Abstract
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has recently emerged throughout the world, resulting in more than 400 million cases and over 6 million deaths worldwide as of January 2022. Coronaviruses subvert or use certain aspects of the unfolded protein response in the endoplasmic reticulum to overcome protein translation shutdown to benefit their replication. New virions use the ER-Golgi intermediate compartment to assemble and gain transportation to the cell membrane. Extensive remodeling of the ER has been demonstrated during SARS-CoV-2 infection. In this review article, we discuss the role of the endoplasmic reticulum secretory pathway in the replication cycle of SARS-CoV-2. Currently, there is a dearth of therapeutic options for intervening with SARS-CoV-2 infection. To accelerate drug development, efforts around the globe have been focusing on repurposing drugs that have already been approved for clinical use by regulatory agencies. Targeting the ERS pathway is reasonable, as prior work has shown that SARS-CoV-2 egress is dependent on this pathway. Here we discuss the feasibility of off-patent, FDA-approved, pharmacological inhibitors of the ERS pathway to suppress the SARS-CoV-2 replication cycle, a promising approach that warrants investigation.
Keywords: SARS-CoV-2, COVID-19, Unfolded protein response, ERS pathway
1. Introduction
Three new human coronaviruses from animal reservoirs have emerged over a relatively recent timeframe. Along with these outbreaks, thousands of additional coronaviruses have been estimated to reside solely in bats, and raise concerns for their zoonotic transmissions and pandemic potential in the future. The COVID-19 manifests itself as a mild illness of the upper respiratory tract. At the time of hospitalisation, COVID-19 patients present with fever and dry cough; occasionally, patients may experience difficulty in breathing, musculoskeletal pain, headache/dizziness, diarrhoea, nausea and haemoptysis (Cucinotta and Vanelli, 2020; Henderson et al., 2020; Spiteri et al., 2020). Severe COVID-19 cases progress to acute respiratory distress syndrome (ARDS), on average around 8, 9 days after symptom onset (Spiteri et al., 2020). SARS-CoV-2 has an estimated mortality of between 1 and 5%, yet human-to-human transmission continues unabated, despite intensifying public health strategies.
Our ability to prevent current and future emergent viruses will be determined by our understanding of the coronavirus replication cycle and virus-host interactions. Cells adapt to endoplasmic reticulum (ER) stress caused by viral infection through the activation of the unfolded protein response (UPR), which expands the capacity to handle the increased protein load in the ER (Kluge et al., 2020). However, when UPR fails to manage the burden of viral proteins in ER, the production of secretory and surface proteins in the host cells is reduced and cellular function is impaired (Hetz and Papa, 2018). Indeed, compromised production of secretory and surface proteins in SARS-CoV-2 infected cells can explain the etiology of COVID-19 patients (Hetz, 2012; Huang et al., 2020). Impaired ER homoeostasis in SARS-CoV-2 infected cells can compromise the assembly and folding of olfactory and taste receptors which can explain the symptoms of abnormal taste and smell in COVID-19 patients (Zhou et al., 2020; Mehta et al., 2020). Similarly, compromised assembly and secretion of pulmonary surfactant in SARS-CoV-2 infected alveolar epithelial cells can contribute to symptoms of ARDS (Zhou et al., 2020; Mehta et al., 2020). Hereafter, we comment on the role of the endoplasmic reticulum secretory pathway as a potential target against SARS-CoV-2.
2. SARS-CoV-2 replication cycle
2.1. Infection
The main mechanism of SARS-CoV-2 transmission is via infected respiratory droplets inhaled by susceptible individuals as a result of close-range contact (Cevik et al., 2020). Although host cellular receptors are primarily found in the respiratory tract, the gastrointestinal tracts and the conjunctiva have also indicated susceptibility to infection (Cevik et al., 2020). SARS-CoV-2 virus can persist for many days on certain smooth surfaces, and this facilitates another route of transmission from unclean hands to the mucosa of the nose, eyes, and mouth (Cevik et al., 2020). The distribution of the ACE2 receptor across the respiratory tract, intestinal and endothelial cells, kidneys, and other locations, may explain the infection sites and account for patient symptoms and complications (Cevik et al., 2020).
Coronavirus infection begins with the specific binding of the SARS-CoV-2 spike (S) protein to the peptidase domain of the angiotensin-converting enzyme 2 (ACE2) host cell receptor (V'kovski et al., 2021). This occurs along with the assistance of other host factors such as the transmembrane protease serine 2 (TMPRSS2) which cleaves the S protein and has significant interaction with ACE2 (Baughn et al., 2020). TMPRSS2 facilitates cell entry via the S protein, leading to membrane fusion and release of the viral genome into the host cytoplasm (Hartenian et al., 2020).
Several host proteins act as cofactors that potentiate entry of SARS-CoV-2 into the cell. Scavenger receptor class B member 1 (SRB1) is a cell-surface receptor that plays a role in high-density lipoprotein (HDL) uptake (Wei et al., 2020). Neuropilin-1 (NRP1) is a transmembrane glycoprotein that serves as a cell surface receptor for semaphorins (Daly et al., 2020; Cantuti-Castelvetri et al., 2020). Both SRB1 and NRP1 can promote SARS-CoV-2 entry in the presence of ACE2, which indicates that they function as cofactors that enhance infection through ACE2 (Baggen et al., 2021). Furthermore, several other host factors that interact with S protein were suggested as candidate SARS-CoV-2 receptors (Baggen et al., 2021). AXL Receptor Tyrosine Kinase (AXL), low-density lipoprotein receptor class A domain–containing protein 3 (LDLRAD3), C-type lectin domain family 4 member G (CLEC4G), and Basigin (BSG) can all interact with SARS-CoV-2 S protein and promote infection in absence of ACE2 (Wang et al., 2021, 2020; Zhu et al., 2021). Interestingly, Glucose-Regulated Protein, 78 kDa (GRP78), a key regulator of UPR, can form a complex with SARS-CoV-2 S protein and ACE2 (Carlos et al., 2021). Treatment with a humanized monoclonal antibody against GRP78 (hMAb159) reduced ACE2 expression on plasma membrane, as well as SARS-CoV-2 infection in vitro (Carlos et al., 2021).
SARS-CoV-2 RNA has not only been detected in throat and nasopharyngeal swabs of infected patients, but also in blood, urine, saliva, and stool samples (Malik, 2020). Studies indicate that the greatest infectiousness potential occurs just prior to or within the initial five days of symptom onset, as indicated by the observation of the peak SARS-CoV-2 viral load in the respiratory tract during the first week of illness or at the time of symptom onset (Cevik et al., 2020). Infected individuals may be asymptomatic or mildly symptomatic if their early immune responses are successful in suppressing viral replication and preventing its entry into the lungs (Tan et al., 2021). However, many of these people may be unaware of their condition and as a result will not self-isolate or seek treatment, perpetuating the transmission of the virus to others (Tan et al., 2021).
Most people display an antibody response to the virus between 10 and 14 days after the infection, with a smaller number of cases requiring extended time for antibody production and others having no antibody response whatsoever (Kellam and Barclay, 2020). People with low antibody titers may be at risk of reinfection with SARS-CoV-2 and should be regularly monitored for evidence of recurring disease (Kellam and Barclay, 2020).
2.2. Replication, assembly, and release
SARS-CoV-2 replicates its RNA to produce full length copies of the virus which are integrated into newly produced viral particles (V'kovski et al., 2021). Once the virus enters the cell, the release of the genomic RNA facilitates viral translation via two open reading frames, ORF1a and ORF1ab (V'kovski et al., 2021). These ORFs permit RNA attachment to the host cell ribosome for the purpose of viral replicase gene translation (Haque et al., 2020). Polyproteins pp1a and pp1ab are the resulting products that are processed into sixteen non-structural proteins (nsp) that subsequently form the viral replication-transcription complex (RTC) and thus participate in viral pathogenesis (V'kovski et al., 2021).
Extensive remodeling of the host ER leads to formation of double-membrane vesicles, within which viral RNA synthesis occurs via its RNA-dependent RNA polymerase (RdRp) (Hartenian et al., 2020; Tay et al., 2020). The RdRp of SARS-CoV-2 also plays a crucial role in the assembly of the viral RTC and is composed of a catalytic subunit (nsp12) and accessory subunits (nsp8 and nsp7) (Hillen et al., 2020). Its role in genome replication and transcription of sub-genomic RNAs (sgmRNAs) that encode viral RNA is essential for coronavirus genome replication (Hillen et al., 2020). Both the nsps and the RdRp holoenzyme are considered as potential targets for the treatment of SARS-CoV-2 (Chen et al., 2020).
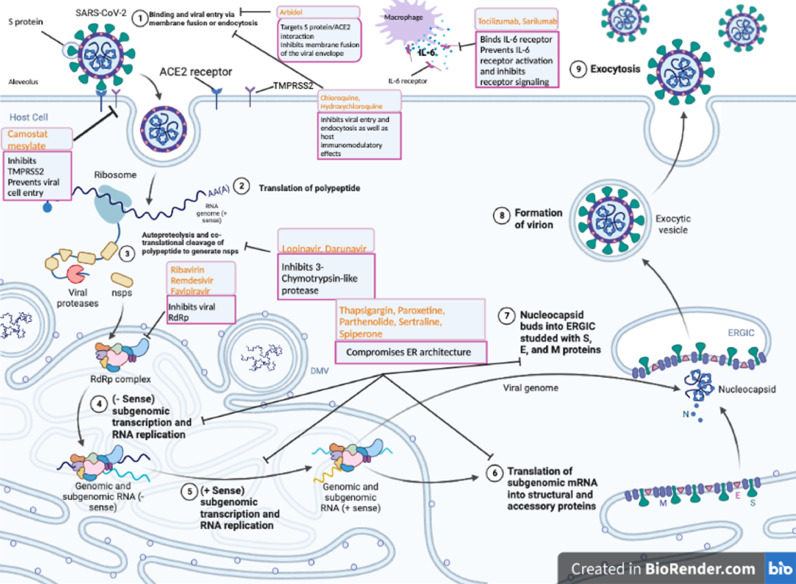
The sgmRNAs transcribed by the RdRp are then translated into structural proteins (M, E, N and S) and accessory proteins (ORF3a, ORF3c, ORF6, ORF7a, ORF7b, ORF8b and ORF9b) (Astuti, 2020). The M, E, and S proteins function together to produce virus-like particles and assemble the virus (Haque et al., 2020). This process is further catalyzed by the N protein, which accounts for the enclosed genome fusion into the endoplasmic-reticulum–Golgi intermediate compartment (ERGIC) (Haque et al., 2020). Nucleocapsids in the ERGIC interact with other structural proteins to form budding vesicles that can be released from the cell as new virions (Astuti, 2020). While the E protein alters the secretory pathways to facilitate viral release, the M protein binds to the nucleocapsid and assists in virion assembly completion (Haque et al., 2020). The virions can then be transported to the cell surface and released through exocytosis (Haque et al., 2020). These viral replication cycle steps present potential targets for drug therapy (Fig. 1 ). A variety of strategies to inhibit SARS-CoV-2 entry are being explored for therapeutic purposes - including ACE2 engagement block, host protease inactivation, and S2-mediated membrane fusion inhibition as well as RdRp inhibition (Tay et al., 2020).
Fig. 1.
Representation of SARS-CoV-2 viral lifecycle and potential drug targets. 1. Binding of coronavirus to cellular receptors such as ACE-2 with host factors (such as TMPRSS2) promote viral entry via membrane fusion and endocytosis. 2. Translation of the viral RNA (polypeptide). 3. Autoproteolysis and co-translational cleavage of polypeptide to generate nsps. 4. (-Sense) subgenomic transcription and RNA replication. 5. (+sense) subgenomic transcription and RNA replication. 6. Translation of subgenomic mRNA into structural and accessory proteins. 7. Nucleocapsid buds into ERGIC studded with S, E, and M proteins. 8, 9. Finally, virions are secreted from the infected cell by exocytosis. Key compounds targeting steps that are attractive therapeutic targets are coloured in orange. ACE2, angiotensin-converting enzyme 2; nsp, non-structural proteins; S protein, spike protein; E protein, envelope protein; M protein, membrane protein; and TMPRSS2, type 2 transmembrane serine protease. Thapsigargin, a non-competitive inhibitor of the sarco/endoplasmic reticulum Ca2+ ATPase (SERCA), blocks the ability of the cell to pump calcium into the sarcoplasmic and endoplasmic reticulum and causes activation of UPR. Paroxetine, Parthenolide, Sertraline, and Spiperone are FDA-approved drugs that compromise ER architecture. Thapsigargin, Paroxetine, Parthenolide, Sertraline, and Spiperone are ERS pathway inhibitors and inhibit steps 4–7.
Several functional genetic screens as well as interactome screens have been performed to identify SARS-CoV-2 host factors. In addition to the receptors and proteases required for virus entry, these screens identified proteins involved in vesicle trafficking, PI3K signaling, lipid homeostasis, translation, protein homeostasis and P-body/stress granule assembly, as crucial host factors for SARS-CoV-2 infection (previously reviewed in 60). The functional relevance of these identified proteins in the SARS-CoV-2 life cycle remains to be demonstrated, but some have known functions in the life cycle of other viruses.
The upper respiratory tract is the primary site of SARS-CoV-2 infection, however, cell lines from different tissues have been used to identify pro-viral host factors. CRISPR-knockout functional screens were based on cell survival, which has the limitation that genes that are essential for cell proliferation are unlikely to be detected. Future studies should utilize bidirectional CRISPR screens using the cell lines derived from the lungs to identify the host factors required for infection. Similarly, screens to identify cellular proteins that interact with a given SARS-CoV-2 protein used ectopic expression of individual viral proteins. This will lead to many false positive hits, due to very high and non-physiologic levels of viral protein expression that will not recapitulate the expression level and structural changes associated with natural infection.
3. Endoplasmic reticulum secretory pathway
The endoplasmic reticulum secretory (ERS) pathway primarily comprises the endoplasmic reticulum (ER) and the Golgi apparatus, as well as the vesicles that travel in between them. It functions as a folding, modification, and quality regulation system for the secretory proteins as well as resident proteins of the ERS pathway (Shikano and Colley, 2013). All proteins entering the ERS pathway possess an ER signal sequence that directs them to the rough ER (Lodish et al., 2000). The proteins then travel from the ER to the Golgi apparatus in vesicular carriers to reach the vacuoles or the cell surface, and this transport is balanced by the retrograde traffic of membrane and luminal proteins in order to maintain the integrity of cellular compartments (Vitale and Denecke, 1999).
79% genome sequence identity as well as a similar pathophysiology is shared between SARS-CoV-2 and SARS-CoV (Tay et al., 2020; Frieman et al., 2008). SARS-CoV depends on the ERS pathway to complete its replication cycle, involving the steps of viral assembly and secretion of virions (Fung et al., 2014). ER stress has been reported in virus infected cells, including severe acute respiratory syndrome coronavirus (SARS-CoV), murine hepatitis virus, avian infectious bronchitis virus (Fung and Liu, 2014; Koseler et al., 2020). The role of the ERS pathway in the replication cycle of SARS-CoV-2 has also been clearly demonstrated (Zhu et al., 2020; Gordon et al., 2020). SARS-CoV-2 modifies and uses the ERS signaling networks to complete its replication cycle (Sureda et al., 2020). Mapping of SARS-CoV-2 proteins and human proteins has shown that SARS-CoV-2 interacts with components of the ERS pathway (Gordon et al., 2020). For example, membrane (M) protein interacts with multiple ER morphology proteins, ORF8 interacts with several ER protein quality control proteins, Spike (S) and nsp13 interacts with multiple Golgi apparatus proteins (Gordon et al., 2020). Furthermore, during SARS-CoV-2 replication, spike (S) protein, membrane (M) protein, and envelope (E) protein are synthesized by the ribosome and inserted into the ER membrane (Ujike and Taguchi, 2015). Mature viral proteins are transported from the ER to the ERGIC, where lipid membrane, nucleocapsid protein, RNA, and these viral proteins help assemble viral particles (Hartenian et al., 2020). Viral particles are then secreted from the cell after being transported to the plasma membrane along the secretory pathway.
4. SARS-CoV-2-induced endoplasmic reticulum stress response
The ER primarily functions to safeguard the synthesis and correct folding of transmembrane proteins as well as proteins that are readily secreted and those involved in cell secretome regulation (Sureda et al., 2020). The UPR consists of three signaling pathways and reduces ER stress by inducing the temporary attenuation of translation, upregulating the production of ER chaperones, and increasing phospholipid biosynthesis (Bechill et al., 2020). There are three ER transmembrane proteins that regulate the UPR pathways, including IRE1 (inositol-requiring protein 1α), ATF6 (activating transcription factor 6α), and PERK (protein kinase R [PKR]-like ER kinase) (Bechill et al., 2020; Lin et al., 2007). These proteins serve as stress sensors by detecting the extent of unfolded protein present in the ER lumen and collectively initiate the UPR (Ron and Walter, 2007). The PERK pathway works as an integrated stress response pathway and modulates apoptosis, while the IRE1 pathway affects both apoptosis induction and ER folding, and the ATF6 pathway enhances ER folding capacity (Fung and Liu, 2014). This permits signal transmission across the ER membrane to the cytosol and nucleus, and encourages the cell to return to homeostasis by increasing ER folding capacity and lowering protein synthesis (Fung and Liu, 2014). If homeostasis is untenable, apoptosis will result.
All three sensors of the UPR have been implicated in SARS-CoV-2 infection. SARS-CoV-2 has the ability to selectively prevent host protein production and take control of the ERS pathway in order to synthesize viral proteins (Ha et al., 2020; Chan et al., 2020). An overload of proteins can skew the balance between ER folding capacity and the demand for protein synthesis (Sureda et al., 2020). Particularly, translation of viral proteins (M, E and S) occurs on ER membranes that increases the protein load on the ER. Consequently, the increased client protein load on the cell's transport and folding machineries can activate UPR (Sicari et al., 2020). GRP78 interacts with the spike protein of SARS-CoV-2 and serves as a host factor for virus entry and infection (Bechill et al., 2020). GRP78, ACE2 and Spike protein form a protein complex in the ER which can titrate out the GRP78 from its normal cellular function in ER (Bechill et al., 2020). Activation of the ER stress sensors is mediated by the disassociation of the ER chaperone GRP78 from the sensor domain of ER stress sensors located in the lumen of ER (Bechill et al., 2020). Furthermore, double membrane vesicles (DMV) that form replication organelles are derived from the ER. The formation of DMVs will shrink the size of ER and increase the local concentration of proteins in ER which can impair protein folding. Taken together, (i) increased synthesis of viral proteins, (ii) titration of GRP78 by ACE2 and S protein and (iii) use of ER membranes to form DMVs will contribute to the activation of UPR during SARS-CoV-2 infection.
Recent studies have reported that overexpression of the ORF8 and S proteins of SARS-CoV-2 was sufficient in activating all three branches of the UPR pathway (Echavarría-Consuegra, 2021). These proteins appear to be divergent from their SARS-CoV counterparts, displaying potential for future investigation (Echavarría-Consuegra, 2021).
The ORF8 protein activates both the IRE1 and ATF6 branches, showing therapeutic promise for simultaneous inhibition of both branches (Echavarría-Consuegra, 2021). Since both ER stress and UPR activation are also involved in fibrotic disease progression, a known complication of COVID-19, these pathways contribute to the respiratory pathophysiology associated with this virus (Echavarría-Consuegra, 2021). Although pharmacological inhibition of the UPR demonstrated a significant negative effect on SARS-CoV-2 replication, this intervention may have an additional therapeutic effect of diminishing COVID-19 associated pathophysiology as well (Echavarría-Consuegra, 2021). Lastly, the activation of ATF4 and CHOP promoter activities by SARS-CoV-2 ORF3a suggests a role of the PERK pathway during infection (Sicari et al., 2020).
In addition to the accumulation of unfolded proteins in the ER, UPR is also activated by aberrant lipid composition of the ER membrane referred to as lipid bilayer stress (Halbleib et al., 2017). The changes in membrane composition associated with lipid bilayer stress promote IRE1 oligomerization (Halbleib et al., 2017). As membrane thickness and molecular packing thickness increases, the resulting lipid bilayer stress increases IRE1 oligomerization which activates UPR (Halbleib et al., 2017). The key proteins in cholesterol homeostasis (SREBF2, SCAP, NPC1, NPC2) and lipid transport (TMEM30A, SUGMAR1) were reported as cellular host factors that play a role in the viral replication cycle (Baggen et al., 2021). Virus infection may lead to lipid bi-layer stress and activate UPR. IRE1 activation during SARS-CoV-2 infection portrays IRE1 as an additional therapeutic target against SARS-CoV-2. A further understanding of the mechanisms through which SARS-CoV-2 proteins affect the activation of ER stress receptors as well as the UPR, would be essential for their potential role as therapeutic targets against SARS-CoV-2 (Echavarría-Consuegra, 2021).
4.1. SARS-CoV-2-infection and autophagic flux
Perturbations in the ER, calcium homeostasis or cellular energy status activate the autophagy pathway in addition to the UPR. Both pathways are induced upon infection with a variety of viruses and have an especially significant association during CoV infections. Autophagy is a reliable mechanism that assists in limiting viral replication and maintaining homeostasis within the cell (Siqueira et al., 2018). Autophagosomes sequester cytosolic targets (cargos) such as defective organelles or protein aggregates for degradation and this process is carried out by approximately 30 conserved ATG proteins (Siqueira et al., 2018). Once autophagosome formation has occurred, the process of autophagic flux is completed upon autophagosome and lysosome fusion facilitating cargo degradation within autophagolysosomes (Siqueira et al., 2018).
CoV replication complexes share some features of autophagosomes, including lysosomal marker acquisition via infection as well as co-localization with multiple organelle markers (Prentice et al., 2003). The use of CoV double membrane vesicles (DMVs) resembling autophagosomes as a platform for viral replication may be an important therapeutic target, since DMV biogenesis often requires components of the autophagic flux. Since all identified CoVs alter the autophagy pathway for their own use in some manner, this displays the necessity for this pathway in the viral replication cycle. Additionally, TMEM41B is an ER-transmembrane protein involved in autophagy and an essential requirement for CoV infection that further highlights this finding (Schneider et al., 2021). The possibility for repurposing pre-approved drugs that modulate the UPR and autophagy as broad-spectrum inhibitors for several respiratory viruses, including CoV, may be an efficient way to limit viral spread (Senger et al., 2020).
5. Inhibitors of endoplasmic reticulum secretory pathway
The pathophysiology of severe COVID-19 is characterized by acute respiratory distress and excessive inflammation with the ability to induce respiratory and multi-organ failure, as well as death (Zhou et al., 2020; Ruscitti et al., 2020). SARS-CoV-2 depends on the ERS pathway to complete its life cycle such as viral assembly and secretion of virions. For the purposes of maintaining the cell's homeostasis and its survival, cells transport a large number of proteins to the extracellular medium by using the ERS pathway. As mentioned previously, the UPR attempts to restore ER homeostasis or cause apoptosis depending on the intensity and duration of stress (Ha et al., 2020). The ER resident chaperone (GRP78) has been identified as a master regulator of the UPR that is upregulated during times of ER stress or protein overload in ER (Ha et al., 2020). By functioning as an essential host factor for viral infection, GRP78 may pose as a drug target for SARS-CoV-2 treatment and shows potential to disrupt various stages of the viral replication cycle including viral entry, protein production, and release. Further investigation into agents targeting GRP78 may indicate a possible approach to avoid viral infection and propagation.
Suppressing CoV replication by reprogramming the ER stress pathways has recently demonstrated therapeutic potential of UPR pathway. Thapsigargin, a chemical activator of the UPR, displayed an extensive antiviral effect on three different CoVs (including SARS-CoV-2) in four different cell types (Shaban et al., 2021). Thapsigargin activated the PERK branch of the ER stress pathway (which is also activated by CoV), blocked the autophagic flux in CoV-infected cells, reversed virus-induced translational shut-down, improved viability of infected cells, and restored GRP78 and IRE1 levels with long-term treatment (Shaban et al., 2021). The latter suggests that the thapsigargin-mediated response may reverse and reprogram the CoV-induced block of inducible host factors and override the suppressive effects of CoVs on ER functions (Shaban et al., 2021).
Its ability to target many central mechanisms essential for CoV replication portray thapsigargin's potential role in future CoV targeted therapeutics. Inhibiting pathways of the UPR also demonstrate significant potential for successful SARS-CoV-2 therapy. The IRE1 axis of the UPR produces spliced XBP1s that help regulate the secretome of the cells (Sureda et al., 2020). SARS-CoV-2 has been known to induce UPR through its ORF8b and also activate NLR family pyrin domain containing 3 (NLRP3) inflammasomes present in macrophages which are involved in cytokine regulation (e.g. IL-1β) (Sureda et al., 2020). Thus, inhibiting the RNAase activity of IRE1 may be an effective approach in treating SARS-CoV-2 infection through the modulation of macrophage secretomes (Sureda et al., 2020). In addition to the IRE1 axis, the PERK pathway is also activated by SARS-CoV-2 infection (Sureda et al., 2020). The recent development of PERK inhibitors function to alleviate the attenuation of IFN responses as well as the innate immunity, and present yet another therapeutic approach for SARS-CoV-2 (Sureda et al., 2020).
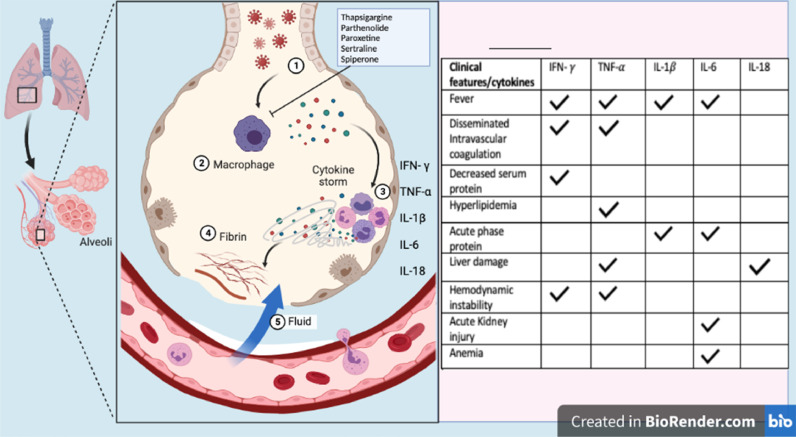
SARS-CoV-2 infection causes injury and death of virus-infected cells and tissues as part of the virus replicative cycle. Viral infection in airway epithelial cells leads to high levels of virus-associated pyroptosis which is an inflammatory form of apoptosis or autophagy. Airway epithelial cell and macrophages detect the pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) leading to an elevated secretion of pro-inflammatory cytokines into the blood of infected patients. This causes a massive production of chemokines and pro-inflammatory cytokines which can result in the development of a cytokine storm (Fig. 2 ). The ERS pathway is required for SARS-CoV-2 morphogenesis as well as the processing of secretory proteins such as cytokines and is a promising therapeutic target for COVID-19 (Sureda et al., 2020; Vaninov, 2020). Previous studies indicate that about 40% of SARS-CoV-2 interacting proteins are associated with the ERS pathway (Gordon et al., 2020). Proteins such as nsp8, ORF8, M and nsp13 may play a role in rearranging the endoplasmic reticulum and Golgi trafficking during coronavirus infection in an effort to promote viral replication (Gordon et al., 2020). The sec61 translocon mediates viral protein insertion into the ER membrane, and since nsp8 interacts with three components of signal recognition particles, the virus may utilize the sec61-mediated protein translocation pathway for entry into the ER (Gordon et al., 2020). Thus, inhibiting co-translational entry into the secretory pathway may assist in preventing viral infection, and this shows tremendous potential as a target for SARS-CoV-2 treatment (Gordon et al., 2020). Notably, an inhibitor of the ERS pathway would simultaneously reduce the production of multiple proinflammatory cytokines and blunt the cytokine storm more effectively than cytokine-blocking antibodies. ERS pathway is an extremely attractive anti-viral target because its inhibition would be a “double strike” against SARS-CoV-2: (i) block SARS-CoV-2 assembly and secretion and (ii) reduce the production of multiple proinflammatory cytokines.
Fig. 2.
Schematic representation of clinical features associated with cytokine response in SARS-CoV-2 infections. 1. Coronavirus infects the cells of the lung. 2. Immune cells such as macrophages produce cytokines in response to the virus. 3. Cytokines attract more immune cells such as monocytes, neutrophils, eosinophils, and other white blood cells which produce more inflammatory cytokines, resulting in hyperinflammation that damages lung cells. 4. Damage can occur through fibrin formation. 5. Weakened blood vessels allow fluid to seep in to the lung cavities, leading to respiratory failure. The activated cytokines result in clinical features involving fever (IFN-γ, TNF-α, IL-1β, IL-6), disseminated intravascular coagulation (IFN-γ, TNF-α), decreased serum protein (IFN-γ), hyperlipidemia (TNF-α), acute phase protein (IL-1β, IL-6), liver damage (TNF-α, IL-18), hemodynamic instability (IFN-γ, TNF-α), acute kidney injury (IL-6), and anemia (IL-6). Thapsigargin, Paroxetine, Parthenolide, Sertraline, and Spiperone are ERS pathway inhibitors that reduce the production of multiple proinflammatory cytokines (Step 2).
A recent screening of a library of off-patent , FDA-approved drugs with known bioavailability and safety in humans has identified potent inhibitors of the ERS pathway (Wang et al., 2020). Based on their ability to inhibit protein secretion and safety, affordability, and documented anti-viral and anti-inflammatory effects, four compounds appear promising: Parthenolide, Paroxetine, Sertraline and Spiperone.
Parthenolide, is a naturally occurring sesquiterpene lactone derived from feverfew (Tanacetum parthenium) and exhibits a variety of antiparasitic, anti-viral, and anti-inflammatory effects. The anti-viral activity of Parthenolide has been demonstrated against Herpes simplex virus 1 (Farhan and Rabouille, 2011). Parthenolide treatment reduces infiltration of inflammatory cells, airway permeability and production of pro-inflammatory cytokines in LPS-induced acute lung injury mouse model (Zhao et al., 2018). Phase 1 dose-escalation study of ACT001, an orally-available Parthenolide derivative showed satisfactory bioavailability and treatment was well tolerated with no dose-limiting toxicities (Clinical trial: ACTRN12616000228482). SARS‐CoV‐2 encodes two proteases 3C-like cysteine protease (3 CLpro) also known as Main protease (Mpro) and papain-like protease (PLpro) (Lakhera et al., 2022). These proteases cleave polyproteins pp1a and pp1ab to produce sixteen non-structural proteins (nsps) (Lakhera et al., 2022). As such both viral proteases are druggable targets against SARS-CoV-2, since nsps form the viral replication-transcription complex (RTC) and participate in viral pathogenesis. In molecular docking and molecular dynamics simulations, Parthenolide showed the best binding affinity and reactivity response against MPro and PLpro (Lakhera et al., 2022). Indeed, Parthenolide interacts with SARS-CoV-2 PLpro and inhibits its deISGylation activity (Zou et al., 2022). These suggest that Parthenolide is a promising candidate against the SARS-CoV-2 that needs further investigation.
Paroxetine and Sertraline are selective serotonin reuptake inhibitors (SSRI) that are clinically used for the treatment of depression in human patients. Paroxetine has been shown to ameliorate the reactive microglia-mediated inflammatory responses in astrocytes (Benassi-Zanqueta et al., 2018). Sertraline has a wide range of anti-inflammatory actions across many body systems, with benefits reported in rat models of rheumatoid arthritis and cerebral anti-inflammatory action (Jang et al., 2016; Zhang et al., 2020). Sertraline has been reported to show strong anti-viral activity in an in vivo murine model of Ebola virus infection (Baharav et al., 2012). The effects of various SSRIs (Paroxetine, Sertraline, Fluoxetine and Escitalopram) have been evaluated on SARS-CoV-2 replication. Fluoxetine inhibits SARS-CoV-2 replication, but neither Paroxetine nor Escitalopram had any effect on viral replication (Zimniak et al., 2021). However, Fluoxetine had no effect on replication of other viruses such as Rabies virus, human respiratory syncytial virus, Human Herpesvirus 8 or Herpes simplex virus (Zimniak et al., 2021). Sertraline has shown antiviral activity against SARS-CoV-2 in Vero E6 cells (Peng et al., 2021). Sertraline reduced viral load, and IL-6 and TNF-α mRNA at day 4 post-infection in hamsters (Peng et al., 2021). Furthermore, sertraline did not induce antiviral drug resistance after 10 serial passages of SARS-CoV-2 in vitro (Peng et al., 2021). It is highly unlikely that inhibition of serotonin reuptake transporters is responsible for the anti-SARS-CoV-2 effect of Fluoxetine and Sertraline, since neither Paroxetine nor Escitalopram interfered with viral replication (Peng et al., 2021). Further studies are warranted to study the mechanism of action of fluoxetine and sertraline for their anti-SARS-CoV-2 activity.
Spiperone is a dopaminergic D2 antagonist that is used as an antipsychotic agent and a psychotropic drug. Spiperone has been shown to reduce pro-inflammatory cytokine expression and NO production in LPS‐stimulated microglia cells (Zheng et al., 2008). Spiperone is reported to show anti-viral activity against the human pathogenic Polyomaviruses (Goodwin et al., 2009). This drug was also recently identified as a candidate molecule that can disrupt the interaction between the S protein and ACE2 in molecular docking experiments utilizing the structure of omicron S protein in complex with ACE2 (Bahadur Gurung et al., 2022). However, activity of Spiperone against SARS-CoV-2 has not yet been tested in wet-laboratory experiments (Bahadur Gurung et al., 2022).
6. Conclusions
Recent studies have demonstrated the potential for the ERS pathway as a drug target for SARS-CoV-2 infection, however, much remains to be discovered regarding the host-virus interaction at this stage. This viral infectious disease can manifest as fever, pneumonia, ARDS in more severe cases, cytokine storm development, multiple organ damage, or even death. While new strategies to target the virus are beginning to surface, the dependency of the virus on the ERS pathway to complete its replication cycle, morphogenesis, and processing of secretory proteins, presents this as an extremely attractive therapeutic target that possesses remarkable variability. Cells are unlikely to develop resistance to ERS inhibitors and the efficacy of ERS inhibitors will not be compromised by current and emerging variants of SARS-CoV-2. Further investigations into the viral exploitation of the ERS and UPR pathways would not only be a pragmatic approach, but seem to be essential for a targeted treatment of the virus.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Maarisha Upadhyay: Writing – original draft, Visualization, Writing – review & editing. Sanjeev Gupta: Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
No data was used for the research described in the article.
References
- Astuti I., Ysrafil Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. 2020;14(4):407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggen J., Vanstreels E., Jansen S., et al. Cellular host factors for SARS-CoV-2 infection. Nat. Microbiol. 2021;6:1219–1232. doi: 10.1038/s41564-021-00958-0. [DOI] [PubMed] [Google Scholar]
- Bahadur Gurung A., Ajmal Ali M., Elshikh M.S., Aref I., Amina M., Lee J. An in silico approach unveils the potential of antiviral compounds in preclinical and clinical trials as SARS-CoV-2 omicron inhibitors. Saudi J. Biol. Sci. 2022;29(6) doi: 10.1016/j.sjbs.2022.103297. Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baharav E., Bar M., Taler M., Gil-Ad I., Karp L., Weinberger A., Weizman A. Immunomodulatory effect of sertraline in a rat model of rheumatoid arthritis. Neuroimmunomodulation. 2012;19(5):309–318. doi: 10.1159/000339109. [DOI] [PubMed] [Google Scholar]
- Baughn L.B., Sharma N., Elhaik E., Sekulic A., Bryce A.H., Fonseca R. Targeting TMPRSS2 in SARS-CoV-2 infection. Mayo Clin. Proc. 2020;95(9):1989–1999. doi: 10.1016/j.mayocp.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechill J., Chen Z., Brewer J.W., Baker S.C. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J. Virol. 2020;82(9):4492–4501. doi: 10.1128/JVI.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benassi-Zanqueta E., Marques C.F., Nocchi S.R., Dias Filho B.P., Nakamura C.V., Ueda Nakamura T. Parthenolide influences herpes simplex virus 1 replication in vitro. Intervirology. 2018;61(1):14–22. doi: 10.1159/000490055. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos A.J., et al. Te chaperone GRP78 is a host auxiliary factor for SARS-CoV- 2 and GRP78 depleting antibody blocks viral entry and infection. J. Biol. Chem. 2021;296:10075. doi: 10.1016/j.jbc.2021.100759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik M., Kuppalli K., Kindrachuk J., Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;23(371):m3862. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- Chan C., Siu K., Chin K., Yuen K., Zheng B., Jin D. Modulation of the unfolded protein response by the severe acute respiratory syndrome coronavirus spike protein. J. Virol. 2020 doi: 10.1128/JVI.00659-06. [online] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Malone B., Llewellyn E., Grasso M., Shelton P.M.M., Olinares P.D.B., Maruthi K., Eng E.T., Vatandaslar H., Chait B.T., Kapoor T.M., Darst S.A., Campbell E.A. Structural basis for helicase-polymerase coupling in the SARS-CoV-2 replication-transcription complex. Cell. 2020;182(6):1560–1573. doi: 10.1016/j.cell.2020.07.033. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cucinotta D., Vanelli M. WHO declares COVID-19 a pandemic. Acta Biomed. 2020;91(1):157–160. doi: 10.23750/abm.v91i1.9397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly J.L., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echavarría-Consuegra L., et al. Manipulation of the Unfolded Protein Response: A Pharmacological Strategy against Coronavirus Infection. PLoS Pathog. 2021;17(6):e1009644. doi: 10.1371/journal.ppat.1009644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhan H., Rabouille C. Signalling to and from the secretory pathway. J. Cell Sci. 2011;124(Pt 2):171–180. doi: 10.1242/jcs.076455. [DOI] [PubMed] [Google Scholar]
- Frieman M., Heise M., Baric R. SARS coronavirus and innate immunity. Virus Res. 2008;133(1):101–112. doi: 10.1016/j.virusres.2007.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Huang M., Liu D.X. Coronavirus-induced ER stress response and its involvement in regulation of coronavirus-host interactions. Virus Res. 2014;194:110–123. doi: 10.1016/j.virusres.2014.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;5:296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung T.S., Liu D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014;17(5):296. doi: 10.3389/fmicb.2014.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin E.C., Atwood W.J., DiMaio D. High-throughput cell-based screen for chemicals that inhibit infection by simian virus 40 and human polyomaviruses. J. Virol. 2009;83(11):5630–5639. doi: 10.1128/JVI.00203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha D.P., Van Krieken R., Carlos A.J., Lee A.S. The stress-inducible molecular chaperone GRP78 as potential therapeutic target for coronavirus infection. J. Infect. 2020;81(3):452–482. doi: 10.1016/j.jinf.2020.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halbleib K., Pesek K., Covino R., Hofbauer H., Wunnicke D., Hänelt I., Hummer G., Ernst R. Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell. 2017;67(4):673–684. doi: 10.1016/j.molcel.2017.06.012. [DOI] [PubMed] [Google Scholar]
- Haque S.M., Ashwaq O., Sarief A., Azad John Mohamed A.K. A comprehensive review about SARS-CoV-2. Future Virol. 2020;15(9):625–648. doi: 10.2217/fvl-2020-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of Coronaviruses. J. Biol. Chem. 2020;11(37):12910–12934. doi: 10.1074/jbc.REV120.01393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson L.A., Canna S.W., Schulert G.S., Volpi S., Lee P.Y., Kernan K.F., Caricchio R., Mahmud S., Hazen M.M., Halyabar O., et al. On the alert for cytokine storm: Immunopathology in COVID-19. Arthritis Rheumatol. 2020;72(7):1059–1063. doi: 10.1002/art.41285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C. The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012;13(2):89–102. doi: 10.1038/nrm3270. [DOI] [PubMed] [Google Scholar]
- Hetz C., Papa F.R. The unfolded protein response and cell fate control. Mol. Cell. 2018;69(2):169–181. doi: 10.1016/j.molcel.2017.06.017. [DOI] [PubMed] [Google Scholar]
- Hillen H.S., Kokic G., Farnung L., et al. Structure of replicating SARS-CoV-2 polymerase. Nature. 2020;584:154–156. doi: 10.1038/s41586-020-2368-8. [DOI] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang Y.J., Back M.J., Fu Z., Lee J.H., Won J.H., Ha H.C., Lee H.K., Jang J.M., Choi J.M., Kim D.K. Protective effect of sesquiterpene lactone parthenolide on LPS-induced acute lung injury. Arch. Pharm. Res. 2016;39(12):1716–1725. doi: 10.1007/s12272-016-0716-x. [DOI] [PubMed] [Google Scholar]
- Kellam P., Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J. Gen. Virol. 2020;101(8):791–797. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluge S., Janssens U., Welte T., Weber-Carstens S., Marx G., Karagiannidis C. German recommendations for critically ill patients with COVID-19. Med. Klin. Intensivmed. Notfmed. 2020;15(3):111–114. doi: 10.1007/s00063-020-00689-w.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseler A., Sabirli R., Goren T., Turkcuer I., Kurt O. Endoplasmic reticulum stress markers in SARS-CoV-2 infection and pneumonia: case-control study. In Vivo. 2020;34(3):1645–1650. doi: 10.21873/invivo.11956. Suppl. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakhera S., Devlal K., Ghosh A., Chowdhury P., Rana M. Modelling the DFT structural and reactivity study of feverfew and evaluation of its potential antiviral activity against COVID-19 using molecular docking and MD simulations. Chem. Zvesti. 2022;76(5):2759–2776. doi: 10.1007/s11696-022-02067-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.H., Li H., Yasumura D., Cohen H.R., Zhang C., Panning B., Shokat K.M., Lavail M.M., Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodish H., Berk A., Zipursky S.L., et al. Molecular Cell Biology. 4th ed. W. H. Freeman; New York: 2000. Overview of the secretory pathway. Section 17.3. [Google Scholar]
- Malik Y.A. Properties of coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020;42(1):3–11. Apr. [PubMed] [Google Scholar]
- Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Hlh Across Speciality Collaboration UK COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395(10229):1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Ding C., Jiang L., Tang W., Liu Y., Zhao L., Yi Z., Ren H., Li C., He Y., et al. Discovery of potential anti-SARS-CoV-2 drugs based on large-scale screening in vitro and effect evaluation in vivo. Sci. China Life Sci. 2021;65:1181–1197. doi: 10.1007/s11427-021-2031-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prentice E., Jerome G.W., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2003;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Ruscitti P., Berardicurti O., Iagnocco A., Giacomelli R. Cytokine storm syndrome in severe COVID-19. Autoimmun. Rev. 2020;19(7):102562. doi: 10.1016/j.autrev.2020.102562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider W.M., Luna J.M., Hoffmann H.H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–132. doi: 10.1016/j.cell.2020.12.006. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger M.R., Evangelista T.C.S., Dantas R.F., da Silva Santana M.V., Gonçalves L.C.S., Neto L.R., de S., et al. COVID-19: molecular targets, drug repurposing and new avenues for drug discovery. Mem. Inst. Oswaldo Cruz. 2020;115 doi: 10.1590/0074-02760200254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaban M.S., Müller C., Mayr-Buro C., et al. Multi-level inhibition of coronavirus replication by chemical ER stress. Nat. Commun. 2021;12(1):5536. doi: 10.1038/s41467-021-25551-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikano S., Colley K. Encyclopedia of Biological Chemistry. Academic Press; 2013. Secretory pathway; pp. 203–209. [Google Scholar]
- Sicari D., Chatziioannou A., Koutsandreas T., Sitia R., Chevet E. Correction: role of the early secretory pathway in SARS-CoV-2 infection. J. Cell Biol. 2020;219(9) doi: 10.1083/jcb.202006005. 08132020c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira M., Ribeiro R., Travassos L.H. Autophagy and its interaction with intracellular bacterial pathogens. Front. Immunol. 2018;9:935. doi: 10.3389/fimmu.2018.00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiteri G., Fielding J., Diercke M., Campese C., Enouf V., Gaymard A., Bella A., Sognamiglio P., Sierra Moros M.J., Riutort A.N., et al. First cases of coronavirus disease\2019 (COVID-19) in the WHO European Region, 24 January to 21 February 2020. Eur. Surveill. 2020;25(9):2000178. doi: 10.2807/1560-7917.ES.2020.25.9.2000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureda A., Alizadeh J., Nabavi S.F., Berindan-Neagoe I., Cismaru C.A., Jeandet P., Los M.J., Clementi E., Nabavi S.M., Ghavami S. Endoplasmic reticulum as a potential therapeutic target for COVID-19 infection management? Eur. J. Pharmacol. 2020;882 doi: 10.1016/j.ejphar.2020.173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C., Xiao Y., Meng X., Huang X., Li C., Wu A. Asymptomatic SARS-CoV-2 infections: what do we need to know? Infect. Control Hosp. Epidemiol. 2021;42(1):114–115. doi: 10.1017/ice.2020.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay M.Z., Poh C.M., Renia L., MacAry P.A., Ng L.F.P. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20(6):363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Taguchi F. Incorporation of spike and membrane glycoproteins into coronavirus virions. Viruses. 2015;7(4):1700–1725. doi: 10.3390/v7041700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaninov N. In the eye of the COVID-19 cytokine storm. Nat. Rev. Immunol. 2020;20(5):277. doi: 10.1038/s41577-020-0305-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A., Denecke J. The endoplasmic reticulum-gateway of the secretory pathway. Plant Cell. 1999;11(4):615–628. doi: 10.1105/tpc.11.4.615. Apr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V'kovski P., Kratzel A., Steiner S., et al. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Xie J., Zhao L., Fei X., Zhang H., Tan Y., Zhou L., Liu Z., Ren Y., Yuan L. et al: Aveolar macrophage activation and cytokine storm in the pathogenesis of severe COVID-19. In.: Research Square; 2020. [DOI] [PMC free article] [PubMed]
- Wang K., et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct. Target. Ther. 2020;5:283. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat. Metab. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- Zhang X., Zhu L.B., He J.H., Zhang H.Q., Ji S.Y., Zhang C.N., Hou N.N., Huang C.P., Zhu J.H. Paroxetine suppresses reactive microglia-mediated but not lipopolysaccharide induced inflammatory responses in primary astrocytes. J. Neuroinflamm. 2020;17(1):50. doi: 10.1186/s12974-020-1712-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Liu P., Boncompain G., Loos F., Lachkar S., Bezu L., Chen G., Zhou H., Perez F., Kepp O., et al. Identification of pharmacological inhibitors of conventional protein secretion. Sci. Rep. 2018;8(1):14966. doi: 10.1038/s41598-018-33378-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L.T., Hwang J., Ock J., Lee M.G., Lee W.H., Suk K. The antipsychotic spiperone attenuates inflammatory response in cultured microglia via the reduction of proinflammatory cytokine expression and nitric oxide production. J. Neurochem. 2008;107(5):1225–1235. doi: 10.1111/j.1471-4159.2008.05675.x. [DOI] [PubMed] [Google Scholar]
- Zhou F., Yu T., Du R., Fan G., Liu Y., Liu Z., Xiang J., Wang Y., Song B., Gu X., et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Wang W., Liu Z., Liang C., Wang W., Ye F., Huang B., Zhao L., Wang H., Zhou W., et al. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 2020;11(1):3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, S. et al. Genome-wide CRISPR activation screen identifes novel receptors for SARS-CoV-2 entry. Preprint at bioRxiv (2021). [DOI] [PMC free article] [PubMed]
- Zimniak M., Kirschner L., Hilpert H., et al. The serotonin reuptake inhibitor fluoxetine inhibits SARS-CoV-2 in human lung tissue. Sci. Rep. 2021;11:5890. doi: 10.1038/s41598-021-85049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Shan H., Sun D., Xia L., Shi Y., Wan J., Zhou A., Wu Y., Xu H., Lei H., Xu Z., Wu Y. Parthenolide reveals an allosteric mode to inhibit the deISGylation activity of SARS-CoV-2 papain-like protease. Acta Biochim. Biophys. Sin. 2022 doi: 10.3724/abbs.2022092. (Shanghai) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.