isolated from cells and tissues using TRIzol reagent (Thermo Fisher Scientific Inc.) according to the manufacturer’s instruction. The concentration and purity of the RNA were determined with NanoDrop Onec (Thermo Fisher Scientific Inc.). 2 μg of template RNA was reverse transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer’s instruction. Quantitative PCR (qPCR) was performed in a 96 well plate using PowerUp SYBR Green Master Mix (Applied Biosystems) in QuantStudio 3 Real-Time PCR System (Applied Biosystems). Primers for qPCR were listed in Table S3. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) for human samples and hypoxanthine phosphoribosyltransferase 1 (Hprt1) for murine samples were used as reference genes for relative quantification.
To assess mRNA stability, Huh7 human liver cancer cells and mouse primary hepatocytes were incubated with actinomycin D (Act D) (5 μg/ml) (Sigma-Aldrich) and Veh (DMSO) or iD1 for the indicated time. mRNA levels were determined by RT-qPCR, normalized to the level of GAPDH or Hprt mRNA, and expressed as a percent change of the initial mRNA level. Before Act D treatment, Huh7 cells were incubated with a PPARα agonist, Wy-14643 (200 μM) (Cayman Chemical) for 6 h, to induce FGF21 mRNA expression. To calculate half-lives of the indicated mRNAs, the first-order degradation kinetics was employed.
For RIP-RT-qPCR, lysates from cells expressing empty, Flag-YTHDF2, or Flag-CNOT6L vector were pre-cleared and incubated with the complex of 1 μg of anti-Flag antibody (F1804, Sigma-Aldrich) and 25 μl of Dynabeads Protein G at 4°C. Beads were washed 5 times with the lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, Roche complete protease inhibitor cocktail) using a Magnetic Separator (Invitrogen). Immunoprecipitated (IPed) and input RNAs were isolated from anti-flag immunoprecipitates and total cell lysates, respectively. Levels of IPed and input mRNAs were determined by RT-qPCR. IPed mRNA levels were normalized to those of input.
For RNA-seq, total RNAs were isolated from livers of mice treated with/without iD1 using TRIzol reagent (Thermo Fisher Scientific Inc.) and purified with an RNeasy Kit (Qiagen) according to the manufacturer’s instruction. The quality of RNA was determined with the Bioanalyzer nanochip (Agilent Technologies, Inc.). mRNA-seq libraries were prepared with TruSeq stranded mRNA-seq kit and read in NextSeq 500 (75bp single read) (Illumina, Inc.). All experiments were done in biological triplicates from livers of mice treated with/without iD1. The data was analyzed by RaNA-seq (Prieto and Barrios, 2019). The result of functional enrichment analysis was shown in Table S2. The complete dataset reported herein has been submitted to the NCBI GEO database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE185851.
Polysome profiling
Polysome profiling and RT-qPCR were carried out as described previously (Morita et al., 2017). Briefly, livers were lysed in 500 μL of hypotonic buffer (5 mM Tris-HCl [pH 7.5], 2.5 mM MgCl2, 1.5 mM KCl, 100 μg/mL cycloheximide, 2 mM DTT, 0.5% Triton X-100, and 0.5% sodium deoxycholate). Lysates were loaded onto 10-50% (wt/vol) sucrose density gradients (20 mM HEPES-KOH [pH 7.6], 100 mM KCl, and 5 mM MgCl2) and centrifuged at 35,000 rpm (SW 40 Ti rotor, Beckman Coulter, Inc.) for 2 h at 4°C. Gradients were fractionated, and optical density at 254 nm was continuously recorded using an ISCO fractionator (Teledyne ISCO). RNAs from each fraction and inputs were isolated using Trizol (Invitrogen) and GlycoBlue (Invitrogen) according to the manufacturer’s instructions. RT-qPCR reactions were carried out using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) and PowerUp SYBR Green Master Mix (Applied Biosystems) in QuantStudio 3 Real-Time PCR System (Applied Biosystems) according to the manufacturer’s instruction. Primers for qPCR were listed in Table S3.
Luciferase assay
For 3’ UTR luciferase assay, human 3’UTRs of GDF15 and FGF21 mRNAs and mouse 3’UTRs of Gdf15 and Fgf21 mRNAs were amplified by PCR from a cDNA pool of Huh7 cells or WT mouse livers and inserted downstream of the Firefly luciferase coding region (pGL3-control, Promega). For promotor luciferase assay, human promotors of GDF15 and FGF21 genes were amplified by PCR from a genome of Huh7 cells. Oligos for cloning of 3’UTRs and promoters were listed in Table S4. Renilla luciferase (pRL-TK, Promega) was used as a control for both assays. The indicated plasmids (50-100 ng) were transfected into Huh7 cells in a 96-well plate using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instruction. Cell extracts were prepared and assayed for Firefly and Renilla luciferase activities using Dual-Glo Luciferase Assay System (Promega) and Varioskan LUX Multimode Microplate Reader (Thermo Fisher Scientific Inc.). The transfection efficacy was normalized to Renilla luciferase activity.
Co-Immunoprecipitation and immunoblotting
HEK293T cells were transiently co-transfected with plasmids encoding Myc-CNOT6L and either Flag only or Flag-YTHDF2 by using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s instruction. Cells were washed with 1 x PBS 5 times and lysed with the lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, and Roche complete protease inhibitor cocktail). After centrifugation, protein concentrations were measured by the Bio-Rad protein assay. Dynabeads Protein G was washed with the lysis buffer 5 times, and cell lysates were pre-cleared with the beads before co-IP. 1 μg of anti-Flag antibody was bound to the beads according to the manufacturer’s instruction. The bead-antibody complex was incubated with lysates at 4°C. Beads were washed with the lysis buffer 5 times before reconstitution with 1 x SDS-PAGE sample buffer. Proteins were resolved by SDS-PAGE, and then transferred to PVDF membranes. Antibodies against Myc (TAG003) and Flag (F1804) were purchased from Bioshop Canada Inc. and Sigma-Aldrich, respectively. Horseradish peroxidase-conjugated anti-mouse IgG (light chain specific) were from Jackson ImmunoResearch.
Mouse livers and Huh7 cells were lysed with the lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% NP-40, and Roche complete protease inhibitor cocktail). After centrifugation, protein concentrations were measured by the Bio-Rad protein assay. Proteins were resolved by SDS-PAGE, and then transferred to PVDF membranes. Antibodies against CNOT1 (#44613), α-tubulin (#2144), and β-actin (#4970) were purchased from Cell Signaling. Antibodies against GDF15 (sc-377195) and FGF21 (sc-81946) were purchased from Santa Cruz. Antibodies against CNOT6L (CNOT6L-B) and CNOT3 (CNOT3-Full) were described previously (Morita et al., 2019). Horseradish peroxidase-conjugated anti-mouse and rabbit IgG (light-chain specific) were from Jackson ImmunoResearch.
Cell culture
Huh7 liver cancer cells, HEK293 cells, and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Corning), supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin/streptomycin (Corning). Primary hepatocytes were isolated from anesthetized 8-week-old male mice by portal vein perfusion (GIBCO) and collagenase (Sigma-Aldrich) digestion. After purification of hepatocytes by differential centrifugation using Percoll (Sigma-Aldrich), cells were plated in Collagen Type I-coated plates (Corning) and cultured in William’s E Medium with supplements (GIBCO). For GDF15 mRNA induction, Huh7 cells were treated with an ER stress inducer, thapsigargin (Tg: 1 μM), and/or CNOT6/6L deadenylase inhibitor, iD1, for 6 h. For FGF21 mRNA induction, Huh7 cells were treated with a PPARα agonist, WY-14643 (WY: 200 μM), and/or CNOT6/6L deadenylase inhibitor, iD1, for 6 h.
Adenovirus/AAV production and injection
Adenovirus infections were carried out according to the manufacturer’s protocol (Takara Bio.). Briefly, shRNA targeting Gdf15, Fgf21 (Morita et al., 2019), Cnot6, or Cnot6l mRNA listed in Table S5 was inserted into pBAsi-mU6 (Takara Bio.), and the region composed of the promoter and shRNA sequence was inserted into pAxcwit2 adenovirus vector (Takara Bio.). pAxcwit2 containing shRNA against Gfp (Takara Bio.) was used as a negative control. Adenovirus was purified from HEK293 cells according to the manufacturer’s instruction (Takara Bio.). Adenovirus expressing shRNA was administered to 8-week-old mice through tail-vain injection (1 x 109 plaque-forming units [pfu] per mouse).
For food intake measurement of shGdf15 and shFgf21 mice, we infected mice with the corresponding adenovirus, administrated iD1 to mice 7 days later, then measured food intake for 3 or 4 days. We separately prepared shGdf15 and shFgf21 mice for serum and tissues isolations and dissected them 14 days after the infection. For food intake measurement of sh6/6l mice, we infected mice with the adenovirus, measured food intake every day, then dissected mice 7 days after infection.
For AAV8-TBG-Cre injection, pAAV.TBG.PI.Cre.rBG was purchased from Addgene (a gift from James M. Wilson; Addgene viral prep #107787-AAV8; http://n2t.net/addgene:107787; RRID:Addgene_107787). For AAV8-TBG-Cont, pAAV.TBG.PI.Null.bGH was purchased from Addgene (a gift from James M. Wilson, Addgene viral prep #105536-AAV8; http://n2t.net/addgene:105536; RRID:Addgene_105536). AAV8-TBG-Cre and AAV8-TBG-Cont were administered into 6/6lf/f mice through tail-vain injection (1.0 x 1011 genome copies per mouse). After AAV8 injection, we measured food intake and body weight every day and dissected the mice 18 days following the tail-vein injection.
Blood tests, GTT, ITT, metabolic studies, histological analysis
Levels of blood glucose, serum triglycerides, serum ketone bodies, serum FGF21, serum GDF15, and serum insulin were measured using a glucose meter (Bayer), LabAssay Triglyceride (Wako), Autokit Total Ketone Bodies (Wako), FGF21 MOUSE ELISA (BioVender Inc.), Mouse/Rat GDF-15 Quantikine ELISA Kit (R&D Systems), and Ultra Sensitive Mouse Insulin ELISA Kit (Crystal Chem Inc.), respectively. Glucose tolerance tests (GTTs) were performed in mice fed or fasted for 16 h by IP injection of 0.5 g glucose/kg of body weight. Insulin tolerance tests (ITTs) were performed in fed mice by IP injection of 1.5 U insulin/kg of body weight. For fasting analysis, mice were housed individually and deprived of food for 16 h.
For in vivo metabolic studies, each mouse was housed in a separate cage and maintained on a 12-h dark-light cycle with access to food and water ad libitum. Food intake was measured during the indicated time. Oxygen consumption was analyzed during a 96 h at 25°C with an Oxymax-CLAMS system (Columbus Instruments) according to the manufacturer’s instruction. Oxygen consumption and respiratory exchange ratio (RER) were calculated according to the manufacturer’s protocol (Columbus Instruments). Oxygen consumption is expressed as the volume of O2 consumed/kg of body weight/h. RER and heat production were calculated using the equations. RER = carbon dioxide production (VCO2)/oxygen consumption (VO2).
For iD1 treatment, mice were intraperitoneally administrated with iD1 (2 mg/kg of body weight) for the indicated time. For poly(I:C) treatment, mice were intraperitoneally administrated with poly(I:C) (10 mg/kg of body weight) for the indicated time. For LY2405319 (LY) treatment, mice were intraperitoneally administrated with LY (5 mg/kg of body weight) for the indicated time. For an HDF-induced obese mouse model, 8-week-old mice were fed on HFD for 12 weeks. For fasting and refeeding, mice fed, fasted for 24 h, or refed for 2 h after 24 h-fasting were prepared. Veh or iD1 was injected 3 h before the dissection.
For forced treadmill running, a 4-lane motorized treadmill (Columbus Instruments) with a slope at 10° and electrical shock (0.25 mA x 163 V and 1 Hz) was used. 8-12 weeks old male mice were familiarized with a 4-lane motorized treadmill (Columbus Instruments) three times (10 min at 10 m/min) one week before the experimental day. These mice were subsequently exposed to a running paradigm of 10 min at 10 m/min, then 40 min at 20 m/min followed by gradually increased speed (2 m/min every min) until exhaustion. Exhaustion was defined when mice fell back to the grid for more than 5 s. Mice were sacrificed before, immediately after, or 3 h after exercise. During the 3 h post-exercise recovery period, mice were returned to cages with food and water. Collected tissues were rapidly frozen in liquid nitrogen and stored at −80°C for later analyses.
After metabolic studies, mice were dissected to collect serum and tissues. Liver triglycerides were extracted from liver tissue homogenates using 2-propanol followed by measurement with LabAssay Triglyceride kit (Wako). Livers, eWAT, and BAT were fixed in 10% formalin overnight. Paraffin-embedded sections were analyzed by hematoxylin and eosin staining.
Immunofluorescence
10% neutral buffered formalin-perfused brains were fixed in 10% neutral buffered formalin for 48 h, then infiltrated in 20% and 30% sucrose, and embedded in O.C.T. compound. 25 μm-thick frozen coronal sections of the hindbrain and the hypothalamus were cut with a cryomicrotome (Leica Biosystems) and stored in cryoprotectant at −20°C until further processing. Free-floating sections were washed with 1 x PBS, and incubated in 3% donkey serum/PBST for 1 h, followed by overnight incubation with rabbit anti-c-Fos antibody (Sigma-Aldrich, F7799). After washing with 1 x PBS, sections were incubated with anti-rabbit Alexa Fluor 568 (Invitrogen, A10042) for 1 h at room temperature. After final washing, sections were mounted onto glass slides and coverslipped with VECTASHIELD® Hardset Antifade Mounting Medium with DAPI (H-1500). c-Fos positive cells in NTS, AP, and PVN were visualized and quantified using fluorescence microscopy (Keyence BZ-X800).
QUANTIFICATION AND STATISTICAL ANALYSIS
All values represent mean ± SEM. Differences among groups were compared using two-way ANOVA followed by between-group comparison with Bonferroni’s post-hoc test, one-way ANOVA with Dunnett’s post-hoc test, or Student’s t-test (two-tailed, unpaired) when there were only two groups. All statistical analyses were performed using GraphPad Prism 7, and the differences were considered significant when p < 0.05. The detailed information about statistical analyses is shown in Table S5.
Supplementary Material
Table S1. List of CNOT6L-interacting mRNAs encoding hepatic secretory proteins. Related to Figure 2.
Table S2. Functional enrichment analysis of RNA-seq data from livers of mice treated with/without iD1. Related to Figure 2.
Table S3. List of qPCR primers. Related to STAR Methods.
Table S4. List of cloning primers and shRNA target sequences. Related to STAR Methods.
Table S5. Information about the statistical analyses. Related to STAR Methods.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal CNOT6L antibody | (Morita et al., 2019) | #CNOT6L-B |
| Mouse monoclonal CNOT6L antibody | (Morita et al., 2019) | #694-3 |
| Rabbit monoclonal CNOT1 antibody | Cell Signaling Technology | #44613 |
| Rabbit polyclonal CNOT3 antibody | (Morita et al., 2019) | #CNOT3-full |
| Sheep polyclonal GDF15 antibody | R&D Systems | AF6385 |
| Mouse monoclonal GDF15 antibody | Santa Cruz Biotechnology | sc-377195 |
| Mouse monoclonal FGF21 antibody | Santa Cruz Biotechnology | sc-81946 |
| Mouse monoclonal α-tubulin antibody | Cell Signaling Technology | #2144 |
| Mouse monoclonal β-actin antibody | Cell Signaling Technology | #4970 |
| Mouse monoclonal Myc antibody | Bioshop Canada Inc. | TAG003 |
| Mouse monoclonal Flag M2 antibody | Sigma-Aldrich | F1804 |
| Rabbit polyclonal c-Fos antibody | Sigma-Aldrich | F7799 |
| HRP-linked anti-mouse IgG (light chain specific) | Jackson ImmunoResearch | 115-035-174 |
| HRP-linked anti-rabbit IgG (light chain specific) | Jackson ImmunoResearch | 211-032-171 |
| HRP-linked anti-mouse IgG | Cell Signaling Technology | #7076 |
| HRP-linked anti-rabbit IgG | Cell Signaling Technology | #7074 |
| Anti-rabbit IgG, Alexa Fluor 568 | Thermo Fisher Scientific Inc. | A10042 |
| Bacterial and virus strains | ||
| AAV8-TBG-Cre | Addgene | 107787-AAV8 |
| AAV8-TBG-Cont | Addgene | 105536-AAV8 |
| Escherichia coli strain BL21 Competent Cell | Thermo Fisher Scientific Inc. | EC0114 |
| Adenovirus | Takara Inc. | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| CNOT6L (residues 158–555 a.a.) | (Wang et al., 2010) | N/A |
| CNOT6 (residues 153-557a.a.) | (Wang et al., 2010) | N/A |
| CNOT7 (Full length) | (Yang et al., 2008) | N/A |
| CNOT8 (Full length) | (Yang et al., 2008) | N/A |
| Glutathione-Sepharose 4B | GenScript | L00206 |
| HRV GST-3C protease | Acrobiosystem | 3CCN31331000U |
| BML-2840 ICCB Known Bioactives Library | Enzo Life Sciences, Inc. | BML-2840-0500 |
| BML-2865 Natural Product Library | Enzo Life Sciences, Inc. | BML-2865-0500 |
| Isopropyl β-D-1-thiogalactopyranoside | Thermo Fisher Scientific Inc. | 34060 |
| Dynabeads Protein G | Thermo Fisher Scientific Inc. | 10004D |
| TRIzol Reagent | Thermo Fisher Scientific Inc. | 15596018 |
| GlycoBlue Coprecipitant | Thermo Fisher Scientific Inc. | AM9516 |
| Actinomycin D | Sigma-Aldrich | A1410 |
| DMSO | Corning | 25950CQC |
| WY-14643 | Cayman Chemical | 7073010 |
| Thapsigargin | Sigma-Aldrich | T9033 |
| Tris | Thermo Fisher Scientific Inc. | BP152 |
| EDTA | Thermo Fisher Scientific Inc. | BP118 |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Sigma-Aldrich | 11873580001 |
| IGEPAL CA-630 (NP-40) | Sigma-Aldrich | 56741 |
| Triton X-100 | Thermo Fisher Scientific Inc. | BP151 |
| Sodium deoxycholate | Thermo Fisher Scientific Inc. | BP349 |
| HEPES | Thermo Fisher Scientific Inc. | BP310 |
| Cycloheximide | Sigma-Aldrich | C7698 |
| Sucrose | Thermo Fisher Scientific Inc. | BP220 |
| NaCl | Thermo Fisher Scientific Inc. | BP358 |
| KCl | Thermo Fisher Scientific Inc. | BP366 |
| MgCl2 | Thermo Fisher Scientific Inc. | M33 |
| DMEM | Corning | 10-013-CV |
| FBS | Sigma-Aldrich | F8067 |
| Penicillin-Streptomycin Solution | Corning | 30-002-Cl |
| Percoll | Sigma-Aldrich | P7828 |
| Perfusion medium | GIBCO | 17701-038 |
| Liver Digest Medium | GIBCO | 17703-034 |
| William’s E Medium | GIBCO | A1217601 |
| Primary Hepatocyte Thawing and Plating Supplements | GIBCO | CM3000 |
| Primary Hepatocyte Maintenance Supplements | GIBCO | CM4000 |
| Collagen type I | Corning | 354236 |
| Insulin | Novo Nordisk | NDC 0169-1833-02 |
| LY2405319 | (Gaich et al., 2013) | N/A |
| Poly(I:C) | InvivoGen | tlrl-pic |
| High-fat diet | Research Diets, Inc. | D12492 |
| Standard diet | Research Diets, Inc. | D12450B |
| 10% Formalin | Thermo Fisher Scientific Inc. | 245-684 |
| 2-propanol | Thermo Fisher Scientific Inc. | A416 |
| O.C.T. compound | Sakura Finetek USA, Inc. | 4583 |
| VECTASHIELD® Hardset Antifade Mounting Medium with DAPI | Thermo Fisher Scientific Inc. | H-1500 |
| Critical commercial assays | ||
| Amicon Ultra-4 10K centrifuge filter | Millipore | Z648027 |
| COSMOSIL 3C18-MS-II | Nacalai Tesque | 05514-01 |
| Magnetic Separator | Invitrogen | 12321D |
| RNeasy Mini Kit | Qiagen | NC9677589 |
| High-Capacity cDNA Reverse Transcription Kit | Applied Biosystems | 4368814 |
| PowerUp SYBR Green Master Mix | Applied Biosystems | A25742 |
| TruSeq Stranded mRNA Library Prep | Illumina, Inc. | 20020594 |
| Lipofectamine 3000 | Invitrogen | L3000-015 |
| Dual-Glo Luciferase Assay System | Promega | PR-E2940 |
| Bio-Rad protein assay | Bio-Rad Laboratories, Inc. | 5000006 |
| 24GA 0.75IN 0.7x19mm yellow, closed IV catheter | BD Medical | BD 383511 |
| 100μm cell strainer | Falcon | 352360 |
| Glucose meter | Bayer | BD15789 |
| LabAssay Triglyceride | Wako | 290-63701 |
| Autokit Total Ketone Bodies | Wako | 415-73301 411-73401 |
| FGF21 MOUSE ELISA | BioVender Inc. | RD291108200R |
| Mouse/Rat GDF-15 Quantikine ELISA Kit | R&D Systems | MGD150 |
| Ultra Sensitive Mouse Insulin ELISA Kit | Crystal Chem Inc. | 90080 |
| Deposited data | ||
| RIP-CHIP raw data | (Morita et al., 2019) | GEO: GSE62365 |
| RNA-seq raw data from livers | This paper | GEO: GSE185851 |
| Experimental models: Cell lines | ||
| Huh7 cells | JCRB | JCRB0403 |
| Mouse primary hepatocytes | This paper | N/A |
| 293T cells | ATCC | CRL-3216 |
| 293 | ATCC | CRL-1573 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Jackson laboratory | 000664 |
| Mouse: Cnot6l−/− | (Morita et al., 2019) | N/A |
| Mouse: Fgf21f/f | Jackson laboratory | 022361 |
| Mouse: Alb-cre | Jackson laboratory | 003574 |
| Mouse: Cnot6ltm1a(EUCOMM)Wts | EMMA | N/A |
| Mouse: Cnot6tm1a(KOMP)Wtsi | MMRRC | N/A |
| Mouse: Cnot6lf/f | This paper | N/A |
| Mouse: Cnot6f/f | This paper | N/A |
| Mouse: ACTB-FLPe | Jackson laboratory | 005703 |
| Mouse: Gdf15tm1a(KOMP)Wtsi | MMRRC | N/A |
| Oligonucleotides | ||
| 6-FAM-labeled poly(A) RNA | Sigma-Aldrich | N/A |
| TAMRA-labeled oligo (dT) DNA | Sigma-Aldrich | N/A |
| FITC-labeled poly(A)20 RNA | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| pGL3-control | Promega | PR-E1741 |
| pGL3-3’UTR of human GDF15 mRNA | This paper | N/A |
| pGL3-3’UTR of mouse Gdf15 mRNA | This paper | N/A |
| pGL3-3’UTR of human FGF21 mRNA | This paper | N/A |
| pGL3-3’UTR of mouse Fgf21 mRNA | This paper | N/A |
| pRL-TK | Promega | PR-E2241 |
| pBAsi-mU6-shGfp | Takara Bio. | 3222 |
| pBAsi-mU6-shGdf15 | This paper | N/A |
| pBAsi-mU6-shFgf21 | This paper | N/A |
| pBAsi-mU6-shCnot6 | This paper | N/A |
| pBAsi-mU6-shCnot6l | This paper | N/A |
| pAxcwit2-shGfp | Takara Bio. | 6170 |
| pAxcwit2-shGdf15 | This paper | N/A |
| pAxcwit2-shFgf21 | This paper | N/A |
| pAxcwit2-shCnot6 | This paper | N/A |
| pAxcwit2-shCnot6l | This paper | N/A |
| pAAV.TBG.PI.Cre.rBG | Addgene | 107787 |
| pAAV.TBG.PI.Null.bGH | Addgene | 105536 |
| pCMV-Myc-CNOT6L | This paper | N/A |
| pcDNA3-flag-YTHDF2 | Addgene | 52300 |
| Software and algorithms | ||
| Molecular Operating Environment | Chemical Computing Group | https://www.chemcomp.com/ |
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/ |
| Endnote | Clarivate | https://endnote.com/ |
| Illustrator 2021 | Adobe | https://www.adobe.com/ |
| Photoshop 2021 | Adobe | https://www.adobe.com/ |
| Other | ||
| iBright FL1000 Imaging System | Thermo Fisher Scientific Inc. | A32752 |
| A Biacore system | GE Healthcare | 28975001 |
| Liquid Chromatograph Mass Spectrometer | Shimadzu | LCMS-8050 |
| NanoDrop Onec | Thermo Fisher Scientific Inc. | 027420P21H2 |
| QuantStudio 3 Real-Time PCR System | Thermo Fisher Scientific Inc. | A28136 |
| Varioskan LUX Multimode Microplate Reader | Thermo Fisher Scientific Inc. | VL0L00D0 |
| 4-lane motorized treadmill | Columbus Instruments | 1012M-4 |
| Fluorescence microscopy | Keyence | BZ-X800 |
Highlights.
Hepatic CNOT6L controls food intake, energy expenditure, and fat utilization
Gdf15 and Fgf21 mRNAs are degraded by CNOT6L deadenylase in response to stimuli
GDF15 and FGF21 mediate the CNOT6L effects on food intake and energy expenditure
Targeting CNOT6L has a therapeutic potential to treat diet-induced metabolic disorders
ACKNOWLEDGMENTS
We thank T. Yamamoto for Cnot6l−/− mice; M. Kato-Murayama for the protein preparation; D.E. Moller for LY2405319; the Animal Facility and the Histology Facility at the University of Texas Health Science Center at San Antonio (UTHSCSA) for mouse experiments and histological analyses; the Genome Sequencing Facility and the Bioanalytics and Single-Cell Core Facility at UTHSCSA for RNA-seq and technical assistance.
This work was supported by the University of Texas Rising Stars Award, Cancer Prevention and Research Institute of Texas (CPRIT) Award (RP220267), Helen F. Kerr Foundation Grant, Shelby Tengg Foundation Grant, Cancer Center Support Grant (P30 CA054174), Grant-in-Aid for Scientific Research (18K07237 and 21K07102), and JST FOREST Program. S.K. was supported by AHA Postdoctoral Fellowship, JSPS Overseas Research Fellowship, and Uehara Memorial Foundation Postdoctoral Fellowship. T.H.-M.H. was supported by NIH grants U54 CA217297 and P30 CA054174 and CPRIT grant RP150600. I.T is a Senior Scholar of the Fonds de la recherche du Québec—Santé (FRQS) and was supported by a grant from the Canadian Institutes of Health Research (CIHR PJT-175050). O.L. is a Wallenberg Academy Fellow and was supported by grants from the Swedish Research Council and the Swedish Cancer Society. N.M. was supported by San Antonio Claude D. Pepper Older Americans Independence Center (P30 AG044271), the San Antonio Nathan Shock Center of Excellence on the Biology of Aging (P30 AG021890), grants from the NIH (R01-DK80157 and R01-DK089229), and the American Diabetes Association.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information containing 7 Figures, 5 Tables, and 1 Data can be found online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Basquin J, Roudko VV, Rode M, Basquin C, Seraphin B, and Conti E (2012). Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Mol Cell 48, 207–218. [DOI] [PubMed] [Google Scholar]
- Bhaskar V, Roudko V, Basquin J, Sharma K, Urlaub H, Seraphin B, and Conti E (2013). Structure and RNA-binding properties of the Not1-Not2-Not5 module of the yeast Ccr4-Not complex. Nat Struct Mol Biol 20, 1281–1288. [DOI] [PubMed] [Google Scholar]
- Boland A, Chen Y, Raisch T, Jonas S, Kuzuoglu-Ozturk D, Wohlbold L, Weichenrieder O, and Izaurralde E (2013). Structure and assembly of the NOT module of the human CCR4-NOT complex. Nat Struct Mol Biol 20, 1289–1297. [DOI] [PubMed] [Google Scholar]
- Chaaban I, Rizk OH, Ibrahim TM, Henen SS, El-Khawass EM, Bayad AE, El-Ashmawy IM, and Nematalla HA (2018). Synthesis, anti-inflammatory screening, molecular docking, and COX-1,2/−5-LOX inhibition profile of some novel quinoline derivatives. Bioorg Chem 78, 220–235. [DOI] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, and Kim VN (2014). TAIL-seq: genome-wide determination of poly(A) tail length and 3' end modifications. Mol Cell 53, 1044–1052. [DOI] [PubMed] [Google Scholar]
- Charles ED, Neuschwander-Tetri BA, Pablo Frias J, Kundu S, Luo Y, Tirucherai GS, and Christian R (2019). Pegbelfermin (BMS-986036), PEGylated FGF21, in Patients with Obesity and Type 2 Diabetes: Results from a Randomized Phase 2 Study. Obesity (Silver Spring) 27, 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll AP, Chen M, Taskar P, Rimmington D, Patel S, Tadross JA, Cimino I, Yang M, Welsh P, Virtue S, et al. (2020). GDF15 mediates the effects of metformin on body weight and energy balance. Nature 578, 444–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart MA (2016). The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip Rev RNA 7, 438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day EA, Ford RJ, Smith BK, Mohammadi-Shemirani P, Morrow MR, Gutgesell RM, Lu R, Raphenya AR, Kabiri M, McArthur AG, et al. (2019). Metformin-induced increases in GDF15 are important for suppressing appetite and promoting weight loss. Nat Metab 1, 1202–1208. [DOI] [PubMed] [Google Scholar]
- Du H, Zhao Y, He J, Zhang Y, Xi H, Liu M, Ma J, and Wu L (2016). YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun 7, 12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson PJ, Wang F, Du Y, Liu Q, Pickard RT, Gonciarz MD, Coskun T, Hamang MJ, Sindelar DK, Ballman KK, et al. (2017). The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nat Med 23, 1215–1219. [DOI] [PubMed] [Google Scholar]
- Feldmann HM, Golozoubova V, Cannon B, and Nedergaard J (2009). UCP1 ablation induces obesity and abolishes diet-induced thermogenesis in mice exempt from thermal stress by living at thermoneutrality. Cell Metab 9, 203–209. [DOI] [PubMed] [Google Scholar]
- Fisher FM, Kim M, Doridot L, Cunniff JC, Parker TS, Levine DM, Hellerstein MK, Hudgins LC, Maratos-Flier E, and Herman MA (2017). A critical role for ChREBP-mediated FGF21 secretion in hepatic fructose metabolism. Mol Metab 6, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher FM, and Maratos-Flier E (2016). Understanding the Physiology of FGF21. Annu Rev Physiol 78, 223–241. [DOI] [PubMed] [Google Scholar]
- Flippo KH, and Potthoff MJ (2021). Metabolic Messengers: FGF21. Nat Metab 3, 309–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler CJ (2021). The endocannabinoid system - current implications for drug development. J Intern Med 290, 2–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Sikstrom K, Alain T, Morita M, McLaughlan S, Larsson O, and Topisirovic I (2014). Polysome fractionation and analysis of mammalian translatomes on a genome-wide scale. J Vis Exp 87, e51455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng L, Lam KSL, and Xu A (2020). The therapeutic potential of FGF21 in metabolic diseases: from bench to clinic. Nat Rev Endocrinol 16, 654–667. [DOI] [PubMed] [Google Scholar]
- Hashimoto O, Funaba M, Sekiyama K, Doi S, Shindo D, Satoh R, Itoi H, Oiwa H, Morita M, Suzuki C, et al. (2018). Activin E Controls Energy Homeostasis in Both Brown and White Adipose Tissues as a Hepatokine. Cell Rep 25, 1193–1203. [DOI] [PubMed] [Google Scholar]
- Hawley JA, Hargreaves M, Joyner MJ, and Zierath JR (2014). Integrative biology of exercise. Cell 159, 738–749. [DOI] [PubMed] [Google Scholar]
- Hsu JY, Crawley S, Chen M, Ayupova DA, Lindhout DA, Higbee J, Kutach A, Joo W, Gao Z, Fu D, et al. (2017). Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature 550, 255–259. [DOI] [PubMed] [Google Scholar]
- Hu X, Yin B, Cappelle K, Swevers L, Smagghe G, Yang X, and Zhang L (2018). Identification of novel agonists and antagonists of the ecdysone receptor by virtual screening. J Mol Graph Model 81, 77–85. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Dutchak P, Zhao G, Ding X, Gautron L, Parameswara V, Li Y, Goetz R, Mohammadi M, Esser V, et al. (2007). Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell Metab 5, 415–425. [DOI] [PubMed] [Google Scholar]
- Inagaki T, Lin VY, Goetz R, Mohammadi M, Mangelsdorf DJ, and Kliewer SA (2008). Inhibition of growth hormone signaling by the fasting-induced hormone FGF21. Cell Metab 8, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsenty G, and Olson EN (2016). Bone and Muscle Endocrine Functions: Unexpected Paradigms of Inter-organ Communication. Cell 164, 1248–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazak L, Chouchani ET, Lu GZ, Jedrychowski MP, Bare CJ, Mina AI, Kumari M, Zhang S, Vuckovic I, Laznik-Bogoslavski D, et al. (2017). Genetic Depletion of Adipocyte Creatine Metabolism Inhibits Diet-Induced Thermogenesis and Drives Obesity. Cell Metab 26, 660–671 e663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke S, Pandya-Jones A, Saito Y, Fak JJ, Vagbo CB, Geula S, Hanna JH, Black DL, Darnell JE Jr., and Darnell RB (2017). m(6)A mRNA modifications are deposited in nascent pre-mRNA and are not required for splicing but do specify cytoplasmic turnover. Genes Dev 31, 990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, and Adams AC (2014). Inventing new medicines: The FGF21 story. Mol Metab 3, 221–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Kim SH, Min YK, Yang HM, Lee JB, and Lee MS (2013). Acute exercise induces FGF21 expression in mice and in healthy humans. PLoS One 8, e63517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Junior LC, Campos A, Niero R, Correa R, Vander Heyden Y, and Filho VC (2020). Xanthones and Cancer: from Natural Sources to Mechanisms of Action. Chem Biodivers 17, e1900499. [DOI] [PubMed] [Google Scholar]
- Klein AB, Nicolaisen TS, Ortenblad N, Gejl KD, Jensen R, Fritzen AM, Larsen EL, Karstoft K, Poulsen HE, Morville T, et al. (2021). Pharmacological but not physiological GDF15 suppresses feeding and the motivation to exercise. Nat Commun 12, 1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinert M, Clemmensen C, Sjoberg KA, Carl CS, Jeppesen JF, Wojtaszewski JFP, Kiens B, and Richter EA (2018). Exercise increases circulating GDF15 in humans. Mol Metab 9, 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, and Mangelsdorf DJ (2019). A Dozen Years of Discovery: Insights into the Physiology and Pharmacology of FGF21. Cell Metab 29, 246–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai KK, Kolippakkam D, and Beretta L (2008). Comprehensive and quantitative proteome profiling of the mouse liver and plasma. Hepatology 47, 1043–1051. [DOI] [PubMed] [Google Scholar]
- Lan T, Morgan DA, Rahmouni K, Sonoda J, Fu X, Burgess SC, Holland WL, Kliewer SA, and Mangelsdorf DJ (2017). FGF19, FGF21, and an FGFR1/beta-Klotho-Activating Antibody Act on the Nervous System to Regulate Body Weight and Glycemia. Cell Metab 26, 709–718 e703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Sonenberg N, and Nadon R (2010). Identification of differential translation in genome wide studies. Proc Natl Acad Sci U S A 107, 21487–21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson O, Sonenberg N, and Nadon R (2011). anota: Analysis of differential translation in genome-wide studies. Bioinformatics 27, 1440–1441. [DOI] [PubMed] [Google Scholar]
- Li X, Morita M, Kikuguchi C, Takahashi A, Suzuki T, and Yamamoto T (2017). Adipocyte-specific disruption of mouse Cnot3 causes lipodystrophy. FEBS Lett 591, 358–368. [DOI] [PubMed] [Google Scholar]
- Luan HH, Wang A, Hilliard BK, Carvalho F, Rosen CE, Ahasic AM, Herzog EL, Kang I, Pisani MA, Yu S, et al. (2019). GDF15 Is an Inflammation-Induced Central Mediator of Tissue Tolerance. Cell 178, 1231–1244 e1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markan KR, Naber MC, Ameka MK, Anderegg MD, Mangelsdorf DJ, Kliewer SA, Mohammadi M, and Potthoff MJ (2014). Circulating FGF21 is liver derived and enhances glucose uptake during refeeding and overfeeding. Diabetes 63, 4057–4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maryati M, Kaur I, Jadhav GP, Olotu-Umoren L, Oveh B, Hashmi L, Fischer PM, and Winkler GS (2014). A fluorescence-based assay suitable for quantitative analysis of deadenylase enzyme activity. Nucleic Acids Res 42, e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meex RCR, and Watt MJ (2017). Hepatokines: linking nonalcoholic fatty liver disease and insulin resistance. Nat Rev Endocrinol 13, 509–520. [DOI] [PubMed] [Google Scholar]
- Morita M, Oike Y, Nagashima T, Kadomatsu T, Tabata M, Suzuki T, Nakamura T, Yoshida N, Okada M, and Yamamoto T (2011). Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3+/− mice. EMBO J 30, 4678–4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita M, Prudent J, Basu K, Goyon V, Katsumura S, Hulea L, Pearl D, Siddiqui N, Strack S, McGuirk S, et al. (2017). mTOR Controls Mitochondrial Dynamics and Cell Survival via MTFP1. Mol Cell 67, 922–935 e925. [DOI] [PubMed] [Google Scholar]
- Morita M, Siddiqui N, Katsumura S, Rouya C, Larsson O, Nagashima T, Hekmatnejad B, Takahashi A, Kiyonari H, Zang M, et al. (2019). Hepatic posttranscriptional network comprised of CCR4-NOT deadenylase and FGF21 maintains systemic metabolic homeostasis. Proc Natl Acad Sci U S A 116, 7973–7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa D, Yanagiya A, Georgiadou E, Wu Y, Stylianides T, Rutter GA, Suzuki T, and Yamamoto T (2020). Loss of beta-cell identity and diabetic phenotype in mice caused by disruption of CNOT3-dependent mRNA deadenylation. Commun Biol 3, 476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugridge JS, Coller J, and Gross JD (2018). Structural and molecular mechanisms for the control of eukaryotic 5'-3' mRNA decay. Nat Struct Mol Biol 25, 1077–1085. [DOI] [PubMed] [Google Scholar]
- Mullican SE, Lin-Schmidt X, Chin CN, Chavez JA, Furman JL, Armstrong AA, Beck SC, South VJ, Dinh TQ, Cash-Mason TD, et al. (2017). GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nat Med 23, 1150–1157. [DOI] [PubMed] [Google Scholar]
- Mullican SE, and Rangwala SM (2018). Uniting GDF15 and GFRAL: Therapeutic Opportunities in Obesity and Beyond. Trends Endocrinol Metab 29, 560–570. [DOI] [PubMed] [Google Scholar]
- Nicholson AL, and Pasquinelli AE (2019). Tales of Detailed Poly(A) Tails. Trends Cell Biol 29, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Yi H, Kim Y, Chang H, and Kim VN (2016). Regulation of Poly(A) Tail and Translation during the Somatic Cell Cycle. Mol Cell 62, 462–471. [DOI] [PubMed] [Google Scholar]
- Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, and Kim YK (2019). Endoribonucleolytic Cleavage of m(6)A-Containing RNAs by RNase P/MRP Complex. Mol Cell 74, 494–507 e498. [DOI] [PubMed] [Google Scholar]
- Passmore LA, and Coller J (2022). Roles of mRNA poly(A) tails in regulation of eukaryotic gene expression. Nat Rev Mol Cell Biol 23, 93–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Alvarez-Guaita A, Melvin A, Rimmington D, Dattilo A, Miedzybrodzka EL, Cimino I, Maurin AC, Roberts GP, Meek CL, et al. (2019). GDF15 Provides an Endocrine Signal of Nutritional Stress in Mice and Humans. Cell Metab 29, 707–718 e708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinzi L, Caporuscio F, and Rastelli G (2018). Selection of protein conformations for structure-based polypharmacology studies. Drug Discov Today 23, 1889–1896. [DOI] [PubMed] [Google Scholar]
- Potthoff MJ, Inagaki T, Satapati S, Ding X, He T, Goetz R, Mohammadi M, Finck BN, Mangelsdorf DJ, Kliewer SA, et al. (2009). FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proc Natl Acad Sci U S A 106, 10853–10858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priest C, and Tontonoz P (2019). Inter-organ cross-talk in metabolic syndrome. Nat Metab 1, 1177–1188. [DOI] [PubMed] [Google Scholar]
- Prieto C, and Barrios D (2019). RaNA-Seq: Interactive RNA-Seq analysis from FASTQ files to functional analysis. Bioinformatics. [DOI] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. (2011). Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol 29, 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz JR, Goldsmith MR, Little SB, and Pasquinelli MA (2008). Computational molecular modeling for evaluating the toxicity of environmental chemicals: prioritizing bioassay requirements. Environ Health Perspect 116, 573–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisch T, Chang CT, Levdansky Y, Muthukumar S, Raunser S, and Valkov E (2019). Reconstitution of recombinant human CCR4-NOT reveals molecular insights into regulated deadenylation. Nat Commun 10, 3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizel Y, Morgan A, Gao L, Lan Y, Manduchi E, Waite EL, Wang AW, Wells A, and Kaestner KH (2020). Collapse of the hepatic gene regulatory network in the absence of FoxA factors. Genes Dev 34, 1039–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, and Larsson O (2007). Improved precision and accuracy for microarrays using updated probe set definitions. BMC Bioinformatics 8, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford JA, Nogiec CD, Lindholm ME, Adkins JN, Amar D, Dasari S, Drugan JK, Fernandez FM, Radom-Aizik S, Schenk S, et al. (2020). Molecular Transducers of Physical Activity Consortium (MoTrPAC): Mapping the Dynamic Responses to Exercise. Cell 181, 1464–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A, Charles ED, Neuschwander-Tetri BA, Loomba R, Harrison SA, Abdelmalek MF, Lawitz EJ, Halegoua-DeMarzio D, Kundu S, Noviello S, et al. (2019). Pegbelfermin (BMS-986036), a PEGylated fibroblast growth factor 21 analogue, in patients with non-alcoholic steatohepatitis: a randomised, double-blind, placebo-controlled, phase 2a trial. Lancet 392, 2705–2717. [DOI] [PubMed] [Google Scholar]
- Scheja L, and Heeren J (2019). The endocrine function of adipose tissues in health and cardiometabolic disease. Nat Rev Endocrinol 15, 507–524. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, and Selbach M (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Sekiyama K, Ushiro Y, Kurisaki A, Funaba M, and Hashimoto O (2019). Activin E enhances insulin sensitivity and thermogenesis by activating brown/beige adipocytes. J Vet Med Sci 81, 646–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefan N, and Haring HU (2013). The role of hepatokines in metabolism. Nat Rev Endocrinol 9, 144–152. [DOI] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, and Bartel DP (2014). Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Adachi S, Morita M, Tokumasu M, Natsume T, Suzuki T, and Yamamoto T (2015). Post-transcriptional Stabilization of Ucp1 mRNA Protects Mice from Diet-Induced Obesity. Cell Rep 13, 2756–2767. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Suzuki T, Soeda S, Takaoka S, Kobori S, Yamaguchi T, Mohamed HMA, Yanagiya A, Abe T, Shigeta M, et al. (2020). The CCR4-NOT complex maintains liver homeostasis through mRNA deadenylation. Life Sci Alliance 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi A, Takaoka S, Kobori S, Yamaguchi T, Ferwati S, Kuba K, Yamamoto T, and Suzuki T (2019). The CCR4-NOT Deadenylase Complex Maintains Adipocyte Identity. Int J Mol Sci 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai VWW, Husaini Y, Sainsbury A, Brown DA, and Breit SN (2018). The MIC-1/GDF15-GFRAL Pathway in Energy Homeostasis: Implications for Obesity, Cachexia, and Other Associated Diseases. Cell Metab 28, 353–368. [DOI] [PubMed] [Google Scholar]
- Tu Y, Yang Y, Li Y, and He C (2021). Naturally occurring coumestans from plants, their biological activities and therapeutic effects on human diseases. Pharmacol Res 169, 105615. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- Ukleja M, Cuellar J, Siwaszek A, Kasprzak JM, Czarnocki-Cieciura M, Bujnicki JM, Dziembowski A, and Valpuesta JM (2016). The architecture of the Schizosaccharomyces pombe CCR4-NOT complex. Nat Commun 7, 10433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya F, Gavalda-Navarro A, Peyrou M, Villarroya J, and Giralt M (2017). The Lives and Times of Brown Adipokines. Trends Endocrinol Metab 28, 855–867. [DOI] [PubMed] [Google Scholar]
- Wang GX, Zhao XY, Meng ZX, Kern M, Dietrich A, Chen Z, Cozacov Z, Zhou D, Okunade AL, Su X, et al. (2014a). The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med 20, 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Morita M, Yang X, Suzuki T, Yang W, Wang J, Ito K, Wang Q, Zhao C, Bartlam M, et al. (2010). Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. EMBO J 29, 2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014b). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe C, Morita M, Hayata T, Nakamoto T, Kikuguchi C, Li X, Kobayashi Y, Takahashi N, Notomi T, Moriyama K, et al. (2014). Stability of mRNA influences osteoporotic bone mass via CNOT3. Proc Natl Acad Sci U S A 111, 2692–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster MW, Chen YH, Stowell JAW, Alhusaini N, Sweet T, Graveley BR, Coller J, and Passmore LA (2018). mRNA Deadenylation Is Coupled to Translation Rates by the Differential Activities of Ccr4-Not Nucleases. Mol Cell 70, 1089–1100 e1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widhalm JR, and Rhodes D (2016). Biosynthesis and molecular actions of specialized 1,4-naphthoquinone natural products produced by horticultural plants. Hortic Res 3, 16046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, Zhao XY, Ji Y, Li C, Guo L, et al. (2019). Landscape of Intercellular Crosstalk in Healthy and NASH Liver Revealed by Single-Cell Secretome Gene Analysis. Mol Cell 75, 644–660 e645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, Walker K, Min X, Hale C, Tran T, Komorowski R, Yang J, Davda J, Nuanmanee N, Kemp D, et al. (2017). Long-acting MIC-1/GDF15 molecules to treat obesity: Evidence from mice to monkeys. Sci Transl Med 9. [DOI] [PubMed] [Google Scholar]
- Yang L, Chang CC, Sun Z, Madsen D, Zhu H, Padkjaer SB, Wu X, Huang T, Hultman K, Paulsen SJ, et al. (2017). GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nat Med 23, 1158–1166. [DOI] [PubMed] [Google Scholar]
- Yang X, Morita M, Wang H, Suzuki T, Yang W, Luo Y, Zhao C, Yu Y, Bartlam M, Yamamoto T, et al. (2008). Crystal structures of human BTG2 and mouse TIS21 involved in suppression of CAF1 deadenylase activity. Nucleic Acids Res 36, 6872–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H, Park J, Ha M, Lim J, Chang H, and Kim VN (2018). PABP Cooperates with the CCR4-NOT Complex to Promote mRNA Deadenylation and Block Precocious Decay. Mol Cell 70, 1081–1088 e1085. [DOI] [PubMed] [Google Scholar]
- Zaccara S, and Jaffrey SR (2020). A Unified Model for the Function of YTHDF Proteins in Regulating m(6)A-Modified mRNA. Cell 181, 1582–1595 e1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccara S, Ries RJ, and Jaffrey SR (2019). Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol 20, 608–624. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Yan D, Guo E, Ding B, Yang W, Liu R, Yamamoto T, and Bartlam M (2016). Structural basis for inhibition of the deadenylase activity of human CNOT6L. FEBS Lett 590, 1270–1279. [DOI] [PubMed] [Google Scholar]
- Zhao BS, Roundtree IA, and He C (2017). Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol 18, 31–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of CNOT6L-interacting mRNAs encoding hepatic secretory proteins. Related to Figure 2.
Table S2. Functional enrichment analysis of RNA-seq data from livers of mice treated with/without iD1. Related to Figure 2.
Table S3. List of qPCR primers. Related to STAR Methods.
Table S4. List of cloning primers and shRNA target sequences. Related to STAR Methods.
Table S5. Information about the statistical analyses. Related to STAR Methods.
Data Availability Statement
The bulk RNA-seq data are publicly available at GEO under accession number (GSE185851). The data generated in this study will be shared by the lead contact upon reasonable request.
This study did not generate new code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.