ABSTRACT
Fluorescence dilution approaches can detect bacterial cell division events and can detect if there are differential rates of cell division across individual cells within a population. This approach typically involves inducing expression of a fluorescent protein and then tracking partitioning of fluorescence into daughter cells. However, fluorescence can be diluted very quickly within a rapidly replicating population, such as pathogenic bacterial populations replicating within host tissues. To overcome this limitation, we have generated two revTetR reporter constructs, where either mCherry or yellow fluorescent protein (YFP) is constitutively expressed and repressed by addition of tetracyclines, resulting in fluorescence dilution within defined time frames. We show that fluorescent signals are diluted in replicating populations and that signal accumulates in growth-inhibited populations, including during nitric oxide (NO) exposure. Furthermore, we show that tetracyclines can be delivered to the mouse spleen during Yersinia pseudotuberculosis infection and defined a drug concentration that results in even exposure of cells to tetracyclines. We then used this system to visualize bacterial cell division within defined time frames postinfection. revTetR-mCherry allowed us to detect slow-growing cells in response to NO in culture; however, this strain had a growth defect within mouse tissues, which complicated results. To address this issue, we constructed revTetR-YFP using the less toxic YFP and showed that heightened NO exposure correlated with heightened YFP signal, indicating decreased cell division rates within this subpopulation in vivo. This revTetR reporter will provide a critical tool for future studies to identify and isolate slowly replicating bacterial subpopulations from host tissues.
KEYWORDS: Yersinia pseudotuberculosis, cell division rate, fluorescence dilution, nitric oxide
INTRODUCTION
Bacterial growth and metabolism have been the focus of decades of research, in part because we need to learn more about the pathways utilized by bacteria for growth to be able to target these essential processes with antibiotics or novel therapeutics. It is also well appreciated that bacteria with decreased metabolic rates are less susceptible to antibiotics (1–4). Earlier studies focused on growth of bacteria and antibiotic susceptibility under different laboratory conditions (1, 5–7); more recently, there has been a focus on studying bacterial growth and metabolism within host tissues to more closely approximate bacterial growth conditions at the point where antibiotics or other therapeutics would be administered (8–10). Importantly, bacterial pathogens may utilize a range of different nutrients for growth within host tissues, and individual bacteria, or subsets of bacteria, may concurrently utilize different metabolites to promote their growth (10–14). Some of these substrates may support higher bacterial replication rates, and others may support lower replication rates. Nutrient availability and the presence of host-derived antimicrobials, in addition to interactions with different immune cells subsets, can also generate complex microenvironments within host tissues, which can result in differences in growth rates across bacterial populations (12, 15–18).
When a bacterial infection is treated, differences in bacterial growth rates can dramatically affect antibiotic efficacy. Slow-growing subsets of bacteria are inherently less susceptible to antibiotic treatment, due to decreased metabolic rates (1, 3, 4). This is termed antibiotic tolerance: a transient, phenotypic change in a subset of bacterial cells within a population, which alters antibiotic susceptibility (19, 20). Many of these experiments have been performed in bacteriological media, so it still remains largely unclear which pathways promote decreased antibiotic susceptibility and slowed growth within host tissues. To identify these pathways, investigators will need to isolate antibiotic-tolerant cells from the host environment for downstream analyses. The specific bacterial pathways linked to tolerance may depend in part on the antibiotic and the mode of action, or the environment around the bacteria during the antibiotic exposure (18, 21, 22).
Molecular tools currently exist to identify and isolate slow-growing bacterial cells (8, 23–26), but there are also limitations with some of these approaches that need to be addressed to apply them within the host environment. One of the main approaches is utilizing fluorescence dilution to identify individual cells that have divided within a certain time frame. Typically, fluorescence dilution approaches involve inducing expression of a fluorescent protein, then removing the inducer, and detecting changes in the amount of fluorescent protein within cells as a readout for cell division events and protein partitioning into daughter cells (27, 28). Many research groups have successfully utilized this approach to identify dividing and nondividing bacterial cells by inducing fluorescent expression prior to introducing bacteria into tissue culture or infection models (27–30). For some bacterial infection models, this approach can easily be applied, because the size of the colonizing population of bacteria is relatively large and the number of cell division events within host tissues is relatively small (24, 25, 27). However, in other infection models, only a few bacterial cells establish infection, and these cells go on to replicate many times, which would quickly dilute out fluorescent signals (17, 31–33). Furthermore, it is difficult to deliver inducer compounds into host tissues at levels sufficient to modulate gene expression and more challenging to remove these compounds to allow for subsequent fluorescence dilution.
Yersinia pseudotuberculosis is an enteric pathogen that spreads systemically from intestinal tissues to colonize deep tissue sites, such as the spleen (34–36). There is a strong population bottleneck within intestinal tissues, and few bacterial cells spread systemically to colonize deep tissue sites (37). Within deep tissues, individual bacteria found clonal clusters of replicating extracellular bacteria, termed microcolonies or pyogranulomas (17, 38, 39). Microcolonies can grow to contain hundreds to thousands of bacteria within a single replication site, despite recruitment of circumscribing layers of neutrophils and monocytes (17, 39, 40). Recent studies have shown that subpopulations of bacteria are present within microcolony structures and that subsets of bacteria stressed by the presence of nitric oxide (NO) preferentially survive treatment with doxycycline, suggesting they may represent a slow-growing subset of cells (17, 22). However, it has been difficult to determine growth rates in this model system, since fluorescence dilution approaches cannot typically be used when the bacterial population contracts and expands so dramatically.
To overcome these limitations, we have generated revTetR reporter constructs, where exposure to tetracyclines will inhibit additional fluorescent gene expression, resulting in fluorescence dilution. These constructs will promote fluorescent signal accumulation in nondividing cells, and fluorescent protein will be diluted into dividing cells, thus allowing us to differentiate between dividing and nondividing subpopulations of bacteria. Tetracyclines can also easily be delivered into our mouse model of Yersinia pseudotuberculosis systemic infection (22), which allows us to visualize fluorescence dilution as a measure of cell division at specific stages of infection and within specific time frames. Here, revTetR constructs allow us to ask if responses to host-derived stresses, specifically nitric oxide (NO), affect in vivo cell division rates. These reporter constructs will be a useful tool for future studies to isolate and characterize bacterial subpopulations with differential growth rates.
RESULTS
The revTetR reporter can be used to visualize differential bacterial growth rates in vitro with mCherry fluorescence dilution.
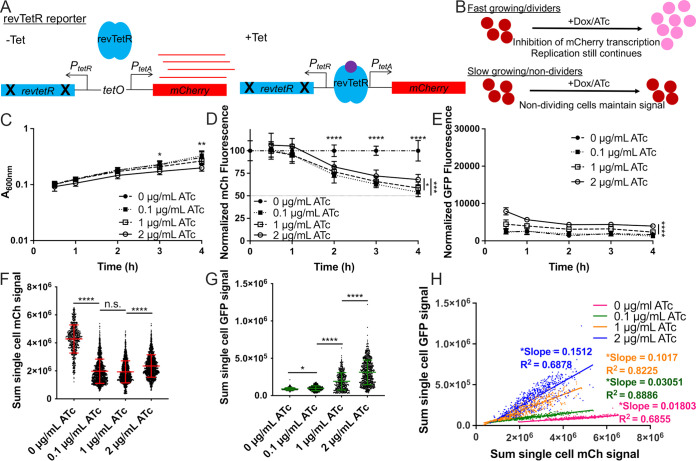
To detect differential cell division rates within host tissues, we generated a fluorescence dilution reporter system by modifying our recently described TetON fluorescent reporter with stable mCherry (tetR::PtetA::mCherry) (22). The revTetR reporter (revtetR::PtetA::mCherry) was constructed by introducing two mutations (E15A and L17G) into the wild-type tetR gene. These two mutations were predicted to cause a reversal of TetR activity resulting in repression of PtetA specifically in the presence of tetracycline and tetracycline derivatives (41, 42). Therefore, revTetR is a tetracycline-responsive reporter that allows cells to constitutively express mCherry in the absence of tetracycline derivatives, and subsequent addition of tetracyclines should inhibit additional mCherry transcription (Fig. 1A). This reporter can be used to compare growth rates of bacteria via a fluorescence dilution approach (27, 28). For example, in cultures treated with tetracycline, fast-growing cells will continue to divide, and the existing mCherry signal will be partitioned between daughter cells with every replication event (Fig. 1B). However, slow-growing or nondividing cells will not go through the same number of replication events and therefore will not undergo fluorescence dilution (Fig. 1B).
FIG 1.
The revTetR reporter can be used to visualize differential bacterial growth rates in vitro with mCherry fluorescence dilution. (A) The revTetR-mCherry reporter was created by introducing two mutations (E15A and L17G) in the wild-type tetR sequence, which results in constitutive mCherry expression in the absence of tetracyclines (−Tet). Tetracycline addition (+Tet) promotes revTetR binding and repression of mCherry transcription. (B) After tetracycline addition (+Dox/ATc), fast-growing/dividing bacteria (top row, red circles) will partition mCherry equally into daughter cells with each cell division event, resulting in dilution. Slow-growing/nondividing cells will retain a high level of mCherry (bottom row). (C to E) Exponential-phase cultures of the revTetR-mCherry dps strain (2-h subculture in LB, 37°C with rotation) were treated with the indicated ATc doses at 0 h and then further incubated at 37°C with rotation. Aliquots were taken at the indicated time points to detect (C) optical density by absorbance (A600), (D) mCh fluorescence (normalized by dividing total fluorescence by OD values, untreated value set to 100%), dotted line depicts one doubling (50%), and (E) GFP fluorescence normalized to OD. (C to E) Data shown from 9 replicates. (F) Cells treated with the indicated doses of ATc for 4 h were fixed for fluorescence microscopy. Sum single-cell mCherry signal is shown. (G) Sum single-cell GFP signal is shown. (H) Correlation plot depicting single-cell sum mCherry and sum GFP signals. (F to H) Data shown from three biological replicates. Mean and standard deviation are shown. Statistics: (C to E) two-way analysis of variance (ANOVA) with Tukey’s multiple-comparison test, comparisons made between 2 μg/mL and other groups; (F and G) Kruskal-Wallis one-way ANOVA with Dunn’s posttest; (H) linear regression with R2 value and slope of best-fit line; significantly nonzero slope indicates values are correlated. *, P < 0.05; **, P < 0.01; ****, P < 0.0001; n.s., not significant.
To confirm that the mCherry fluorescence from the revTetR reporter dilutes into actively dividing cells, we generated a reporter strain containing the revTetR reporter alongside a plasmid containing dps::gfp-ssrA (revTetR dps) (16). Dps, a ferritin-like iron-sequestering protein, accumulates within bacteria in stationary phase in response to multiple different stresses and protects DNA from oxidative damage (43–45). Slow-growing cells have increased levels of the dps (GFP) reporter signal (16). If the revTetR reporter is functioning as predicted, we would expect to detect heightened revTetR (mCherry) in cells expressing dps, identifying these cells as slow-growing, nondividing cells within the population. revTetR dps cells were grown into exponential phase and then treated with increasing doses of anhydrotetracycline (ATc), a tetracycline derivative that binds TetR or revTetR but has limited ribosomal targeting (46, 47). Aliquots of each sample were measured for optical density or absorbance (A600) and fluorescence using a microplate reader. The absorbance values increased across all treatment groups, and ATc did not have a detrimental effect upon growth rate with either 0.1 μg/mL or 1 μg/mL treatment (Fig. 1C). However, 2 μg/mL ATc treatment did slightly inhibit growth, resulting in significantly lower optical density (OD) values at 3 h and 4 h posttreatment. mCherry fluorescence values were normalized to cell number by dividing the background-subtracted fluorescence value from each well by the corresponding OD value. The resulting value was further normalized to the untreated sample value for each time point (set at 100%). Following 2 h of ATc treatment, all treated groups had significantly reduced mCherry fluorescence relative to that of untreated cells (Fig. 1D). At 4 h posttreatment, the fluorescent signal for all treated groups decreased by approximately 50% relative to the signal of the untreated group, showing that ATc successfully induces dilution of mCherry with each replication event (Fig. 1D). A 50% dilution in fluorescent signal should correlate with one cell division event in all the cells within the population, suggesting the doubling time was approximately 4 h. This is slow for Y. pseudotuberculosis strains and was the first indication that the high levels of mCherry expression associated with this revTetR construct may affect bacterial growth. We noted that 2 μg/mL ATc treatment resulted in significantly higher mCherry (Fig. 1D) and dps reporter signal (Fig. 1E) than 0.1 μg/mL and 1 μg/mL treatment, again suggesting that the 2 μg/mL dose may slightly inhibit growth and providing evidence that slowed growth can be detected by an increase in mCherry signal. Cells from the final time point (4 h) were fixed and imaged using fluorescence microscopy to quantify single-cell fluorescence using the sum single-cell signal for each channel. The average signal of the untreated group (0 μg/mL) was significantly higher than the average signal of the group treated with the lowest dose of ATc (0.1 μg/mL), again showing that signal dilution was successful (Fig. 1F). Cells treated with 1 μg/mL ATc had mCherry fluorescence similar to that of the 0.1 μg/mL-treated group. However, at the highest dose of ATc (2 μg/mL), the average mCherry signal significantly increased (Fig. 1F), and we found that there were also significant increases in dps reporter expression with increasing doses of ATc (Fig. 1G), suggesting that high concentrations of ATc may have an inhibitory effect on bacterial cells. Slower-growing cells with heightened dps reporter expression should also exhibit heightened revTetR-mCherry signal, and we confirmed that these signals were correlated under all treatment conditions, based on significantly nonzero slopes of linear regression best-fit lines (Fig. 1H). Collectively, these data indicate that revTetR fluorescence accumulates in slow-growing cells and that actively growing cells partition mCherry into daughter cells, resulting in significant decreases in mCherry signal.
Growth inhibition with doxycycline promotes mCherry fluorescence accumulation.
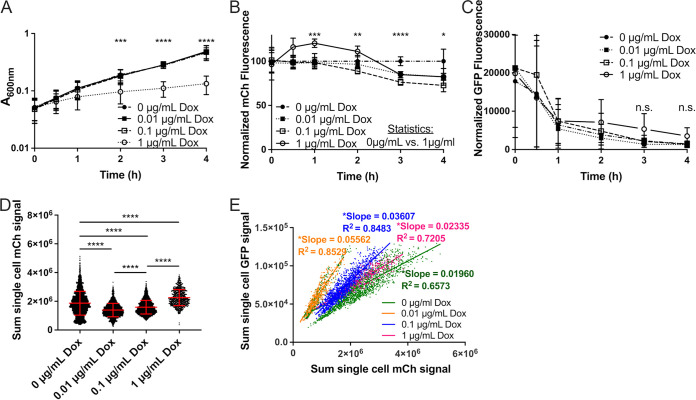
To further test whether growth-inhibited cells exhibit heightened revTetR-mCherry fluorescence, cells of the revTetR dps strain were subcultured to exponential phase and treated with increasing doses of the antibiotic, doxycycline (Dox). Aliquots of each sample were measured for optical density (OD) by absorbance (A600) and fluorescence at hourly time points using a microplate reader. Cells exposed to 1 μg/mL Dox had significantly decreased growth rates; however, the doses lower than 1 μg/mL (0.01 to 0.1 μg/mL) were subinhibitory and did not affect growth (Fig. 2A). mCherry fluorescence was initially heightened with 1 μg/mL Dox treatment (Fig. 2B), and dps reporter signal was slightly, but not significantly, elevated (Fig. 2C), consistent with growth inhibition at this treatment concentration. Three hours after treatment, the mCherry signal in all treatment groups was significantly below untreated values, including the 1 μg/mL treatment group (Fig. 2B), suggesting that growth may resume after initial growth inhibition. Single-cell analysis was performed by fluorescence microscopy, and the sum single-cell fluorescence values were graphed for each treatment group. Dilution of mCherry signal could be seen with treatment of 0.01 μg/mL Dox, but as the dosage increased, the mCherry signal within cells increased, indicating signal accumulation and stalled growth (Fig. 2D). The mCherry fluorescent signal was significantly higher in cells treated with 1 μg/mL Dox than in untreated cells (0 μg/mL), suggesting that growth inhibition resulted in significant revTetR-mCherry fluorescence accumulation. Again, we observed a correlation between dps reporter signal and revTetR-mCherry signal under all treatment conditions, confirming that slower-growing cells have heightened revTetR fluorescence (Fig. 2E).
FIG 2.
Growth inhibition with doxycycline promotes mCherry fluorescence accumulation. Exponential-phase cultures of the revTetR-mCherry dps strain were treated with the indicated doses of doxycycline (Dox) at 0 h, aliquots were taken at the indicated time points to detect (A) optical density by absorbance (A600), (B) mCh fluorescence (normalized by dividing total fluorescence by OD values, untreated value set to 100%), and (C) GFP fluorescence normalized to OD. (A to C) Data shown from 9 replicates. (D) Cells treated with the indicated doses of Dox for 4 h were fixed for fluorescence microscopy. Sum single-cell mCherry signal is shown. (E) Correlation plot depicting single-cell sum mCherry and sum GFP signals. (D and E) Data shown from three biological replicates. Mean and standard deviation are shown. Statistics: (A to C) two-way ANOVA, Tukey’s multiple-comparison test, comparisons shown between untreated (0 μg/mL) and 1 μg/mL Dox; (D) Kruskal-Wallis one-way ANOVA with Dunn’s posttest; (E) linear regression with R2 value and slope of best-fit line; significantly nonzero slope indicates that values are correlated. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; n.s., not significant.
Nitric oxide-stressed cells divide at a lower rate than unstressed cells.
It has been shown recently that exposure to nitric oxide (NO) results in cell division arrest, through collapse of the FtsZ cell division machinery (48). Our results have also suggested that NO-stressed Yersinia pseudotuberculosis preferentially survives doxycycline treatment during growth in the mouse spleen (22), which is consistent with a hypothesis that NO causes cell division arrest within host tissues and subsequently promotes decreased antibiotic susceptibility. To test this hypothesis, we generated a strain containing the revTetR-mCherry reporter and Phmp::gfp, which can be used to mark NO-stressed cells (16, 17).
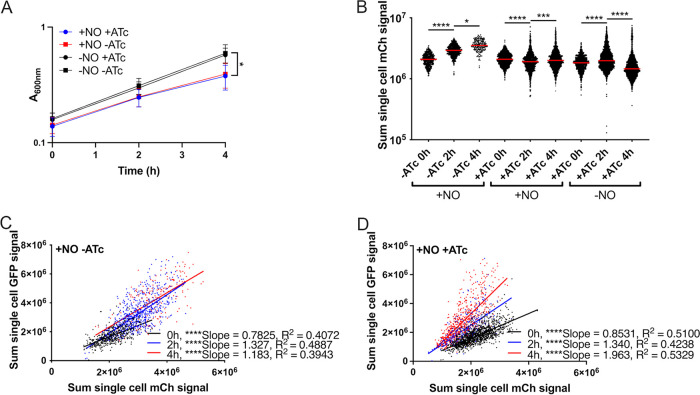
The revTetR-mCherry Phmp::gfp strain was treated with the slow-releasing NO donor compound DETA-NONOate for 2 h and then further treated with ATc (1 μg/mL). The Phmp::gfp reporter was used to detect NO exposure and to mark the level of stress experienced by individual cells, which can be variable across the population in response to DETA-NONOate (16, 17). Growth inhibition during treatment with the NO donor, DETA-NONOate, was confirmed based on a significantly lower absorbance value in treated cultures at the 4 h time point (Fig. 3A). Induction of hmp in response to the NO donor was also confirmed across all treatment groups (Fig. S1). In the absence of ATc treatment, NO-exposed cells increased in mCherry signal over time, which is consistent with growth inhibition and decreased protein turnover (Fig. 3B). In cells treated with NO and ATc, the revTetR-mCherry fluorescence decreased between 0 h and 2 h, indicating some cell division within this time frame, but signal slightly increased between 2 h and 4 h ATc treatment, indicating cell division arrest (Fig. 3B). This timing was consistent with the growth inhibition observed at 4 h in Fig. 3A. In contrast, cells grown in the absence of NO treatment had a significant drop in mCherry fluorescence between 2 h and 4 h, suggesting significant levels of cell division events within this time frame (Fig. 3B). Consistent with previous literature, these results indicate that NO treatment results in growth arrest, and reduced levels of cell division events (48), as detected by revTetR-mCherry.
FIG 3.
Nitric oxide-stressed cells divide at a lower rate than do unstressed cells. Exponential-phase cultures of the revTetR-mCherry Phmp::gfp strain were cultured 2 h in the presence (+NO) or absence (−NO) of NO, then split, and either treated with 1 μg/mL ATc (+ATc) or left untreated (−ATc). Time points are shown relative to ATc addition. (A) Aliquots were taken at the indicated time points to detect optical density by absorbance (A600). Mean and standard deviation are shown. (B) Aliquots of cells were fixed at the indicated time points for fluorescence microscopy. Sum single-cell mCherry signal is shown. Median value/group is highlighted. (C to D) Correlation plots depicting single-cell sum mCherry and mean GFP signals. Comparisons were made for NO-treated cells (+NO) in the presence (+ATc) and absence (−ATc) of ATc, and cells were imaged at the indicated time points. Values were normalized to the max signal intensity detected in samples, represented by a value of 1.0. All data shown represent three biological replicates. Number of cells analyzed is shown. Statistics: (A) two-way ANOVA, Tukey’s multiple-comparison test, comparisons shown between untreated (−NO) and NO donor treated (+NO) samples; (B) Kruskal-Wallis one-way ANOVA with Dunn’s posttest; (C and D) linear regression with R2 value and slope of best-fit line; significantly nonzero slope indicates values are correlated. *, P < 0.05; ****, P < 0.0001; n.s., not significant.
To determine whether heightened NO stress correlated with a reduced level of cell division events, single-cell mCherry (revTetR) and green fluorescent protein (GFP; Hmp) values from NO-treated cells were compared in the absence and presence of ATc treatment using correlation plots and linear regression analyses. In the absence of ATc treatment, the distributions of cells within the NO-stressed bacterial populations were very similar in fluorescence across all time points (Fig. 3C). In the presence of ATc treatment, there was a clear shift in the NO-treated population toward reduced mCherry signal over time (Fig. 3D). This suggested that some cell division still occurred in the presence of NO treatment, which was consistent with Fig. 3B. However, the correlation between the two fluorescent signals strengthened over the time course based on increased R2 values (Fig. 3D). These data support the conclusion that cells experiencing higher levels of NO stress divide more slowly and retain higher levels of revTetR-mCherry fluorescence.
Low doses of doxycycline and anhydrotetracycline diffuse evenly across microcolonies and do not inhibit bacterial growth.
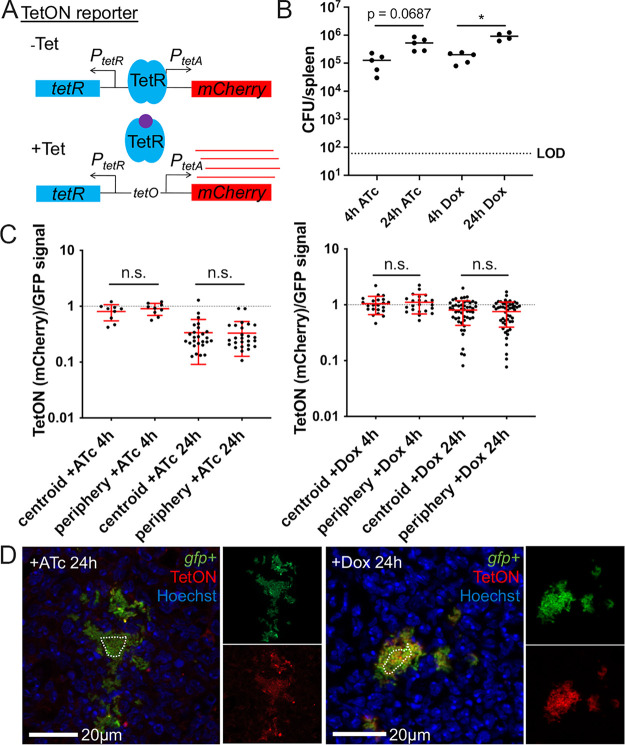
One of the major goals for generating the revTetR reporter was to set up a system to detect bacterial cell division events within host tissues over defined periods of time. To do this, we would need to administer tetracyclines during infection, ensure that the dosage used would not affect bacterial growth, and also diffuse evenly across microcolonies to allow for accurate detection of signal dilution and cell division. We previously developed a model of doxycycline treatment of Y. pseudotuberculosis infection and found that a single 40 mg/kg injection of doxycycline significantly reduces bacterial CFU and also accumulates at a higher concentration at the periphery of microcolonies (22). We hypothesized that lowering the tetracycline dose may allow us to administer subinhibitory levels of antibiotic that would still modulate the revTetR reporter, while allowing for more even diffusion of the antibiotic across microcolonies. Our rationale was that lower antibiotic concentrations would be more readily eliminated from circulation, so antibiotic movement into the spleen would occur within a more defined time frame, limiting continued accumulation.
To identify a tetracycline concentration that results in equal diffusion across microcolonies, a previously characterized reporter, PtetA::mCherry-ssrA (TetON), was used for tetracycline detection (22). The TetON reporter contains the wild-type tetR gene; addition of tetracyclines induces the production of mCherry, and therefore levels of mCherry can be used as a readout for antibiotic exposure (Fig. 4A). C57BL/6 mice were inoculated intravenously with the TetON gfp+ strain, which has constitutive expression of GFP alongside the TetON reporter, allowing for visualization of relative reporter expression. A single dose of 4 mg/kg Dox or 4 mg/kg ATc was administered intraperitoneally at 48 h postinfection. This time point was chosen for antibiotic administration since we can detect subsets of bacteria responding to host-derived nitric oxide at this time point, thus allowing us to ask whether stressed subpopulations of bacteria divide more slowly than unstressed bacteria.
FIG 4.
Low doses of doxycycline and anhydrotetracycline diffuse evenly across microcolonies and do not inhibit bacterial growth. C57BL/6 mice were infected intravenously with the TetON gfp+ strain, and a single dose of Dox (4 mg/kg) or ATc (4 mg/kg) was administered intraperitoneally at 48 h postinfection. Spleens were harvested at the indicated time points after treatment. (A) Schematic of the TetON reporter: addition of tetracycline relieves TetR repression and allows for mCherry transcription. (B) CFU/spleen quantification at the indicated time points posttreatment. Dotted line, limit of detection; dots, individual mice. (C) Reporter signal was detected with fluorescence microscopy and is shown as the ratio of TetON (mCherry)/GFP signal at the centroid and periphery of microcolonies. Time points indicate time posttreatment with ATc (left panel) or Dox (right panel). Dotted line, equivalent levels of mCherry and GFP signal. Each dot, individual microcolony. Mean and standard deviation are shown. (D) Representative images. Merged images and individual channels are shown. Cells outside the dotted line are defined as peripheral. Data set represents 4 to 5 mice per group, collected from two independent experiments; 2 to 8 microcolonies were analyzed per spleen from a single cross-section of tissue. Statistics: (B) Kruskal-Wallis one-way ANOVA with Dunn’s posttest; (C) Wilcoxon matched-pairs; *, P < 0.05; n.s., not significant.
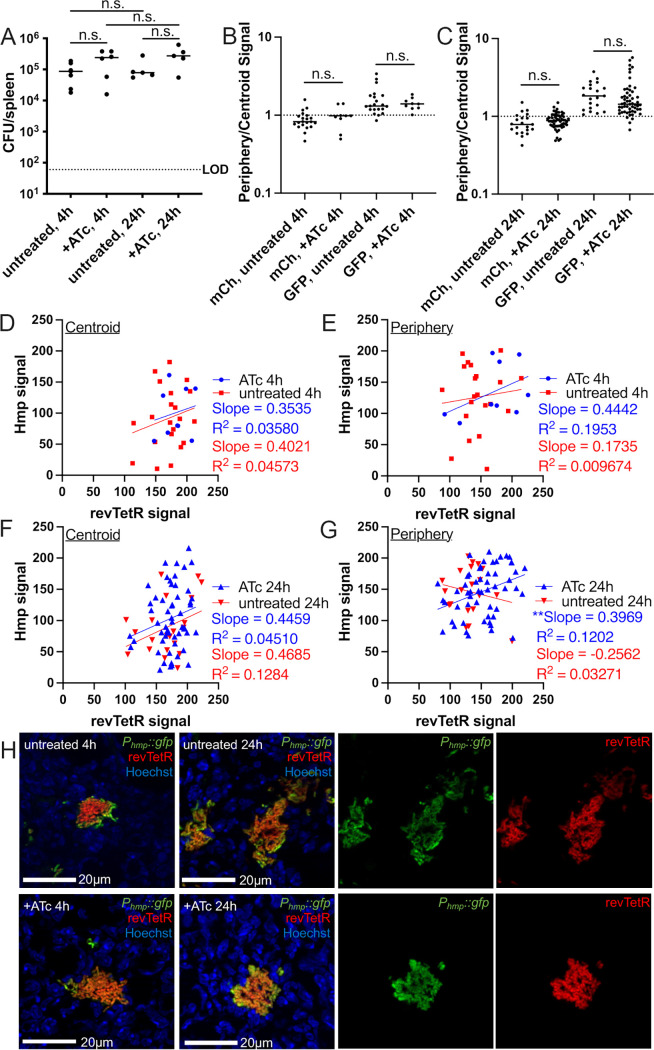
At 4 h or 24 h posttreatment, spleens were harvested to quantify CFU and quantify reporter expression by fluorescence microscopy. Bacterial numbers continued to increase between 4 h and 24 h posttreatment with 4 mg/kg doses of either ATc or Dox, confirming that this dosage does not interfere with bacterial growth (Fig. 4B). Microcolonies were then visualized by fluorescence microscopy to determine if ATc and Dox diffuse evenly across microcolonies at this dosage. The centroid and periphery of each microcolony were quantified for mCherry and GFP signal, and the ratio of mCherry/GFP was graphed. There were no significant differences in mCherry/GFP ratios between the centroid and periphery of microcolonies treated with either ATc or Dox, suggesting that each drug diffused evenly across microcolonies (Fig. 4C). The mCherry/GFP ratios remained comparable at the centroid and peripheries even at 24 h after treatment, showing that throughout the infection time course, there was even diffusion of the drugs (Fig. 4C and D). These data suggest that lowering the tetracycline dosage 10-fold results in even diffusion of a subinhibitory dose of tetracyclines, which will allow us to modulate revTetR reporter expression in all cells across the microcolony without affecting bacterial growth.
revTetR signal appears to increase in both the centroid and periphery of microcolonies.
To determine if there are slowly dividing subpopulations of bacteria within microcolonies, we infected mice with the revTetR-mCherry gfp+ strain, which constitutively expressed gfp, and treated mice with ATc at 48 h postinfection or left mice untreated. Mice were sacrificed at either 4 h or 24 h posttreatment, and spleens were harvested to quantify CFU and visualize reporter expression by fluorescence microscopy. Bacterial CFU were comparable between ATc-treated and untreated mice at both time points, suggesting that ATc treatment did not impair bacterial growth (Fig. 5A). However, we did not see an increase in CFU between 4 h and 24 h, which suggests that there was limited, or variable, bacterial growth within this selected time frame for this bacterial strain. To determine if there were differences in reporter expression, mCherry signal was quantified relative to the constitutive GFP signal at the periphery and centroid of individual microcolonies. Since we have previously shown that bacteria experience heightened levels of NO stress and express higher levels of the T3SS at the periphery of microcolonies, we were expecting to see heightened levels of mCherry signal relative to those of GFP at the periphery of microcolonies. At 4 h posttreatment, we did not observe any differences in mCherry signal at the periphery of microcolonies compared to that at the centroid, in the absence or presence of ATc treatment (Fig. 5B). This suggested that there may not have been many cell division events within the first 4 h of treatment, which is why tissue was also harvested 24 h posttreatment. At 24 h posttreatment, untreated mice had significantly lower levels of mCherry signal at the periphery of microcolonies, and ATc-treated mice also had significantly lower mCherry signal at the periphery, indicating that we were not seeing increased mCherry signal at the periphery of microcolonies as expected (Fig. 5C and D). It did appear that ATc-treated microcolonies had higher mCherry signals at both the centroid and periphery relative to those of untreated microcolonies, although this was not statistically significant. These results suggest that revTetR signal remained high after ATc treatment, indicating that there may be slowly dividing cells present within microcolonies. Without much bacterial growth across these time points in general, it was difficult to assess whether slow-growing cells were present in a specific spatial location or part of a subpopulation without an additional reporter alongside revTetR.
FIG 5.
revTetR-mCherry signal appears to increase in both the centroid and periphery of microcolonies. C57BL/6 mice were infected intravenously with the revTetR-mCherry gfp+ strain, and a single dose of ATc (4 mg/kg) was administered intraperitoneally at 48 h postinfection. Spleens were harvested at the indicated time points after treatment to quantify CFU and prepare tissue for fluorescence microscopy. (A) CFU/spleen quantification at the indicated time points posttreatment. Dotted line, limit of detection; dots, individual mice; horizontal lines, median values. (B) Reporter signal at 4 h quantified using the ratio of revTetR (mCherry)/GFP signal at the centroid and periphery of microcolonies. Dotted line, equivalent levels of mCherry and GFP signal. Each dot, individual microcolony. Mean and standard deviation are shown. (C) Reporter signal at 24 h quantified using ratio of revTetR (mCherry)/GFP signal at the centroid and periphery of microcolonies. Mean and standard deviation are shown. (D) Representative images. Data set represents 4 to 6 mice per group, collected from three independent experiments; 2 to 13 microcolonies were analyzed per spleen from a single cross-section of tissue. Statistics: (A) Kruskal-Wallis one-way ANOVA with Dunn’s posttest; (B and C) Wilcoxon matched-pairs; *, P < 0.05; **, P < 0.01; n.s., not significant.
NO stress correlates with heightened mCherry signal accumulation and decreased cell division rates.
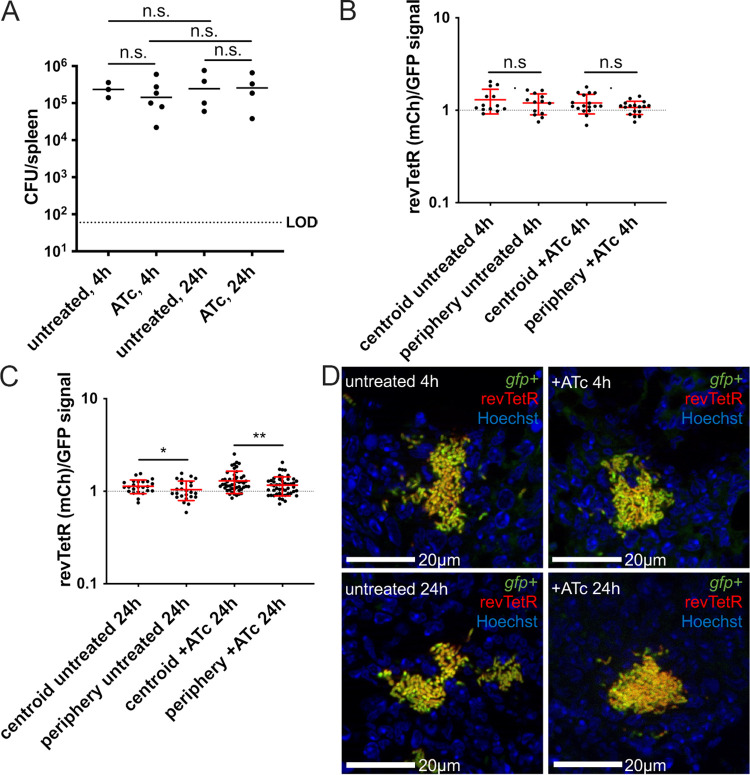
Hmp+ bacteria responding to NO stress preferentially survive doxycycline treatment, suggesting that they may represent a slow-growing subpopulation (22). Consistent with this, in culture, NO-stressed cells accumulated heightened mCherry signal, suggesting that they were dividing more slowly than unstressed cells (Fig. 3). However, in our mouse model, it was difficult to determine if peripheral cells of the microcolony had heightened revTetR signal without marking the peripheral subpopulation with the hmp reporter. To determine if NO-stressed bacteria are dividing more slowly than unstressed cells within host tissues, we infected mice with the revTetR-mCherry Phmp::gfp strain and treated mice with ATc at 48 h postinfection or left mice untreated. Spleens were harvested at 4 h and 24 h posttreatment to quantify CFU and visualize reporter expression by fluorescence microscopy. CFU within ATc-treated mice did not differ significantly from CFU within untreated mice; again, we noted that there was not significant growth based on total CFU between 4 h and 24 h but anticipated that there would be sufficient replication at the level of individual microcolonies to assess differences in revTetR signal (Fig. 6A).
FIG 6.
NO stress correlates with heightened mCherry signal accumulation and decreased cell division rates. Mice were infected with the revTetR-mCherry Phmp::gfp strain and a single dose of ATc (4 mg/kg) was administered intraperitoneally at 48h postinfection. Spleens were harvested at the indicated time points after treatment and prepared for fluorescence microscopy. (A) CFU/spleen quantification. Dotted line: limit of detection, dots: individual mice. (B) Reporter signal at 4h quantified using ratio of periphery/centroid signal for either mCherry (mCh) or GFP. Dotted line: equivalent levels of mCherry and GFP signal. Each dot: individual microcolony. (C) Reporter signal at 24 h quantified using ratio of periphery/centroid signal for either mCh or GFP. (D to G) Correlation plots depicting single-cell revTetR signal (mCherry) and Hmp signal (GFP) at the indicated time points, comparing +/− ATc treatment. Graphs represent either centroid or peripheral values; dots, individual microcolonies. (H) Representative images with the green and red channels shown alongside merged images for 24 h posttreatment images. Data set represents 5 to 6 mice per group, collected from three independent experiments; 3 to 8 microcolonies were analyzed per spleen from a single cross-section of tissue. Median values are shown with horizontal lines. Statistics: (A to C) Kruskal-Wallis one-way ANOVA with Dunn’s posttest (D to G) linear regression with R2 value and slope of best-fit line; significantly nonzero slope indicates that values are correlated. **, P < 0.01; n.s., not significant.
Periphery and centroid measurements were taken from each microcolony image and combined into a single periphery/centroid ratio for each microcolony to allow for comparison across treatment groups. At 4 h post ATc treatment, the average ratio for the hmp reporter signal was above a value of 1, indicating higher expression in the peripheral cells (Fig. 6B). The average ratio for revTetR-mCherry values was close to 1 in both ATc-treated and untreated mice, suggesting equal expression in peripheral and centroid cells and very little cell division in this time frame (Fig. 6B). At 24 h post ATc treatment, GFP ratios remained above 1, indicating high peripheral expression of the hmp reporter (Fig. 6C). The revTetR-mCherry ratio appeared to increase slightly but remained below a value of 1 and was not significantly different from that of untreated mice (Fig. 6C). This again suggested that there was little cell division within this time frame, since we could not detect much impact of the ATc treatment. We then used linear regressions to determine if there was a correlation in fluorescent signals at distinct spatial locations within the microcolony and overall saw little impact of the ATc treatment on the fluorescence of the cells (Fig. 6D to H). The exception was the analysis at the periphery 24 h posttreatment, where ATc treatment resulted in a slight positive correlation, suggesting that Hmp+ cells were less likely to divide (Fig. 6G and H). The limited levels of cell division with this strain in vivo made these results difficult to interpret, and we then sought to modify the reporter to generate a new revTetR construct with normal growth kinetics in vivo.
Dilution of the revTetR-YFP signal coincides with the replication of fast-growing cells.
To address the growth issues in the revTetR-mCherry strain evident in previous experiments, we modified the revTetR reporter to express YFP instead of mCherry (revtetR::PtetA::yfp). We surmised that the growth issues were due to the more toxic and disruptive nature of constant mCherry overexpression in the absence of tetracycline derivatives, and YFP is known to be less toxic for cells (49, 50).
To determine if revTetR-YFP strains exhibit normal growth kinetics, and confirm that NO stress is sufficient to slow growth and promote YFP signal accumulation, we generated a strain containing the revTetR-YFP reporter and Phmp::mCherry (revtetR::PtetA::yfp Phmp::mCherry). Cells were grown in the presence and absence of the NO donor compound DETA-NONOate for 2 h and then treated with ATc (1 μg/mL) as described above. Aliquots were taken at the indicated time points after ATc addition to quantify single-cell fluorescence by microscopy. Growth inhibition due to the NO donor was confirmed based on increased signal between 0 h and 2 h post ATc treatment (Fig. S2A), and all treated cultures responded to the NO donor based on induction of hmp (Fig. S2B). In contrast, cells grown in the absence of NO showed dramatic YFP signal dilution between 0 to 2 h (Fig. S2A). This suggested that the revTetR-YFP construct behaves similarly to revTetR-mCherry and can be used to mark growth-arrested cells. However, unstressed cells (−NO, +ATc) no longer had signal dilution between 2 to 4 h post ATc treatment, suggesting that growth was slowing in these cultures. Since cells were incubated with NO for 2 h before ATc treatment, this indicates that the cells were beginning to enter stationary phase by the 4 h time point (6 h of growth). These changes in growth kinetics indicate that the revTetR-YFP strain may grow faster than the revTetR-mCherry strain in culture. Additionally, the signal dilution between 2 to 4 h in the +NO +ATc group (Fig. S2A) suggests that cells are recovering from NO exposure and are starting to replicate after 6 h of exposure. This is consistent with our previous work (16) and consistent with the presence of some Hmplow/mCherrylow cells in the +NO +ATc group at 4 h (Fig. S2B).
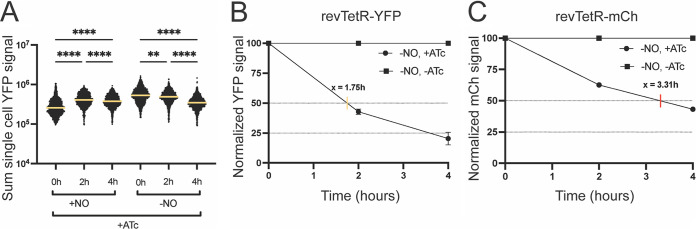
To ensure we were capturing the time frame of growth arrest, we modified the in vitro NO donor experiment to shorten the NO exposure. Cells containing the revTetR-YFP reporter and Phmp::mCherry were either treated with the NO donor compound or left untreated for only 30 min before ATc addition, and single-cell fluorescence was quantified via microscopy. Cells grown in the absence of ATc treatment continued to accumulate YFP signal, which was unsurprising (Fig. S3A). Heightened hmp induction in the +NO groups compared to that in the −NO groups confirmed NO exposure throughout the time course (Fig. S3B). With this experimental protocol, cells exposed to the NO donor and ATc treatment increased in revTetR-YFP signal between 0 to 2 h, with only a slight decline between 2 to 4 h, indicating overall growth arrest (Fig. 7A). Cells grown in the absence of the NO donor with ATc treatment showed continuous signal dilution, indicating continuous growth and replication (Fig. 7A). These results suggest that in this faster-growing strain, YFP signal dilution does correspond with bacterial cell growth, and signal accumulation corresponds with growth arrest.
FIG 7.
Dilution of the revTetR-YFP signal coincides with replication of fast-growing cells. The revTetR-YFP Phmp::mCherry strain was grown in the presence (+NO) or absence (−NO) of NO for 30 min. Cultures were then treated with 1 μg/mL ATc (+ATc) or left untreated. Aliquots were taken at time points after ATc addition. (A) Sum single-cell YFP signal determined by microscopy. Bars: mean. At least 1,500 individual cells were quantified per group. (B and C) Replication rates of unstressed (−NO) cells were calculated by first normalizing to fluorescent signal at 0 h, and then values of treated cells (+ATc) were further normalized to the untreated values (−ATc, set to 100%) to depict signal dilution over time. Linear regressions were used to calculate the time it takes for 100% of the cells in culture to undergo one full replication (50% dilution of the signal). This was done for (B) revTetR-YFP (yellow tick, 1.75 h for full replication cycle) and (C) revTetR-mCherry (red tick, 3.31 h). Statistics: (A) Kruskal-Wallis one-way ANOVA with Dunn’s posttest; **, P < 0.001; ****, P < 0.0001.
Since our results above indicate that the growth kinetics are different between the revTetR-YFP and revTetR-mCherry strains, we estimated the length of time it takes for 100% of the cells in culture to undergo one full replication cycle, as indicated by a 50% dilution of the revTetR signals. Calculations were completed using data from unstressed cells: −NO +ATc and −NO −ATc groups. YFP signals from our single-cell microscopy data were normalized to 100% at 0 h, and then the ATc-treated group (−NO +ATc) data were normalized to the untreated control group (−NO −ATc) to account for signal dilution over time. Using a linear regression equation, the revTetR-YFP strain was estimated to undergo one replication cycle, or doubling, in 1.75 h (Fig. 7B). This is consistent with normal growth kinetics of Y. pseudotuberculosis at 37°C (16). We then confirmed that the revTetR-mCherry strain does indeed grow slower, with an estimated 3.31 h for one replication cycle (Fig. 7C). Together, these data indicate that the revTetR-YFP reporter can be used to differentiate between dividing and nondividing cells and confirm that the growth issues seen in the revTetR-mCherry strains have been resolved with revTetR-YFP.
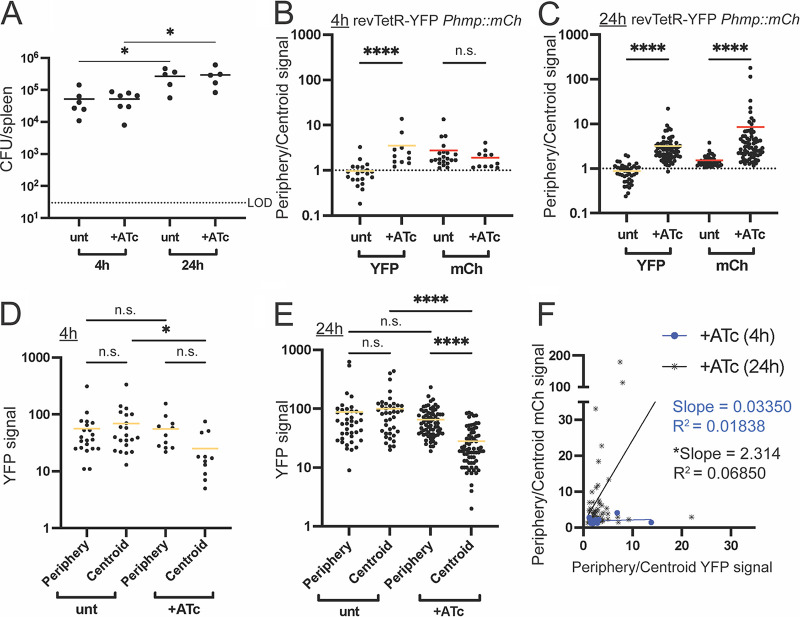
Cells that lack NO stress rapidly dilute revTetR-YFP signal, indicating higher cell division rates.
Our previous work indicates that cells at the periphery of the microcolony respond to NO stress and may be slow-growing, since Hmp+ cells had reduced susceptibility to a ribosome-targeting antibiotic (22). While our in vitro data support this (Fig. 7), our previous study using a modified TIMER protein (DsRed42) to detect slow-growing bacteria suggested that NO stress is transient within host tissues (16). At late time points during infection, Hmp− cells at the centroid of microcolonies were instead slow-growing due to nutrient limitation (16). In our mouse infections with the revTetR-mCherry Phmp::gfp strain described above, it appeared that cells responding to NO stress (Hmp+, GFP+) may have higher mCherry levels (Fig. 6G). Due to growth issues with the strain, it was difficult to interpret these results. To clarify if the subset of NO-exposed cells are truly growing at a slower rate than unstressed cells within host tissues, we infected mice with the revTetR-YFP Phmp::mCherry strain and then treated them with ATc (+ATc) at 48h postinfection or left mice untreated (unt). We confirmed that the revTetR-YFP Phmp::mCherry strain was growing well within host tissues based on significant increases in CFU in the spleen, comparing between 4 h and 24 h post ATc treatment (Fig. 8A). Spleens were processed for fluorescence microscopy, and periphery and centroid measurements were taken from each microcolony and combined into a single periphery/centroid ratio to allow for comparison across treatment groups. At 4 h posttreatment, the average ratio for the hmp (mCh) reporter signal was above a value of 1 in both untreated (unt) and treated mice (+ATc), indicating higher expression in the peripheral cells, as expected. The average ratio of revTetR-YFP signal was close to 1 in untreated mice, suggesting equal expression in peripheral and centroid cells (Fig. 8B). In contrast, ATc-treated mice had a ratio above 1, suggesting that there is higher YFP expression in the periphery than in the centroid (Fig. 8B). Similar trends held true at 24 h posttreatment, although the hmp (mCh) reporter signal was higher in treated mice for unknown reasons (Fig. 8C).
FIG 8.
Cells that lack NO stress rapidly dilute revTetR-YFP signal, indicating higher cell division rates. Mice were infected with the revTetR-YFP Phmp::mCherry strain and then either injected with 4 mg/kg ATc (+ATc) or left untreated (unt) at 48 h postinfection. (A) CFU/spleen was quantified for each mouse at the indicated time points post ATc treatment. Dotted line indicates limit of detection (LOD), and each dot represents one mouse. (B and C) The ratio of periphery/centroid signals was quantified with revTetR (YFP) and Hmp reporter (mCherry) signals at (B) 4 h and (C) 24 h post ATc treatment, along with the raw YFP signals at (D) 4 h and (E) 24 h posttreatment. (F) Correlation plots are shown for periphery/centroid ratios for revTetR (YFP) and Hmp (mCherry) reporter signals, comparing treated groups at each time point. (B to F) Each dot represents a microcolony. Mean values are highlighted. Data represent 4 to 5 mice per group per time point, with 11 to 71 microcolonies analyzed for each group and time point. Statistics: (A to E) Kruskal-Wallis one-way ANOVA with Dunn’s posttest; (F) linear regression was used to determine correlation between values, and a significant deviation from zero indicates that the values are correlated; *, P < 0.05; ****, P < 0.0001.
An increased periphery/centroid ratio can mean that either the periphery signal increased or, conversely, the centroid signal decreased. Because of the nature of fluorescence dilution, we would expect that this means the centroid dilutes revTetR-YFP signal more rapidly than the periphery, suggesting that the centroid divides more rapidly. To assess this, raw YFP signal values were compared across treatment groups and time points. At 4 h post ATc treatment, YFP signals for untreated mice were not significantly different between the centroid and periphery (Fig. 8D). The YFP signal in the centroid of ATc-treated mice trended lower, but not significantly lower, than that in the periphery (Fig. 8D). However, the centroids of ATc-treated mice were significantly lower than the centroids of untreated mice, suggesting cell division and signal dilution at the centroid (Fig. 8D). At 24 h post ATc treatment, untreated mice had similar levels of YFP signal at the centroid and periphery of microcolonies, while centroids of treated mice had levels of YFP significantly lower than those of treated peripheries or untreated centroids (Fig. 8E). These results suggest significant cell division within the centroid of microcolonies and relative growth arrest at the periphery, since peripheral cells were not changing in YFP signal (Fig. 8E). These conclusions are corroborated by the correlation of revTetR-YFP and mCherry (Hmp) periphery/centroid ratios, where revTetR and Hmp exhibit a weak correlation at 24 h posttreatment, based on a significantly nonzero slope (Fig. 8F). It is important to note that while cells in the periphery tend to have higher YFP signals in the treated mice at 24 h (Fig. 8F), some subsets of peripheral cells do not exhibit this high YFP signal (Fig. 9). It is, however, striking how much YFP dilution occurs at the centroid of microcolonies, which strongly supports the conclusion that the centroid divides rapidly (Fig. 8E and Fig. 9). Overall, this microcolony phenotype is consistent with our previous finding that only small subsets of peripheral cells actively respond to NO stress at any given time point (16), which appears to correlate with detection of growth arrest with revTetR-YFP.
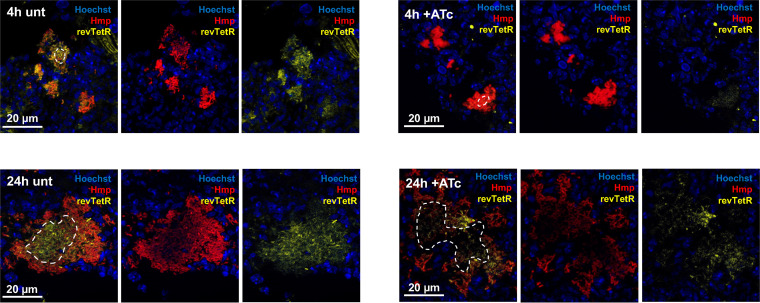
FIG 9.
Representative images of spleens during revTetR-YFP Phmp::mCherry infection. Representative images are shown from spleens of mice infected with the revTetR-YFP Phmp::mCherry strain, as described and quantified in Fig. 8. Merged images are shown alongside red and yellow single channels images at the indicated time points after ATc treatment. Bacterial cells outside the white dotted lines (see merged images) are considered peripheral, and interior cells at the geometric centroid are considered centroid.
DISCUSSION
In this study, we have developed revTetR constructs that can be utilized to detect bacterial cell division events within host tissues using a fluorescence dilution approach. We have shown that both revTetR-mCherry and revTetR-YFP constructs result in heightened fluorescent signal in growth-arrested cells in culture and have also shown that nitric oxide (NO) stress is sufficient to promote increased fluorescent signal in culture (Fig. 3 and 7). This was consistent with previous studies showing that NO exposure results in an arrest of cell division (48). Our in vivo results in the mouse model of infection were initially less clear; our revTetR-mCherry strains had notable slowed growth within host tissues (Fig. 5A and Fig. 6A), which made it difficult to differentiate dividing and nondividing cells. Our results with revTetR-YFP were generally consistent with those with revTetR-mCherry; however, expression of the less toxic YFP allowed bacteria to replicate within host tissues during the chosen time frame postinfection, and we detected clear differences in cell division rates across microcolonies. Bacterial cells at the centroid clearly replicate more quickly and dilute YFP signal, while we saw that YFP signal was maintained within peripheral cells (Fig. 7D and E). Some Hmp+ cells retained YFP signal while others did not, which we believe reflects transient NO exposures within host tissues (16).
The Hmp+ subpopulation was a focus of this study primarily because we knew that this subpopulation preferentially survives doxycycline treatment in the Y. pseudotuberculosis mouse model of infection. We hypothesized that this was due to increased levels of growth arrest as a result of NO exposure (22, 48). However, it is important to note that the Hmp+ subpopulation is a complex mixture of cells that also includes subsets expressing high levels of the type-III secretion system (T3SS) due to direct neutrophil contact (17). Based on the pattern of revTetR-YFP signal within host tissues (Fig. 9), revTetR-YFP signal is heightened in a subpopulation of peripheral cells that is larger than the subset of bacteria in direct neutrophil contact. Additional experiments would be needed to directly compare revTetR-YFP signal with T3SS expression. We also chose to mark the Hmp+ subpopulation here with a stable fluorescent protein to detect NO exposure. However, we know that only subsets of this population are actively responding to NO at any given time point (16), which adds further complexity to this subpopulation. It will be very interesting to utilize this revTetR-YFP construct in future experiments to isolate bacterial cells with decreased division rates from the host environment and further characterize the pathways that are associated with slowed bacterial growth within host tissues.
Importantly, we believe that the cell division rates within microcolonies vary over the course of infection, and revTetR-YFP will allow us to probe this in more detail by selecting different time points over the course of infection for ATc treatment and infection endpoints. Here, we chose to compare between 52 h and 72 h postinfection, which equates to +ATc 4 h and 24 h, respectively. These were time points where we expected rapid expansion within the bacterial population, allowing us to differentiate between slowly dividing and quickly dividing cells. These time points worked well for distinguishing differential growth rates with revTetR-YFP and showed that the centroid of microcolonies was dividing rapidly and diluting fluorescent signal (Fig. 9). However, this does seem to contrast with some of our other results that indicate the centroid is slow-growing (16). In Patel et al., slowed growth was detected in the centroid of microcolonies with a slow-folding DsRed variant, DsRed42, specifically at 72 h postinfection (16). We hypothesize that the time point is key here, and with the time points we have chosen in this study, we are capturing a period of rapid growth that precedes nutrient depletion at later time points. By switching from revTetR-mCherry to revTetR-YFP, we have generated strains without growth attenuation and also generated a construct where revTetR-YFP fluorescence could be directly compared with DsRed42 to better understand bacterial growth dynamics over the course of infection.
Experiments with the revTetR-mCherry constructs have shown us that to truly detect differences in cell division rates in vivo, we need to sample time points where there is rapid expansion within the bacterial population. Based on our estimates in Fig. 7, revTetR-YFP strains double approximately every 1.75 h during culture at 37°C, while revTetR-mCherry strains double approximately every 3.3 h and may take as long as 4 h to divide, based on Fig. 1D. The doubling time, or replication rate, thus essentially establishes the sensitivity of the system and needs to be taken into account when choosing the time points for experiments. Conversely, by completing full time courses, researchers would be able to utilize these reporters to approximate the growth rate of strains grown under different conditions. We believe that this application could be particularly useful in approximating the bacterial cell division rates within host tissues over the course of infection by preparing samples at multiple different time points posttreatment and also by varying when ATc treatment is administered postinfection. We plan to explore this in future experiments.
In this study, we compared revTetR-mCherry reporter signal during treatment with two tetracycline derivatives, the active antibiotic, doxycycline (Dox), and the inactive derivative, anhydrotetracycline (ATc). Both tetracycline derivatives bind to TetR and revTetR; however, ATc is predicted to bind these proteins with higher affinity and has poor affinity to bacterial ribosomes (46, 47). This allowed us to modulate revTetR with ATc without significantly inhibiting translation; however, we would expect to see translational inhibition during Dox treatment and did see evidence of this when detecting dps reporter signal alongside revTetR-mCherry. Because Dox inhibits translation, the amount of dps signal did not significantly increase with Dox treatment due to translational inhibition (Fig. 2C). However, we were able to conclude that slow-growing or growth-arrested cells had higher levels of revTetR-mCherry signal using ATc and NO treatments.
We recently showed with a slow-folding fluorescent protein derivative (DsRed42) that NO stress was sufficient to promote fluorescence accumulation but also found that there was limited overlap in slow-growing and NO-stressed populations within host tissues (16). Destabilized and stable versions of fluorescent proteins were used to show that NO stress was transient within host tissues and that few bacteria were actively responding and detoxifying NO at any given time point (16). However, the fact that Hmp+ bacteria preferentially survive doxycycline treatment suggests that, while NO exposure may be transient, there could also be sustained changes within cells exposed to high levels of NO (22). Although Yersinia repairs iron-sulfur cluster-containing proteins in response to this stress (51), there may be long-term damage or even genetic changes that occur following NO exposure. It would be very interesting to determine if any genetic changes occur within microcolonies over the course of infection, especially given that these bacterial populations are clonal, founded by a single bacterium, and presumed to be genetically homogenous (17).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Y. pseudotuberculosis IP2666 was used as the wild-type strain throughout. For in vitro experiments, overnight cultures were grown in LB at 26°C with rotation for 16 h. Exponential-phase cells for in vitro experiments were generated by subculturing overnight cultures by diluting 1:100 into fresh LB and rotating at 37°C 2 h. All in vitro experiments were performed by culturing at 37°C with rotation. For all mouse infections, bacterial inocula were grown overnight (16 h) to postexponential phase in 2× YT broth (LB, with 2× concentration of yeast extract and tryptone) at 26°C with rotation.
revTetR reporter construction.
The revTetR reporter was generated by introducing specific point mutations into our previously described tetR::PtetA::mCherry reporter construct in pMMB67EH (22). tetR was mutated by site-directed mutagenesis at two sites, E15A (GAG→GCU) and L17G (CUU→GGU), using 4 point mutations to generate the revTetR construct. An additional L25V mutation could not be successfully introduced. Primers were designed that contained the desired mutations and were used to PCR amplify around the pMMB67EH plasmid using PfuUltra II high-fidelity polymerase (Agilent). PCR products were purified (Qiagen) and transformed into electrocompetent DH5αλpir Escherichia coli. Colonies were screened based on mCherry fluorescence in the absence of tetracyclines, and plasmids were then isolated using the PureYield Plasmid Midiprep kit (Promega) and sequenced to confirm mutations. revtetR::PtetA::mCherry plasmids were transformed into Y. pseudotuberculosis strains as described previously (52) to generate the following reporter strains: revtetR::PtetA::mCherry Pdps::gfp-ssrA, revtetR::PtetA::mCherry gfp+, and revtetR::PtetA::mCherry Phmp::gfp. Pdps::gfp-ssrA, gfp+, and Phmp::gfp reporter constructs have been described previously (16) and are all inserted into the pACYC184 plasmid. revtetR::PtetA::yfp was constructed by PCR amplifying revtetR::PtetA and fusing this to yfp using overlap extension PCR. revtetR::PtetA::yfp was inserted into the pACYC184 plasmid and transformed into a Y. pseudotuberculosis strain containing the previously described Phmp::mCherry plasmid (17).
In vitro reporter characterization.
Once subcultures reached exponential phase (2 h of growth), the indicated doses of doxycycline (Dox; 0.01 μg/mL, 0.1 μg/mL, or 1 μg/mL) or anhydrotetracycline (ATc; 0.1 μg/mL, 1 μg/mL, or 2 μg/mL) were added to samples to promote mCherry dilution, with one sample left untreated as a control. Samples were incubated at 37°C with rotation throughout the time course, and time points of 0, 0.5, 1, 2, 3, and 4 h postantibiotic treatment were taken. For each time point, 600 μL from each sample was pelleted and resuspended in 600 μL of phosphate-buffered saline (PBS). Two hundred microliters of each sample was then pipetted into 3 wells of a black-walled, clear-bottom 96-well plate and analyzed using a Synergy microplate reader (Biotek Instruments) for OD600 and fluorescence signal measurements. Optical density (A600) was used to approximate cell number, and mCherry fluorescence was detected with 560 nm excitation/610 nm emission. All experiments were repeated in triplicate to generate a total of three independent data sets, or nine replicates. In strains of Y. pseudotuberculosis also containing a GFP reporter, GFP fluorescence was detected with 480 nm excitation/520 nm emission.
Induction of hmp using nitric oxide donor compound.
Overnight cultures were diluted 1:50 into fresh LB with the nitric oxide donor compound DETA-NONOate (2.5 mM dose) or were left untreated. After either 2 h or 30 min of incubation, each culture was evenly split into two separate tubes. Samples were then either treated further with ATc (1 μg/mL dose) or left untreated, for a total of four different treatment groups: +NO/+ATc, +NO/−ATc, −NO/+ATc, and −NO/−ATc. Aliquots were taken every 2 h after ATc treatment to quantify absorbance and fluorescence, and cells were prepared for fluorescence microscopy, as described below. Fixed cells were imaged and quantified for single-cell fluorescence measurements.
Fluorescence microscopy: bacteria.
To visualize individual bacterial cells, 500 μL from each sample was pelleted, resuspended in 4% paraformaldehyde (PFA), and incubated overnight at 4°C for fixation. Agarose pads were prepared by placing two pieces of tape approximately 1 cm apart on slides, pipetting 25 μL of 1% agarose in PBS, and placing a glass coverslip over the area. After leaving the agarose to solidify for 20 min, the coverslips and tape were peeled off and the agarose was trimmed to 1-cm2 squares. PFA-fixed bacteria were pelleted and resuspended in 50 μL PBS, and 5 μL of sample was pipetted onto each agarose pad. Bacteria were imaged with the 63× oil immersion objective, using a Zeiss Axio Observer.Z1 (Zeiss) fluorescence microscope with Colibri.2 LED light source and an ORCA-R2 digital CCD camera (Hamamatsu). Five images were taken of each sample in distinct fields of view, using differential interference contrast (DIC), GFP, and mCherry channels. Volocity image analysis software was used to specifically select individual bacterial cells and quantify the fluorescent signal associated with each cell.
Murine model of systemic infection.
The following strains were used for mouse infections: Y. pseudotuberculosis tetR::PtetA::mCherry-ssrA gfp+ (22), revtetR::PtetA::mCherry gfp+, revtetR::PtetA::mCherry Phmp::gfp, and revtetR::PtetA::yfp Phmp::mCherry. Six- to 8-week-old female C57BL/6 mice were obtained from Jackson Laboratories (Bar Harbor, ME). All animal studies were approved by the Institutional Animal Care and Use Committee of Johns Hopkins University. Mice were injected intravenously with 103 bacteria for all experiments. After 48 h, mice were treated with the indicated dose (4 mg/kg, equates to approximately 72 μg in 100 μL sterile PBS) of Dox or ATc or left untreated. At the indicated time points, either 4 h or 24 h posttreatment, mice were euthanized via both lethal dose of isoflurane and cervical dislocation, and spleens were removed, halved, and processed for histology or homogenized to quantify CFU.
Histology.
Harvested spleen halves were fixed in 4% PFA overnight at 4°C and were embedded using Sub Xero OCT compound (Mercedes Medical), frozen on dry ice, and stored at −80°C. Spleens were cross-sectioned into 10-μm sections using a cryostat microtome; one representative section was imaged per mouse. To visualize reporters, sections were thawed in PBS, stained with Hoechst at a 1:10,000 dilution, and washed in PBS, and coverslips were mounted using ProLong Gold (Life Technologies). Images were taken of all microcolonies within each section with a 63× oil immersion objective, using a Zeiss Axio Observer.Z1 (Zeiss) fluorescence microscope with Colibri.2 LED light source, an Apotome.2 (Zeiss) for optical sectioning, and an ORCA-R2 digital CCD camera (Hamamatsu).
Image analysis.
For in vitro experiments, Volocity image analysis software was used to select single bacterial cells in each image with the “find objects” function. The mean and sum signal intensity were calculated for each object (single cell) for mCherry, GFP, and YFP channels. Sum signal intensity values were used for revTetR reporter quantification. For in vivo experiments, either Volocity or Fiji image analysis software was used to select regions of interest at the centroid and periphery of each microcolony and to quantify the fluorescent signal intensity of each channel to generate relative signal intensities of fluorescent reporters. A small representative point (~0.01 pixels2) was selected at the centroid and periphery of each microcolony, which generated a “signal intensity” value at that point that correlates with the sum. Centroid bacteria were defined as surrounded only by other bacteria. Peripheral bacteria were defined based on contact with host cells (Fig. 4D). All data were analyzed and graphed, and all statistical analyses were performed, using GraphPad Prism 9.
ACKNOWLEDGMENTS
We thank the members of the Davis lab, who provided feedback and suggestions during the final steps of manuscript preparation. We also thank the members of the Pekosz and Klein lab for their helpful feedback throughout this project. This work was supported by NIAID grants 1K22AI123465-01 and 1R21AI154116-01A1 to K.M.D. R.E.B. is also supported by training grant 2T32AI007417-26 through NIAID. We declare no conflicts of interest.
Conceptualization: B.L., R.K.D., K.M.D.; formal analysis: B.L., R.E.B., K.M.D.; funding acquisition and supervision: K.M.D.; investigation: B.L., R.E.B., K.L.C., K.M.D.; methodology: B.L., R.E.B., K.M.D.; writing—original draft preparation: B.L., R.E.B., K.M.D.; writing—review & editing: B.L., R.E.B., K.L.C., R.K.D., K.M.D.
Footnotes
Supplemental material is available online only.
Contributor Information
Kimberly M. Davis, Email: kdavi140@jhu.edu.
Denise Monack, Stanford University.
REFERENCES
- 1.Balaban N, Merrin J, Chait R, Kowalik L, Leibler S. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622–1625. 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- 2.Goerke C, Wolz C. 2010. Adaptation of Staphylococcus aureus to the cystic fibrosis lung. Int J Med Microbiol 300:520–525. 10.1016/j.ijmm.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Harms A, Maisonneuve E, Gerdes K. 2016. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science 354:aaf4268. 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- 4.Lopatkin A, Stokes J, Zheng E, Yang J, Takahashi M, You L, Collins J. 2019. Bacterial metabolic state more accurately predicts antibiotic lethality than growth rate. Nat Microbiol 4:2109–2117. 10.1038/s41564-019-0536-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnoldini M, Vizcarra I, Peña-Miller R, Stocker N, Diard M, Vogel V, Beardmore R, Hardt W, Ackermann M. 2014. Bistable expression of virulence genes in Salmonella leads to formation of an antibiotic-tolerant subpopulation. PLoS Biol 12:e1001928. 10.1371/journal.pbio.1001928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan B, Singh P. 2011. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334:982–986. 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sánchez-Romero M, Casadesús J. 2014. Contribution of phenotypic heterogeneity to adaptive antibiotic resistance. Proc Natl Acad Sci USA 111:355–360. 10.1073/pnas.1316084111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claudi B, Spröte P, Chirkova A, Personnic N, Zankl J, Schürmann N, Schmidt A, Bumann D. 2014. Phenotypic variation of Salmonella in host tissues delays eradication by antimicrobial chemotherapy. Cell 158:722–733. 10.1016/j.cell.2014.06.045. [DOI] [PubMed] [Google Scholar]
- 9.Kaiser P, Regoes R, Dolowschiak T, Wotzka S, Lengefeld J, Slack E, Grant A, Ackermann M, Hardt W. 2014. Cecum lymph node dendritic cells harbor slow-growing bacteria phenotypically tolerant to antibiotic treatment. PLoS Biol 12:e1001793. 10.1371/journal.pbio.1001793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manina G, Dhar N, McKinney J. 2015. Stress and host immunity amplify Mycobacterium tuberculosis phenotypic heterogeneity and induce nongrowing metabolically active forms. Cell Host Microbe 17:32–46. 10.1016/j.chom.2014.11.016. [DOI] [PubMed] [Google Scholar]
- 11.Perry W, Spraggins J, Sheldon J, Grunenwald C, Heinrichs D, Cassat J, Skaar E, Caprioli R. 2019. Staphylococcus aureus exhibits heterogeneous siderophore production within the vertebrate host. Proc Natl Acad Sci USA 116:21980–21982. 10.1073/pnas.1913991116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunrath O, Bumann D. 2019. Host resistance factor SLC11A1 restricts Salmonella growth through magnesium deprivation. Science 366:995–999. 10.1126/science.aax7898. [DOI] [PubMed] [Google Scholar]
- 13.Rosenthal A, Qi Y, Hormoz S, Park J, Li S, Elowitz M. 2018. Metabolic interactions between dynamic bacterial subpopulations. Elife 7:e33099. 10.7554/eLife.33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stewart P, White B, Boegli L, Hamerly T, Williamson K, Franklin M, Bothner B, James G, Fisher S, Vital-Lopez F, Wallqvist A. 2019. Conceptual model of biofilm antibiotic tolerance that integrates phenomena of diffusion, metabolism, gene expression, and physiology. J Bacteriol 201:e00307-19. 10.1128/JB.00307-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton N, Schürmann N, Casse O, Steeb A, Claudi B, Zankl J, Schmidt A, Bumann D. 2014. Disparate impact of oxidative host defenses determines the fate of Salmonella during systemic infection of mice. Cell Host Microbe 15:72–83. 10.1016/j.chom.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Patel P, O’Hara B, Aunins E, Davis K. 2021. Modifying TIMER to generate a slow-folding DsRed derivative for optimal use in quickly-dividing bacteria. PLoS Pathog 17:e1009284. 10.1371/journal.ppat.1009284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis K, Mohammadi S, Isberg R. 2015. Community behavior and spatial regulation within a bacterial microcolony in deep tissue sites serves to protect against host attack. Cell Host Microbe 17:21–31. 10.1016/j.chom.2014.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rowe S, Wagner N, Li L, Beam J, Wilkinson A, Radlinski L, Zhang Q, Miao E, Conlon B. 2020. Reactive oxygen species induce antibiotic tolerance during systemic Staphylococcus aureus infection. Nat Microbiol 5:282–290. 10.1038/s41564-019-0627-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brauner A, Fridman O, Gefen O, Balaban N. 2016. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol 14:320–330. 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- 20.Fisher R, Gollan B, Helaine S. 2017. Persistent bacterial infections and persister cells. Nat Rev Microbiol 15:453–464. 10.1038/nrmicro.2017.42. [DOI] [PubMed] [Google Scholar]
- 21.Stapels D, Hill P, Westermann A, Fisher R, Thurston T, Saliba A, Blommestein I, Vogel J, Helaine S. 2018. Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362:1156–1160. 10.1126/science.aat7148. [DOI] [PubMed] [Google Scholar]
- 22.Raneses J, Ellison A, Liu B, Davis K. 2020. Subpopulations of stressed Y. pseudotuberculosis preferentially survive doxycycline treatment within host tissues. mBio 11:e00901-20. 10.1128/mBio.00901-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henry T, Brynildsen M. 2016. Development of persister-FACSeq: a method to massively parallelize quantification of persister physiology and its heterogeneity. Sci Rep 6:25100. 10.1038/srep25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luk C, Valenzuela C, Gil M, Swistak L, Bomme P, Chang Y, Mallet A, Enninga J. 2021. Salmonella enters a dormant state within human epithelial cells for persistent infection. PLoS Pathog 17:e1009550. 10.1371/journal.ppat.1009550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulte M, Olschewski K, Hensel M. 2021. Fluorescent protein-based reporters reveal stress response of intracellular Salmonella enterica at level of single bacterial cells. Cell Microbiol 23:e13293. 10.1111/cmi.13293. [DOI] [PubMed] [Google Scholar]
- 26.Zalis E, Nuxoll A, Manuse S, Clair G, Radlinski L, Conlon B, Adkins J, Lewis K. 2019. Stochastic variation in expression of the tricarboxylic acid cycle produces persister cells. mBio 10:e01930-19. 10.1128/mBio.01930-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Helaine S, Thompson J, Watson K, Liu M, Boyle C, Holden D. 2010. Dynamics of intracellular bacterial replication at the single cell level. Proc Natl Acad Sci USA 107:3746–3751. 10.1073/pnas.1000041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roostalu J, Jöers A, Luidalepp H, Kaldalu N, Tenson T. 2008. Cell division in Escherichia coli cultures monitored at single cell resolution. BMC Microbiol 8:68. 10.1186/1471-2180-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helaine S, Holden D. 2013. Heterogeneity of intracellular replication of bacterial pathogens. Curr Opin Microbiol 16:184–191. 10.1016/j.mib.2012.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Saliba A, Li L, Westermann A, Appenzeller S, Stapels D, Schulte L, Helaine S, Vogel J. 2017. Single-cell RNA-seq ties macrophage polarization to growth rate of intracellular Salmonella. Nat Microbiol 2. 10.1038/nmicrobiol.2016.206. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez R, Lane M, Wagner N, Weening E, Miller V. 2015. Dissemination of a highly virulent pathogen: tracking the early events that define infection. PLoS Pathog 11:e1004587. 10.1371/journal.ppat.1004587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McVicker G, Prajsnar T, Williams A, Wagner N, Boots M, Renshaw S, Foster S. 2014. Clonal expansion during Staphylococcus aureus infection dynamics reveals the effect of antibiotic intervention. PLoS Pathog 10:e1003959. 10.1371/journal.ppat.1003959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pollitt E, Szkuta P, Burns N, Foster S. 2018. Staphylococcus aureus infection dynamics. PLoS Pathog 14:e1007112. 10.1371/journal.ppat.1007112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Capron J, Delamarre J, Delcenserie R, Gineston J, Dupas J, Lorriaux A. 1981. Liver absess complicating Yersinia pseudotuberculosis ileitis. Gastroenterology 81:150–152. 10.1016/0016-5085(81)90665-X. [DOI] [PubMed] [Google Scholar]
- 35.Hubbert W, Petenyi C, Glasgow L, Uyeda C, Creighton S. 1971. Yersinia pseudotuberculosis infection in the United States. Septicemia, appendicitis, and mesenteric lymphadenitis. Am J Trop Med Hyg 20:679–684. 10.4269/ajtmh.1971.20.679. [DOI] [PubMed] [Google Scholar]
- 36.Paff J, Triplett D, Saari T. 1976. Clinical and laboratory aspects of Yersinia pseudotuberculosis infections, with a report of two cases. Am J Clin Pathol 66:101–110. 10.1093/ajcp/66.1.101. [DOI] [PubMed] [Google Scholar]
- 37.Barnes P, Bergman M, Mecsas J, Isberg R. 2006. Yersinia pseudotuberculosis disseminates directly from a replicating bacterial pool in the intestine. J Exp Med 203:1591–1601. 10.1084/jem.20060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peterson L, Philip N, DeLaney A, Wynosky-Dolfi M, Asklof K, Gray F, Choa R, Bjanes E, Buza E, Hu B, Dillon C, Green D, Berger S, Gough P, Bertin J, Brodsky I. 2017. RIPK1-dependent apoptosis bypasses pathogen blockage of innate signaling to promote immune defense. J Exp Med 214:3171–3182. 10.1084/jem.20170347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Khairallah C, Sheridan B, van der Velden A, Bliska J. 2018. CCR2+ inflammatory monocytes are recruited to Yersinia pseudotuberculosis pyogranulomas and dictate adaptive responses at the expense of innate immunity during oral infection. Infect Immun 86:e00782-17. 10.1128/IAI.00782-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Green E, Clark S, Crimmins G, Mack M, Kumamoto C, Mecsas J. 2016. Fis is essential for Yersinia pseudotuberculosis virulence and protects against reactive oxygen species produced by phagocytic cells during infection. PLoS Pathog 12:e1005898. 10.1371/journal.ppat.1005898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Resch M, Striegl H, Henssler E, Sevvana M, Egerer-Sieber C, Schiltz E, Hillen W, Muller Y. 2008. A protein functional leap: how a single mutation reverses the function of the transcription regulator TetR. Nucleic Acids Res 36:4390–4401. 10.1093/nar/gkn400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scholz O, Henssler E, Bail J, Schubert P, Bogdanska-Urbaniak J, Sopp S, Reich M, Wisshak S, Köstner M, Bertram R, Hillen W. 2004. Activity reversal of Tet repressor caused by single amino acid exchanges. Mol Microbiol 53:777–789. 10.1111/j.1365-2958.2004.04159.x. [DOI] [PubMed] [Google Scholar]
- 43.Almiron M, Link A, Furlong D, Kolter R. 1992. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev 6:2646–2654. 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 44.Nair S, Finkel S. 2004. Dps protects cells against multiple stresses during stationary phase. J Bacteriol 186:4192–4198. 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karas V, Westerlaken I, Meyer A. 2015. The DNA-binding protein from starved cells (Dps) utilizes dual functions to defend cells against multiple stresses. J Bacteriol 197:3206–3215. 10.1128/JB.00475-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Degenkolb J, Takahashi M, Ellestad G, Hillen W. 1991. Structural requirements of tetracycline-Tet repressor interaction: determination of equilibrium binding constants for tetracycline analogs with the Tet repressor. Antimicrob Agents Chemother 35:1591–1595. 10.1128/AAC.35.8.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rasmussen B, Noller H, Daubresse G, Oliva B, Misulovin Z, Rothstein D, Ellestad G, Gluzman Y, Tally F, Chopra I. 1991. Molecular basis of tetracycline action: identification of analogs whose primary target is not the bacterial ribosome. Antimicrob Agents Chemother 35:2306–2311. 10.1128/AAC.35.11.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jones-Carson J, Yahashiri A, Kim J, Liu L, Fitzsimmons L, Weiss D, Vázquez-Torres A. 2020. Nitric oxide disrupts bacterial cytokinesis by poisoning purine metabolism. Sci Adv 6:eaaz0260. 10.1126/sciadv.aaz0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barbier M, Damron F. 2016. Rainbow vectors for broad-range bacterial fluorescence labeling. PLoS One 11:e0146827. 10.1371/journal.pone.0146827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen Y, Chen Y, Wu J, Shaner N, Campbell R. 2017. Engineering of mCherry variants with long Stokes shift, red-shifted fluorescence, and low cytotoxicity. PLoS One 12:e0171257. 10.1371/journal.pone.0171257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Davis K, Krupp J, Clark S, Isberg R. 2019. Iron-sulfur cluster repair contributes to Yersinia pseudotuberculosis survival within deep tissues. Infect Immun 87:e00533-19. 10.1128/IAI.00533-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson R, Davis K. 2020. Yersinia pseudotuberculosis: cultivation, storage, and methods for introducing DNA. Curr Protoc Microbiol 59:e122. 10.1002/cpmc.122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download iai.00167-22-s0001.pdf, PDF file, 0.3 MB (322.7KB, pdf)