Abstract
Background: TP53 protein is lost or mutated in about half of all types of human cancers and small molecules to regulate mutant p53 repair, or interrupt ubiquitination degradation of p53 induced by E3-ubiquitin ligase Mdm2 have a potential application in clinical application. Methods: To inhibit the deubiquitinase activity of 19S proteasome and restore the p53 protein level, in this study, we utilized p53 knockout mice to test the anti-cancer effect of a specific USP14 and UCH37 inhibitor b-AP15. Results: Our results show that UCHL5, USP14 and COPS5 are upregulated in p53-related tumors, and higher expression of these genes results in a shorter overall survival in patients with p53 deficiency. Treatment with b-AP15, a UCHL5 and USP14 deubiquitinating activity inhibitor in 19S regulatory subunit, induces tumor regression and prolong the survival period of tumor-loaded mice through down-regulation of COPS5 and its downstream AP-1 and E2F1, and up-regulation of the cell cycle-related proteins p27 and Cyclin E1. Conclusions: Thus, our results suggested that inhibition of UCHL5 and USP14 deubiquitinating activity in 19S proteasome may contribute an extensive approach to preventing tumor progress due to p53 deficiency.
Keywords: therapy, p53, cancer, inhibitor, UCHL5, USP14
Introduction
Wild-type p53 protein is crucial for maintaining genome integrity and cell physiological function. 1 The transcriptional activity of p53 activate downstream target genes, including miR-34a, MDM2, BAX, GADD45, PCNA, NOXA, and CDKN1A, which are culpable for DNA repair, cell-cycle control, apoptosis, or senescence. 2 Therefore, p53 protein reduction, mutational inactivation or loss-of-function existent in over 50% of human cancers, brings about incontrollable cell cycle check-point, senescence and apoptosis. 3
A mouse strain bearing deletion of p53 gene was constructed using gene targeting technology in which the role of p53 for malignant transformation was analysis and the anti-cancer effect of small molecule inhibitors was tested. 4 Homozygous p53-deficient mice develop and reproduce normally; however, adult mice in 4-6 months are highly susceptible to cancer, 5 due to definition of programmed cell death or cell-cycle arrest and increase of damaged cells following DNA damage. 6 Previous studies have confirmed that thymic lymphoma was the most common tumor found in mice with p53 deficiency, and osteosarcoma and soft tissue sarcoma were also detected. 7 Therefore, p53-deficient mouse strains were widely used for research on gene function and mechanism, drug target research, and development of new p53 targeted drugs.
As reported, small molecule inhibitors was researching for functional restoration of mutant p53 to wild-type, inducing p53 level increase, disruption of the binding between murine double minute-2 (Mdm2) and p53 to prevent p53 ubiquitination and degradation. 8 However, modesty of 19S proteasome deubiquitinating activity to prevent p53 ubiquitination degradation and induce tumor regression is not reported.
In this study, we utilized p53 knockout mice, in which p53 deficiency induced genesis and development of neoplasm in adult mice, to test the anti-cancer effect of a small molecule inhibitor. Our results show that treatment with b-AP15, an ubiquitin C-terminal hydrolase L5 (UCHL5) and ubiquitin specific peptidase 14 (USP14) deubiquitinating activity inhibitor, in 19S regulatory subunit, results in tumor regression and prolong the survival period of tumor-loaded mice through down-regulation of COPS5 and its downstream AP-1 and E2F1, and up-regulation of the cell cycle-related proteins p27 and Cyclin E1. Thus, our results suggested that inhibition of UCHL5 and USP14 deubiquitinating activity in 19S proteasome may contribute an extensive approach to preventing tumor progress due to p53 deficiency.
Materials and Methods
Data Processing and Analysis of p53 Deletion-Related Tumors
The expression levels of p53 and three p53-related genes (UCHL5, USP14 and COPS5) were investigated in tumor samples and normal tissue samples in the TCGA and GEO dataset. We downloaded the transcriptome sequencing data of 42, 258 and 86 patients with p53 deletion-related tumors (including lymphoid lymphoma, sarcoma and osteosarcoma, respectively) through TCGA data portal (https://portal.gdc.cancer.gov/). The clinic pathological characteristics were recorded including the patient's clinical information, tumor characteristics, and overall survival (OS) for all patients. Overall survival was calculated from the first day of diagnosis until the time of last follow-up or death and evaluated by the Kaplan–Meier curves and differences between survival rates were examined using the log-rank test.
Additionally, two datasets of diffuse large B-cell lymphoma (DLBC) (GSE26725) and osteosarcoma (GSE28425) patients from Gene Expression Omnibus (GEO) dataset were used for gene expression analysis between normal tissues and cancer tissues: the gene expression profiling from 12 B-cell chronic lymphocytic leukemia (B-CLL) patient peripheral blood samples in comparison with 5 healthy donor peripheral blood samples were analyzed using GEO dataset GSE26725 and 19 osteosarcoma cell line and 4 human bone samples were selected for GEO dataset GSE28425.
Animal Studies
The experimental procedure was approved by the Institutional Animal Care and Use Committee of Shanghai Tenth People's Hospital (SY-2019-0215). Three-month-old wild-type and p53−/− mice of either sex were used in this study. We made efforts to minimize the number of animals utilized and to decrease their suffering. All procedures in the experiments were in compliance with Guide for the Care and Use of Laboratory Animals (eighth Edition) 9 and agreed by the Institutional Animal Care and Use Committee guidelines at Shanghai Tenth People's Hospital, Tongji University School of Medicine (SYDW-20-125). The reporting of this study conforms to ARRIVE 2.0 guidelines. 10
Genotype of mice was tested using polymerase chain reaction (PCR). About 10 ng template genomic DNA from tail biopsies was employed per reaction. Sequences of PCR primers for p53 genotyping and sequencing analysis are as follows: Forward, 5′-AGTTCTGCCACGTGGTTGGT-3; Reverse 1, 5′-GTCTCCTGGCTCAGAGGGAG-3′; Reverse 2, 5′-CAGAGGCCACTTGTGTAGCG-3′. Amplifying under the following PCR conditions: denaturation at 95 °C for 4 min, followed by 36 cycles at 95 °C for 40 s, 62.5 °C for 30 s, and 72 °C for 30 s Reaction products were analyzed by DNA agarose gel electrophoresis and then sequenced. For all analyses performed on transgenic mice, age-matched wild-type mice of the same genetic background were used as control animals.
b-AP15 is reported as an UCHL5 and USP14 inhibitor. 11 b-AP15 (5mg/kg, 1%DMSO + 30%PEG300 + 1%Tween80 + ddH2O) was given i.p. twice a week for the number of days indicated. b-AP15 was purchased from SelleckChem. Mice without any treatment were used as the mock, and mice treated with vehicle (1%DMSO + 30%PEG300 + 1%Tween80 + ddH2O) were used as the control. Mice were observed by magnetic resonance imaging, X-ray, or micro-CT diagnosis for tumor phenotypes once a week.
H&E Staining and Immunohistochemistry (IHC)
Standard H&E staining and IHC analysis were employed to analyze protein expression levels in wild-type and p53−/− mice. For IHC analysis, tissues were blocked with phosphate-buffered saline containing bovine serum albumin (BSA) and treated with 2% H2O2 for endogenous peroxidases inactivation. Antigen retrieval was performed at 97 °C for 40 min in citrate buffer (Dako, Carpinteria, Calif). Proliferative cell nuclear antigen (PCNA) was tested using a mouse monoclonal antibody diluted 1:50 (Clone PC 10, Dako). Staining for P53 used a monoclonal antibody (diluted 1:100) specifically reacting with the P53 (Clone ab90363; Abcam, Cambridge, MA, USA). Cleaved Caspase-3 was stained with a mouse monoclonal antibody diluted 1:500 (Clone ab214430; Abcam). COPS5 monoclonal antibody (clone sc-13157; Santa Cruz Biotechnology, Santa Cruz, Calif), p27 mouse monoclonal (Clone ab193379; Abcam) and AP-1 rabbit monoclonal antibody (Clone ab40766; Abcam) were used at a dilution of 1:150, 1:150 and 1:200, respectively. E2F1 antibody (Clone SAB4500682; Sigma, St Louis, Mo) and Cyclin E1 antibody (Clone SAB4503516; Sigma, St Louis, Mo) was used at a dilution of 1:50. Primary antibodies were labeled with streptavidin–horseradish peroxidase kits. Negative controls were included that were treated identically but received no primary antibody to analyze specific IHC staining.
Statistical Analysis
Means ± standard deviations were used for data expression. Categorical data were recounted using numbers and percentages. Statistical significance was assessed by one-way ANOVA with Tukey test or Mann-Whitney U test, and the results was comparing with two independent sample Wilcoxon rank sum test using SPSS 20.0 or Prism 6.0 GraphPad software. P < .05 indicates the level of significance.
Results
Expression Level and Clinical Significance of USP14 in Tumors with p53 Deficiency
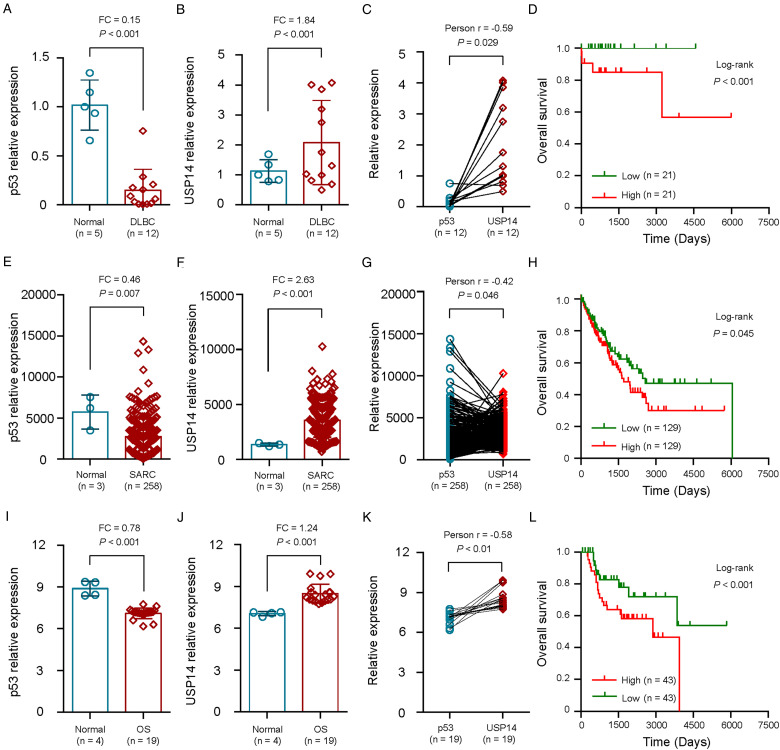
Considering the close association of 19S regulatory subunit deubiquitinases UCHL5 and USP14 with COPS5 in tumors with p53 deficiency, we firstly studied the expression levels and correlation of USP14 and p53 in normal or p53-related tumors. Our results indicated that the levels of p53 expression were significantly lower in DLBCs compared with normal samples in the GSE26725 dataset (Fold change (FC) = 0.15, P < .001; Figure 1a).
Figure 1.
Expression and clinical significance of USP14 in tumors with p53 deficiency. a-c, The expression level of p53 (a) and USP14 (b) and their correlation (c) in 12 DLBC tissues and 5 noncancerous tissues from GEO dataset GSE26725. d, The Kaplan-Meier method was used to evaluate the relationship between USP14 expression and overall survival of 42 DLBC patients from TCGA dataset. e-g, The expression level of p53 (e) and USP14 (f) and their correlation (g) in 258 SARC tissues and 3 noncancerous tissues from TCGA dataset. h, The Kaplan-Meier method was used to evaluate the relationship between USP14 expression and overall survival of 258 SARC patients from TCGA dataset. i-l, The expression level of p53 (i) and USP14 (j) and their correlation (k) in 19 OS tissues and 4 noncancerous tissues from GEO dataset GSE28425. l, The Kaplan-Meier method was used to evaluate the relationship between USP14 expression and overall survival of 86 OS patients from TCGA dataset. DLBC, diffuse large b-cell lymphoma; SARC, sarcoma; OS, osteosarcoma.
However, USP14 over-expressed in DLBCs compare with normal tissues (FC = 1.84, P < .001; Figure 1b). Besides, we analyzed the correlation between the expression level of p53 and USP14 in 12 DLBC samples from GEO dataset (GSE26725). As showed in Figure 1c, the relationship between p53 expression level and USP14 expression level presented inverse correlation (r = −0.59, P = .029; Figure 1c).
We then evaluated the prognostic value of USP14 expression levels for 42 patients with DLBC from TCGA dataset. The OS in DLBC patients with a low expression of USP14 was prolonged when compared to patients with high expression of USP14 (P < .001; Figure 1d), which indicated that USP14 expression was positively correlated with lower OS in DLBC patients.
Similarly, p53 expression was significantly lower in sarcomas when compared with normal samples in the TCGA dataset (FC = 0.46, P = .007; Figure 1e) and USP14 expression was significantly higher in sarcomas when compared with normal samples (FC = 2.63, P < .001; Figure 1f). The relationship between p53 expression level and USP14 expression level presented inverse correlation in 258 sarcoma patients (r = −0.42, P = .046; Figure 1g). We then explored the clinical significance of USP14 in sarcoma patients and the results showed that high USP14 expression was correlated with lower OS (P = .045; Figure 1h) in 258 sarcoma patients.
Besides, we found that p53 expression was significantly down-regulated in 19 osteosarcoma cell lines when compared with normal bone cell lines in the GEO dataset (GSE28425) (FC = 0.78, P < .001; Figure 1i) and USP14 expression significantly up-regulates in osteosarcoma cell lines when compared with normal bone cell lines (FC = 1.24, P < .001; Figure 1j). The relationship between p53 expression level and USP14 expression level was also negatively correlated correlation (P < .01; Figure 1k). The clinical significance of USP14 in 86 osteosarcoma patients from TCGA dataset showed that high USP14 expression was correlated with lower OS (P < .001; Figure 1I).
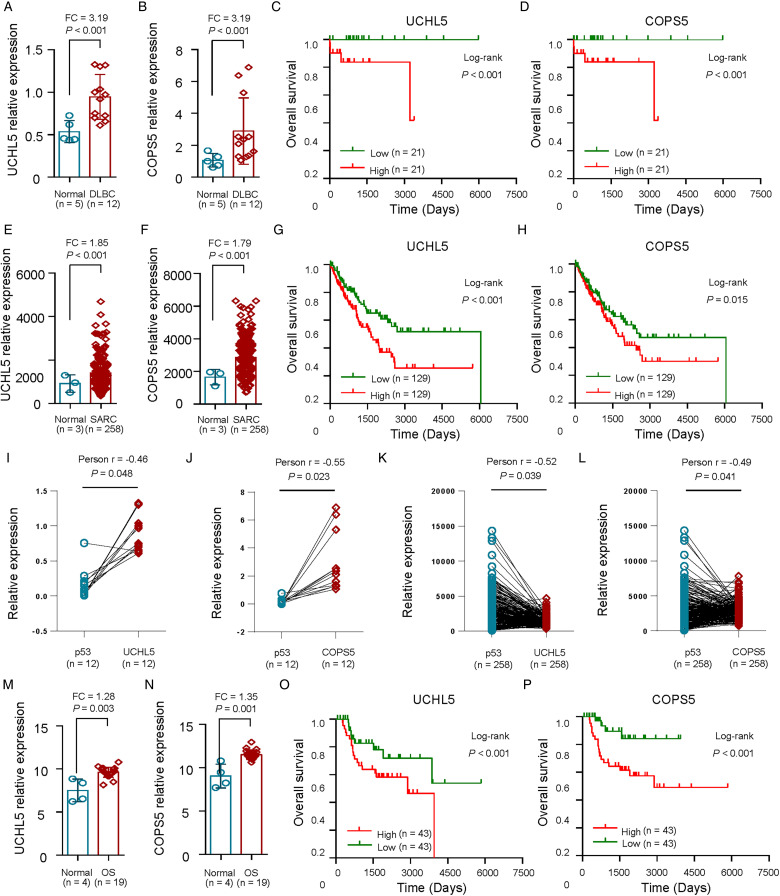
Expression and Clinical Significance of UCHL5 and COPS5 in Tumors with p53 Deficiency
Since our previous data had suggested that UCHL5 and COPS5 expression level was negatively correlated with p53 in p53-deficient cancer samples, and COPS5 was a direct target of UCHL5 and USP14, we further examined the expression and prognostic value of UCHL5 expression together with COPS5 levels using multivariate analysis of OS.
The results showed that DLBC patients with high UCHL5 and COPS5 expression (Figure 2a, b) and high UCHL5 and COPS5 levels had significantly decreased OS (P < .001) (Figure 2c, d). Similarly, sarcoma samples over-expressed UCHL5 and COPS5 (Figure 2e, f) and sarcoma patients with high UCHL5 and COPS5 expression had significantly decreased OS (P < .001) (Figure 2g, h), which suggested that UCHL5 and COPS5 might have potential prognostic value for sarcoma patients. As showed in Figure 2i-j, the relationship between p53 expression level with UCHL5 or COPS5 expression level presented inverse correlation in 12 DLBC tissues from GEO dataset GSE26725 (P < .05). Moreover, the relationship between p53 expression level with UCHL5 or COPS5 expression level presented inverse correlation in 258 sarcoma patients (P < .05; Figure 2k-l).
Figure 2.
Expression and clinical significance of UCHL5 and COPS5 in tumors with p53 deficiency. a-b, Relative levels of UCHL5 (a) and COPS5 (b) in 12 DLBC tissues and 5 noncancerous tissues from GEO dataset GSE26725. c-d, The Kaplan-Meier method was used to evaluate the relationship between UCHL5 (c) and COPS5 (d) expression and overall survival of 42 DLBC patients from TCGA dataset. e-f, Relative levels of UCHL5 (e) and COPS5 (f) in 258 SARC tissues and 3 noncancerous tissues from TCGA dataset. g-h, The Kaplan-Meier method was used to evaluate the relationship between UCHL5 (g) and COPS5 (h) expression and overall survival of 258 SARC patients from TCGA dataset. i-j, The expression correlation of p53 with UCHL5 (i) or COPS5 (j) in 12 DLBC tissues from GEO dataset GSE26725. k-l, The expression correlation of p53 with UCHL5 (k) or COPS5 (l) in 258 SARC tissues from TCGA dataset. m-n, Relative levels of UCHL5 (m) and COPS5 (n) in 19 OS tissues and 4 noncancerous tissues from GEO dataset GSE28425. o-p, The Kaplan-Meier method was used to evaluate the relationship between UCHL5 (o) and COPS5 (p) expression and overall survival of 86 OS patients from TCGA dataset. DLBC, diffuse large b-cell lymphoma; SARC, sarcoma; OS, osteosarcoma.
Moreover, osteosarcoma cell lines exhibited enhanced UCHL5 and COPS5 expression compared with the normal bone cell lines in the GEO dataset (GSE28425) (P < .01; Figure 2m, n). As expected, the clinical significance of UCHL5 and COPS5 in 86 osteosarcoma patients from TCGA dataset showed that UCHL5 and COPS5 expression was positively correlated with lower OS (P < .001; Figure 2o, p).
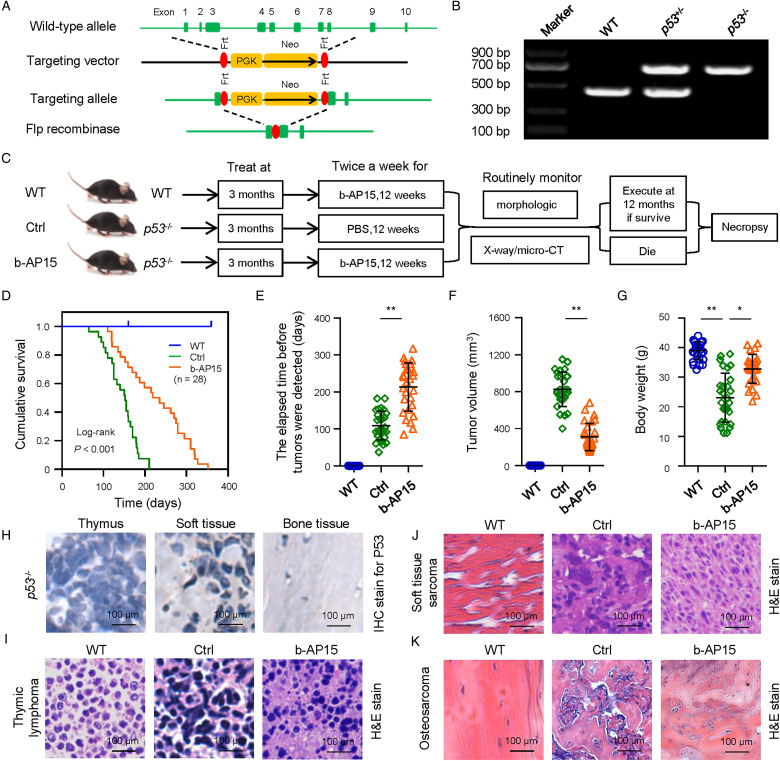
Inhibition of UCHL5 and USP14 Activity Leads to Tumor Suppression in Vivo
Adult p53−/− mice was generated to investigate the role of p53 in tumor formation and evaluate drug effect on tumors with p53 deficiency (Figure 3a-c). Our results showed that the homozygous mice with p53 deficiency fell on cancer death due to malignant lymphomas of thymus at an early age (about in 4 and 6 months) and soft tissue sarcoma and osteosarcoma (Figure 3d). Here, we investigated the effect of a specific UCHL5 and USP14 inhibitor b-AP15 on tumor growth in p53-deficient model in vivo (Figure 3c). We found a significantly prolonged OS in p53−/− mice with b-AP15 administration (Figure 3d). Delayed onset of observable tumor (Figure 3e) and tumor volume (Figure 3f) were shown in mice with b-AP15 treatment compared to the mice of the vehicle-treated group. Weight of control mice was found to significantly decrease. On the contrary, mice with b-AP15 treatment demonstrated normal body weight (Figure 3g) and main organ weights (including lung and liver).
Figure 3.
The effect of b-AP15 on tumor regressions in mice with p53 deficiency. a, Scheme for generation of primary tumors with Flp recombinase in FRT-flanked p53 mice. b, Genotypic detecting of offspring from a p53 heterozygous cross. Tail biopsies were collected at weaning and offspring were screened for the p53 mutation. c, Experimental design of generation of primary tumors and treatment with b-AP15 for tumors with p53 deficiency. e-g, Responses to treatment with b-AP15 as assessed by the elapsed time before tumors were detected (e), tumor volume (f), and body weight (g). h, IHC staining analysis and quantitative analysis. i-k, H&E staining analysis of normal or primary tumors in thymus (i), soft tissue (j) and bone (k). * P < .05 and ** P < .01 versus control.
The tissue morphology of b-AP15–treated p53−/− mice was restored (Figure 3h), which were malignant phenotypes of three main types of cancer, including lymphomas of thymus (Figure 3i), soft tissue sarcoma (Figure 3j) and osteosarcoma (Figure 3k), in the control group.
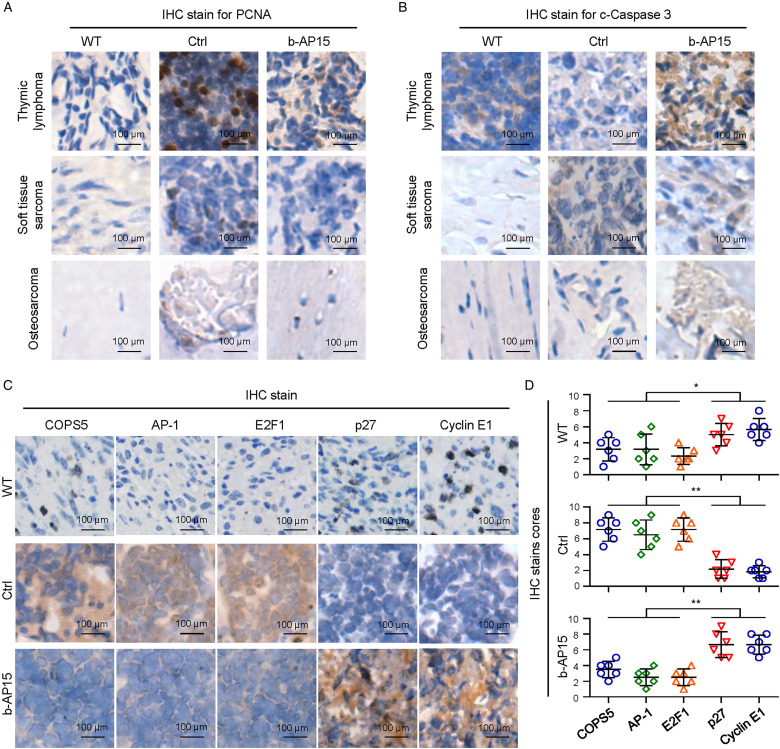
Treatment with b-AP15 Resulted in Durable Tumor Regression Through Cell Cycle and Apoptosis Regulation Mechanism in p53−/− Mice
The mitosis index and PCNA positive cells from b-AP15–treated mice markedly decreased (Figure 4a). We next tested whether cell apoptosis marker c-Caspase-3 was similarly induced in p53−/− mice in vivo and found that apoptotic cells also increased (Figure 4b), which suggests the molecular mechanisms of tumor suppress and apoptosis induction by b-AP15.
Figure 4.
The inhibitory effect of b-AP15 on COPS5-dependent regulation mechanism. a-b, IHC staining analysis for PCNA (a) and c-Caspase-3 (b) in wild-type (WT) or p53-deficient mice with or without treatment with b-AP15. c-d, The effect of b-AP15 in homozygous p53−/− mice on COPS5, its downstream factors AP-1 and E2F1, and G1/S phase marker proteins Cyclin E1and p27 was analyzed by IHC staining analysis and quantitative analysis in WT or p53-deficient mice with or without treatment with b-AP15. Ctrl, control; WT, wild type. Data showed as means ± SDs. * P < .05 and ** P < .01 versus control.
We previous observed that b-AP15 induced obvious G2/M phase arrest. In accordance with these findings, we further studied the inhibitory effect of b-AP15 on G1/S phase marker proteins Cyclin E1 and p27. Our results showed that the protein levels of Cyclin E1 and p27 were significantly down-regulated in p53−/− mice, however, b-AP15 treatment rescued Cyclin E1 and p27 levels (Figure 4c), suggesting that b-AP15 inhibit tumor progress in vivo and molecular mechanisms of regulation in cell cycle are involved in tumors with p53 deficiency.
b-AP15 Treatment Induce Down-Regulation of COPS5 and its Downstream Factors
We previous explored the interacting proteins and probable substrates of UCHL5 and USP14 through mass spectrometry and yeast two-hybrid system. We demonstrated that a critical negative regulator COPS5 was recognizing as an interacting substrate of UCHL5 and USP14. 10
In this study, COPS5 protein was found to significantly upregulated (Figure 4c, d); in contrast, treatment with b-AP15 significantly decreased COPS5 protein level (Figure 4c, d). Moreover, deletion of p53 increased the COPS5 downstream factors AP-1 and E2F1 level; in contrast, treatment with b-AP15 significantly decreased AP-1 and E2F1 protein level in vivo in p53−/− mice (Figure 4c, d). Thus, it is possible that b-AP15 act as a small molecule anticancer component for tumors with p53 deficiency by decreasing AP-1 and E2F1 in the COPS5 pathway.
Discussion
Cancer cells feature uncontrolled cell cycle, apoptosis evading and resistance developing, and insensitivity to anti-growth signal.12,13 Thus, induction of control of cell cycle progression and apoptosis would be promising strategies for cancer therapy p53 mutation or functions loss leads to genomic instability and induces tumorigenesis. 14 Thus, modulation of p53 level and function or regulation of the components in p53 signaling pathways were encouraging since p53 plays an important role in cellular control and response mechanism.15–17
Genetically engineered mice with p53 deficiency was proved to be associated with a high frequency of spontaneous cancers, 5 in which p53 is nonfunctional as a transcription factor, or through ubiquitination and degradation by E3 ubiquitin ligase Mdm2 or CSN5 in ubiquitin 26S proteasome pathway.13,18,19 CSN5, also known as Jab1 or COPS5, is found to regulate degradation of p53 proteins by ubiquitination and is confirmed to overexpress in a variety of types of tumors.20–22 However, the precise roles of 19S proteasome regulator subunit in the p53 regulation and in tumorigenesis are not well characterized.
In this study, we utilized p53−/− mice to investigate the role of p53 in tumor formation and evaluate drug effect on tumors with p53 deficiency. Our results show that b-AP15, a selected inhibitor specifically and selectively blocks UCHL5 and USP14 deubiquitylating activity in 19S regulatory particle can inhibit tumor growth and induce cell apoptosis. Additionally, b-AP15 can antagonize the COPS5 and it downstream factors AP-1 and E2F1, which suggested that UCHL5 and USP14 are pivotal regulators for p53 and COPS5. Therefore, our current research may allow targeting UCHL5 and USP14 in 19S regulatory particle for anti-cancer drug development.
b-AP15 was reported to inhibit tumor cell growth and induce cell apoptosis, and displays anti-tumor role in multiple tumor cell models 23 without inhibiting 26S proteasomes proteolytic activities. 24 More important, in this study, our in vivo studies indicated that b-AP15 is well tolerated. Mice with b-AP15 treatment demonstrated normal body weight and main organ weights, suggesting that b-AP15 is not genotoxic. It is worth noting that, Gubat et al showed that the dienone compound b-AP15 inhibits proteasomal degradation of long-lived proteins. Deletion of genes encoding mitochondrial proteins decreased the sensitivity to b-AP15, suggesting that mitochondrial dysfunction is coupled to cell death induced by b-AP15 and likely also other dienone compounds of the same class to affect protein degradation and proteasome function at more than one level. 25 Besides, Demmers et al observed distinct effects on the global ubiquitinome upon removal of either USP14 or UCH37, while the simultaneous removal of both DUBs suggested less functional redundancy than previously anticipated. When comparing treatment of wild-type versus USP14/UCH37 double-knockout cells with small molecule inhibitor b-AP15, they showed that broad and severe off-target effects were observed, questioning the alleged specificity of this inhibitor. 26
These findings shed lights that inhibition of UCHL5 and USP14 deubiquitinating activity in 19S proteasome may contribute an extensive approach to preventing tumor progress due to p53 deficiency. However, this study are limited by small sample size and animal model in providing a comprehensive overview of p53 deficiency. Further validation using human clinical trial and functional characterization are needed to delineate the exact treatment effect and mechanistic details.
Conclusions
Thus, our research displayed a promising efficacy for p53-deficient disease models through targeting DUBs in 19S regulatory particle and a clinical application prospect to improve outcome for p53-deficient patients.
Acknowledgements
We thank to the Central Laboratory for Medical Research, Shanghai Tenth People's Hospital, Tongji University School of Medicine for the technical assistance.
Abbreviations
- B-CLL
B-cell chronic lymphocytic leukemia
- BSA
bovine serum albumin
- DLBC
diffuse large B-cell lymphoma
- FC
Fold change
- GEO
Gene Expression Omnibus
- IHC
immunohistochemistry
- Mdm2
murine double minute-2
- OS
overall survival
- PCR
polymerase chain reaction
- PCNA
Proliferative cell nuclear antigen
- UCHL5
ubiquitin C-terminal hydrolase L5
- USP14
ubiquitin specific peptidase 14.
Author Contribution: ZYJ, JH, YSM, DF and YGO designed the study. ZYJ, JH, JWZ, XFW, YSM, ZXX, HRS, CC, BZX and DF performed the animal experiments. ZYJ, XFW, YSM, JBL, DF and YGO performed the data analyses. ZYJ, YSM, YGO and DF wrote the manuscript. ZYJ, JH, JWZ, XFW and YSM contributed equally to this work. All the authors approved the final manuscript.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethics Approval and Consent to Participate: All experimental procedures were approved by the Institutional Animal Care and Use Committee (IACUC) guidelines at Tongji University School of Medicine (SYDW-19-215).
Consent for Publication: This manuscript has not been submitted simultaneously to another journal in whole or in part. The manuscript is not under simultaneous consideration by any other publication. There is no conflict of interests for any of the authors, and all of the named authors have agreed to submit the paper in its present form.
Availability of Data and Material: The datasets supporting the conclusions of this article are included within the article.
Funding: This study was supported partly by grants from 100 talents pool of teachers in Affiliated Hospital of Shanghai Health Medical College (ZPBRK-19-01), and Key program of Hunan Provincial Department of Science and Technology (2020WK2020).
ORCID iD: Da Fu https://orcid.org/0000-0002-0878-2575
References
- 1.Loureiro JB, Abrantes M, Oliveira PA, Saraiva L. P53 in skin cancer: from a master player to a privileged target for prevention and therapy. Biochim Biophys Acta Rev Cancer. 2020;1874:188438. 10.1016/j.bbcan.2020.188438. [DOI] [PubMed] [Google Scholar]
- 2.Wu C, Guo E, Ming Jet al. Radiation-Induced DNMT3B promotes radioresistance in nasopharyngeal carcinoma through methylation of p53 and p21. Mol Ther Oncolytics. 2020;17:306-319. 10.1016/j.omto.2020.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie J, Zhang W, Liang Xet al. et al. RPL32 Promotes lung cancer progression by facilitating p53 degradation. Mol Ther Nucleic Acids. 2020;21:75-85. 10.1016/j.omtn.2020.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song J, Yu J, Jeong LS, Lee SK. A novel cytarabine analog evokes synthetic lethality by targeting MK2 in p53-deficient cancer cells. Cancer Lett. 2020;197:54-65. 10.1016/j.canlet.2020.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Jacks T, Remington L, Williams BOet al. et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1-7. 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 6.Singh A, Bhattacharyya N, Srivastava Aet al. et al. MicroRNA-215-5p treatment suppresses mesothelioma progression via the MDM2-p53-signaling axis. Mol Ther. 2019;27:1665-1680. 10.1016/j.ymthe.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura A, Kirsch DG, McLaughlin MEet al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661-665. 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 8.Si W, Zhou B, Xie Wet al. Angiogenic factor AGGF1 acts as a tumor suppressor by modulating p53 post-transcriptional modifications and stability via MDM2. Cancer Lett. 2021;497:28-40. 10.1016/j.canlet.2020.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals. 8th edition. National Academies Press (US); 2011. [PubMed] [Google Scholar]
- 10.Percie du Sert N, Hurst V, Ahluwalia A, et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. Br J Pharmacol. 2020;177(16):3617-3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma YS, Wang XF, Yu F, et al. Inhibition of USP14 and UCH37 deubiquitinating activity by b-AP15 as a potential therapy for tumors with p53 deficiency. Signal Transduct Target Ther. 2020;5:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18:176. 10.1186/s12943-019-1109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang M, Yu F, Chen X, Li P, Wang K. The underlying mechanisms of noncoding RNAs in the chemoresistance of hepatocellular carcinoma. Mol Ther Nucleic Acids. 2020;21:13-27. 10.1016/j.omtn.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Yu J, Wang Qet al. Tartrate-Resistant acid phosphatase 5/ACP5 interacts with p53 to control the expression of SMAD3 in lung adenocarcinoma. Mol Ther Oncolytics. 2020;16:272-288. 10.1016/j.omto.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garufi A, Baldari S, Pettinari Ret al. A ruthenium(II)-curcumin compound modulates NRF2 expression balancing the cancer cell death/survival outcome according to p53 status. J Exp Clin Cancer Res 2020;39:122. 10.1186/s13046-020-01628-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raimundo L, Ramos H, Loureiro JB, Calheiros J, Saraiva L. BRCA1/P53: two strengths in cancer chemoprevention. Biochim Biophys Acta Rev Cancer. 2020;1873:188339. 10.1016/j.bbcan.2020.188339. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Yan B, Meng Set al. et al. Theaflavin induces apoptosis of A375 human melanoma cells and inhibits tumor growth in Xenograft zebrafishes through P53- and JNK-related mechanism. Front Pharmacol. 2020;11:1317. 10.3389/fphar.2020.01317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng X, Li S, Kong Fet al. Long noncoding RNA PiHL regulates p53 protein stability through GRWD1/RPL11/MDM2 axis in colorectal cancer. Theranostics. 2020;10:265-280. 10.7150/thno.36045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng J, Guo J, North BJet al. et al. Functional analysis of deubiquitylating enzymes in tumorigenesis and development. Biochim Biophys Acta Rev Cancer. 2019;1872:188312. 10.1016/j.bbcan.2019.188312. [DOI] [PubMed] [Google Scholar]
- 20.Danielpour D, Purighalla S, Wang E, Zmina PM, Sarkar A, Zhou G. JAB1/COPS5 Is a putative oncogene that controls critical oncoproteins deregulated in prostate cancer. Biochem Biophys Res Commun. 2019;518:374-380. 10.1016/j.bbrc.2019.08.066. [DOI] [PubMed] [Google Scholar]
- 21.Wang S, Oh DY, Leventaki Vet al. MicroRNA-17 acts as a tumor chemosensitizer by targeting JAB1/CSN5 in triple-negative breast cancer. Cancer Lett. 2019;465:12-23. 10.1016/j.canlet.2019.08.016. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Jia Y, Zhang Y, Zhou D, Sun H, Ma X. alpha5-nAChR contributes to epithelial-mesenchymal transition and metastasis by regulating Jab1/Csn5 signalling in lung cancer. J Cell Mol Med. 2020;24:2497-2506. 10.1111/jcmm.14941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hillert EK, Brnjic S, Zhang Xet al. Proteasome inhibitor b-AP15 induces enhanced proteotoxicity by inhibiting cytoprotective aggresome formation. Cancer Lett. 2019;448:70-83. 10.1016/j.canlet.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 24.D'Arcy P, Brnjic S, Olofsson MHet al. et al. Inhibition of proteasome deubiquitinating activity as a new cancer therapy. Nat Med. 2011;17:1636-1640. 10.1038/nm.2536. [DOI] [PubMed] [Google Scholar]
- 25.Gubat J, Selvaraju K, Sjöstrand L, et al. Comprehensive target screening and cellular profiling of the cancer-active compound b-AP15 indicate abrogation of protein homeostasis and organelle dysfunction as the primary mechanism of action. Front Oncol. 2022;12:852980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Wal L, Bezstarosti K, Demmers JAA. A ubiquitinome analysis to study the functional roles of the proteasome associated deubiquitinating enzymes USP14 and UCH37. J Proteomics. 2022;262:104592. [DOI] [PubMed] [Google Scholar]