Abstract
Mechanisms underlying breast cancer brain metastasis (BCBM) are still unclear. In this study, we observed that extracellular vesicles (EVs) secreted from breast cancer cells with increased expression of tGLI1, a BCBM-promoting transcription factor, strongly activated astrocytes. EV-derived microRNA/miRNA microarray revealed tGLI1-positive breast cancer cells highly secreted miR-1290 and miR-1246 encapsulated in EVs. Genetic knockin/knockout studies established a direct link between tGLI1 and both miRNAs. Datamining and analysis of patient samples revealed that BCBM patients had more circulating EV-miRs-1290/1246 than those without metastasis. Ectopic expression of miR-1290 or miR-1246 strongly activated astrocytes whereas their inhibitors abrogated the effect. Conditioned media from miR-1290- or miR-1246-overexpressing astrocytes promoted mammospheres. Furthermore, miRs-1290/1246 suppressed expression of FOXA2 transcription repressor, leading to CNTF cytokine secretion and subsequent activation of astrocytes. Finally, we conducted a mouse study to demonstrate that astrocytes overexpressing miR-1290, but not miR-1246, enhanced intracranial colonization and growth of breast cancer cells. Collectively, our findings demonstrate, for the first time, that breast cancer EV-derived miR-1290 and miR-1246 activate astrocytes in the brain metastatic microenvironment and that EV-derived miR-1290 promotes progression of brain metastases through the novel EV-miR-1290→FOXA2→CNTF signaling axis.
Keywords: Extracellular vesicles, microRNAs, Breast cancer brain metastasis, Brain microenvironment, tGLI1
1. Introduction
In 2022, an estimated 287,850 women will be diagnosed with breast cancer with 43,250 breast cancer cases resulting in death [1]. Metastasis to distant organs accounts for 90% of breast cancer deaths [2]. Breast cancer brain metastases (BCBM) occur in approximately 10–30% of patients with metastatic breast cancer, and BCBM is associated with poor prognosis with a median survival of only 6–18 months [3]. This poor survival rate may be attributed to limited knowledge of the mechanisms contributing to BCBM.
Truncated glioma-associated oncogene homolog 1 (tGLI1) was discovered by our lab as a gain-of-function GLI1 zinc finger transcription factor [4]. tGLI1, an alternatively spliced variant of GLI1, is a terminal effector of the Sonic hedgehog (SHH) signaling pathway [4]. We have demonstrated that tGLI1 expression is tumor-specific as it is expressed in breast cancer and glioblastoma (GBM), but not in normal breast or brain tissue [4–7]. tGLI1 regulates GLI1 target genes and also has the ability to upregulate its own eight target genes including VEGF-A, VEGF-C, VEGFR2, TEM7, HPSE, CD24, CD44, and OCT4 contributing to increased growth, migration, invasion, and stemness of cancer cells [4, 8–10]. We recently reported that tGLI1-positive breast cancer cells preferentially metastasize to the brain through mediating metastasis-initiating cancer stem cells [11]. Additional mechanisms by which tGLI1-positive breast cancer cells communicate with the brain microenvironment to promote breast cancer colonization and metastasis remain unclear.
Primary breast tumors can prime distant organs prior to colonization and astrocytes are activated before breast cancer cells extravasate into the brain [12,13]. Primary tumors can communicate with distant organs through the secretion of extracellular vesicle (EV)-derived microRNAs (miRNAs). Many EVs originate from the endosome, range in size from 30 to 150 nm in diameter, and play critical roles in intercellular communication through the transport of many bioactive molecules including growth factors, DNA, and miRNAs [14]. MiRNAs are small, 18–23 nucleotide, endogenous noncoding RNAs that regulate gene expression in many physiological processes by targeting mRNAs [15]. Tumor-derived EV-miRNAs promote the destruction of the blood-brain barrier leading to brain metastasis [16]. Importantly, EVs can fuse with astrocytes resulting in the release of oncogenic miRNAs, which prime astrocytes to secrete factors that promote tumor growth and invasion in the brain [17].
In the present study, we investigated whether tGLI1-positive breast cancer cells primed brain microenvironmental cells to promote BCBM. We observed that tGLI1-positive breast cancer cells activate astrocytes through secretion of EVs containing increased miR-1290 and miR-1246 levels. miR-1290 and miR-1246 expression was higher in sera of metastatic breast cancer patients and BCBM tumors compared to sera from patients without metastasis and matched primary tumors, respectively. Analysis of serum samples showed more circulating EV-miR-1290 and miR-1246 in BCBM patients than those without metastasis. Both miRNAs were associated with poor clinical outcomes in breast cancer patients. Overexpression and knockdown studies linked both miRNAs to astrocyte activation and mammosphere formation. Furthermore, we identified Forkhead Box A2 (FOXA2), a miR-1290 and miR-1246 target, as a direct transcriptional repressor of ciliary neurotrophic factor (CNTF). Moreover, astrocytes overexpressing miR-1290, but not miR-1246, promoted growth of co-implanted breast cancer cells in the brain in an intracranial mouse model. In this study, we uncovered a novel role that EV-derived miR-1290 and miR-1246 play in activating astrocytes, and identified the novel miR-1290-FOXA2-CNTF signaling axis and its important role in promoting BCBM.
2. Materials and methods
2.1. Cell lines, lentiviruses, and expression plasmids
MDA-MB-231 and SKBR3 cells were purchased from ATCC. MDA-231-BRM and CN34 cells were a kind gift from Dr. Joan Massagué at the Sloan Kettering Institute [18]. SKBRM cells, a brain metastatic human breast cancer cell line derived from HER2-enriched SKBR3 cells, were established by Drs. Fei Xing and Kounosuke Watabe [19]. The E6/E7/hTERT immortalized human astrocytes were a gift from Dr. Russell O. Pieper at University of California, San Francisco. Isogenic MDA-MB-231 and SKBRM cell lines with vector, GLI1 or tGLI1 were previously developed in our laboratory [4,11]. Astrocytes stably expressing vector, miR-1290, or miR-1246 were generated using lentiviral constructs (SBI; CD511B-1) followed by GFP sorting. SKBRM-tGLI1 cells with stable knockdown (miROff) vector, miR-1290 (Off-1290), and miR-1246 (Off-1246) were established using lentiviral constructs (ABM; mh30080) followed by GFP sorting. All cell lines have been authenticated using standard methods, tested for mycoplasma, treated if necessary (Sigma-Aldrich; 10-799-050-001), and tested again prior to use. Extracellular vesicle tracking CD63-GFP lentivirus was obtained from Dr. Xandra Breakefield’s laboratory [20]. Overexpression lentiviruses for miR-1290, miR-1246, and miR-1270 were obtained from Dr. Yin-Yuan Mo at University of Tennessee.
2.2. EV isolation and nanoparticle tracking analysis (NTA)
EV-free FBS was prepared by diluting FBS 1:3 in culture media, centrifuged at 4 °C for 24 h at 25,000×g, and passed through a 0.22 μm cellulose acetate filter. Cells were cultured in media containing 10% EV-free FBS for 48–72 h. Media was harvested and centrifuged at 4 °C for 10 min at 300×g. Supernatant was collected and centrifuged for 20 min at 2000×g. Supernatant was collected and subjected to centrifugation for 30 min at 16,500×g and passed through a 0.22 μm cellulose acetate filter. EVs were then isolated through three methods: 1) ultracentrifugation method pellets EVs by centrifugation at 120,000×g for 70 min; 2) size exclusion chromatography method loads 500 μL of supernatant onto a qEV column from iZon (Cambridge, MA) and fractions were collected as outlined in the manufacturer’s manual; and 3) ExoQuick Kit EV isolation uses a polymer-based method following the System Biosciences manufacturer’s protocol (Palo Alto, CA; ExoQuick-TC). EVs from human and mouse sera were isolated using the ExoQuick Kit from System Biosciences. Size distribution was measured by NTA using Nanosight NS50 (Malvern Instruments, UK).
2.3. Breast cancer mammosphere assay
Cells were harvested and seeded in 24-well ultra-low attachment plates (Corning) at 1000–4000 cells per well in either a 1:1 ratio of mammosphere medium and CM from astrocytes overexpressing each miRNA or in mammosphere media only. Astrocyte-CM was generated by culturing cells in serum-free medium for 24–48 h. Mammosphere medium contained Dulbecco’s modified Eagle’s medium/F12 (Gibco), B27 (Gibco; 2%), insulin (Sigma; 4 μg/mL), SHH (Sigma; 100 ng/mL) and EGF (Sigma; 20 ng/mL). Mammospheres were supplemented with fresh sphere medium every other day and cultured from 7 to 14 days. Mammospheres were counted and imaged under a 5× objective.
2.4. Glial fibrillary acidic protein (GFAP) and FOXA2 immunofluorescence staining (IF)
Human astrocytes were seeded in treated chamber slides for 24 h, then washed with warm PBS and stimulated with conditioned media (CM) overnight. Cells were washed in PBS, fixed in 4% formaldehyde in PBS for 15 min, washed again and blocked in 1% goat serum in PBST for 1 h at room temperature. GFAP (Cell Signaling; 3656) 1° antibody was incubated overnight in 1% BSA/PBST at 4 °C. Cells were washed with PBS, incubated in DAPI (1:1000 in PBS), washed with PBS, and were placed on slides with aqueous mounting medium and sealed. Slides were imaged at 10x using an Olympus FV1200 spectral laser scanning confocal microscope or at 20x using the ImageXpress Pico microscope. IF staining for FOXA2 (R&D; AF2400) followed the same protocol as described above with frozen tissue on slides.
2.5. Cytokine array
Human astrocytes overexpressing miR-1290, miR-1246, or vector control were used. Cells were grown in serum-free medium for 24–48 h, centrifuged for 5 min at 300×g, and added to the cytokine array membranes. Procedures were followed as outlined in the Human Cytokine Array C2000 kit from RayBiotech (Norcross, GA).
2.6. Intracranial inoculation mouse model
All animal experiments were conducted as approved by the Wake Forest Institutional Animal Care and Use Committee. Intracranial implantations were performed as previously described [6]. Briefly, athymic female mice 5–6 weeks of age were anesthetized with ketamine/xylazine and isogenic SKBRM cells and astrocytes were stereotactically implanted into the right frontal lobe of the brain. Isogenic cell lines were implanted at a 1:1 ratio of SKBRM to astrocytes (total of 2 × 104 cells/5 μL PBS). Tumor growth was monitored 1x weekly using IVIS imaging and mice were euthanized at humane endpoint or end of study. Blood serum was collected, all organs imaged ex vivo, and mouse brains were stored in OCT for further analysis.
2.7. Statistical analyses
Log-Rank (Mantle-Cox) test, one-way ANOVA, two-way ANOVA, Pearson correlation, and student’s t-test were performed using Graphpad Prism 5 and 9 and described previously [11]. N = 3 experimental replicates unless otherwise indicated. Results are represented as ± SE.
See the supplementary information for a complete description of the methods used.
3. Results
3.1. Breast cancer-derived miR-1290- and miR-1246-containing EVs activate astrocytes
We previously observed that tGLI1-positive breast cancer stem cells (BCSCs) strongly activate and interact with astrocytes [11], the most abundant glial cell type in the brain, and reactive astrocytes promote metastasis and tumor growth in the brain [13,21]. To determine how tGLI1-expressing breast cancer cells activate astrocytes, we aimed to identify which secreted factors induced astrocyte activation. To this end, we stimulated human astrocytes with conditioned media (CM) from isogenic MDA-MB-231 cells overexpressing vector (231-Vector), GLI1 (231-GLI1), or tGLI1 (231-tGLI1). GFAP expression, a marker of astrocyte activation, was evaluated by immunofluorescence [13]. CM from 231-tGLI1 cells significantly activated astrocytes compared to control (FBS-free media) and CM from 231-Vector and 231-GLI1 cells (Fig. 1A–B). Since 231-tGLI1 cells activated astrocytes without cellular contact, we investigated whether 231-tGLI1 cells secrete proteins or EVs to activate astrocytes. We performed size exclusion chromatography to isolate protein and EV fractions from 231-tGLI1 CM (Fig. 1C). To confirm proper isolation of EVs, we performed NTA and found the EVs were the appropriate diameter (130–200 nm; Fig. 1D). To examine whether astrocytes could take up EVs secreted from cancer cells, we overexpressed GFP-labeled CD63, a tetraspanin used to characterize EVs, in breast cancer cells, collected CM, extracted EVs by ultracentrifugation, stimulated astrocytes with purified EVs, and then performed confocal microscopy. The secreted EVs were readily taken up by astrocytes as indicated by intracellular GFP signal (Fig. 1E). Next, we examined whether secreted proteins or EVs from 231-tGLI1 cells activated astrocytes and found that the EV fractions, but not the protein fractions, significantly increased GFAP expression in astrocytes (Fig. 1F–G). Since EVs contain a variety of bioactive molecules that are released into target cells including miRNAs [14], we investigated whether 231-tGLI1 cells contained increased EV-derived miRNAs, and thereby activated astrocytes. We conducted a miRNA microarray of EV-miRNAs isolated from 231-tGLI1 and 231-vector cells (Fig. 1H). Analysis of significantly differentially expressed miRNAs identified three miRNAs (miR-1290, miR-1246, and miR-1270) that were upregulated in EVs from 231-tGLI1 cells by at least two-fold and p-value < 0.01 (Fig. 1I). We confirmed these findings with RT-qPCR and observed significant enrichment of all three miRNAs in EVs-derived from 231-tGLI1 cells (Fig. 1J). Collectively, these data demonstrate that tGLI1-positive breast cancer cells secrete EV-derived miRNAs that are taken up by astrocytes and lead to astrocyte activation.
Fig. 1. Breast cancer-derived miR-1290- and miR-1246-containing extracellular vesicles activate astrocytes.
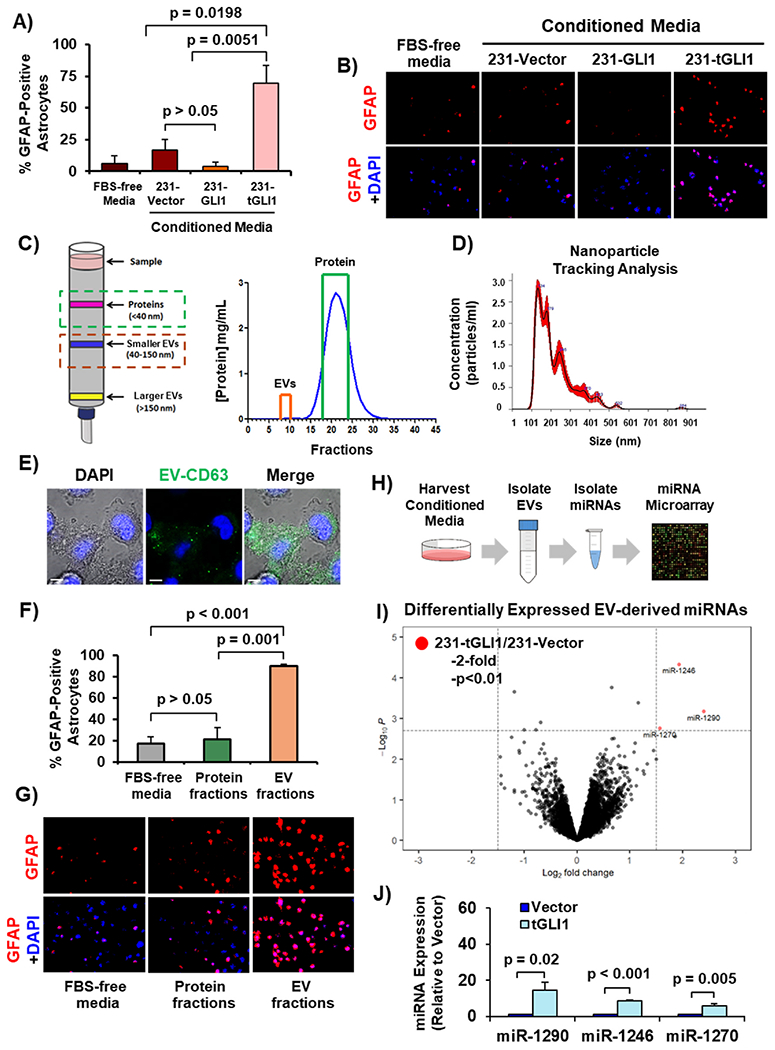
(A–B) Astrocyte activation by CM from 231-tGLI1 cells, as detected by GFAP immunofluorescence. Control astrocytes were incubated with FBS-free media. (A) Quantification of percent GFAP positive cells. (B) Representative images with 10× magnification. (C–D) Size exclusion chromatography of CM from 231-tGLI1 cells. (C) Schematic of extracellular vesicle (EV) or protein elution from size exclusion column and protein quantification of each fraction. Orange box indicates pooled EV fractions and green box indicates pooled protein fractions. (D) NTA of isolated EVs. (E) EV uptake assay. Image of astrocytes after overnight stimulation with CD63-GFP labeled EVs. 100× magnification. Scale bar indicates 5 μm. (F–G) Astrocyte activation, indicated by GFAP immunofluorescence, using EV or protein fractions. (F) Quantification of percent GFAP-positive astrocytes. (G) Representative images in 10× magnification. (H) Schematic of the miRNA microarray of isolated EV-derived miRNAs from 231-tGLI1 and 231-Vector cells. (I) Differentially expressed miRNAs identified from the miRNA microarray. Microarray data was analyzed using the limma package and visualized using the Enhanced Volcano package in R. (J) RT-qPCR of secreted EV-derived miRNAs from 231-Vector and 231-tGLI1 cells. Student’s t-test was used in Panels A, F, and J to compute p-values. N = 3 experimental replicates.
3.2. EV-derived miR-1290 and miR-1246 are highly expressed in metastatic breast cancer patient sera, associated with poor clinical outcomes, enriched in BCBM, and upregulated by tGLI1
To help narrow the list of these miRNAs, we analyzed publicly available patient datasets obtained from Gene Expression Omnibus (GEO). Analysis of a large cohort of circulating miRNAs from serum of 2686 non-cancer and 1269 breast cancer patients revealed significant enrichment of all three miRNAs in patients with breast cancer (Fig. 2A; GSE73002). Analysis of a cohort with subtype information showed that all three miRNAs were expressed at higher levels in HER2-enriched breast cancer and triple-negative breast cancer (TNBC) than the luminal subtypes (Fig. 2B; GSE86278). Furthermore, we found miR-1290 and miR-1246, but not miR-1270, were more highly expressed in sera from breast cancer patients with metastases compared to those with no metastases (Fig. 2C; GSE68373). Most notably, miR-1290 and miR-1246, but not miR-1270, levels in brain metastases were significantly higher compared to matched primary breast tumors (Fig. 2D; GSE37407). miR-1290 and miR-1246 expression was also correlated with worse overall survival in TNBC patients (Fig. 2E) and metastasis-free survival (MFS; Fig. 2F; GSE40267). Expression of miR-1270 was not included in this dataset. These findings directed us to further investigate miR-1290 and miR-1246 for their roles in activating astrocytes and promoting BCBM.
Fig. 2. EV-derived miR-1290 and miR-1246 are highly expressed in metastatic breast cancer patient sera, associated with poor clinical outcomes, enriched in BCBM, and upregulated by tGLI1.
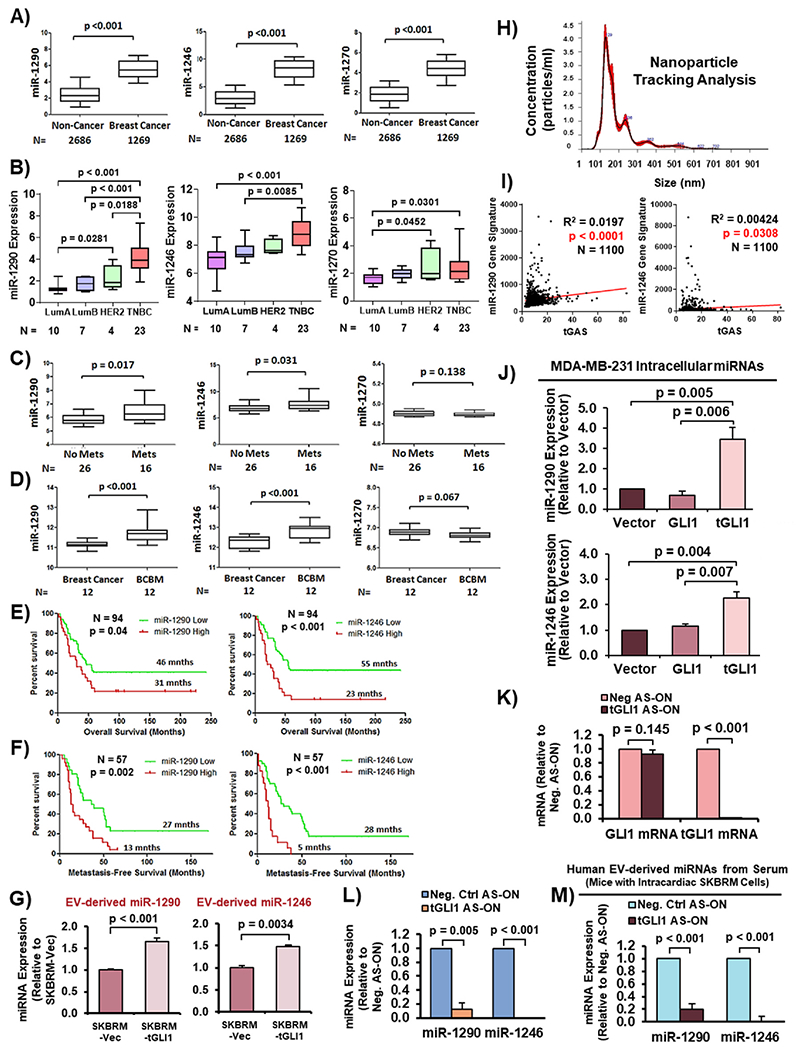
(A) MiRNA expression in sera of healthy individuals (non-cancer) compared to breast cancer patients (GSE73002). (B) Expression of miRNAs in breast cancer patient tumors with known subtype information (GSE86278). LumA, luminal A. LumB, luminal B. TNBC, triple-negative breast cancer. (C) MiRNA expression in sera of breast cancer patients without metastases (No Mets) compared to those with metastases (Mets; GSE68373). (D) MiRNA expression in primary breast tumors and matched brain metastases (GSE37407). (E–F) Kaplan-Meier curves showing overall survival and metastasis-free survival (MFS) of breast cancer patients with low versus high miRNA expression (GSE40267). Log-Rank (Mantle-Cox) test was used to compute p-values. (G) MiRNA expression in EVs from SKBRM-Vector and SKBRM-tGLI1 cells. (H) NTA of isolated EVs. (I) Correlation of tGAS and miR-1290/miR-1246 gene signatures. Pearson correlation was used to calculate p-values. (J) Intracellular expression of miR-1290 and miR-1246 in isogenic MDA-MB-231 cell lines. (K) RT-qPCR to validate effective and selective tGLI1 knockdown by tGLI1 AS-ON in SKBRM cells. (L) tGLI1 knockdown reduced miR-1290 and miR-1246 expression. (M) Expression of EV-derived miR-1290 and miR-1246 in sera of mice carrying SKBRM BCBM with or without tGLI1 knockdown. Serum was harvested from mice 25 days post inoculation. Students t-test was used in Panels A–D, G, and J-M. N = 3 experimental replicates.
To further examine the association of miR-1290 and miR-1246 with tGLI1-expressing BCBM, we isolated EVs via ultracentrifugation from the CM of SKBRM cells, a brain-tropic subline of SKBR3 cells, that stably express vector (SKBRM-Vector) or tGLI1 (SKBRM-tGLI1), and determined miRNA expression. We found expression of both miRNAs was significantly increased in SKBRM-tGLI1 cells compared to SKBRM-Vector cells (Fig. 2G). EV fractions were validated using NTA (Fig. 2H). Furthermore, we confirmed the link between both miRNAs and tGLI1 as indicated by the positive correlations between miR-1290/miR-1246 gene signatures [22] and the tGLI1 activation signature [5–8, 10] (tGAS; TCGA; Fig. 2I). Moreover, we found significantly increased intracellular miR-1290 and miR-1246 expression in 231-tGLI1 cells compared to 231-Vector and 231-GLI1 cells (Fig. 2J). Next, we selectively knocked down tGLI1 with previously validated tGLI1 antisense-oligonucleotides (AS-ON) [6] and found tGLI1, not GLI1, mRNA expression was significantly decreased (Fig. 2K). Furthermore, we found significantly decreased miR-1290 and miR-1246 expression in SKBRM cells with tGLI1 knockdown (Fig. 2L). We validated these in vitro findings by isolating EV-derived miRNAs from sera of mice that were intracardially implanted with tGLI1 AS-ON SKBRM cells in our recent study [11], which showed that mice in the tGLI1 AS-ON group presented with smaller brain metastases compared to the control group. Circulating EV-derived miR-1290 and miR-1246 levels were significantly decreased in mice carrying SKBRM with tGLI1 knockdown (Fig. 2M). Taken together, these results showed, for the first time, that miR-1290 and miR-1246 may play an important role in BCBM and that both miRNAs are upregulated by tGLI1 in vitro and in vivo.
3.3. Breast cancer-derived miR-1290 and miR-1246 activate astrocytes, which promote breast cancer stem cells
Analysis of EV-derived miR-1290 and miR-1246 levels in sera from breast cancer patients without metastases (No Met; N = 11) versus those with only brain metastases (Brain Met; N = 9) revealed an increase in expression of both miRNAs in the serum samples from patients with only brain metastases (Fig. 3A). EVs isolated using the ExoQuick Kit were validated via NTA (Fig. 3B) and western blotting for biomarkers (Supplementary Fig. 1A; CD-9 for smaller EVs, CD-154 for larger EVs, Annexin V for apoptotic bodies). Furthermore, we found miR-1290 and miR-1246 levels were elevated in EVs of brain-tropic lines (231-BRM and SKBRM) than those of their parental cells (MDA-MB-231 and SKBR3; Fig. 3C–D).
Fig. 3. Breast cancer-derived miR-1290 and miR-1246 activate astrocytes, which promote breast cancer stem cells.
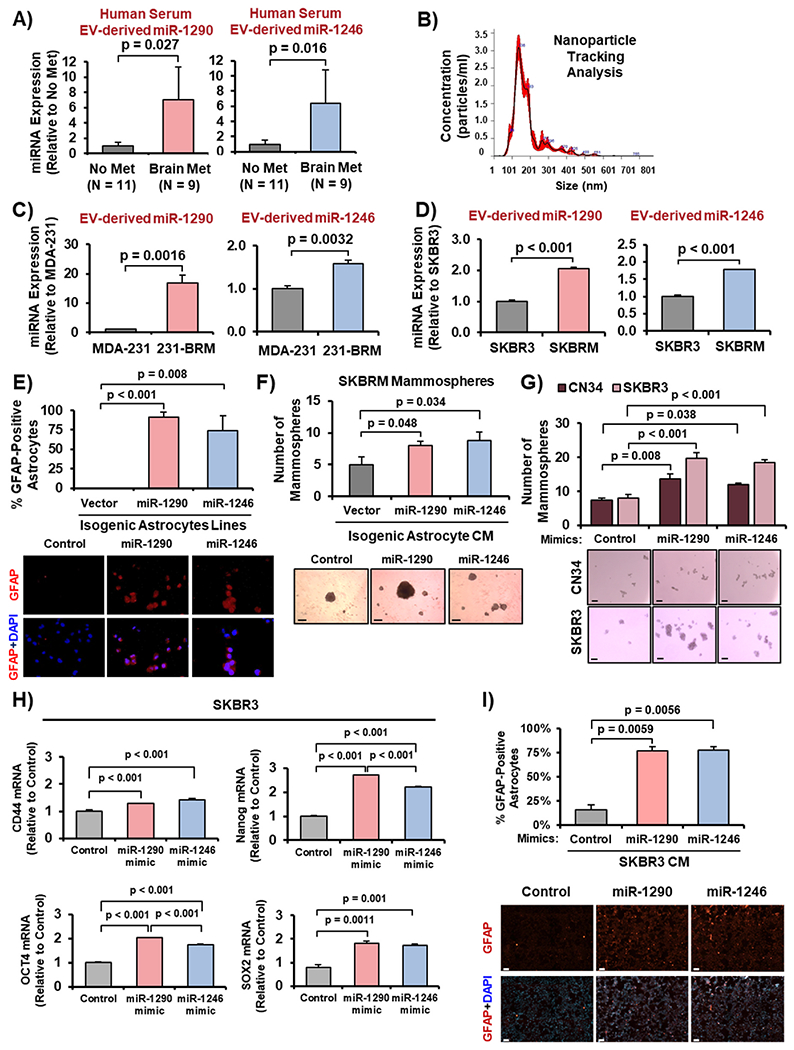
(A) Higher levels of EV-miR-1290 and miR-1246 in the serum of human breast cancer patients with only brain metastases (Brain Met; N = 9) compared to those with no metastases (No Met; N = 11). (B) NTA of isolated EVs. (C–D) miR-1290 and miR-1246 expression in EVs from CM of parental lines (MDA-231 and SKBR3) relative to brain-tropic lines (231-BRM and SKBRM). (E) Activation of astrocytes by miR-1290 and miR-1246 as indicated by GFAP immunofluorescence. (F) CM of miR-1290- and miR-1246-activated astrocytes promotes SKBRM mammosphere formation. 5× magnification. Scale bar indicates 100 μm. (G) Ectopic expression of miR-1290- and miR-1246 mimics promotes SKBR3 and CN34 mammosphere development. 5× magnification. Scale bar indicates 100 μm. (H) Ectopic expression of miR-1290 and miR-1246 increased mRNA expression of four stemness genes in SKBR3 cells as indicated by RT-qPCR. (I) CM from SKBR3 cells overexpressing miR-1290 or miR-1246 activated astrocytes as indicated by GFAP immunofluorescence. Scale bar indicates 100 μm. Student’s t-test was used in Panels A, C-D, H to derive p-values. One-way ANOVA was used in Panels E–G, I to compute p-values. N = 3 experimental replicates.
Next, we investigated whether miR-1290 and miR-1246 activate astrocytes and found that astrocytes stably expressing miR-1290 or miR-1246 were significantly activated compared to the control astrocytes (Fig. 3E). Further, we examined whether astrocytes activated by miR-1290 and miR-1246 could promote breast cancer stemness, and observed that CM from miR-1290- or miR-1246-activated astrocytes significantly increased mammosphere formation of SKBRM cells (Fig. 3F). We confirmed this finding by transfecting breast cancer cells with miR-1290 and miR-1246 mimics, which are chemically-modified molecules designed to mimic mature miRNAs. We found that the miRNA mimics promoted mammosphere development in SKBR3 cells and CN34 TNBC cells (Fig. 3G). Since mammospheres enrich the BCSC population, we determined whether miR-1290 and miR-1246 increased expression of cancer stemness genes. Ectopic expression of miR-1290 and miR-1246 upregulated expression of four known stemness genes, CD44, Nanog, OCT4, and SOX2 (Fig. 3H). Additionally, we observed that CM from SKBR3 cells overexpressing miR-1290 or miR-1246 significantly activated astrocytes compared to control (Fig. 3I). Overexpression of miR-1290 and miR-1246 were validated with RT-qPCR (Supplementary Fig. 1B). Collectively, these results demonstrate that EV-derived miR-1290 and miR-1246 are enriched in BCBM in vitro, and that breast cancer-derived miR-1290 and miR-1246 activate astrocytes, which promote BCSCs.
3.4. miR-1290 and miR-1246 inhibition decreases the ability of breast cancer cells to activate astrocytes and promote mammosphere formation
To complement our overexpression studies, we inhibited each miRNA using lentiviral anti-miRNA constructs in SKBRM-tGLI1 cells and determined the impact on astrocyte activation. The lentivirus encodes an antisense miRNA which binds to mature miRNAs in a base-pair specific manner to prevent miRNAs from regulating target mRNAs. CM from SKBRM-tGLI1-miROff-1290 (Off-1290) and SKBRM-miROff-1246 (Off-1246) cells lost the ability to significantly activate astrocytes (Fig. 4A). Moreover, SKBRM-tGLI1-miROff-1290 and SKBRM-miROff-1246 cells displayed reduced ability to form mammospheres compared to SKBRM-tGLI1-miROff-Vector cells (Fig. 4B). To confirm effective miR-1290 and miR-1246 inhibition, we determined whether the miRNA knockdown increased expression of known miR-1290 and miR-1246 target genes [23–30]. SKBRM-tGLI1-miROff-1290 expressed significantly higher levels of miR-1290 targets, p27, IRF2, SCAI, and NAT1, than SKBRM-tGLI1-miROff-Vector cells (Fig. 4C). Similarly, SKBRM-tGLI1-miROff-1246 expressed significantly higher levels of miR-1246 targets, DYRK1A, PRKAR1A, and PPP2CB, compared to SKBRM-tGLI1-miROff-Vector cells (Fig. 4D). Interestingly, LIG4 and IL6ST mRNA expression was significantly decreased in SKBRM-tGLI1 cells with miR-1246 inhibition, suggesting that miR-1246 may maintain the ability to regulate certain genes in a cancer or tissue-specific manner or that regulation of LIG4 and IL6ST gene expression is more complex (Fig. 4D). Taken together, these data demonstrate that miR-1290 and miR-1246 inhibition reduces the ability of breast cancer cells to activate astrocytes and promote mammosphere formation.
Fig. 4. miR-1290 and miR-1246 inhibition decreases astrocyte activation and mammosphere formation.
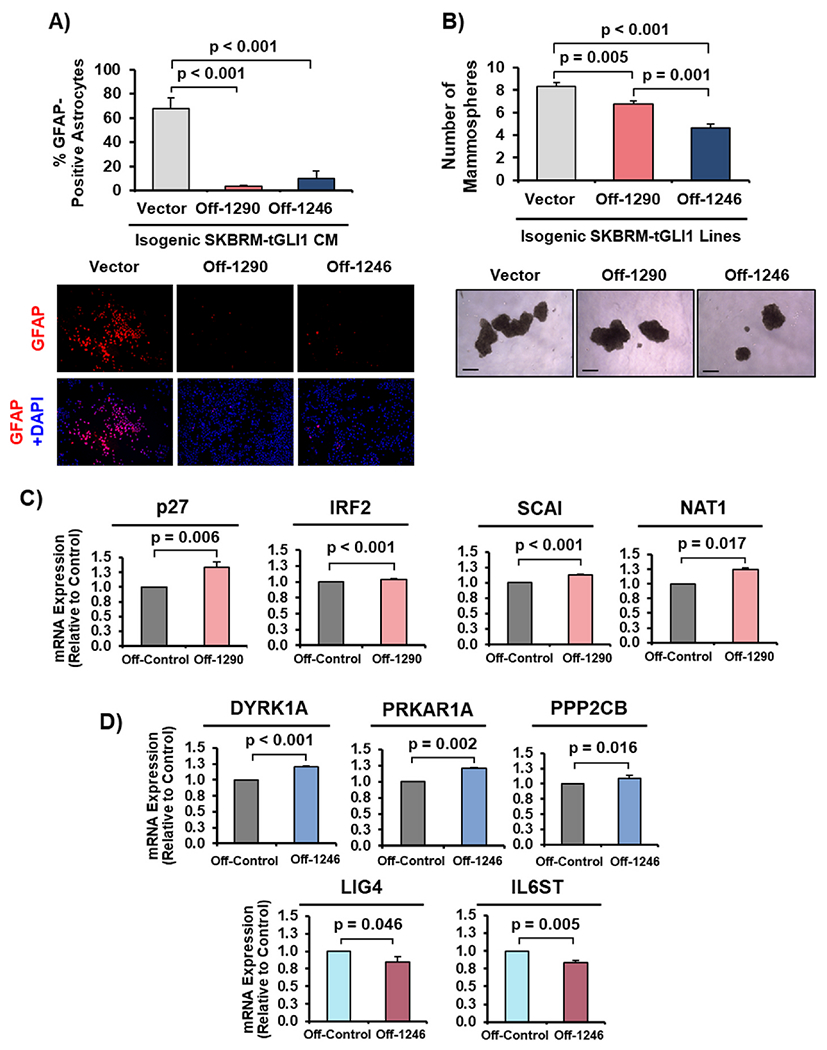
(A) GFAP expression in astrocytes stimulated by CM from SKBRM cells overexpressing tGLI1 with miR-1290 or miR-1246 inhibitors (SKBRM-tGLI1-miR-Off-1290 or -1246). 10× magnification. (B) Mammosphere assay of SKBRM-tGLI1-miR-Off-Control, −1290, or −1246 cells. 5× magnification. Scale bar indicates 100 μm. (C–D) RT-qPCR of miR-1290 and miR-1246 target genes, respectively, in SKBRM-tGLI1-miROff-1290 (Off-1290) and miR-1246 (Off-1246). One-way ANOVA was used in Panel A–B to compute p-values. Student’s t-test (two-tailed) was used in Panels C–D to derive p-values. N = 3 experimental replicates.
3.5. miR-1290- and miR-1246-activated astrocytes secrete CNTF to promote breast cancer stem cells
Given that CM from astrocytes overexpressing miR-1290 and miR-1246 promoted mammosphere formation of SKBRM cells (Fig. 3F), we hypothesized that these astrocytes secrete cytokines to promote breast cancer cells. To identify these cytokines, we subjected CM from astrocytes stably overexpressing control vector, miR-1290 or miR-1246 to a cytokine array comprised of 174 cytokines. We identified a number of cytokines and growth factors that were significantly differentially expressed, such as ciliary neurotrophic factor (CNTF), which was identified to be highly secreted by both miR-1290- and miR-1246-activated astrocytes (Fig. 5A; 71.82- and 172.82-fold increase, respectively). CNTF is secreted by reactive astrocytes and is a potent survival factor in the brain [31,32]; CNTF receptor-alpha (CNTFR-α) is highly expressed in breast cancer [33]. To validate the cytokine array results, we stimulated astrocytes with CM from isogenic SKBRM cell lines and then measured secreted CNTF using an ELISA. CM from SKBRM-tGLI1 cells significantly increased the ability of astrocytes to secrete CNTF compared to CM from SKBRM-Vector and SKBRM-GLI1 cells (Fig. 5B). Astrocytes overexpressing miR-1290 and miR-1246 showed increased CNTF mRNA and protein expression compared to control (Fig. 5C–D). Furthermore, we found that CNTF significantly activated astrocytes (Fig. 5E) and promoted SKBRM mammosphere formation (Fig. 5F). CNTFR-α expression is more highly expressed in brain metastases compared to normal breast tissues and primary breast tumors (Fig. 5G). Using an established 12-gene activation signature of CNTF [31] to divide a patient cohort, we observed that patients with high tumoral CNTF activation presented with shortened brain MFS (Fig. 5H). Consistent with these findings, we found that high CNTFR gene signature [31] correlated with worse brain MFS (Fig. 5I). Collectively, these novel findings demonstrate that miR-1290- and miR-1246-activated astrocytes secrete CNTF to promote BCSCs, and the CNTF-CNTFR pathway is associated with BCBM.
Fig. 5. miR-1290- and miR-1246-activated astrocytes secrete CNTF to promote breast cancer stem cells.
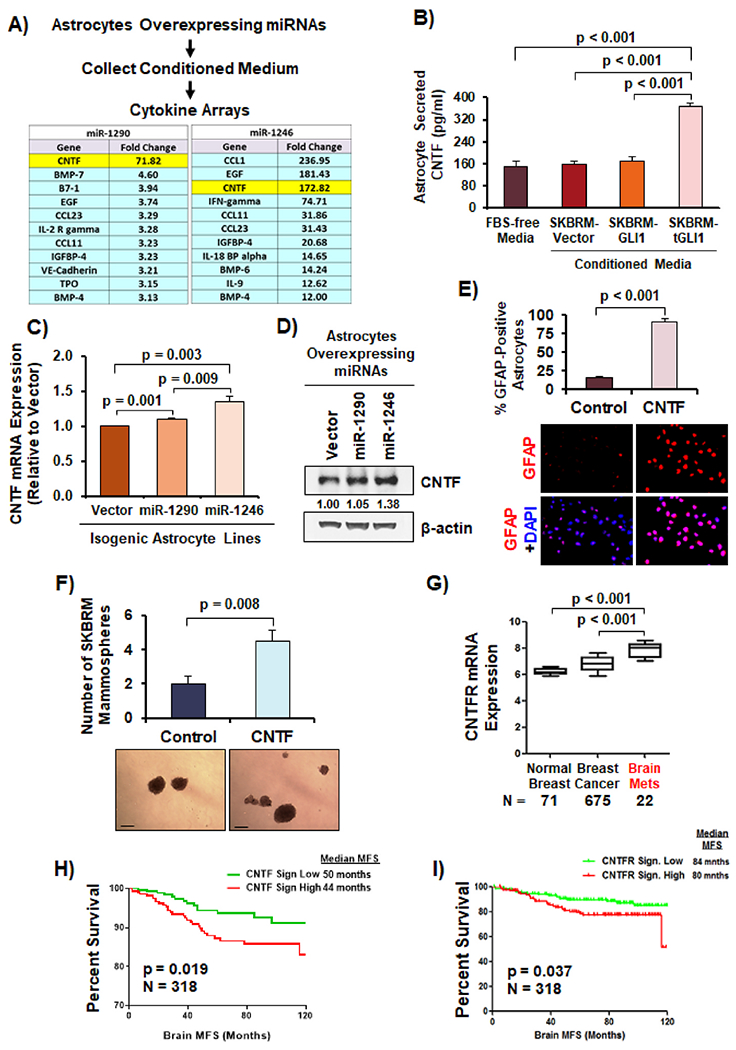
(A) Schematic of cytokine array assay. Table of top secreted cytokines and growth factors from astrocytes overexpressing miR-1290 and -1246 compared to control astrocytes. (B) Astrocytes secreted more CNTF in the presence of CM from SKBRM-tGLI1 cells as indicated by CNTF ELISA. (C) CNTF mRNA expression in astrocytes overexpressing vector, miR-1290, and miR-1246 using RT-qPCR. (D) CNTF protein expression in astrocytes overexpressing vector, miR-1290, and miR-1246 as indicated by Western blot analysis with quantification. (E) CNTF activated astrocytes as indicated by GFAP immunofluorescence. Human astrocytes were stimulated with or without 100 ng/mL of CNTF. 10× magnification. (F) CNTF increased mammosphere formation of SKBRM cells. 5× magnification. Scale bar indicates 100 μm. (G) Expression of CNTFR-α in normal breast tissue, breast cancer, and BCBM. (H–I) Kaplan-Meier plot of brain MFS using with CNTF and CNTFR gene signatures in breast cancer GEO datasets (GSE3740/40267/68373/73002/86278/158309). Log-Rank (Mantle-Cox) test was used in Panels H–I. Student’s t-test was used in Panels B–G to compute p-values. N = 3 experimental replicates.
3.6. miR-1290 and miR-1246 upregulate CNTF expression through reducing expression of FOXA2 in astrocytes
Next, we aimed to determine how miR-1290 and miR-1246 upregulate CNTF expression by identifying a transcriptional repressor: (1) whose expression can be suppressed by miR-1290 and miR-1246, and (2) can transcriptionally repress CNTF expression. We searched for potential transcriptional repressors through GeneCards [34] and found forkhead box A2 (FOXA2) to be a predicted repressor of CNTF expression. To determine whether FOXA2 is negatively regulated by miR-1290 and miR-1246, we used Targetscan [35], a miRNA-target search engine, to identify predicted binding sites for miR-1290 and miR-1246 in the 3′-untranslated regions (3′-UTRs) of FOXA2 and found two potential miR-1290 binding sites and one potential miR-1246 binding site (Fig. 6A). To determine whether miR-1290 and miR-1246 suppress FOXA2 expression, we performed RT-qPCR and found significantly decreased FOXA2 mRNA expression in astrocytes overexpressing miR-1290 or miR-1246 compared to vector (Fig. 6B). We also observed a decrease in FOXA2 protein expression in astrocytes transfected with miR-1290 and miR-1246 mimics (Fig. 6C). Furthermore, using a luciferase reporter under the control of the FOXA2 3′-UTR, we found that miR-1290 and miR-1246 expression significantly reduced activity of the FOXA2 3′-UTR reporter compared to vector (Fig. 6D), suggesting that miR-1290 or miR-1246 target the FOXA2 3′-UTR. Next, we determined whether FOXA2 represses CNTF expression and found that ectopic FOXA2 expression significantly suppressed CNTF mRNA and protein expression in astrocytes (Fig. 6E–F). To determine whether FOXA2 binds to the human CNTF promoter, we conducted a ChIP assay followed by qPCR and observed that FOXA2 directly binds to two regions within the human CNTF promoter (Fig 6G). To determine whether FOXA2 could suppress the CNTF promoter, we performed luciferase reporter assays and found that overexpression of FOXA2 suppressed CNTF promoter activity in all three isogenic astrocytes cell lines (Fig. 6H). In summary, these results reveal a novel mechanism in which breast cancer EV-derived miR-1290 and miR-1246 negatively regulate expression of FOXA2 in astrocytes, FOXA2 transcriptionally represses the cytokine CNTF, and reduced FOXA2 leads to increased CNTF expression and secretion, which results in astrocyte activation and breast cancer aggressiveness (Fig. 6I).
Fig. 6. miR-1290 and 1246 upregulate CNTF expression through reducing expression of FOXA2 in astrocytes.
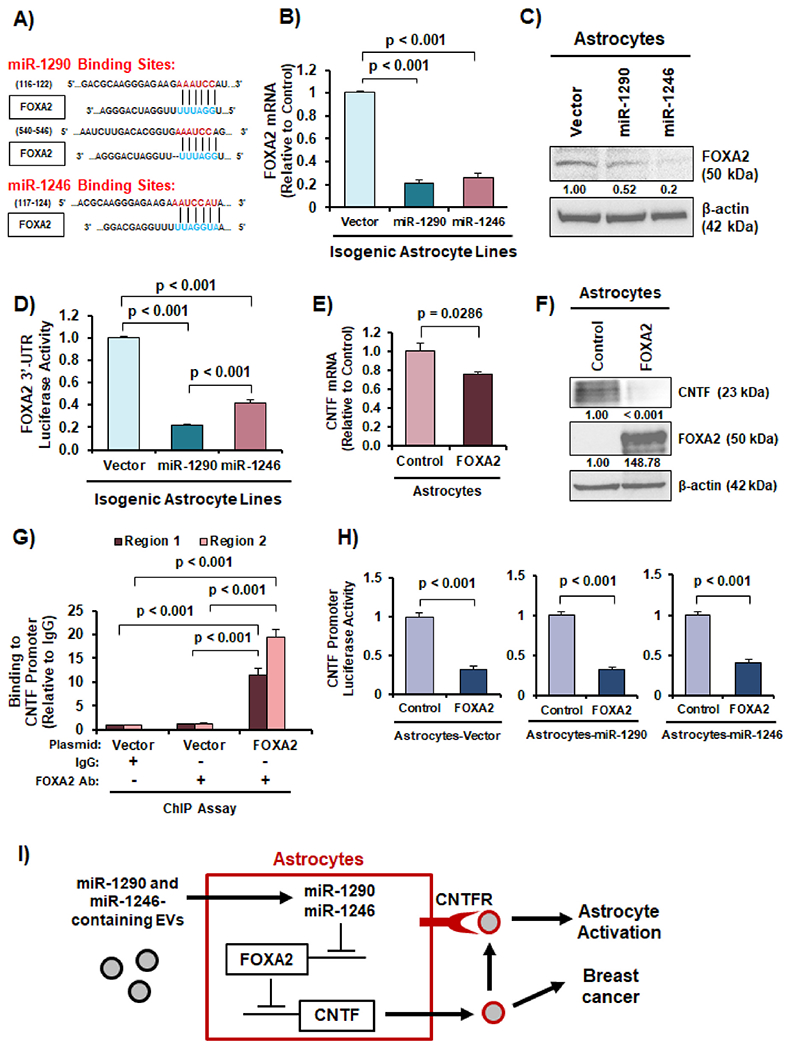
(A) Predicted miR-1290 and miR-1246 binding sites in the 3′-UTR of FOXA2 from Targetscan. (B) FOXA2 mRNA expression was reduced in astrocytes overexpressing miR-1290 and miR-1246. (C) FOXA2 protein expression was decreased in astrocytes transfected with miR-1290 and miR-1246 mimics as indicated by western blotting. (D) FOXA2 3′-UTR luciferase reporter activity was significantly lower in astrocytes overexpressing miR-1290 or miR-1246. (E) Ectopic expression of FOXA2 reduced CNTF mRNA expression in astrocytes as indicated by RT-qPCR. (F) Ectopic expression of FOXA2 decreases CNTF protein expression in astrocytes as indicated by western blotting. (G) FOXA2 binds to the human CNTF gene promoter in astrocytes as shown by ChIP-qPCR. (H) CNTF gene promoter activity was suppressed by miR-1290 or miR-1246 in astrocytes as indicated by dual luciferase reporter assay. (I) Schematic illustration of the novel EV-derived miR-1290/1246→FOXA2→CNTF signaling axis that leads to astrocyte activation and breast cancer aggressiveness. Students t-test was in Panels B, D-E, G-H to compute p-values. N = 3 experimental replicates.
3.7. miR-1290-expressing astrocytes promote intracranial colonization and growth of brain-tropic breast cancer cells in vivo
Given that miR-1290- and miR-1246-expressing astrocytes promote brain-tropic breast cancer cells, we devised a model to examine whether astrocytes activated by miR-1290 and miR-1246 could promote breast cancer cell colonization and growth in the brain. Whether miR-1290 and miR-1246 can promote breast cancer growth in the brain in vivo has not been studied. We intracranially co-implanted (1) SKBRM + Astrocytes-Vector, (2) SKBRM + Astrocytes-miR-1290, or (3) SKBRM + Astrocytes-miR-1246 into 5–6 week old, athymic female mice (N = 10 per group). Notably, SKBRM cells maintain endogenous levels of tGLI1 and express luciferase so IVIS imaging was utilized to measure bioluminescence throughout the study (Fig. 7A). We observed that Astrocytes-miR-1290 significantly enhanced the ability of SKBRM cells to colonize and grow in the mouse brain (Group 2 vs Group 1; p = 0.02) (Fig. 7B–C). In contrast, Astrocytes-miR-1246 did not significantly promote the growth of SKBRM cells in the brain (Group 3 vs Group 1; p > 0.05). EV-derived miR-1290 was significantly higher in the serum of mice with SKBRM + Astrocytes-miR-1290 compared to the other two groups (Fig. 7D; top). While circulating EV-miR-1246 levels in mice with Astrocytes-miR-1246 was higher, the difference did not reach statistical significance (Fig. 7D; bottom). Next, we examined mouse brain sections for tumor cell proliferation using immunohistochemical analysis (IHC; Ki-67) and found that Astrocytes-miR-1290, but not Astrocytes-miR-1246, significantly enhanced SKBRM cell proliferation in the mouse brain (Fig. 7E,G). Additionally, we conducted GFAP IHC using a human GFAP antibody to determine the extent of astrocyte activation within the tumors and observed that GFAP-positive Astrocytes-miR-1290 were more tumor infiltrative than GFAP-positive Astrocytes-miR-1246 or Astrocytes-Vector (Fig. 7F–G). CNTF IHC revealed that Astrocytes-miR-1290, but not Astrocytes-miR-1246, significantly increased CNTF expression compared to Astrocytes-Vector (Fig. 7H–I). To examine FOXA2 expression in astrocytes, we performed IF co-staining for FOXA2 and GFAP on the mouse brain sections. Of the GFAP-positive (red) astrocytes, we found significantly decreased FOXA2 expression (green) in Astrocytes-miR-1290 compared to Astrocytes-miR-1246 or Astrocytes-Vector (Fig. 7J–K). In summary, we report, for the first time, that tGLI1-expressing brain-metastatic breast cancer cells secrete EV-derived miR-1290, which activates astrocytes in the brain metastatic niche through the novel miR-1290-FOXA2-CNTF signaling axis, and that astrocyte-secreted CNTF promotes breast cancer cell growth in the brain (Fig. 7L).
Fig. 7. miR-1290-expressing astrocytes promote intracranial colonization and growth of brain-tropic breast cancer cells in vivo.
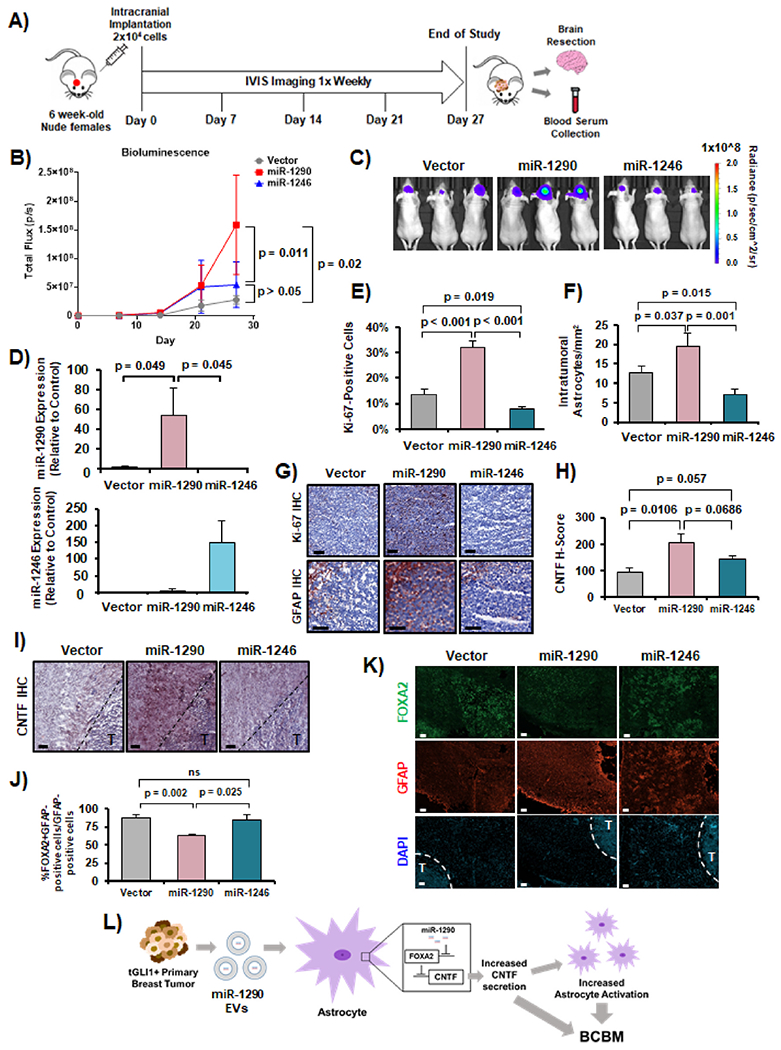
Isogenic SKBRM breast cancer lines and isogenic astrocyte lines were co-implanted into the mouse brain and imaged weekly via an IVIS imager. Brain resection and blood serum collection occurred at humane endpoint or end of study. Ten mice per group were used. Group 1: SKBRM + Astrocytes-Vector (Vector); Group 2: SKBRM + Astrocytes-miR-1290 (miR-1290); and Group 3: SKBRM + Astrocytes-miR-1246 (miR-1246). (A) Experimental design. (B) Bioluminescence measuring tumor growth throughout the study. Two-way ANOVA was used to compute p-values. (C) Representative IVIS images at Day 27. (D) Circulating EV-miR-1290 and miR-1246 isolated from mouse serum using RT-qPCR. (E) Ki-67 IHC. (F) Tumor-infiltrating, activated astrocytes as indicated by GFAP IHC. (G) Representative IHC images at 20× magnification. Scale bar indicates 100 μm. (H) CNTF IHC of mouse brain sections. (I) Representative CNTF IHC images at 20× magnification. Scale bar indicates 100 μm. (J) IF co-staining of FOXA2 and GFAP in mouse brain sections. (K) Representative FOXA2 and GFAP IF images at 20× magnification. “T” indicates intracranial tumor. Scale bar indicates 100 μm. (L) Schematic of described mechanism by which tGLI1 upregulates EV-derived miR-1290 to activate astrocytes, increase CNTF, and promote breast cancer cell growth in the brain. Students t-test was used in Panels D–F, H, and J to calculate p-values. N = 5 experimental replicates.
4. Discussion
In the present study, we made the following novel observations: 1) tGLI1-positive breast cancer cells secrete EVs containing miR-1290 and miR-1246 that activate astrocytes; 2) EV-derived miR-1290 and miR-1246 are highly enriched in BCBM, associated with poor clinical outcomes, and upregulated by tGLI1; 3) ectopic expression of miR-1290 and miR-1246 activates astrocytes and promotes BCSC populations, while miRNA knockdown abrogates these effects; 4) miR-1290 and miR-1246-primed astrocytes secrete CNTF to promote mammospheres; 5) miR-1290 and miR-1246 in astrocytes repress FOXA2 expression, a negative regulator of CNTF, resulting in increased expression of CNTF and astrocyte activation; and 6) astrocytes activated by miR-1290 are more tumor-infiltrative than the control astrocytes, downregulate FOXA2 expression, upregulate CNTF expression, and promote intracranial SKBRM xenograft growth in vivo. Together, these findings provide novel insights into the molecular mechanisms that underlie BCBM.
We observed that tGLI1-positive breast cancer cells and tumors express higher levels of miR-1290 and miR-1246 than tGLI1-negative/low breast cancer. We also found that tGLI1-expressing breast cancer cells secrete increased levels of EV-derived miR-1290 and miR-1246 to activate astrocytes. The mechanism by which tGLI1 promotes expression/secretion of miR-1290 and miR-1246 remains unclear. Given that tGLI1 overexpression induced intracellular expression of miR-1290 and miR-1246 whereas tGLI1 knockdown reduced expression (Fig. 2J–L), tGLI1 transcription factor likely directly binds to the promoter regions of miR-1290 and miR-1246, leading to transactivation. It is challenging to identify the mechanism by which a miRNA is upregulated by a transcription factor for multiple reasons. About 50% of miRNA genes are located in intergenic (non-protein coding) regions and a transcription start site can be localized at 200-20,000 bases upstream of the miRNA coding region [36]. Expression of intronic miRNAs can be regulated by the gene promoter or promoter regions independent of the gene [36].
tGLI1-positive breast cancer cells may preferentially sort and package miR-1290 and miR-1246 into EVs. Of note, whether tGLI1 regulates miRNA sorting and packaging into EVs in any cell or tumor type is currently unknown. Previous work has indicated that the Snail transcription factor regulates expression of several miRNAs through two different mechanisms, by directly binding to their promoter regions and by altering miRNA sorting and packaging into EVs in colorectal adenocarcinoma [37]. Additionally, miR-1246 was shown to be highly secreted by malignant mammary cells (Luminal B, HER2-enriched, and TNBC) compared to the normal mammary epithelial cells, regardless of the intracellular abundance of the miRNAs [38]. These findings suggest tumor cells may regulate miRNA expression and regulate the sorting and release of EV-derived miRNAs. Whether tGLI1 regulates miRNA sorting and packaging into EVs warrants further investigation.
Another potential mechanism includes tGLI1’s ability to regulate miRNA processing machinery by simultaneously upregulating factors that preferentially load miR-1290 and miR-1246 into EVs. Whether tGLI1 regulates the miRNA processing machinery in any cell or tumor type to alter miRNA expression is unknown. Several transcription factors increase miRNA expression by modifying the miRNA processing machinery. For example, the oncogenic transcription factor c-Myc not only regulates the transcription of several miRNAs, but also the expression of Drosha, the RNAse III endoribonuclease that processes pre-miRNAs, thereby mediating miRNA expression [39]. The regulatory function of miRNAs requires Argonaute (AGO) proteins which form the miRNA-RNA-induced silencing complexes that regulate gene expression by targeting mRNAs [14]. tGLI1 or its targets may also increase expression of any of the four AGO proteins in the human genome (AGO1-4) [40]. Previously, AGO2, the most predominant isoform in humans, is regulated by both the epidermal growth factor receptor and mitogen-activated protein kinase pathways in multiple breast cancer cell lines [41]. Further, IHC analysis of AGO2 in breast tumor tissues revealed significantly increased positive staining in the basal-like subtype breast tumors compared to other breast cancer subtypes, suggesting upregulated AGO2 expression may contribute to more aggressive breast cancer phenotypes [42]. The role of tGLI1 in AGO regulatory proteins has not been studied and should be explored in further studies.
Finally, tGLI1 may indirectly upregulate expression of miR-1290 and miR-1246, by increasing expression of transcription factors that activate miR-1290 and miR-1246 expression. Previous work has identified that OCT4 can directly regulate the expression of miR-1246 [43]. Since the Oct4 gene is a transcriptional target of tGLI1 [11], it is possible that tGLI1 indirectly upregulates miR-1290 and miR-1246 through OCT4. The role of tGLI1 in regulating miR-1290 and miR-1246 expression and secretion warrants further investigation. Above-mentioned potential mechanisms underlying tGLI1 upregulation of miR-1290 and miR-1246 may occur concurrently.
Our results indicate that miR-1290 and miR-1246 are highly expressed in BCBM and are important for both BCSC growth and astrocyte activation. While these observations are novel, previous work has reported that miR-1290 and miR-1246 contribute to lung cancer stem cells and metastasis [44]. Further, EVs from pediatric glioma stem cells were highly enriched in miR-1290 and miR-1246 compared to EVs from normal (neural) stem cells, suggesting an important role of these miRNAs in gliomas [45]. miR-1290 has also been shown to be associated with malignant phenotypes in multiple cancers including colorectal carcinoma [23], glioma [46], oral squamous cell carcinoma (OSCC) [47], and HCC [48]. Interestingly, miR-1290 was found to be down-regulated in estrogen receptor (ER)-positive breast tumors [49]. This is consistent with our findings (Fig. 2B) showing lower levels of miR-1290 in less aggressive subtypes of breast cancer. These observations suggest that ER may inhibit miR-1290 in these ER-positive breast cancers.
We reported the novel role of miR-1246 in activating astrocytes through the FOXA2-CNTF signaling axis. miR-1246 has been implicated in breast cancer proliferation [50], invasion and drug resistance [50], promotion of metastasis in colorectal carcinoma [51], and invasion in OSCC [52] and lung cancer [53]. In contrast, overexpression of miR-1246 inhibited tumor growth and promoted apoptosis in a prostate cancer mouse xenograft model [54]. The roles of miR-1246 in different tumor types may vary and should be examined further.
CNTF is secreted by astrocytes during neuron injury to help with survival and recovery [55]. Of note, tumor-associated astrocytes secrete specific cytokines that drive brain- and lung-specific metastases of TNBC cells [56]. We discovered that astrocytes stimulated by tGLI1-positive breast cancer cells secrete CNTF, leading to astrocyte activation (an autocrine effect). Since CNTFR-α is highly enriched in patient breast tumors and in BCBM (Fig. 5G), astrocyte-secreted CNTF likely promotes the intracranial growth and progression of BCBM as a paracrine effect. The CNTF-CNTFR-α pathway has been shown to play an important role in HCC; CNTFR-α was highly expressed in HCC and CNTFR-α activation by CNTF led to increased glucose uptake and reduced cell-cycle arrest to promote cell growth [57]. Furthermore, CNTF was shown to protect neuroblastoma cells from oxidative stress-induced apoptosis [58].
There are currently no published reports with FOXA2 directly repressing expression of CNTF in any cancer type. In normal CNS homeostasis, CNTF is expressed at low levels [59]. However, following injury such as brain injury [60] or stroke [61], CNTF has been shown to be upregulated in activated, GFAP-positive astrocytes. FOXA2 has important roles in the differentiation of dopaminergic neurons in the brain and is expressed in normal astrocytes [62]. Given that we observed upregulated CNTF to activate astrocytes and promote mammospheres, miR-1290 suppression of FOXA2 may be an important regulatory mechanism in reactive astrocytes.
In this study, we uncovered the novel roles of FOXA2, a miR-1290 and miR-1246 target, and a transcriptional repressor of CNTF, suggesting it functions as a tumor suppressor. FOXA2 negatively regulates the epithelial-to-mesenchymal transition in several cancers, including breast [58], endometrial [63], and pancreatic cancers [64]. FOXA2 is associated with decreased metastasis in lung cancer [65], gastric cancer [66], and HCC [67]. Low FOXA2 expression in GBM is predictive of poor survival and associated with adverse events in patients who received chemotherapy or radiotherapy [68]. In contrast, several studies reported that FOXA2 can play a tumor-promoting role. FOXA2 has been correlated with TNBC proliferation and stemness and is associated with increased relapse [69]. In the brain, FOXA2 is important for development and maintenance of dopaminergic neurons [70] and mediates the SHH pathway by negatively or positively regulating SHH pathway components [71]. These mixed results suggest that the functionality of FOXA2 is complex and likely tumor type- and cell context-dependent.
In summary, this study demonstrates the novel role that EV-derived miR-1290 and miR-1246 play in activating astrocytes in the brain through the novel miR-1290/1246→FOXA2→CNTF signaling axis. Additionally, astrocytes activated by miR-1290 have the ability to promote progression of established breast cancer brain metastases. Together, our results shed new light on the functionality of EV-derived miR-1290 and miR-1246, and mechanisms by which brain-metastatic breast cancer primes astrocytes in the brain to facilitate breast cancer brain metastasis.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Dr. Yin-Yuan Mo for the miR-1290, miR-1246, and miR-1270 overexpression lentiviruses. Additionally, we would like to acknowledge Dr. Xandra Breakefield’s laboratory for the EV tracking CD63-GFP, and Dr. Russell O. Pieper for the immortalized human astrocytes.
Funding
This research was funded by United States National Institutes of Health grants R01NS087169 (HWL), 1T32CA247819-01 (ATR), P30CA012197 (BP), 1R01CA228137-01A1 (HWL, KW), as well as, United States Department of Defense grants, W81XWH-17-1-0044 (HWL), W81XWH-19-1-0072 (HWL), W81XWH-19-1-0753 (HWL), and W81XWH-20-1-0044 (HWL).
Declaration of competing interest
A. Thomas declares research support (paid to the institution) from Sanofi; stock ownership in Johnson and Johnson, Bristol Myers Squibb, Pfizer, and Gilead; consulting for Eli Lilly, Genentech; participation in DSMB for BeyondSpring Pharmaceuticals; and royalties from Up-to-Date.
Footnotes
CRediT authorship contribution statement
Sherona R. Sirkisoon: Conceptualization, Data curation, Formal analysis, Methodology, Writing – review & editing, and. Grace L. Wong: Conceptualization, Data curation, Formal analysis, Methodology, Writing – review & editing, Visualization, Writing – original draft. Noah R. Aguayo: Data curation, Formal analysis, Methodology, Writing – review & editing. Daniel L. Doheny: Data curation, Formal analysis, Methodology, Writing – review & editing. Dongqin Zhu: Data curation, Formal analysis, Methodology. Angelina T. Regua: Methodology, Writing – review & editing. Austin Arrigo: Writing – review & editing. Sara G. Manore: Writing – review & editing. Calvin Wagner: editing. Alexandra Thomas: Resources, Investigation, Writing – review & editing. Ravi Singh: Data curation, Formal analysis, Methodology, Writing – review & editing. Fei Xing: Data curation, Formal analysis, Methodology, Writing – review & editing. Guangxu Jin: Formal analysis, Methodology, Writing – review & editing. Kounosuke Watabe: Funding acquisition, Methodology, Writing – review & editing. Hui-Wen Lo: Conceptualization, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing, All authors have read and agreed to the published version of the manuscript.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.canlet.2022.215726.
References
- [1].Siegel RL, Miller KD, Fuchs HE, Jemal A, Cancer stat. 72 (2022) (2022) 7–33. [DOI] [PubMed] [Google Scholar]
- [2].Chaffer CL, Weinberg RA, A Perspect. Cancer Cell Metastasis 331 (2011) 1559–1564. [DOI] [PubMed] [Google Scholar]
- [3].Ekici K, Temelli O, Dikilitas M, Halil Dursun I, Bozdag Kaplan N, Kekilli E, Survival and prognostic factors in patients with brain metastasis: single center experience, J. B.U.ON. : Off. J. Balkan Union Oncol 21 (2016) 958–963. [PubMed] [Google Scholar]
- [4].Lo HW, Zhu H, Cao X, Aldrich A, Ali-Osman F, A novel splice variant of GLI1 that promotes glioblastoma cell migration and invasion, Cancer Res. 69 (2009) 6790–6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cao X, Geradts J, Dewhirst MW, Lo HW, Upregulation of VEGF-A and CD24 gene expression by the tGLI1 transcription factor contributes to the aggressive behavior of breast cancer cells, Oncogene 31 (2012) 104–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rimkus TK, Carpenter RL, Sirkisoon S, Zhu D, Pasche BC, Chan MD, Lesser GJ, Tatter SB, Watabe K, Debinski W, Lo H-W, Truncated Glioma-Associated Oncogene Homolog 1 (tGLI1) Mediates Mesenchymal Glioblastoma via Transcriptional Activation of CD44 78, 2018, pp. 2589–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sirkisoon SR, Carpenter RL, Rimkus T, Anderson A, Harrison A, Lange AM, Jin G, Watabe K, Lo H-W, Interaction between STAT3 and GLI1/tGI1 oncogenic transcription factors promotes the aggressiveness of triple-negative breast cancers and HER2-enriched breast cancer, Oncogene 37 (2018) 2502–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Carpenter RL, Paw I, Zhu H, Sirkisoon S, Xing F, Watabe K, Debinski W, Lo HW, The gain-of-function GLI1 transcription factor TGLI1 enhances expression of VEGF-C and TEM7 to promote glioblastoma angiogenesis, Oncotarget 6 (2015) 22653–22665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC, Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer, Cancer Res. 65 (2005) 338–348. [PubMed] [Google Scholar]
- [10].Zhu H, Carpenter RL, Han W, Lo HW, The GLI1 splice variant TGLI1 promotes glioblastoma angiogenesis and growth, Cancer Lett. 343 (2014) 51–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sirkisoon SR, Carpenter RL, Rimkus T, Doheny D, Zhu D, Aguayo NR, Xing F, Chan M, Ruiz J, Metheny-Barlow LJ, Strowd R, Lin J, Regua AT, Arrigo A, Anguelov M, Pasche B, Debinski W, Watabe K, Lo H-W, TGLI1 transcription factor mediates breast cancer brain metastasis via activating metastasis-initiating cancer stem cells and astrocytes in the tumor microenvironment, Oncogene 39 (2020) 64–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hoshino A, Costa-Silva B, Shen T-L, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Jørgen Labori K, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D, Tumour exosome integrins determine organotropic metastasis, Nature 527 (2015) 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lorger M, Felding-Habermann B, Capturing changes in the brain microenvironment during initial steps of breast cancer brain metastasis, Am. J. Pathol 176 (2010) 2958–2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wong GL, Abu Jalboush S, Lo HW, Exosomal MicroRNAs and organotropism in breast cancer metastasis, Cancers 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ambros V, microRNAs: tiny regulators with great potential, Cell 107 (2001) 823–826. [DOI] [PubMed] [Google Scholar]
- [16].Tominaga N, Kosaka N, Ono M, Katsuda T, Yoshioka Y, Tamura K, Lotvall J, Nakagama H, Ochiya T, Brain metastatic cancer cells release microRNA-181c-containing extracellular vesicles capable of destructing blood-brain barrier, Nat. Commun 6 (2015) 6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, Ochiya T, Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis, J. Biol. Chem 288 (2013) 10849–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massagué J, Genes that mediate breast cancer metastasis to the brain, Nature 459 (2009) 1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Xing F, Liu Y, Sharma S, Wu K, Chan MD, Lo HW, Carpenter RL, Metheny-Barlow LJ, Zhou X, Qasem SA, Pasche B, Watabe K, Activation of the c-met pathway mobilizes an inflammatory network in the brain microenvironment to promote brain metastasis of breast cancer, Cancer Res. 76 (2016) 4970–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lai CP, Kim EY, Badr CE, Weissleder R, Mempel TR, Tannous BA, Breakefield XO, Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters, Nat. Commun 6 (2015) 7029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang L, Cossette SM, Rarick KR, Gershan J, Dwinell MB, Harder DR, Ramchandran R, Astrocytes directly influence tumor cell invasion and metastasis in vivo, PLoS One 8 (2013), e80933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Chen Y, Wang X, miRDB: an online database for prediction of functional microRNA targets, Nucleic Acids Res. 48 (2020) D127–d131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ma Q, Wang Y, Zhang H, Wang F, miR-1290 contributes to colorectal cancer cell proliferation by targeting INPP4B, Oncol. Res 26 (2018) 1167–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jin JJ, Liu YH, Si JM, Ni R, Wang J, Overexpression of miR-1290 contributes to cell proliferation and invasion of non small cell lung cancer by targeting interferon regulatory factor 2, Int. J. Biochem. Cell Biol 95 (2018) 113–120. [DOI] [PubMed] [Google Scholar]
- [25].Li M, He XY, Zhang ZM, Li S, Ren LH, Cao RS, Feng YD, Ji YL, Zhao Y, Shi RH, MicroRNA-1290 promotes esophageal squamous cell carcinoma cell proliferation and metastasis, World J. Gastroenterol 21 (2015) 3245–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Endo Y, Yamashita H, Takahashi S, Sato S, Yoshimoto N, Asano T, Hato Y, Dong Y, Fujii Y, Toyama T, Immunohistochemical determination of the miR-1290 target arylamine N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast cancer, BMC Cancer 14 (2014) 990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang Y, Liao JM, Zeng SX, Lu H, p53 downregulates Down syndrome-associated DYRK1A through miR-1246, EMBO Rep. 12 (2011) 811–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Bott A, Erdem N, Lerrer S, Hotz-Wagenblatt A, Breunig C, Abnaof K, Wörner A, Wilhelm H, Münstermann E, Ben-Baruch A, Wiemann S, miRNA-1246 induces pro-inflammatory responses in mesenchymal stem/stromal cells by regulating PKA and PP2A, Oncotarget 8 (2017) 43897–43914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mo LJ, Song M, Huang QH, Guan H, Liu XD, Xie DF, Huang B, Huang RX, Zhou PK, Exosome-packaged miR-1246 contributes to bystander DNA damage by targeting LIG4, Br. J. Cancer 119 (2018) 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xie K, Liu L, Chen J, Liu F, Exosomal miR-1246 derived from human umbilical cord blood mesenchymal stem cells attenuates hepatic ischemia reperfusion injury by modulating T helper 17/regulatory T balance, IUBMB Life 71 (2019) 2020–2030. [DOI] [PubMed] [Google Scholar]
- [31].Dutta R, McDonough J, Chang A, Swamy L, Siu A, Kidd GJ, Rudick R, Mirnics K, Trapp BD, Activation of the ciliary neurotrophic factor (CNTF) signalling pathway in cortical neurons of multiple sclerosis patients, Brain : J. Neurol 130 (2007) 2566–2576. [DOI] [PubMed] [Google Scholar]
- [32].Kahn MA, Ellison JA, Speight GJ, de Vellis J, CNTF regulation of astrogliosis and the activation of microglia in the developing rat central nervous system, Brain Res. 685 (1995) 55–67. [DOI] [PubMed] [Google Scholar]
- [33].Douglas AM, Goss GA, Sutherland RL, Hilton DJ, Berndt MC, Nicola NA, Begley CG, Expression and function of members of the cytokine receptor superfamily on breast cancer cells, Oncogene 14 (1997) 661–669. [DOI] [PubMed] [Google Scholar]
- [34].Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D, The GeneCards suite: from gene data mining to disease, Genome Sequence Anal. 54 (2016) 1, 30.31–31.30.33. [DOI] [PubMed] [Google Scholar]
- [35].Agarwal V, Bell GW, Nam JW, Bartel DP, Predicting effective microRNA target sites in mammalian mRNAs, Elife (2015) 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE, Chromatin structure analyses identify miRNA promoters, Genes Dev. 22 (2008) 3172–3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Przygodzka P, Papiewska-Pająk I, Bogusz-Koziarska H, Sochacka E, Boncela J, Kowalska MA, Regulation of miRNAs by Snail during epithelial-to-mesenchymal transition in HT29 colon cancer cells, Sci. Rep 9 (2019) 2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pigati L, Yaddanapudi SC, Iyengar R, Kim DJ, Hearn SA, Danforth D, Hastings ML, Duelli DM, Selective release of microRNA species from normal and malignant mammary epithelial cells, PLoS One 5 (2010) e13515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wang X, Zhao X, Gao P, Wu M, c-Myc modulates microRNA processing via the transcriptional regulation of Drosha, Sci. Rep 3 (2013) 1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gebert LFR, MacRae IJ, Regulation of microRNA function in animals, Nat. Rev. Mol. Cell Biol 20 (2019) 21–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Adams BD, Claffey KP, White BA, Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells, Endocrinology 150 (2009) 14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Casey MC, Prakash A, Holian E, McGuire A, Kalinina O, Shalaby A, Curran C, Webber M, Callagy G, Bourke E, Kerin MJ, Brown JA, Quantifying Argonaute 2 (Ago2) expression to stratify breast cancer, BMC Cancer 19 (2019) 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chai S, Ng KY, Tong M, Lau EY, Lee TK, Chan KW, Yuan YF, Cheung TT, Cheung ST, Wang XQ, Wong N, Lo CM, Man K, Guan XY, Ma S, Octamer 4/microRNA-1246 signaling axis drives Wnt/β-catenin activation in liver cancer stem cells, Hepatology (Baltimore, Md 64 (2016) 2062–2076. [DOI] [PubMed] [Google Scholar]
- [44].Zhang WC, Chin TM, Yang H, Nga ME, Lunny DP, Lim EK, Sun LL, Pang YH, Leow YN, Malusay SR, Lim PX, Lee JZ, Tan BJ, Shyh-Chang N, Lim EH, Lim WT, Tan DS, Tan EH, Tai BC, Soo RA, Tam WL, Lim B, Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression, Nat. Commun 7 (2016) 11702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Tűzesi Á, Kling T, Wenger A, Lunavat TR, Jang SC, Rydenhag B, Lötvall J, Pollard SM, Danielsson A, Carén H, Pediatric brain tumor cells release exosomes with a miRNA repertoire that differs from exosomes secreted by normal cells, Oncotarget 8 (2017) 90164–90175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yan L, Cai K, Sun K, Gui J, Liang J, MiR-1290 Promotes Proliferation, Migration, and Invasion of Glioma Cells by Targeting LHX6 233, 2018, pp. 6621–6629. [DOI] [PubMed] [Google Scholar]
- [47].Qin WJ, Wang WP, Wang XB, Zhang XT, Du JD, MiR-1290 targets CCNG2 to promote the metastasis of oral squamous cell carcinoma, Eur. Rev. Med. Pharmacol. Sci 23 (2019) 10332–10342. [DOI] [PubMed] [Google Scholar]
- [48].Yu Y, Han S, Li M, Song Y, Qi F, Circ_0004913 sponges miR-1290 and regulates FOXC1 to inhibit the proliferation of hepatocellular carcinoma, Cancer Cell Int. 20 (2020) 431. [Google Scholar]
- [49].Endo Y, Toyama T, Takahashi S, Yoshimoto N, Iwasa M, Asano T, Fujii Y, Yamashita H, miR-1290 and its potential targets are associated with characteristics of estrogen receptor α-positive breast cancer, Endocr. Relat. Cancer 20 (2013) 91–102. [DOI] [PubMed] [Google Scholar]
- [50].Li XJ, Ren ZJ, Tang JH, Yu Q, Exosomal MicroRNA MiR-1246 promotes cell proliferation, invasion and drug resistance by targeting CCNG2 in breast cancer, Cell. Physiol. Biochem 44 (2017) 1741–1748. [DOI] [PubMed] [Google Scholar]
- [51].Wang SAI, Zeng YA, Zhou J-M, Nie S-L, Peng Q, Gong J, Huo J-R, MicroRNA-1246 promotes growth and metastasis of colorectal cancer cells involving CCNG2 reduction, Mol. Med. Rep 13 (2016) 273. [DOI] [PubMed] [Google Scholar]
- [52].Sakha S, Muramatsu T, Ueda K, Inazawa J, Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma, Sci. Rep 6 (2016) 38750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yang F, Xiong H, Duan L, Li Q, Li X, Zhou Y, MiR-1246 promotes metastasis and invasion of A549 cells by targeting GSK-3β–mediated Wnt/β-catenin pathway, Cancer Treat Res. : Off. J. Kor. Cancer Assoc 51 (2019) 1420–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Bhagirath D, Yang TL, Bucay N, Sekhon K, Majid S, Shahryari V, Dahiya R, Tanaka Y, Saini S, microRNA-1246 is an exosomal biomarker for aggressive prostate, Cancer 78 (2018) 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Albrecht PJ, Enterline JC, Cromer J, Levison SW, CNTF-activated astrocytes release a soluble trophic activity for oligodendrocyte progenitors, Neurochem. Res 32 (2007) 263–271. [DOI] [PubMed] [Google Scholar]
- [56].Sierra A, Price JE, García-Ramirez M, Méndez O, López L, Fabra A, Astrocyte-derived cytokines contribute to the metastatic brain specificity of breast cancer cells, Laboratory investigation, J. Tech. Methods Pathol 77 (1997) 357–368. [PubMed] [Google Scholar]
- [57].Hu X, Zhao Y, He X, Li J, Wang T, Zhou W, Wan D, Wang H, Gu J, Ciliary neurotrophic factor receptor alpha subunit-modulated multiple downstream signaling pathways in hepatic cancer cell lines and their biological implications, Hepatology (Baltimore, Md 47 (2008) 1298–1308. [DOI] [PubMed] [Google Scholar]
- [58].Wang K, Xie M, Zhu L, Zhu X, Zhang K, Zhou F, Ciliary neurotrophic factor protects SH-SY5Y neuroblastoma cells against Aβ1-42-induced neurotoxicity via activating the JAK2/STAT3 axis, Folia Neuropathol. 53 (2015) 226–235. [DOI] [PubMed] [Google Scholar]
- [59].Stöckli KA, Lottspeich F, Sendtner M, Masiakowski P, Carroll P, Götz R, Lindholm D, Thoenen H, Molecular cloning, expression and regional distribution of rat ciliary neurotrophic factor, Nature 342 (1989) 920–923. [DOI] [PubMed] [Google Scholar]
- [60].Asada H, Ip NY, Pan L, Razack N, Parfitt MM, Plunkett RJ, Time course of ciliary neurotrophic factor mRNA expression is coincident with the presence of protoplasmic astrocytes in traumatized rat, striatum 40 (1995) 22–30. [DOI] [PubMed] [Google Scholar]
- [61].Kang SS, Keasey MP, Cai J, Hagg T, Loss of Neuron-Astroglial Interaction Rapidly Induces Protective CNTF Expression After Stroke in Mice 32, 2012, pp. 9277–9287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ma K, Deng X, Xia X, Fan Z, Qi X, Wang Y, Li Y, Ma Y, Chen Q, Peng H, Ding J, Li C, Huang Y, Tian C, Zheng JC, Direct conversion of mouse astrocytes into neural progenitor cells and specific lineages of neurons, Transl. Neurodegener 7 (2018) 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shi W, Wang X, Ruan L, Fu J, Liu F, Qu J, MiR-200a promotes epithelial-mesenchymal transition of endometrial cancer cells by negatively regulating FOXA2 expression, Pharmazie 72 (2017) 694–699. [DOI] [PubMed] [Google Scholar]
- [64].Vorvis C, Hatziapostolou M, Mahurkar-Joshi S, Koutsioumpa M, Williams J, Donahue TR, Poultsides GA, Eibl G, Iliopoulos D, Transcriptomic and CRISPR/Cas9 technologies reveal FOXA2 as a tumor suppressor gene in pancreatic cancer, Am. J. Physiol. Gastrointest. Liver Physiol 310 (2016) G1124–G1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Tang Y, Shu G, Yuan X, Jing N, Song J, FOXA2 functions as a suppressor of tumor metastasis by inhibition of epithelial-to-mesenchymal transition in human lung cancers, Cell Res. 21 (2011) 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Li C, Lu S, Shi Y, MicroRNA-187 promotes growth and metastasis of gastric cancer by inhibiting FOXA2, Oncol. Rep 37 (2017) 1747–1755. [DOI] [PubMed] [Google Scholar]
- [67].Wang J, Zhu CP, Hu PF, Qian H, Ning BF, Zhang Q, Chen F, Liu J, Shi B, Zhang X, Xie WF, FOXA2 suppresses the metastasis of hepatocellular carcinoma partially through matrix metalloproteinase-9 inhibition, Carcinogenesis 35 (2014) 2576–2583. [DOI] [PubMed] [Google Scholar]
- [68].Robertson E, Perry C, Doherty R, Madhusudan S, Transcriptomic profiling of Forkhead box transcription factors in adult glioblastoma multiforme, CANCER GENOMICS PROTEOMICS 12 (2015) 103–112. [PubMed] [Google Scholar]
- [69].Perez-Balaguer A, Ortiz-Martínez F, García-Martínez A, Pomares-Navarro C, Lerma E, Peiró G, FOXA2 mRNA expression is associated with relapse in patients with Triple-Negative/Basal-like breast carcinoma, Breast Cancer Res. Treat 153 (2015) 465–474. [DOI] [PubMed] [Google Scholar]
- [70].Pristerà A, Lin W, Kaufmann A-K, Brimblecombe KR, Threlfell S, Dodson PD, Magill PJ, Fernandes C, Cragg SJ, Ang S-L, Transcription Factors FOXA1 and FOXA2 Maintain Dopaminergic Neuronal Properties and Control Feeding Behavior in Adult Mice 112, 2015, pp. E4929–E4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Metzakopian E, Lin W, Salmon-Divon M, Dvinge H, Andersson E, Ericson J, Perlmann T, Whitsett JA, Bertone P, Ang SL, Genome-wide characterization of Foxa2 targets reveals upregulation of floor plate genes and repression of ventrolateral genes in midbrain dopaminergic progenitors, Development (Camb.) 139 (2012) 2625–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


