ABSTRACT
Wilms’ tumor is the most common type of renal tumor in children. MicroRNAs (miRNAs) are small noncoding RNAs that play crucial regulatory roles in tumorigenesis. We aimed to study the expression profile and function of miR-27a-5p in Wilms’ tumor. miR-27a-5p expression was downregulated in human Wilms’ tumor tissues. Functionally, overexpression of miR-27a-5p promoted cell apoptosis of Wilms’ tumor cells. Furthermore, upregulated miR-27a-5p delayed xenograft Wilms’ tumor tumorigenesis in vivo. Bioinformatics analysis predicted that miR-27a-5p directly targeted the 3′-untranslated region (3′-UTR) of PBOV1, and luciferase reporter assay confirmed the interaction between miR-27a-5p and PBOV1. The function of PBOV1 in Wilms’ tumor was evaluated in vitro, and knockdown of PBOV1 dampened cell migration. In addition, overexpression of PBOV1 antagonized the tumor-suppressive effect of miR-27a-5p in Wilms’ tumor cells. Collectively, our findings reveal the regulatory axis of miR-27a-5p/PBOV1 in Wilms’ tumor, and miR-27a-5p might serve as a novel therapeutic target in Wilms’ tumor.
KEYWORDS: microRNA-27a-5p, Wilms’ tumor cell, PBOV1, biomarker
INTRODUCTION
Wilms’ tumor is the most common renal carcinoma, mostly occurring in children under age 5 (1). The survival of Wilms’ tumor patients has been significantly improved from less than 30% to more than 90% in the last decade due to modern therapeutic strategies and technology (2, 3). However, current therapies, such as radiotherapy and chemotherapy, have serious side effects with poor efficacy in patients with tumor metastasis (4–6). Meanwhile, large-scale next-generation sequencing has identified multiple mutations of the candidate driver in Wilms’ tumor (7). Thus, it is important to further our understanding of the molecular mechanisms of Wilms’ tumor oncogenesis and metastasis and develop new treatment strategies.
MicroRNAs (miRNAs) are small noncoding RNAs that play crucial regulatory roles in various biological processes, including tumorigenesis (8, 9). miRNAs posttranscriptionally regulate their target gene expression via binding to the 3′-untranslated region (3′-UTR) of target mRNAs (8). The expression and function of miRNAs in Wilms’ tumor have also been investigated (10). miRNA microarray profiling results from 36 Wilms’ tumors of different subtypes and normal kidney tissues demonstrated that various miRNAs were dysregulated in blastema Wilms’ tumors and a regressive subtype of Wilms’ tumor (11). Those miRNAs function as oncogenes or tumor suppressors or mediate the chemosensitivity in Wilms’ tumor (11). Watson et al. also reported that miRNAs could predict the chemoresponsiveness in Wilms’ tumor blastema, with 29 miRNAs identified to be markedly differentially expressed in posttreatment high-risk and intermediate-risk patients (12). Among their identified miRNAs, miR-27a-5p was reported as a significantly downregulated miRNA in resistant blastemal cells from high-risk cases compared to those from intermediate-risk cases. The downregulation of miR-27a-5p has been reported to increase the expression of P-glycoprotein, which further contributes to chemoresistance and relapse in leukemia patients (13). However, the detailed functional role and molecular mechanisms of miR-27 in Wilms’ tumor are not fully understood.
In this study, we evaluated the expression profile and function of miR-27a-5p in Wilms’ tumor and cell lines. The results demonstrated that miR-27a-5p was downregulated in human Wilms’ tumor tissues and cells. Overexpression of miR-27a-5p inhibited cell proliferation, cell migration, and invasion and promoted cell apoptosis. Moreover, our data revealed that miR-27a-5p suppressed tumorigenesis via negatively regulating PBOV1. In summary, our findings suggest that miR-27a-5p might serve as a novel therapeutic target in Wilms’ tumor.
RESULTS
miR-27a-5p is downregulated in human Wilms’ tumor tissues and cells.
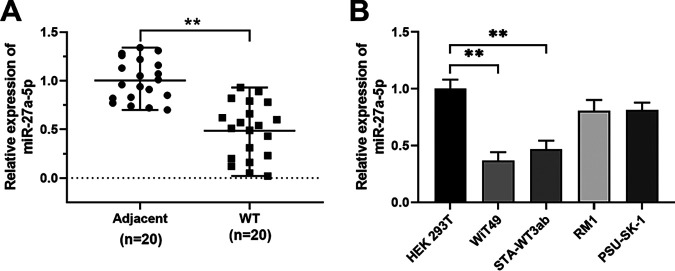
To determine the expression of miR-27a-5p in Wilms’ tumor, we performed quantitative PCR (qPCR) to examine the miR-27a-5p expression in 20 pairs of Wilms’ tumor tissues and adjacent control tissues (Fig. 1A). miR-27a-5p expression was significantly decreased in human Wilms’ tumor tissues (Fig. 1A). Consistently, miR-27a-5p expression was measured in different Wilms’ tumor cell lines, and the results showed that Wilms’ tumor cell lines (WiT49 and STA-WT3ab) had markedly lower levels of miR-27a-5p than that in control cell line HEK 293T (Fig. 1B).
FIG 1.
miR-27a-5p expression in Wilms’ tumor tissues and cell lines. (A) Relative expression of miR-27a-5p was determined in 20 pairs of Wilms’ tumor tissues and adjacent normal control tissues by qPCR. (B) Relative expression of miR-27a-5p was determined in Wilms’ tumor cell lines (WiT49, STA-WT3ab, RM1, and PSU-SK-1) and control cell line HEK 293T by qPCR. **, P < 0.01.
miR-27a-5p inhibits proliferation, migration, and invasion and promotes apoptosis in Wilms’ tumor cells.
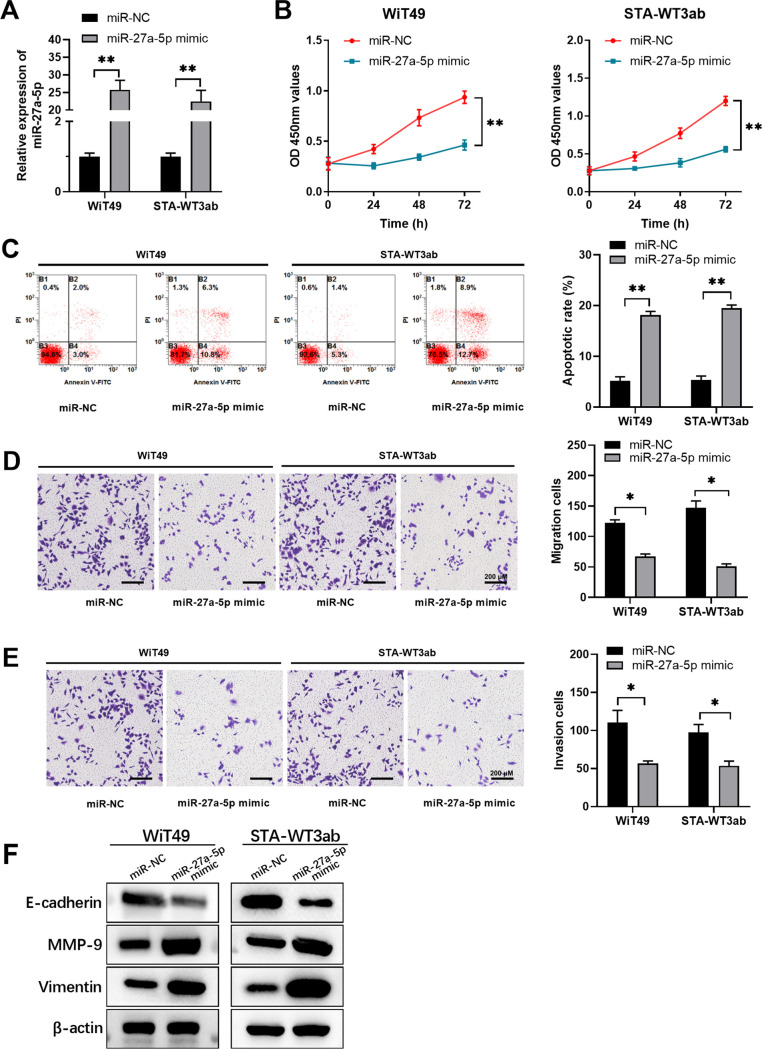
Then, we performed functional assays to assess the role of miR-27a-5p in Wilms’ tumor cells. WiT49 and STA-WT3ab cells, which had relatively underexpressed miR-27a-5p, were transfected with miR-27a-5p mimic to overexpress miR-27a-5p (Fig. 2A). Overexpression of miR-27a-5p remarkably repressed cell proliferation (Fig. 2B) and promoted cell apoptosis (Fig. 2C) in WiT49 and STA-WT3ab cells. Furthermore, compared with the negative control, miR-27a-5p mimic transfection significantly suppressed cell migration and invasion (Fig. 2D and E). To validate the suppressive function of miR-27a-5p overexpression on migration and invasion, we further examined the expression level of migration- and invasion-related proteins, such as MMP-9, vimentin, and E-cadherin. We found that after transfection with miR-27a-5p mimics, MMP-9 and vimentin were increased while E-cadherin was decreased (Fig. 2F), which indicates that the miR-27a-5p overexpression could decrease migration and invasion capability of both WiT49 and STA-WT3ab cells. These findings suggest that miR-27a-5p negatively regulates Wilms’ tumor development.
FIG 2.
miR-27a-5p inhibits proliferation, migration, and invasion and promotes apoptosis in Wilms’ tumor cells. WiT49 or STA-WT3ab cells were transfected with miRNA negative control (miR-NC) or miR-27a-5p mimic. (A) The relative expression of miR-27a-5p in WiT49 or STA-WT3ab cells was analyzed by qPCR. (B) Cell proliferation was analyzed at indicated time points by CCK8 assay. (C) Cell apoptosis was analyzed by annexin V/PI staining and flow cytometry. (D, E) Cell migration and invasion were assessed by transwell assay. (F) Migration- and invasion-related proteins were examined by Western blotting. *, P < 0.05; **, P < 0.01 versus miR-NC.
miR-27a-5p inhibits oncogenesis and metastasis of Wilms’ tumor in vivo.
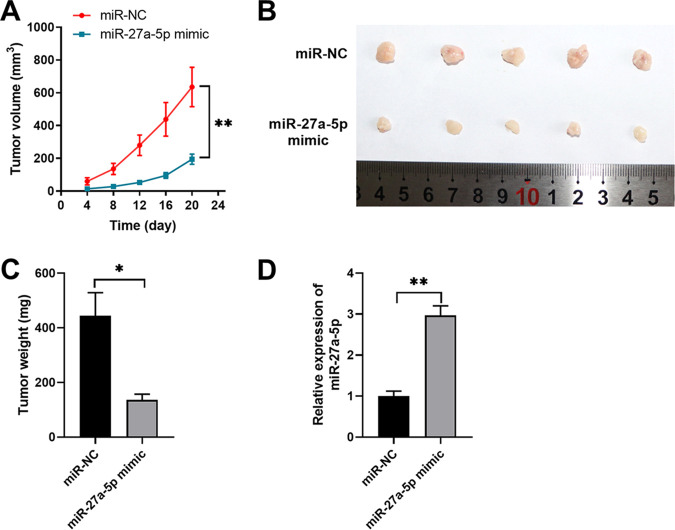
To verify the tumor suppressor role of miR-27a-5p, an in vivo xenograft model was established in nude mice by subcutaneous injection of WiT49 cells transfected with miR-27a-5p mimic or control miRNA. The results demonstrated the tumor inhibitory function of miR-27a-5p in vivo. miR-27a-5p overexpression significantly inhibited Wilms’ tumor development (Fig. 3A). Tumors developed from the miR-27a-5p mimic group showed a much smaller size with a lower tumor weight compared with those developed from control cells (Fig. 3B and C). The upregulated miR-27a-5p expression was confirmed in tumors from the miR-27a-5p mimic group (Fig. 3D).
FIG 3.
miR-27a-5p inhibits Wilms’ xenograft tumor growth in vivo. WiT49 cells were transfected with negative control miRNA (miR-NC) or miR-27a-5p mimic and then inoculated into BALB/c nude mice to develop the xenograft Wilms’ tumor. (A) Tumor growth was monitored and measured at indicated time points. (B) Tumors were extracted and recorded on day 22. (C) Tumor weights of xenograft were analyzed. (D) The relative expression of miR-27a-5p in xenograft tumors was analyzed by qPCR. *, P < 0.05; **, P < 0.01 versus miR-NC.
PBOV1 is a direct target of miR-27a-5p in Wilms’ tumor cells.
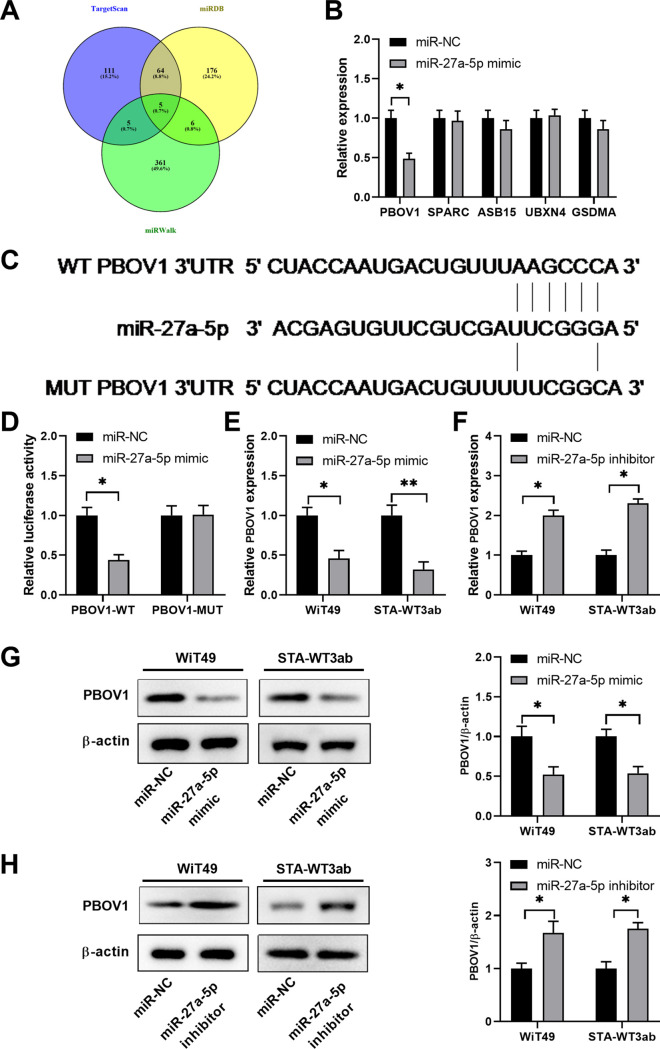
To explore the potential targets regulated by miR-27a-5p, we performed bioinformatics analysis using different online databases (TargetScan, miRDB, and miRWalk). As shown in Fig. 4A, five genes, including PBOV1, SPARC, ASB15, UBXN4, and GSDMA, were predicted to be the potential targets of miR-27a-5p. WiT49 cells were transfected with miRNA negative control (miR-NC) or miR-27a-5p mimic, and the relative expression of these potential targets was analyzed. PBOV1 was markedly downregulated by miR-27a-5p mimic (Fig. 4B). In addition, miR-27a-5p had the putative binding sequences against 3′-UTR of the PBOV1 gene (Fig. 4C). Luciferase reporter assay further validated the interaction between miR-27a-5p and wild-type (WT) 3′-UTR of PBOV1, as miR-27a-5p markedly inhibited the luciferase activity of reporter vector containing WT 3′-UTR of PBOV1 (Fig. 4D). Moreover, we demonstrated that miR-27a-5p mimics significantly inhibited the mRNA and protein levels of PBOV1 in WiT49 or STA-WT3ab cells (Fig. 4E and G). In contrast, inhibition of miR-27a-5p enhanced the expression of PBOV1 in WiT49 or STA-WT3ab cells (Fig. 4F and H).
FIG 4.
PBOV1 is a direct target of miR-27a-5p in Wilms’ tumor cells. (A) Bioinformatics analysis was performed to predict the potential targets of miR-27a-5p using online databases TargetScan, miRDB, and miRWalk. (B) WiT49 cells were transfected with miR-NC or miR-27a-5p mimic. The relative expression of PBOV1, SPARC, ASB15, UBXN4, and GSDMA was analyzed by qPCR 48 h later. (C) The predicted binding sequences between miR-27a-5p and WT or mutated 3′-UTR of PBOV1. (D) WiT49 cells were transfected with miR-NC or miR-27a-5p, together with luciferase reporter vectors containing WT or mutated 3′-UTR of PBOV1. The relative luciferase activity was analyzed 48 h later. (E) WiT49 or STA-WT3ab cells were transfected with miR-NC or miR-27a-5p mimic. The relative expression of PBOV1 mRNA was analyzed by qPCR 48 h later. (F) WiT49 or STA-WT3ab cells were transfected with miR-NC or miR-27a-5p inhibitor. The relative expression of PBOV1 mRNA was analyzed by qPCR 48 h later. (G) WiT49 or STA-WT3ab cells were transfected with miR-NC or miR-27a-5p mimic. The relative expression of PBOV1 protein was analyzed by Western blotting 48 h later. (H) WiT49 or STA-WT3ab cells were transfected with miR-NC or miR-27a-5p inhibitor. The relative expression of PBOV1 protein was analyzed by Western blotting 48 h later. *, P < 0.05; **, P < 0.01 versus miR-NC.
Knockdown of PBOV1 suppresses cell migration and invasion and promotes cell apoptosis of Wilms’ tumor cells.
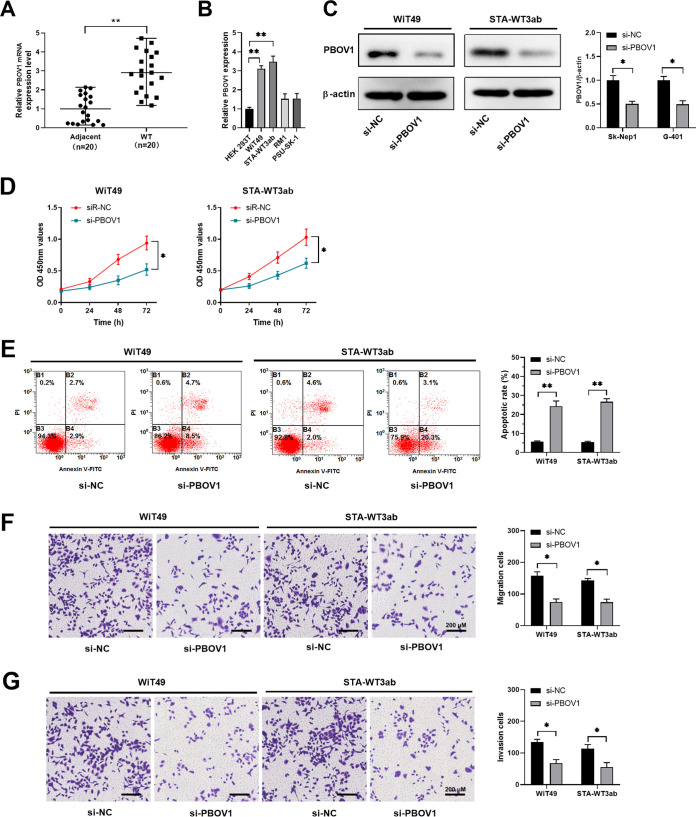
We found that Wilms’ tumor tissues had a much higher expression level of PBOV1 compared with adjacent normal tissues (Fig. 5A). Similarly, we detected higher expression of PBOV1 in Wilms’ tumor cells (WiT49 and STA-WT3ab) compared with that in control HEK 293T cells (Fig. 5B). To study the function of PBOV1, small interfering RNA (siRNA)-targeting PBOV1 was used to suppress the expression of PBOV1 in WiT49 or STA-WT3ab cells. The knockdown efficiency was evaluated by Western blotting (Fig. 5C). Functionally, we demonstrated that knockdown of PBOV1 suppressed cell proliferation and enhanced apoptosis in WiT49 or STA-WT3ab cells (Fig. 5D and E), while transwell assay revealed that inhibition of PBOV1 decreased the capability of cell migration and invasion in WiT49 or STA-WT3ab cells (Fig. 5F and G). Thus, the results suggested that PBOV1 acted as an oncogene in Wilms’ tumor.
FIG 5.
Knockdown of PBOV1 suppresses cell migration and invasion and promotes cell apoptosis of Wilms’ tumor cells. (A) The relative expression of PBOV1 mRNA in 20 pairs of Wilms’ tumor tissues and adjacent normal tissues was analyzed by qPCR. (B) The relative expression of PBOV1 mRNA in Wilms’ tumor cell lines (WiT49, STA-WT3ab, RM1, and PSU-SK-1) and control cell line HEK 293T was analyzed by qPCR. (C to G) WiT49 or STA-WT3ab cells were transfected with negative control (si-NC) or si-PBOV1. (C) The protein level of PBOV1 was analyzed by Western blotting 48 h posttransfection. (D) Cell proliferation was analyzed by CCK8 assay. (E) Cell apoptosis was analyzed by annexin V/PI staining and flow cytometry. (F, G) Cell migration and invasion were assessed by transwell assay. *, P < 0.05; **, P < 0.01 versus si-NC.
Overexpression of PBOV1 antagonizes the tumor suppressor function of miR-27a-5p in Wilms’ tumor cells.
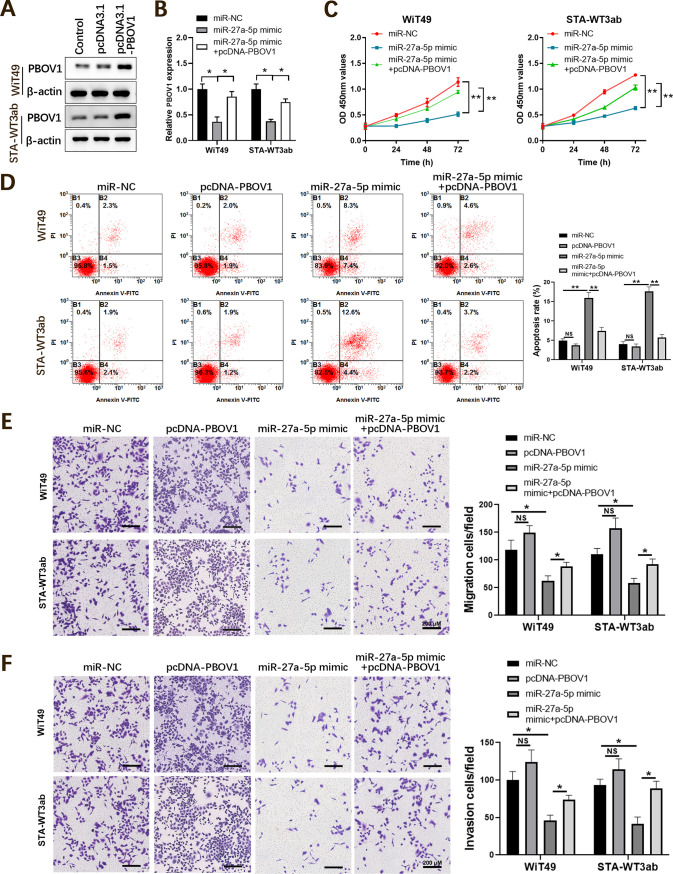
To validate the functional relationship between miR-27a-5p and PBOV1, rescue experiments were performed. A Western blotting assay confirmed that PBOV1 was markedly increased in WiT49 or STA-WT3ab cells (Fig. 6A). Following this, WiT49 or STA-WT3ab cells were cotransfected with miR-27a-5p mimic and PBOV1, thereby divided into the miR-NC, miR-27a-5p mimic, or miR-27a-5p mimic+pcDNA-PBOV1 group. Whereas miR-27a-5p overexpression significantly decreased the expression of PBOV1, overexpression of PBOV1 together with miR-27a-5p rescued PBOV1 expression in Wilms’ tumor cells (Fig. 6B). Functionally, miR-27a-5p mimic inhibited cell proliferation, and PBOV1 overexpression abrogated the inhibitory effect of miR-27a-5p (Fig. 6C). Conversely, miR-27a-5p enhanced cell apoptosis of WiT49 or STA-WT3ab cells. Overexpression of PBOV1 together with miR-27a-5p mimic showed reduced cell apoptosis and was comparable to that in cells transfected with the miR-NC control (Fig. 6D). In addition, overexpression of PBOV1 antagonized the inhibitory effect on cell migration and invasion of miR-27a-5p in WiT49 or STA-WT3ab cells (Fig. 6E and F). Taken together, the data suggested miR-27a-5p exerted its tumor-suppressive role in Wilms’ tumor cells via downregulating PBOV1.
FIG 6.
Overexpression of PBOV1 antagonizes the tumor suppressor effect of miR-27a-5p in Wilms’ tumor cells. WiT49 or STA-WT3ab cells were transfected with negative control (miR-NC), miR-27a-5p mimic, or miR-27a-5p mimic + pcDNA-PBOV1. (A) Confirmation of overexpression of PBOV1 in WiT49 or STA-WT3ab cells by Western blotting. (B) The relative mRNA level of PBOV1 in WiT49 or STA-WT3ab cells was analyzed by qPCR 48 h posttransfection. (C) Cell proliferation was analyzed by CCK8 assay at indicated time points. (D) Cell apoptosis was analyzed by annexin V/PI staining and flow cytometry. (E, F) Cell migration and invasion were assessed by transwell assay. *, P < 0.05; **, P < 0.01.
DISCUSSION
Studies have shown that miRNAs play essential roles in tumorigenesis and metastasis (14, 15). miRNAs also exert the regulatory function in Wilms’ tumor and could be used as diagnostic markers and predictors for chemoresponsiveness (12, 16). Here, we reported that miR-27a-5p was underexpressed in Wilms’ tumor, and miR-27a-5p overexpression suppressed Wilms’ tumor cell growth and metastasis. PBOV1 was demonstrated to be the direct target of miR-27a-5p, and overexpression of PBOV1 abrogated the tumor-suppressive function of miR-27a-5p. Thus, our findings suggest a potential therapeutic target of miR-27a-5p in Wilms’ tumor patients.
miR-27a-5p has been reported to suppress the tumorigenesis of multiple cancers, including prostate cancer, small-cell lung cancer, and cervical adenocarcinoma (17–19). Networks analysis showed that miR-27a-5p was dysregulated in Wilms’ tumor (20). In another study, Watson et al. showed that downregulation of miR-27a was found in high-risk Wilms’ tumors, which might be a predictor of chemoresponsiveness (12). We confirmed that miR-27a-5p was underexpressed in Wilms’ tumor. Consistent with the published data, the tumor-suppressive function of miR-27a-5p was validated both in vitro and in vivo, as overexpression of miR-27a-5p inhibited the growth and metastasis of Wilms’ tumor cells and promoted cell apoptosis.
Bioinformatics analysis predicted multiple potential targets of miR-27a-5p, while we verified that miR-27a-5p mimics specifically inhibited the expression of PBOV1. PBOV1 was first identified as a human tumor-specific gene and is associated with the clinical outcome of cancer patients (21, 22). The high-expression level of PBOV1 promoted G1/S transition and enhanced cell proliferation in prostate cancer (23). The function of PBOV1 was also elucidated in hepatocellular carcinoma (HCC), and overexpression of PBOV1 was correlated with poor prognosis of HCC patients, indicating PBOV1 as a prognostic biomarker of HCC (24). The subcellular localization and the exact regulatory mechanism of PBOV1 is currently unclear. Further investigations regarding the mechanism of PBOV1-related tumorigenesis are required. For example, analysis of gene expression data sets of prostate cancer and breast cancer revealed that PBOV1 expression was highly correlated to the expression level of sonic hedgehog pathway, indicating that the hedgehog pathway could be one of the regulators of PBOV1 expression in those cancer types (21). Zhang et al. demonstrated that PBOV1 was regulated by miR-203 in fracture healing (25). In rheumatoid arthritis, monocyte differentiation was controlled by the lncRNA NTT/PBOV1 axis (26). In this study, we reported for the first time that PBOV1 was directly regulated by miR-27a-5p and PBOV1 overexpression antagonized the function of miR-27a-5p.
Though we demonstrated that the miR-27a-5p/PBOV1 axis regulated Wilms’ tumor development and progression with both in vitro and in vivo evidence, there are several limitations in this study. First, it is of great importance to further study whether high expression of miR-27a-5p mediated the chemoresistance in Wilms’ tumor or not. Second, whether there are other potential miRNAs involved in PBOV1 regulation remains unknown. Additionally, the signaling pathways involved in the miR-27a-5p/PBOV1 axis in Wilms’ tumor need further investigation.
In conclusion, miR-27a-5p acts as a tumor suppressor via negatively regulating PBOV1 in Wilms’ cancer. Our data suggest that miR-27a-5p/PBOV1 might be utilized as a novel therapeutic target in Wilms’ tumor.
MATERIALS AND METHODS
Patient specimens.
Twenty pairs of Wilms’ tumor and adjacent normal kidney tissues were obtained from patients who underwent surgery at the Second Affiliated Hospital of Xi’an Jiaotong University (Xibei Hospital) between March 2019 and September 2019. All Wilms’ tumor tissues were histopathologically confirmed and classified based on typing from the American National Wilms’ Tumor Study 5 and TNM staging system. Tissues were snap-frozen and stored in liquid nitrogen until further analysis. The study was reviewed and approved by the Ethics Committee of Xibei Hospital, and all patients provided written informed consent. The ethics committee of the Second Affiliated Hospital of Xi'an Jiaotong University approved the study. The investigation conforms to the principles outlined in the Declaration of Helsinki, and written informed consent was obtained from all participants.
Cell culture.
Wilms’ tumor-derived renal cancer cell lines WiT49, STA-WT3ab, RM1, and PSU-SK-1 and control HEK 293T cells were purchased from the American Type Culture Collection (ATCC) (USA). Cryopreserved cells were recovered from liquid nitrogen using a cell recovery kit (EXINNO, China) and cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Gibco, USA) and 1% penicillin/streptomycin (Gibco, USA) at 37°C in a 5% CO2 incubator.
Transfection.
Transfection was conducted using Lipofectamine 2000 (Invitrogen, USA) following manufacturer’s instructions. miR-27a-5p mimic or negative control (NC), miR-27a-5p inhibitor, and NC were obtained from GenePharma (Shanghai, China). PBOV1 siRNA and scramble control were purchased from Rubibio Company (Guangzhou, China). pcDNA 3.1-PBOV1 was obtained from GeneCopoeia (MD, USA).
Lentivirus infection and generation of stable cell line.
To generate stable cell line overexpressing miR-27a-5p or miR-NC, WiT49 cells were infected with lentivirus-miR-27a-5p mimic or lentivirus-miR-NC lentivirus particles. Lentivirus-miR-NC vector or lentivirus-miR-27a-5p mimic vector, together with the helper plasmids pHelper 1.0 (Gag and Pol) and pHelper 2.0 (VSVG plasmid) were transiently cotransfected into HEK 293T cells using Lipofectamine 2000 (Invitrogen, USA) to package lentivirus particles, respectively. When WiT49 cells were grown in the logarithmic growth phase, cells were infected with lentivirus particles with a multiplicity of infection of 70. Then, limited dilution method was used, and cells were cultured with puromycin (2 μg/mL) and screened for 14 days to select the stable cell lines overexpressing miR-27a-5p or miR-NC.
Reverse transcriptase quantitative PCR.
To analyze the relative expression levels of miR-27a-5p and PBOV1, RNA was purified from Wilms’ tumor tissues or cultured cells using a total RNA extraction kit (EXINNO) and then reverse transcribed into cDNA using miR-X miRNA synthesis kit (Clontech, USA) or SuperScript Reverse Transcriptase III (Invitrogen, USA) following the manufacturers’ instructions. Quantitative PCR was conducted using the SYBR green mix (TaKaRa, Japan) on a Bio-Rad real-time CFX96 system. U6 snRNA and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were used as the internal control. The gene expression was calculated using the 2−ΔΔCT method. The following primers were used: hsa-miR-27a-5p, 5′-AGGGCTTAGCTGCTTGTGAGC-3′; hsa-PBOV1-Forward, 5′-AGTTCGAGACCAGCCTGACCAG-3′, hsa-PBOV1-Reverse, 5′-TTCAAGCAATTCTCCGCCTCAGC-3′; hsa-SPARC-Forward, 5′-CAAGAAGCCCTGCCTGATGAGAC-3′, hsa-SPARC-Reverse, 5′-TTCCGCCACCACCTCCTCTTC-3′; hsa-GSDMA-Forward, 5′-ACGCTGCTGGATGTGCTTGAG-3′, hsa-GSDMA-Reverse, 5′-AGAGCCCTGCCGTTCCCTTC-3′; hsa-ASB15-Forward, 5′-GCCTGGACATTAGGTTTGACC-3′, hsa-ASB15-Reverse, 5′-GTCAGGGGAGCCAGATAAGC-3′; and hsa-UBXN4-Forward, 5′-CCTTCTGATGCTCCTCTAGAAG-3′, hsa-UBXN4-Reverse, 5′-GGAAACATGGTTGCTAACGAAA-3′.
Western blotting.
Protein was prepared from tumor tissues or cultured cells using radioimmunoprecipitation assay (RIPA) buffer (Beyotime, China) and quantified with a bicinchoninic acid (BCA) protein quantification kit (EXINNO). Protein samples were then diluted in loading buffer (EXINNO), and 20 μg protein was loaded into each well and separated by 10% sodium dodecyl sulfate-polyacrylamide gel followed by transfer onto a polyvinylidene difluoride membrane (Millipore, USA). The membranes were incubated with primary antibody against PBOV1 (Abcam; ab216045) and β-actin (Abcam; ab8227), followed by incubation with horseradish peroxidase conjugated secondary antibody. Protein bands were visualized using an ECL detection kit (AccuRef Scientific, China). β-actin was used as a loading control.
Luciferase reporter assay.
A 214-bp fragment containing the predicted interaction site of the 3′-UTR of wild-type (WT) PBOV1 was synthesized by Tsingke Biotechnology Co., Ltd. (Xi’an, Shaanxi, China), and the mutated fragment was also synthesized. Luciferase reporter vectors containing WT or mutated 3′-UTR of PBOV1 were constructed into the backbone vector pGL3-Luc downstream of the renilla luciferase reporter gene. WiT49 cells were cotransfected with reporter vectors and miR-NC or miR-27a-5p mimic Lipofectamine 3000 (Thermo Fisher, USA) according to the manufacturer’s instructions. After 48 h, renilla and firefly luciferase activity was measured with a Dual-Luciferase reporter assay kit (Promega, USA).
CCK-8 assay.
Transfected WiT49 or STA-WT3ab cells were seeded in 96-well plates at a density of 4,000 cells/well for 24 h. Ten microliters of Cell Counting Kit-8 (CCK-8, Dojindo, Japan) reagent was added into each well at 24, 48, and 72 h posttransfection. After incubating at 37°C and 5% CO2 for 2 h, the absorbance (450 nm) was recorded using a plate reader (Pulang New Technology, Beijing, China).
Cell apoptosis assay.
Transfected WiT49 or STA-WT3ab cells were collected and stained with a BD apoptosis analysis kit (BD Bioscience, USA) following the manufacturer’s instructions. After staining, cells were analyzed using the CytoFlex flow cytometry machine (Beckman Coulter) and annexin V + propidium iodide (PI)—cells were defined as apoptotic cells. The results were analyzed by Flowjo v10.7.1 (TreeStar, USA).
Transwell assay.
Transfected WiT49 or STA-WT3ab cells were resuspended in serum-free medium and seeded to the top chamber (Corning, USA) with or without precoating of Matrigel (BD Bioscience, USA). Complete medium with 10% FBS was added to the bottom chamber. After culture for 24 h at 37°C and 5% CO2, the invaded or migrated cells at the bottom side of transwell membrane were fixed with 4% paraformaldehyde for 10 min and stained with 0.1% crystal violet (Solarbio, China) at room temperature for 30 min. After washing 3 times with phosphate-buffered saline (PBS), the number of migrated cells were counted under a phrase contrast microscope (Nikon, Japan).
Xenograft tumor model.
Ten male BALB/c nude mice (5 to 6 weeks old) were purchased from SLAC animal center (Shanghai, China) and randomly divided into 2 groups. WiT49 cells with stable overexpressing miR-27a-5p mimic or miR-NC were inoculated subcutaneously into nude mice, respectively. Tumor growth was recorded at indicated time points and calculated as follows: volume = length × width2/2. Mice were sacrificed and analyzed on day 22. The experiments were approved by the Animal Care Committee of Xibei Hospital.
Statistical analysis.
Results were shown as mean ± standard deviation (SD) from three independent experiments and analyzed by using GraphPad Prism v7.0 (Prism, USA). Student's t test and one-way analysis of variance (ANOVA) were conducted where necessary. A P value of <0.05 was defined as statistically significant.
Data availability.
The data sets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
ACKNOWLEDGMENTS
Zheng-Tuan Guo and Qiang Yu conceived and designed these experiments. Chunlin Miao, Wenan Ge, and Peng Li performed these experiments. Qiang Yu and Wenan Ge analyzed and interpreted the data. Chunlin Miao and Peng Li wrote the manuscript. Zheng-Tuan Guo and Qiang Yu revised the manuscript. All authors read and approved the final manuscript.
We declare that we have no competing interests.
REFERENCES
- 1.Rivera MN, Haber DA. 2005. Wilms' tumour: connecting tumorigenesis and organ development in the kidney. Nat Rev Cancer 5:699–712. 10.1038/nrc1696. [DOI] [PubMed] [Google Scholar]
- 2.Szychot E, Apps J, Pritchard-Jones K. 2014. Wilms' tumor: biology, diagnosis and treatment. Transl Pediatr 3:12–24. 10.3978/j.issn.2224-4336.2014.01.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopes RI, Lorenzo A. 2017. Recent advances in the management of Wilms' tumor. F1000Res 6:670. 10.12688/f1000research.10760.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pritchard-Jones K. 2002. Controversies and advances in the management of Wilms' tumour. Arch Dis Child 87:241–244. 10.1136/adc.87.3.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akakin A, Yılmaz B, Ekşi MŞ, Yapıcıer Ö, Kılıç T. 2016. Relapsed Wilms' tumor with multiple brain metastasis. Korean J Pediatr 59:S96–S98. 10.3345/kjp.2016.59.11.S96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrlich PF, Ferrer FA, Ritchey ML, Anderson JR, Green DM, Grundy PE, Dome JS, Kalapurakal JA, Perlman EJ, Shamberger RC. 2009. Hepatic metastasis at diagnosis in patients with Wilms tumor is not an independent adverse prognostic factor for stage IV Wilms tumor: a report from the Children's Oncology Group/National Wilms Tumor Study Group. Ann Surg 250:642–648. 10.1097/SLA.0b013e3181b76f20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gadd S, Huff V, Walz AL, Ooms AHAG, Armstrong AE, Gerhard DS, Smith MA, Auvil JMG, Meerzaman D, Chen Q-R, Hsu CH, Yan C, Nguyen C, Hu Y, Hermida LC, Davidsen T, Gesuwan P, Ma Y, Zong Z, Mungall AJ, Moore RA, Marra MA, Dome JS, Mullighan CG, Ma J, Wheeler DA, Hampton OA, Ross N, Gastier-Foster JM, Arold ST, Perlman EJ. 2017. A Children's Oncology Group and TARGET initiative exploring the genetic landscape of Wilms tumor. Nat Genet 49:1487–1494. 10.1038/ng.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.O'Brien J, Hayder H, Zayed Y, Peng C. 2018. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 9:402. 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng Y, Croce CM. 2016. The role of microRNAs in human cancer. Signal Transduct Target Ther 1:15004. 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu X, Li Z, Chan MTV, Wu WKK. 2016. The roles of microRNAs in Wilms' tumors. Tumor Biol 37:1445–1450. 10.1007/s13277-015-4514-8. [DOI] [PubMed] [Google Scholar]
- 11.Ludwig N, Werner TV, Backes C, Trampert P, Gessler M, Keller A, Lenhof H-P, Graf N, Meese E. 2016. Combining miRNA and mRNA expression profiles in Wilms tumor subtypes. Int J Mol Sci 17:475. 10.3390/ijms17040475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watson JA, Bryan K, Williams R, Popov S, Vujanic G, Coulomb A, Boccon-Gibod L, Graf N, Pritchard-Jones K, O'Sullivan M. 2013. miRNA profiles as a predictor of chemoresponsiveness in Wilms' tumor blastema. PLoS One 8:e53417. 10.1371/journal.pone.0053417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Che X, Jian F, Chen C, Liu C, Liu G, Feng W. 2020. PCOS serum-derived exosomal miR-27a-5p stimulates endometrial cancer cells migration and invasion. J Mol Endocrinol 64:1–12. 10.1530/JME-19-0159. [DOI] [PubMed] [Google Scholar]
- 14.Wegert J, Ishaque N, Vardapour R, Geörg C, Gu Z, Bieg M, Ziegler B, Bausenwein S, Nourkami N, Ludwig N, Keller A, Grimm C, Kneitz S, Williams RD, Chagtai T, Pritchard-Jones K, van Sluis P, Volckmann R, Koster J, Versteeg R, Acha T, O'Sullivan MJ, Bode PK, Niggli F, Tytgat GA, van Tinteren H, van den Heuvel-Eibrink MM, Meese E, Vokuhl C, Leuschner I, Graf N, Eils R, Pfister SM, Kool M, Gessler M. 2015. Mutations in the SIX1/2 pathway and the DROSHA/DGCR8 miRNA microprocessor complex underlie high-risk blastemal type Wilms tumors. Cancer Cell 27:298–311. 10.1016/j.ccell.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 15.Si W, Shen J, Zheng H, Fan W. 2019. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics 11:25. 10.1186/s13148-018-0587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitt J, Backes C, Nourkami-Tutdibi N, Leidinger P, Deutscher S, Beier M, Gessler M, Graf N, Lenhof H-P, Keller A, Meese E. 2012. Treatment-independent miRNA signature in blood of Wilms tumor patients. BMC Genomics 13:379. 10.1186/1471-2164-13-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizuno K, Mataki H, Arai T, Okato A, Kamikawaji K, Kumamoto T, Hiraki T, Hatanaka K, Inoue H, Seki N. 2017. The microRNA expression signature of small cell lung cancer: tumor suppressors of miR-27a-5p and miR-34b-3p and their targeted oncogenes. J Hum Genet 62:671–678. 10.1038/jhg.2017.27. [DOI] [PubMed] [Google Scholar]
- 18.Fang F, Huang B, Sun S, Xiao M, Guo J, Yi X, Cai J, Wang Z. 2018. miR-27a inhibits cervical adenocarcinoma progression by downregulating the TGF-betaRI signaling pathway. Cell Death Dis 9:395. 10.1038/s41419-018-0431-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barros-Silva D, Costa-Pinheiro P, Duarte H, Sousa EJ, Evangelista AF, Graça I, Carneiro I, Martins AT, Oliveira J, Carvalho AL, Marques MM, Henrique R, Jerónimo C. 2018. MicroRNA-27a-5p regulation by promoter methylation and MYC signaling in prostate carcinogenesis. Cell Death Dis 9:167. 10.1038/s41419-017-0241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He J, Guo X, Sun L, Wang K, Yao H. 2016. Networks analysis of genes and microRNAs in human Wilms' tumors. Oncol Lett 12:3579–3585. 10.3892/ol.2016.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samusik N, Krukovskaya L, Meln I, Shilov E, Kozlov AP. 2013. PBOV1 is a human de novo gene with tumor-specific expression that is associated with a positive clinical outcome of cancer. PLoS One 8:e56162. 10.1371/journal.pone.0056162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krukovskaia LL, Samusik ND, Shilov ES, Polev DE, Kozlov AP. 2010. Tumor-specific expression of PBOV1, a new gene in evolution. Vopr Onkol 56:327–332. (In Russian.) [PubMed] [Google Scholar]
- 23.Pan T, Liu B, Wen H, Tu Z, Guo J, Yang J, Shen G, Wu R. 2016. PBOV1 promotes prostate cancer proliferation by promoting G1/S transition. Onco Targets Ther 9:787–795. 10.2147/OTT.S92682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xue C, Zhong Z, Ye S, Wang Y, Ye Q. 2018. Association between the overexpression of PBOV1 and the prognosis of patients with hepatocellular carcinoma. Oncol Lett 16:3401–3407. 10.3892/ol.2018.9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang S-Y, Gao F, Peng C-G, Zheng C-J, Wu M-F. 2018. Hsa-miR-203 inhibits fracture healing via targeting PBOV1. Eur Rev Med Pharmacol Sci 22:5797–5803. 10.26355/eurrev_201809_15905. [DOI] [PubMed] [Google Scholar]
- 26.Yang C-A, Li J-P, Yen J-C, Lai I-L, Ho Y-C, Chen Y-C, Lan J-L, Chang J-G. 2018. lncRNA NTT/PBOV1 axis promotes monocyte differentiation and is elevated in rheumatoid arthritis. Int J Mol Sci 19:2806. 10.3390/ijms19092806. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.