ABSTRACT
Here, we report three near-full-length genome sequences of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) obtained in Mexico City, Mexico, during the pandemic of coronavirus disease 19 (COVID-19) in 2020, representing a zooanthroponotic transmission event between humans and a dog. All three genomes belong to the B.1.189 lineage based on the pangolin classification.
ANNOUNCEMENT
Considered the biggest sanitary event of the century, the coronavirus disease (COVID-19) pandemic is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a member of the Coronaviridae family within the Betacoronavirus genus. To date (8 May 2022), the cases and deaths produced by this virus have been 513,955,910 and 6,249,700, respectively. Mexico has been one of the countries with the highest number of deaths (324,334) during this pandemic (https://covid19.who.int/), representing 5.18% of the total mortality worldwide.
The remarkable genome plasticity displayed by SARS-CoV-2 (1) leads to the divergence of multiple phylogenetic clades and the consequent emergence of different viral variants of concern (2). Therefore, the control of this pandemic has represented a major challenge (3, 4).
Recent reports documented the zooanthroponotic spillover of variants of concern like Delta (cats, dogs, pumas, lions, and hamsters) and Omicron (white-tailed deer) in wild and domestic animals (5, 6). Thus, documented infections produced by human-to-animal transmission are increasing (7, 8).
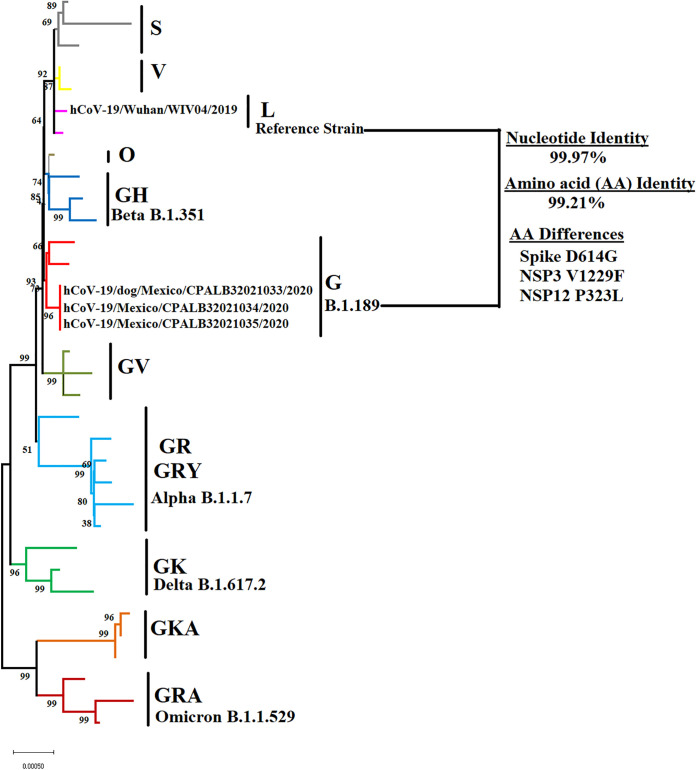
Here, we report three near-full-length genome sequences of SARS-CoV-2 strains obtained from nasopharyngeal swab specimens recovered during a zooanthroponotic spillover event between humans and a dog in Mexico City, Mexico, in 2020. All sequences were classified as part of the pangolin lineage B.1.189 (Fig. 1). Interestingly, no changes were observed in the consensus sequence obtained from the dog, showing the apparent genetic stability of this lineage after infection in different species.
FIG 1.
Phylogenetic tree of SARS-CoV-2 from a zooanthroponotic spillover case in Mexico City during 2020. The zooanthroponotic event described here involved two humans and a dog (Canis lupus familiaris) living in a household. The phylogenetic analysis was conducted by maximum likelihood and the general time reversible model, showing the genetic relationship of sequences reported in this study with different divergent clades of SARS-CoV-2 (GISAID classification) and multiple variants described during the pandemic. In addition, results of the comparison between sequences and the reference strain sequence are shown. The analysis involved a total of 31 representative sequences of different clades obtained from the GISAID database (9).
Viral isolation was performed in Vero cells (ATCC C1008). Subsequently, RNA from the three viral isolates was extracted using the high pure viral RNA kit (Roche), following the manufacturer’s protocol. Next-generation sequencing (NGS) of amplicons was conducted to obtain the SARS-CoV-2 sequences reported in this announcement. For this purpose, a set of 15 primers were developed to cover the genome of SARS-CoV-2 (Table 1). Reverse transcriptase PCR (RT-PCR) reactions were conducted using the SuperScript III one-step RT-PCR system with Platinum Taq DNA polymerase kit, following the manufacturer’s instructions. Libraries were prepared using the Nextera XT DNA library preparation kit following the manufacturer’s protocol. Sequencing and analyses were conducted on the MiSeq system (Illumina). Raw data of samples identified as hCoV-19/dog/Mexico/CPALB32021033/2020, hCoV-19/Mexico/CPALB32021034/2020, and hCoV-19/Mexico/CPALB32021035/2020 consisting of 3,348,413, 8,248,286, and 9,425,298 reads, respectively, with an average read length of 200 bp were analyzed. All analyses were performed in CLC Genomics Workbench v11.0. The paired reads were quality trimmed using default parameters. Reads were then mapped to the reference strain sequence (GenBank accession number NC045512.2). Consensus sequences were obtained using default parameters and annotated based on a comparison with the reference strain. All work conducted in humans and animals was approved by bioethics committee Escuela Nacional de Medicina y Homeopatía (ENMH) number CBE/006/2020 on the project “Zoonosis Virales Emergentes en Tiempos de Circulación de COVID-19 en México.”
TABLE 1.
Sequencing considerationsa
| Amplicon no. | Primer IDb | Primer sequence (5′–3′) | Location in reference sequence | Size (bp) | Annealing temp (°C) |
|---|---|---|---|---|---|
| 1 | 1FCOVID | GCC TTC CCA GGT AAC AAA CCA ACC | 15–1931 | 1,916 | 58 |
| 2RCOVID | GAG CAG TTT CAA GAG TGC GGG AG | ||||
| 2 | 3FCOVID | GCA TTT GCA TCA GAG GCT GCT CG | 1868–4148 | 2,280 | 56 |
| 4RCOVID | CAC CCT CTT GAA CAA CAT CAC CCA C | ||||
| 3 | 5FCOVID | GGC AAT CTT CAT CCA GAT TCT GCC | 4046–6371 | 2,325 | 58 |
| 6RCOVID | TTC CCT GCG CGT CCT CTG ACT TC | ||||
| 4 | 7FCOVID | GTA CCA AAC CAA CCA TAT CCA AAC GC | 6008–8372 | 2,364 | 56 |
| 8RCOVID | CCT GCG CAT TAA TAT GAC GCG CAC | ||||
| 5 | 9FCOVID | CAG CAG CTC GGC AAG GGT TTG TTG | 8169–10209 | 2,040 | 58 |
| 10RCOVID | GGG TTA AGC ATG TCT TCA GAG GTG C | ||||
| 6 | 11FCOVID | CCA CAA ACC TCT ATC ACC TCA GCT G | 10022–12261 | 2,239 | 56 |
| 12RCOVID | CGT TGC ATG GCT GCA TCA CG | ||||
| 7 | 13FCOVID | GGG CAA CCT TAC AAG CTA TAG CC | 12078–14333 | 2,255 | 55 |
| 14RCOVID | CAA TTT GGG TGG TAT GTC TGA TCC C | ||||
| 8 | 15FCOVID | CTG CAG AGT CAC ATG TTG ACA CTG | 14195–16411 | 2,216 | 55 |
| 16RCOVID | CTG TGA CAT CAC AAC CTG GAG C | ||||
| 9 | 17FCOVID | CAC ACC GCA TAC AGT CTT ACA GGC | 16215–18466 | 2,251 | 58 |
| 18RCOVID | CAG GCG GTG GTT TAG CAC TAA C | ||||
| 10 | 19FCOVID | CGA TGT CGA GGG GTG TCA TGC TAC | 18306–20099 | 1,793 | 57 |
| 20RCOVID | GCT TGT TTG GGA CCT ACA GAT GG | ||||
| 11 | 21FCOVID | GGG TGT GGA CAT TGC TGC TAA TAC | 19845–22446 | 2,601 | 56 |
| 22RCOVID | GGG TCA AGT GCA CAG TCT ACA GC | ||||
| 12 | 23FCOVID | GTT GGA CAG CTG GTG CTG CA | 22332–24239 | 1,907 | 58 |
| 24RCOVID | CAG CAC CTG CAC CAA AGG TCC AAC | ||||
| 13 | 25FCOVID | GCC ACC TTT GCT CAC AGA TGA AAT G | 24145–26353 | 2,208 | 55 |
| 26RCOVID | GCG CAG TAA GGA TGG CTA GTG | ||||
| 14 | 27FCOVID | CGA CGA CGA CTA CTA GCG TGC | 26192–28375 | 2,183 | 60 |
| 28RCOVID | CCC ACT GCG TTC TCC ATT CTG G | ||||
| 15 | 29FCOVID | GCA CCC CGC ATT ACG TTT GGT G | 28307–29798 | 1,491 | 60 |
| 30RCOVID | CTT CCA TAT AGG CAG CTC TCC CTA GC |
Description of multiple sets of primers developed in this study to conduct the NGS amplicon sequencing described in this study. The location of the primers corresponds to the nucleotide positions in the reference sequence of SARS-CoV-2 under the accession no. NC045512.2.
ID, identification.
The information from these events is useful for defining the potential role of dogs as reservoirs or intermediate hosts of SARS-CoV-2. In addition, future studies may help evaluate the possible differences in the transmission in animal species among SARS-CoV-2 lineages.
Data availability.
Sequences are available in the Global Initiative on Sharing All Influenza Data (GISAID) database under the following accession numbers: EPI_ISL_11991713 (hCoV-19/dog/Mexico/CPALB32021033/2020), EPI_ISL_11988443 (hCoV-19/Mexico/CPALB32021034/2020), and EPI_ISL_11988444 (hCoV-19/Mexico/CPALB32021035/2020). The raw sequencing data of this project are available in the NCBI Sequence Read Archive (SRA) under the BioProject number PRJNA827138.
ACKNOWLEDGMENTS
This study was supported by grants from SIP-IPN 20196739, 20200873, and 20202442.
Sequencing was carried out following standard procedures at sequencing facilities of the Mexican Agriculture Ministry (SADER).
Contributor Information
Lauro Velazquez-Salinas, Email: lauro.velazquez@usda.gov.
J. Guillermo Estrada-Franco, Email: jestradaf@ipn.mx.
John J. Dennehy, Queens College CUNY
REFERENCES
- 1.Velazquez-Salinas L, Zarate S, Eberl S, Gladue DP, Novella I, Borca MV. 2020. Positive selection of ORF1ab, ORF3a, and ORF8 genes drives the early evolutionary trends of SARS-CoV-2 during the 2020 COVID-19 pandemic. Front Microbiol 11:550674. doi: 10.3389/fmicb.2020.550674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tian D, Sun Y, Zhou J, Ye Q. 2022. The global epidemic of SARS-CoV-2 variants and their mutational immune escape. J Med Virol 94:847–857. doi: 10.1002/jmv.27376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schafer EA, Eberl S, Velazquez-Salinas L. 2022. Beyond the biology: evaluating the role of political, economic, and social factors associated with the incidence and mortality of SARS-CoV-2 during the first seven months of the COVID-19 pandemic. arXiv Preprint arXi v:2204.13995. [Google Scholar]
- 4.Shafer AE, Velazquez-Salinas L. 2021. Controlling the COVID-19 pandemic: the complex epidemiological triad of SARS-CoV-2. Int Jr Infect Dis & Epidemlgy 2:41–42. [Google Scholar]
- 5.Koeppel KN, Mendes A, Strydom A, Rotherham L, Mulumba M, Venter M. 2022. SARS-CoV-2 reverse zoonoses to Pumas and Lions, South Africa. Viruses 14:120. doi: 10.3390/v14010120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandegrift KJ, Yon M, Surendran-Nair M, Gontu A, Amirthalingam S, Nissly RH, Levine N, Stuber T, DeNicola AJ, Boulanger JR, Kotschwar N, Aucoin SG, Simon R, Toal K, Olsen RJ, Davis JJ, Bold D, Gaudreault NN, Richt JA, Musser JM, Hudson PJ, Kapur V, Kuchipudi SV. 2022. Detection of SARS-CoV-2 Omicron variant (B.1.1.529) infection of white-tailed deer. bioRxiv [Preprint]. doi: 10.1101/2022.02.04.479189. [DOI]
- 7.Mahdy MAA, Younis W, Ewaida Z. 2020. An overview of SARS-CoV-2 and animal infection. Front Vet Sci 7:596391. doi: 10.3389/fvets.2020.596391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prince T, Smith SL, Radford AD, Solomon T, Hughes GL, Patterson EI. 2021. SARS-CoV-2 infections in animals: reservoirs for reverse zoonosis and models for study. Viruses 13:494. doi: 10.3390/v13030494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khare S, Gurry C, Freitas L, Schultz MB, Bach G, Diallo A, Akite N, Ho J, Lee RT, Yeo W, Curation Team GC, Maurer-Stroh S. 2021. GISAID's role in pandemic response. China CDC Wkly 3:1049–1051. doi: 10.46234/ccdcw2021.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Sequences are available in the Global Initiative on Sharing All Influenza Data (GISAID) database under the following accession numbers: EPI_ISL_11991713 (hCoV-19/dog/Mexico/CPALB32021033/2020), EPI_ISL_11988443 (hCoV-19/Mexico/CPALB32021034/2020), and EPI_ISL_11988444 (hCoV-19/Mexico/CPALB32021035/2020). The raw sequencing data of this project are available in the NCBI Sequence Read Archive (SRA) under the BioProject number PRJNA827138.