Abstract
One group of penicillin target enzymes, the class A high-molecular-weight penicillin-binding proteins (PBPs), are bimodular enzymes. In addition to a central penicillin-binding–transpeptidase domain, they contain an N-terminal putative glycosyltransferase domain. Mutations in the genes for each of the three Streptococcus pneumoniae class A PBPs, PBP1a, PBP1b, and PBP2a, were isolated by insertion duplication mutagenesis within the glycosyltransferase domain, documenting that their function is not essential for cellular growth in the laboratory. PBP1b PBP2a and PBP1a PBP1b double mutants could also be isolated, and both showed defects in positioning of the septum. Attempts to obtain a PBP2a PBP1a double mutant failed. All mutants with a disrupted pbp2a gene showed higher sensitivity to moenomycin, an antibiotic known to inhibit PBP-associated glycosyltransferase activity, indicating that PBP2a is the primary target for glycosyltransferase inhibitors in S. pneumoniae.
Penicillin-binding proteins (PBPs), membrane-associated proteins that catalyze late steps in murein biosynthesis, are the classic targets for β-lactam antibiotics. They are multidomain proteins, and according to their domain structure, function, and relatedness in peptide sequence, they are classified as multimodular high-molecular-weight PBPs of classes A and B and monofunctional low-molecular-weight PBPs (9). A common feature is a penicillin-binding domain responsible for the enzymatically catalyzed interaction with β-lactam antibiotics that involves a covalent acyl-enzyme intermediate via an active-site serine residue. The critical penicillin-sensitive reaction is a transpeptidation reaction cross-linking the muropeptide side chains of different glycan strands. Class A PBPs, the only PBPs which have been shown to be bifunctional enzymes in vitro, possess an N-terminal glycosyltransferase domain and catalyze transpeptidation as well as glycosyltransferase reaction in vitro (19). We have used the term glycosyltransferase throughout this paper in order to avoid confusion with the transglycosylases as glycan-degrading enzymes as suggested previously by Di Berardino et al. (7). The function of the N-terminal domain of class B PBPs is unknown (1).
Attempts to isolate deletion mutant class B PBPs, such as Escherichia coli PBP2 and PBP3 and Streptococcus pneumoniae PBP2x and PBP2b, failed (3, 14). In contrast, deletion constructs of the class A PBPs E. coli PBP1a and -1b have been obtained, but double mutants could not be obtained, indicating that the cell requires the function of at least one of these PBPs (23). The third class A PBP, PBP1c, in E. coli has not been investigated genetically, since its existence was revealed essentially via genome analysis. There are three class A PBPs in S. pneumoniae: PBP1a, PBP1b, and PBP2a (13), and each is a member of a different subgroup of gram-positive PBPs (9). So far, only PBP1a mutants have been isolated (12, 14). In order to explore the roles of these proteins in S. pneumoniae and especially their N-terminal domain in more detail, we constructed mutant versions of each of the three PBP genes and investigated whether double PBP mutants could also be isolated.
PBP1a, PBP1b, and PBP2a single mutants.
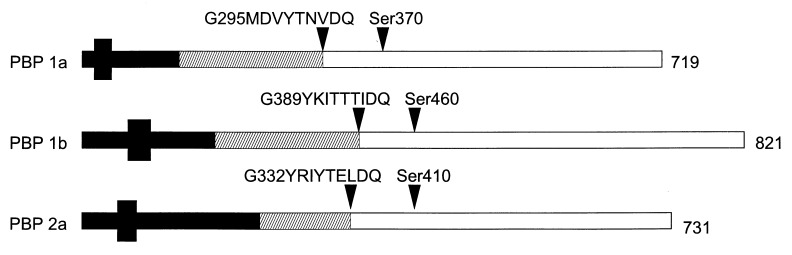
Mutations in each of the three class A PBP genes were obtained via insertion duplication mutagenesis of the laboratory strain R6 (2), using an internal gene fragment cloned into a vector that cannot replicate in S. pneumoniae but that carries an antibiotic resistance marker selectable upon integration into the chromosome by homologous recombination. Two different constructs were made in order to be able to obtain double mutants, using plasmid pJDC9 with an erythromycin resistance gene (6) and pUC19C, a derivative of pUC19 (New England Biolabs) with the cat gene from pC194 (4) cloned into the HincII site. The internal fragments were designed such that the peptide being transcribed after successful recombination of the plasmid into the chromosomal gene terminated within the glycosyltransferase domain (Fig. 1). This domain is defined by six conserved motifs (13). A seventh conserved motif (GxxxxTxxQ [x is any amino acid]) is homologous to motif 4 in class B PBPs, which according to the structure of PBP2x in S. pneumoniae represents the link between the two domains, and the derivatives obtained after mutagenesis terminated well before this motif (21).
FIG. 1.
S. pneumoniae class A PBPs and mutant PBPs used in this study. The structures of PBP1a, PBP1b, and PBP2a are shown schematically, and their length (in amino acid residues) is indicated on the right. The small black box indicates the putative membrane-spanning domain. The conserved motif at the putative transition between the N-terminal glycosyltransferase domain (hatched area) and the penicillin-binding–transpeptidase domain, as well as the active-site serine, are indicated by black triangles. The length of the potentially produced peptide after insertion-duplication mutagenesis is represented by the black bar.
Internal gene fragments were obtained by PCR performed essentially as described previously (15) in a 100-μl volume using 2.5 U of Taq polymerase (Perkin-Elmer, Norwalk, Conn.). The oligonucleotides used for amplification of different genes were as follows: for pbp1a, C10CAACGATTCTGCGCCTAATC30 and G357AGGGAATTGCTTTGCAGATT337; for pbp1b, C326CTATTCGGACGGGACGG343 and G492GTCGCACGAATCACCGCC474; and for pbp2a, G56TGAACTAGAGGACTCTG73 and C660GCATCTTCTACACCCC644. The numbers indicate the position in the genes according to their published sequence (11). After purification of the DNA fragments with the Bio 101 Geneclean II kit (Dianova, Hamburg, Germany), they were first cloned into the PCR II vector by using E. coli INVαF′ (TA cloning kit; Invitrogen, Leek, The Netherlands) prior to cloning into the EcoRI site of pJDC9 and pUC19C.
Purified plasmid DNA was used as donor DNA in transformation experiments with competent S. pneumoniae R6 as acceptor. Pneumococci were grown in C medium throughout (16). Transformation experiments were performed essentially by the published procedure (17) by 30 min of incubation in the presence of DNA at 30°C followed by a 2-h phenotypic expression period at 37°C and growth in agar plates under selective conditions (1 μg of erythromycin per ml or 2 μg of chloramphenicol per ml). Transformants were readily obtained in all cases, and disruption of the respective PBP gene could be confirmed by PCR analysis and Southern hybridization (not shown). Since the transformants should not contain a penicillin-binding domain in the mutated PBP, they could also be verified by analysis of their PBP profiles (Fig. 2). PBPs in cell lysates were labeled with [3H]benzylpenicillin (2 μCi per sample; Amersham Buchler, Braunschweig, Germany) as described and detected by fluorography after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (13). In order to obtain clear separation between PBP1a and PBP1b, SDS-polyacrylamide gels with 10% acrylamide (acrylamide-bisacrylamide [30:0.8]) were used, and for clear separation of PBP2x, -2a, and -2b, 7.5% acrylamide (acrylamide-bisacrylamide [30:1.1]) was used (11) (Fig. 2A and B).
FIG. 2.
Profiles of single and double PBP mutants. PBPs were visualized on fluorograms after labeling of cell lysates with [3H]propionylampicillin. The disrupted PBP(s) of the mutants or of the parent strain (R6) is indicated above the lanes. 1a∗ refers to transformants isolated after the attempt to disrupt pbp2a in a pbp1a mutant. Gels with 10% acrylamide (acrylamide-bisacrylamide [30:0.8]) (A and C) or 7.5% acrylamide (acrylamide-bisacrylamide [30:1.1]) (B) were used. The positions of PBPs are indicated to the left of the fluorograms.
Construction of double mutants.
Double mutants were constructed from pUC19C-derived single mutants, and chromosomal DNA from the pJDC9 derivatives was used in subsequent transformations to introduce the second PBP mutation. Under standard transformation conditions, several hundred transformants, pbp1a pbp1b mutants or pbp1b pbp2a mutants, were obtained, corresponding to a transformation efficiency of 4 × 10−5 to 5 × 10−5, and all showed the expected PBP profiles (Fig. 2). In the pbp1b pbp2a double mutant, the PBP1a band appeared somewhat smeared on the gel (Fig. 2B). PBP1a has an unusual mobility on SDS-polyacrylamide gels in wild-type cells, showing a much higher apparent molecular mass of more than 92 kDa compared to a deduced molecular mass of 79.7 kDa. It is possible that this is due to either modification of PBP1a or its interaction with another component and that this property is affected in the mutants. In contrast, the transformation efficiency dropped by >10-fold when disruption of pbp2a was attempted in a pbp1a mutant used as recipient. Eleven transformants were tested, but none showed a defect in PBP2a on fluorograms (Fig. 2A and B) or contained an insert in the pbp2a gene when investigated by PCR analysis. The transformation was repeated another two times with the same result. Thus, although a negative experiment is not definite proof, the data strongly suggest that simultaneous deletion of both pbp1a and pbp2a is lethal in S. pneumoniae.
Cellular growth of the mutants.
All single mutants and the two types of double mutants obtained grew slower than the R6 strain, with generation times between 43 and 49 min compared to 36 min for the R6 strain (Fig. 3). Different mutants isolated from the same transformation experiments occasionally had slightly different generation times, similar to the results for strains with mutant PBP1a or PBP3 as reported previously (18, 22). They also did not reach the same cell density as the parental strain, and they lysed earlier after reaching stationary-phase growth, suggesting that the lack of each of these enzymes causes some defects related to cell wall biochemistry (Fig. 3). The differences in generation time were less pronounced when the cells were grown in the absence of erythromycin, but the early onset of stationary-phase lysis especially in mutants with a disrupted pbp2a was still clear (data not shown).
FIG. 3.
Growth of S. pneumoniae R6 and class A PBP double mutants. Cells of an exponentially growing culture were diluted in prewarmed C medium supplemented with erythromycin (1 μg/ml), and growth was monitored by nephelometry (in nephelometry units [N]) over time (in hours). Symbols: ●, S. pneumoniae R6; ■, pbp1a pbp1b mutant; ▴, pbp1b pbp2a mutant.
Morphology.
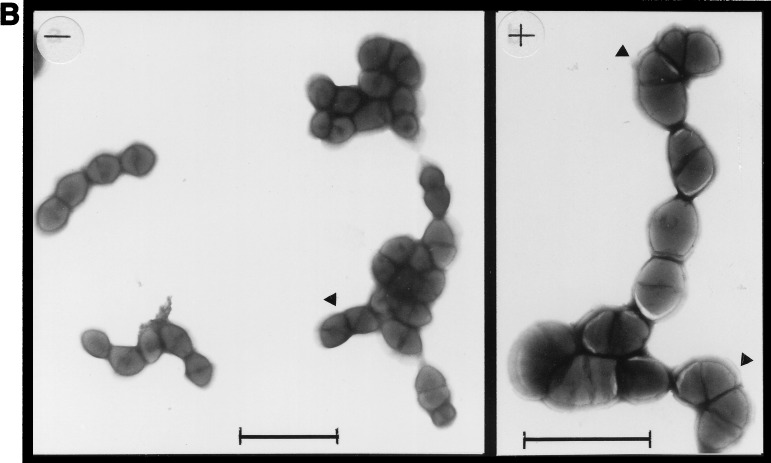
When observed under the phase-contrast microscope, the single mutants did not show any obvious phenotype, whereas the double mutants appeared deformed and grew in small clumps (not shown). Cells of the double mutants were examined in more detail in the electron microscope. Exponentially growing cells were harvested by centrifugation and prepared on carbon film by negative staining according to published procedures, using 1% sodium tungstophosphoric acid (pH 7.0) or 2% ammonium molybdate (24) and observed in an Philips CM100 electron microscope (Fig. 4). They were grown in C medium with or without the addition of 2% choline. The addition of choline prevents cell separation but not cell division, and morphological changes can be observed more easily under these conditions (5). In both the pbp1a pbp1b and pbp1b pbp2a mutants, adjacent septa appeared frequently at odd angles rather than parallel to each other as in the parent R6 strain, resulting in a corkscrew-like growth within the cell chains (Fig. 4).
FIG. 4.
Electron microscopy of S. pneumoniae class A PBP double mutants. Cells are shown after negative staining of exponentially grown cultures of the parent strain S. pneumoniae R6 (A) and the pbp1a pbp1b (B) and pbp1b pbp2a (C) double mutants. Cells were grown in C medium with (+) or without (−) the addition of 2% choline. Arrowheads indicate odd division septa. Bars, 2 μm.
Antibiotic susceptibilities.
The E. coli PBP1b contains a moenomycin-sensitive glycosyltransferase activity (25, 26). Despite the homology of the N-terminal domains of class A PBPs, they are generally not targets for this antibiotic, and the activity of the E. coli monodomain glycosyltransferase that consists of just this module is also not affected by this drug (7). We investigated the susceptibilities of S. pneumoniae single and double mutants to moenomycin by using a narrow range of antibiotic concentrations on blood agar plates (3% sheep blood; moenomycin concentrations used were 0.25, 0.3, 0.5, 0.75, 1, 1.25, and 1.5 μg/ml). The following MICs (in micrograms per milliliter) were obtained for the different strains: R6, 1; pbp1a and pbp1a pbp1b mutants, 0.75; pbp1b mutant, 1.25; pbp2a mutant, 0.5; and pbp1b pbp2a mutant, 0.3. Thus, all mutants with a defective pbp2a gene clearly showed a higher susceptibility to moenomycin than the parental strain did, suggesting that this protein functions as a moenomycin-sensitive glycosyltransferase. This was similar to pbp1a mutants, but here the effect on moenomycin susceptibility was less pronounced, although in vitro data suggested an interaction between the glycosyltransferase domains of PBP1a and moenomycin (8). No difference was found in the MICs of β-lactam antibiotics (cefotaxime, oxacillin, and penicillin G were tested) except that a slightly higher oxacillin MIC was detected for the pbp1a mutant and for both of the double mutants (0.07 to 0.08 μg/ml versus 0.03 μg/ml for the R6 strain). These mutants grew poorly on agar plates, and 48 h instead of the routinely used 24 h for MIC determination was required; therefore, the significance of the MIC changes is difficult to evaluate.
Concluding remarks.
S. pneumoniae is the first organism for which the roles of all class A PBPs were investigated genetically. The importance of these proteins has been deduced from studies on penicillin-resistant strains. The fact that mutations in S. pneumoniae PBP2x and PBP2b are required for primary, low resistance to these drugs has been used as an argument to confirm their essential function (10, 17). Variants with a reduced affinity to penicillins were also observed in all class A PBPs in high-level-resistant strains, and experimental evidence that they can function as resistance determinants was obtained for both PBP1a and PBP2a (13, 20). This shows that inhibition of these PBPs also cannot be tolerated by the cell, at least under certain conditions. In fact, changes in all PBPs including the low-molecular-weight PBP3 have been associated with resistance to β-lactams (15), documenting that the genetic background and function of other PBPs are important parameters that define the indispensable nature of a PBP. However, even a slightly slower growth rate, such as that shown for all class A PBP mutants, may be a handicap in vivo, and an early onset of stationary-phase lysis evident at least in the double mutants will decrease the chances of surviving. Therefore, it seems unlikely that class A PBP mutants can be found outside the laboratory. Taken together, the results suggest that the N-terminal glycosyltransferase domain remains an important target for antimicrobial compounds.
REFERENCES
- 1.Adam M, Fraipont C, Rhazi N, Nguyen-Distèche M, Lakaye B, Frère J-M, Devreese B, Van Beeumen J, van Heijenoort Y, Van Heijenoort J, Ghuysen J-M. The bimodular G57-V577 polypeptide chain of the class B penicillin-binding protein 3 of Escherichia colicatalyzes peptide bond formation from thioesters and does not catalyze glycan chain polymerization from lipid II intermediates. J Bacteriol. 1997;179:6005–6009. doi: 10.1128/jb.179.19.6005-6009.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avery O T, MacLeod C M, McCarty M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types. J Exp Med. 1944;79:137–158. doi: 10.1084/jem.79.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayala J A, Garrido T, De Pedro M A, Vicente M. Molecular biology of bacterial septation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 73–101. [Google Scholar]
- 4.Ballester S, Alonso J, Lopez P, Espinoza M. Comparative expression of the pC194 cat gene in Streptococcus pneumoniae, Bacillus subtilis, and Escherichia coli. Gene. 1990;86:71–79. doi: 10.1016/0378-1119(90)90115-8. [DOI] [PubMed] [Google Scholar]
- 5.Briese T, Hakenbeck R. Interaction between choline and the N-acetyl-muramyl-L-alanine-amidase of Streptococcus pneumoniae. In: Hakenbeck R, Höltje J-V, Labischinski H, editors. The target of penicillin. Berlin, Germany: Walter de Gruyter & Co.; 1983. pp. 173–178. [Google Scholar]
- 6.Chen J-D, Morrison D A. Construction and properties of a new insertion vector, pJDC9, that is protected by transcriptional terminators and useful for cloning of DNA from Streptococcus pneumoniae. Gene. 1988;64:155–164. doi: 10.1016/0378-1119(88)90489-1. [DOI] [PubMed] [Google Scholar]
- 7.Di Berardino M, Dijkstra A, Stüber D, Keck W, Gubler M. The monofunctional glycosyltransferase of Escherichia coliis a member of a new class of peptidoglycan-synthesising enzymes: overexpression and determination of the glycan-polymerising activity. FEBS Lett. 1996;392:184–188. doi: 10.1016/0014-5793(96)00809-5. [DOI] [PubMed] [Google Scholar]
- 8.Di Guilmi A M, Mouz N, Andrieu J P, Hoskins J, Jaskunas S R, Gagnon J, Dideberg O, Vernet T. Identification, purification, and characterization of transpeptidase and glycosyltransferase domains of Streptococcus pneumoniaepenicillin-binding protein 1a. J Bacteriol. 1998;180:5652–5659. doi: 10.1128/jb.180.21.5652-5659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goffin C, Ghuysen J-M. Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol Mol Biol Rev. 1998;62:1079–1093. doi: 10.1128/mmbr.62.4.1079-1093.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grebe T, Hakenbeck R. Penicillin-binding proteins 2b and 2x of Streptococcus pneumoniaeare primary resistance determinants for different classes of β-lactam antibiotics. Antimicrob Agents Chemother. 1996;40:829–834. doi: 10.1128/aac.40.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hakenbeck R, Briese T, Ellerbrok H, Laible G, Martin C, Metelmann C, Schier H-M, Tornette S. Targets of β-lactams in Streptococcus pneumoniae. In: Actor P, Daneo-Moore L, Higgins M L, Salton M R J, Shockman G D, editors. Antibiotic inhibition of bacterial cell surface assembly and function. Washington, D.C: American Society for Microbiology; 1988. pp. 390–399. [Google Scholar]
- 12.Hakenbeck R, Ellerbrok H, Martin C, Morelli G, Schuster C, Severin A, Tomasz A. Penicillin-binding protein 1a and 3 in Streptococcus pneumoniae: what are essential PBP’s. In: De Pedro M A, Höltje J-V, Löffelhardt W, editors. Bacterial growth and lysis metabolism and structure of the bacterial sacculus. New York, N.Y: Plenum Press; 1993. pp. 335–340. [Google Scholar]
- 13.Hakenbeck R, König A, Kern I, van der Linden M, Keck W, Billot-Klein D, Legrand R, Schoot B, Gutmann L. Acquisition of five high-Mr penicillin-binding protein variants during transfer of high-level β-lactam resistance from Streptococcus mitis to Streptococcus pneumoniae. J Bacteriol. 1998;180:1831–1840. doi: 10.1128/jb.180.7.1831-1840.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kell C M, Sharma U K, Dowson C G, Town C, Balganesh T S, Spratt B G. Deletion analysis of the essentiality of penicillin-binding proteins 1A, 2B and 2X of Streptococcus pneumoniae. FEMS Microbiol Lett. 1993;106:171–175. doi: 10.1111/j.1574-6968.1993.tb05954.x. [DOI] [PubMed] [Google Scholar]
- 15.Krauß J, Hakenbeck R. Mutations in PBP3 of a cefotaxime-resistant laboratory mutant C604 and penicillin-resistant clinical isolates of Streptococcus pneumoniae. Antimicrob Agents Chemother. 1997;41:936–942. doi: 10.1128/aac.41.5.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lacks S A, Hotchkiss R D. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 17.Laible G, Hakenbeck R, Sicard M A, Joris B, Ghuysen J-M. Nucleotide sequences of the pbpX genes encoding the penicillin-binding protein 2x from Streptococcus pneumoniaeR6 and a cefotaxime-resistant mutant, C506. Mol Microbiol. 1989;3:1337–1348. doi: 10.1111/j.1365-2958.1989.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 18.Martin C. Molekulargenetische Untersuchungen des Penicillin-bindenden Proteins (PBP) 1a von Streptococcus pneumoniae: Verwandtschaft von PBP 1a Mosaikgenen in Penicillin resistenten klinischen Stämmen. Thesis. Berlin, Germany: Freie Universität Berlin; 1992. [Google Scholar]
- 19.Matsuhashi M. Utilization of lipid-linked precursors and the formation of peptidoglycan in the process of cell growth and division: membrane enzymes involved in the final steps of peptidoglycan synthesis and the mechanism of their regulation. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 55–71. [Google Scholar]
- 20.Muñóz R, Dowson C G, Daniels M, Coffey T J, Martin C, Hakenbeck R, Spratt B G. Genetics of resistance to third-generation cephalosporins in clinical isolates of Streptococcus pneumoniae. Mol Microbiol. 1992;6:2461–2465. doi: 10.1111/j.1365-2958.1992.tb01422.x. [DOI] [PubMed] [Google Scholar]
- 21.Pares S, Mouz N, Pétillot Y, Hakenbeck R, Dideberg O. X-ray structure of Streptococcus pneumoniaePBP2x, a primary penicillin target enzyme. Nat Struct Biol. 1996;3:284–289. doi: 10.1038/nsb0396-284. [DOI] [PubMed] [Google Scholar]
- 22.Schuster C, Dobrinski B, Hakenbeck R. Unusual septum formation in Streptococcus pneumoniae mutants with an alteration in the d,d-carboxypeptidase penicillin-binding protein 3. J Bacteriol. 1990;172:6499–6505. doi: 10.1128/jb.172.11.6499-6505.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spratt B G, Jobanputra V. Mutants of Escherichia coliwhich lack a component of penicillin binding protein 1a are viable. FEMS Lett. 1977;79:374–378. doi: 10.1016/0014-5793(77)80824-7. [DOI] [PubMed] [Google Scholar]
- 24.Steven A C, Trus B L, Maizel J V, Unser M, Parry D A D, Wall J S. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki H, van Heijenoort Y, Tamura T, Mizoguchi J, Hirota Y, Van Heijenoort J. In vitro peptidoglycan polymerization catalysed by penicillin-binding protein 1b of Escherichia coliK 12. FEBS Lett. 1980;110:245–249. doi: 10.1016/0014-5793(80)80083-4. [DOI] [PubMed] [Google Scholar]
- 26.van Heijenoort Y, Derrien M, Van Heijenoort J. Polymerization by transglycosylation in the biosynthesis of the peptidoglycan of Escherichia coliK 12 and its inhibition by antibiotics. FEBS Lett. 1979;89:141–144. doi: 10.1016/0014-5793(78)80540-7. [DOI] [PubMed] [Google Scholar]