The past several years have seen dramatic progress in our understanding of the reactions taking place in the early events of photosynthesis. This has been in large part due to research involving purple photosynthetic bacteria (16, 28, 34, 35, 46, 56). These anaerobic photosynthetic prokaryotes have been and continue to be excellent model organisms in which to investigate the basic mechanisms of photosynthetic light-harvesting and reaction center (RC) photochemistry.
In this minireview, we describe what is currently known about the structure of the bacterial photosynthetic unit (PSU) and then outline the series of reactions which take place between the absorption of a “green” photon by the carotenoids in the antenna system and the charge separation across the membrane by the reaction centers. Much of the background to this topic can be found in the following excellent books (5, 10, 15).
The major light-absorbing pigments in purple bacteria are bacteriochlorophyll (bacteriochlorophyll a [Bchla] or Bchlb) and carotenoids. These pigments are noncovalently bound to two types of integral membrane proteins, forming either reaction centers or antenna complexes (15, 70, 75, 76). All species of purple bacteria contain “core” antenna complexes (light-harvesting complex 1 [LH1]), which surround the reaction centers (7, 12, 24, 75). LH1 complexes typically have a single strong near-infrared absorption band between 870 and 890 nm (Bchla) or at 1,020 nm (Bchlb) (70, 75). Most species also contain a second type of antenna complex (LH2), which is arranged more peripherally (70, 75). LH2 complexes typically have two strong absorption bands in the near-infrared, e.g., at 800 and 820 or 850 nm. The exact ratio of LH2/LH1 complexes present in the photosynthetic membrane is controlled by a variety of environmental factors (1) such as light intensity (for recent reviews of this topic, see references 4 and 58). The structure of the PSU is, therefore, very variable.
Antenna complexes have evolved to increase the effective cross-sectional area for light absorption of each reaction center. Mutant photosynthetic bacteria which lack antenna complexes have been made (30). These mutants will still grow photosynthetically but only at very high, incident light intensities. Antenna complexes supply each reaction center with enough photons to allow photosynthesis to occur at reasonable rates over a wide range of incident light intensities. They also absorb light over a broader spectral range than the reaction centers alone and so allow more of the incident solar spectrum to be used productively.
PRESTRUCTURAL STUDIES
Prior to the determination of the structure of the first LH2 complex in 1995 (46), purple bacterial antenna complexes had been extensively studied by a range of biochemical and molecular biological techniques (8, 9, 30, 31, 37, 72, 73). Antenna complexes had been prepared and characterized from a range of different species (75, 76). In addition, molecular genetic systems were developed in the case of Rhodobacter capsulatus and Rhodobacter sphaeroides, which allowed a wide range of site-directed mutations to be generated (8, 9, 30, 74).
Largely due to work carried out by the research group of Zuber (75, 76), it was established that the LH1 and LH2 complexes are constructed on the same modular principle (75). Each complex is an oligomer of a basic unit which consists of a pair of small, hydrophobic apoproteins (named α and β). Hydropathy analysis of these apoproteins showed that they all contained polar N and C termini and a central hydrophobic region of between 20 and 24 amino acids. This then led to the idea that these apoproteins span the photosynthetic membrane with the central hydrophobic region folded into a single membrane-spanning α-helix. This basic topology was confirmed by experiments with proteases and inside-out and right-side-out membrane vesicles (6, 7, 57). Comparative sequence analysis also pinpointed the role of two conserved histidine residues (α-His 30 and β-His 31 in Rhodopseudomonas acidophila 10050) as being likely fifth ligands to the Mg2+ at the center of the bacteriochlorin rings of the 850- and 875-nm-wavelength-absorbing Bchlas in LH2 and LH1, respectively (75, 76). This was confirmed by resonance Raman spectroscopy (64). Based on these considerations, together with a range of other spectroscopic observations, several structural models of both LH1 and LH2 were proposed (37, 54, 75).
The energy transfer reactions occurring within isolated antenna complexes and intact photosynthetic membranes were studied by picosecond flash photolysis (for reviews, see references 31 and 72). The time of energy transfer for the B800→B850 step in LH2 was put at less than 1 ps, while transfer from LH2 to LH1 was rather multiexponential with the main time being 3 to 5 ps, but with some slower components also visible. The energy transfer step from LH1 to the reaction center took 30 to 50 ps. Once the excitation energy reaches the reaction center, the primary photochemical redox reactions are initiated. As a result of this, cyclic electron transport occurs, a transmembrane proton motive force is generated, and ATP is synthesized.
STRUCTURE OF LH2
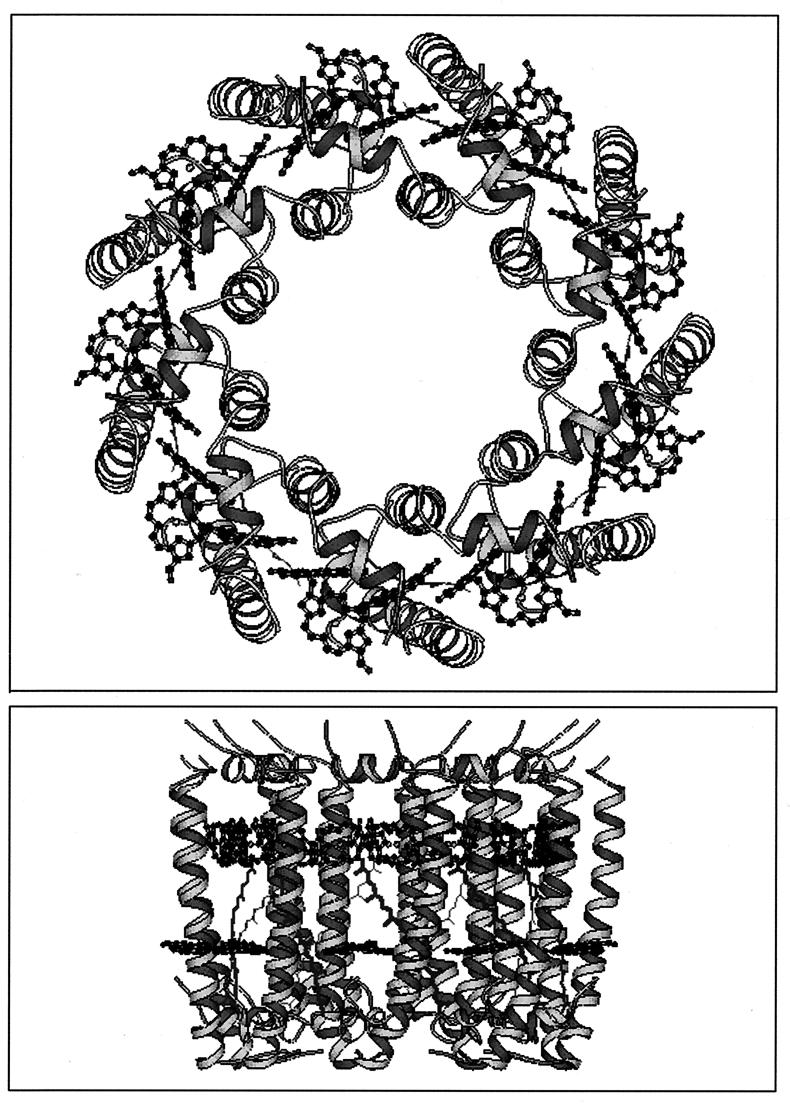
Spurred on by the success of Deisenhofer et al. in crystallizing the purple bacterial reaction center from Rhodopseudomonas viridis (16), we set about trying to crystallize and determine the structure of an LH2 complex. A long struggle then ensued, which finally, after 12 years, was successful, and in 1995, we reported the structure of the LH2 complex from Rhodopseudomonas acidophila 10050 (46). The original structure was described at a resolution of 2.5 Å, but we have now improved this to 2.0 Å with cryocooling (59a). The overall structure is a α9β9 nonamer (Fig. 1). It is rather like an elongated, hollow cylinder (readers should note this is not a pore, as the hole in the middle is filled with lipids). The inner walls of the cylinder are formed from the transmembrane α-helices of the α-apoproteins, and the outer walls are formed from the α-helices of the β-apoproteins. The pigments are all located within these walls of protein. The structure is “capped” top and bottom by the N and C termini of the apoproteins which fold over and interact with each other. For the purposes of this minireview, we will discuss only the detailed arrangement of the pigments. Readers wishing to have more details of the structure of the apoproteins should refer to Prince et al. (59).
FIG. 1.
Schematic representation of the LH2 holocomplex from Rhodopseudomonas acidophila. (Top) View from the cytoplasmic side of the membrane, looking down the central ninefold axis of symmetry. (Bottom) View from within the membrane. The organization of the two rings of Bchla molecules, arranged between the transmembrane α-helices, are shown; the 9 B800 Bchla molecules parallel to the plane of the membrane and the second ring of 18 B850 Bchla molecules, with their bacteriochlorin rings perpendicular to the plane of the membrane. The bottom view also shows the carotenoid (rhodopin-glucoside) which spans the membrane and comes into van der Waal’s contact with both groups of Bchla. Only the chromophoric portions of the pigments are shown. This and most of the other figures in this minireview were produced with the Molscript program (38).
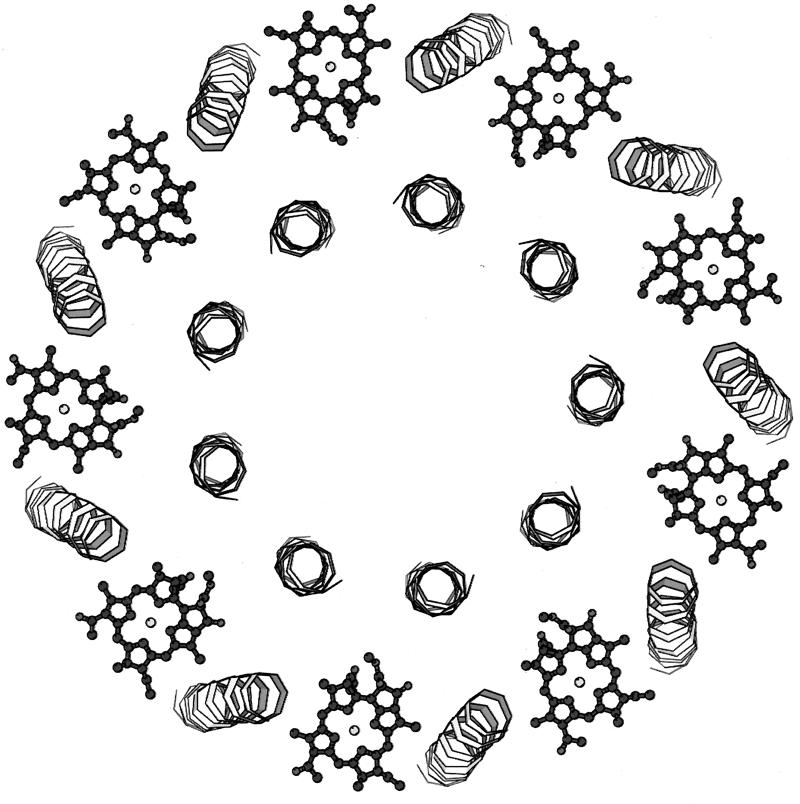
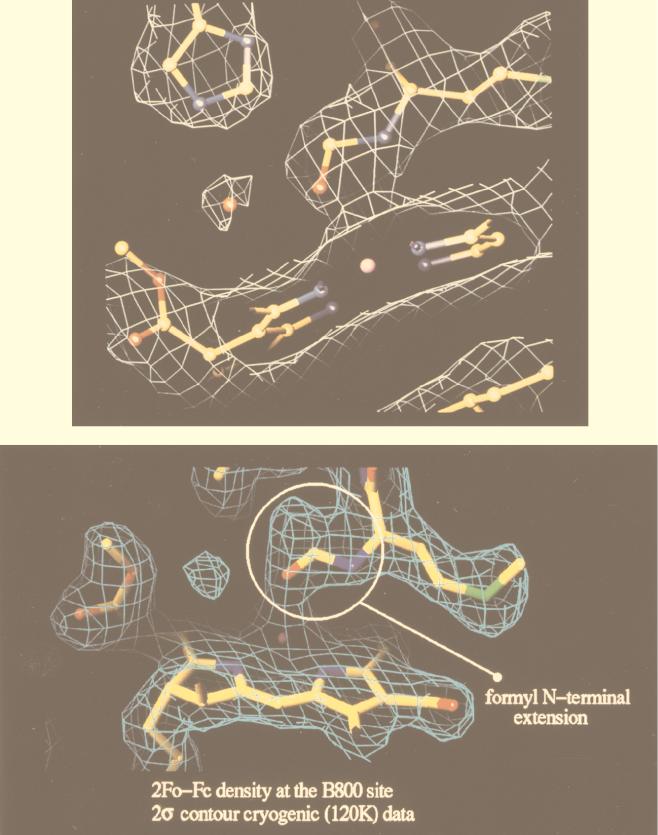
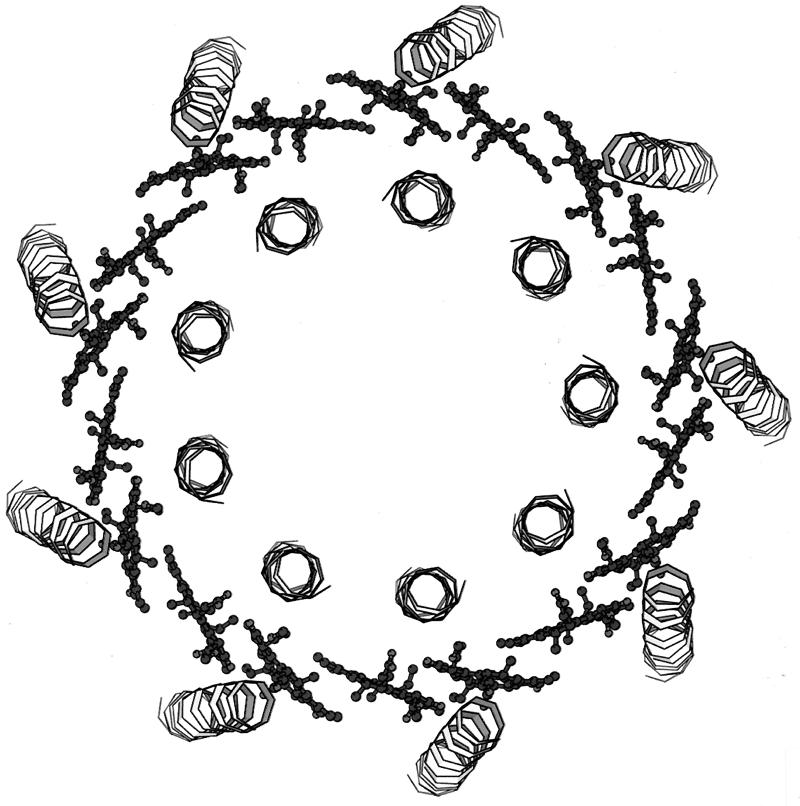
Beginning at the N-terminal (cytoplasmic) side of the complex, the first pigments encountered are a group of monomeric Bchla molecules (Fig. 2). There are nine of these, one per αβ-apoprotein pair. They lie flat, parallel to the putative membrane surface, between the β-apoprotein α-helices. They are separated by 21.2 Å from center to center, and their central Mg2+ ions are complexed to an extension of the N-terminal methionine residue of the α-apoproteins. In the original description of the structure, this extension was modelled as an N-formyl group. With the improved resolution of 2 Å, however, this is clearly seen to be incorrect (Fig. 3). At present we do not know what this extension is. These Bchla molecules have been assigned to those which give rise to the 800-nm-wavelength absorption band (B800). Their binding site is rather polar, which probably explains why their Qy transition (Qy is the Bchla absorption band at 800 or 850 nm) is only slightly redshifted from that of free, monomeric Bchla in organic solvent (∼770 nm). Moving on, further down through the structure toward the C-terminal side, a second group of Bchla molecules are encountered about two-thirds of the way across the transmembrane domain. There are 18 Bchlas in this group (2 per αβ-apoprotein pair), and they are liganded via their central Mg2+ ions to the two conserved histidine residues described above. Their bacteriochlorin rings lie perpendicular to the putative plane of the membrane, parallel to the transmembrane α-helices. They form a closely interacting ring and have a center-to-center separation of 9.5 Å within a αβ pair and 8.9 Å between the next one in the neighboring αβ-apoprotein pairs (Fig. 4). Going round this “ring” of Bchlas, the Mg2+ ions are complexed alternately to the α-apoprotein and then the β-apoprotein. These Bchlas have been assigned as those which give rise to the 850-nm-wavelength absorption band (B850).
FIG. 2.
Location and organization of the B800 Bchlas in the LH2 from Rhodopseudomonas acidophila. The B800 Bchlas (nine Bchla molecules) can be seen arranged peripherally between the β-apoprotein α-helices.
FIG. 3.
Comparison of the electron density in the region of the B800 Bchla binding pocket of LH2 from Rhodopseudomonas acidophila at a resolution of 2.5 Å (top) and 2.0 Å (bottom). (Top) With a resolution of 2.5 Å, the extension of the N-terminal methionine residue of the α-apoprotein is clearly seen in the electron density map, together with the “modelled” formyl group. (Bottom) At this improved resolution of 2.0 Å, the N-terminal extension is also clearly seen. Now, however, it can be seen to bifurcate. The modelled formyl group no longer gives a satisfactory fit to this higher-resolution data. The electron density is shown as the white or blue cage.
FIG. 4.
Organization of B850 Bchlas in the LH2 from Rhodopseudomonas acidophila. The 18 Bchla molecules, which from the B850 ring can be seen, edge on, are arranged between the transmembrane α-helices of the α-apoprotein (inner) and β-apoprotein (outer).
Apart from the overall very hydrophobic binding pocket of the 850-nm-wavelength-absorbing Bchlas, it is also worthwhile pointing out certain of the most important residues which are hydrogen bonded to the Bchla macrocycles. In the intact PSU, energy transfer proceeds down an energy gradient because LH2 absorbs to the blue (high energy) of the LH1 (low energy). This energy gradient results in energy transfer being directed or funnelled toward the reaction center. Indeed, this funnelling is essential for efficient energy transfer from the periphery of the PSU to the reaction center. It is important, therefore, to try to understand the structural factors which control the position of the Qy absorption band of the Bchlas in antenna complexes. Some species of purple bacteria contain naturally occurring wild-type spectral variants of LH2 in which the 850-nm-wavelength absorption band is shifted to 820 nm (11, 75). Careful comparison of their LH2 sequences identified several key, C-terminally located, aromatic residues, the presence or absence of which strongly correlated with this shift in absorbance (76). In Rhodopseudomonas acidophila, for example, residues at positions α44 and α45 were shown to be critical in determining the position of the B850 Qy absorption band. When these residues are Tyr and Typ, the Qy band is at 863 nm, while when they are replaced by Phe and Leu, respectively, the Qy band is blueshifted to 820 nm. Resonance Raman spectroscopy suggested that these residues in the B800-850 complex were hydrogen bonded to the C-9 acetyl group of Bchla, while in the B800-820 complex, these hydrogen bonds were absent (17, 18, 64). The crystallographic structure shows that the B850 C-9 acetyl groups are indeed, hydrogen bonded to α-Tyr 44 and α-Trp 45. Very recently, we have succeeded in determining the three-dimensional (3-D) structure of the B800-B820 complex from Rhodopseudomonas acidophila 7050 (49). The initial indications are that the loss of these hydrogen bonds results in a reorientation of the C-9 acetyl group which twists out of the plane of the bacteriochlorin ring and which may explain the majority of the spectral shift (49). Gudowska-Nowak and coworkers (25) have carried out a detailed analysis of the structure and the absorption properties of the different Bchla molecules present in the water-soluble, FMO antenna complex. They showed that as the C-9 acetyl group twists out of the plane of the bacteriochlorin ring (i.e., moves the carbonyl group out of conjugation with the bacteriochlorin ring), the Qy absorption band is blueshifted compared with its position when the C-9 acetyl group is parallel to the bacteriochlorin ring (i.e., adds another double band into the conjugated system of the macrocycle).
Each αβ-apoprotein pair also contains a single well-resolved carotenoid molecule (rhodopin-glucoside). It has an extended S-shaped conformation ( all trans) and spans the whole depth of the complex. The glucosyl ring is located in a polar pocket on the N-terminal side of the complex. The conjugated chain then runs perpendicular to the edge of the B800 bacteriochlorin ring (closest approach, 3.4 Å) and then crosses over into the next αβ-apoprotein pair before running over the face of the α-bound B850 bacteriochlorin ring (closest approach, 3.68 Å). It is important to point out here that this carotenoid interlinks two αβ-apoprotein pairs and appears to play an important structural role. This may explain why carotenoid deletion mutants fail to assemble LH2 (40, 77) and why in the absence of carotenoids the LH2 apoproteins are synthesized but rapidly degraded (40).
In 1996, the structure of a second LH2 complex from Rhodospirillum molischianum was described by Koepke et al. (35). Its structure is very similar to that from Rhodopseudomonas acidophila, but its oligomerization state differs. It is an octamer rather than a nonamer. The other major difference is the organization of the B800 Bchlas. In the Rhodospirillum molischianum complex, the Mg2+ ions from these Bchlas are liganded to the γ oxygen atom of α-Asp 6. This change results in the plane of the bacteriochlorin ring being dipped into the membrane at an angle of about 20°, and the orientation of the ring being rotated by 90° relative to that of the B800 Bchlas in Rhodopseudomonas acidophila. A more-extensive comparison of the two structures can be found in Cogdell et al. (13). Low-resolution two-dimensional (2D) projection maps of LH2 complexes from Rhodopseudomonas sulphidophilus and Rhodobacter sphaeroides have also been reported (53, 73). These are both nonamers. What controls the size of the ring remains to be determined. It has also been suggested that the ring size may vary in certain species (51a). These are still open questions.
STRUCTURE OF LH1
LH1-RC “core” complexes were first clearly visualized in electron microscopy studies on membranes from Rhodopseudomonas viridis (52, 68). This Bchlb-containing species has only LH1 complexes. It lacks LH2. Its intracytoplasmic membranes are lamellar and contain large regions of quasicrystalline 2D arrays of LH1-RC core complexes. These core structures are circular with a diameter of ∼120 Å. Image processing of these structures suggested that they had sixfold symmetry. Early determinations of stochiometry indicated a Bchla/reaction center ratio of 24:1 (52, 68). Together, this suggested that the reaction center was surrounded by an α12β12 LH1 ring. This idea was given further strong support by Meckenstock et al. (50, 51). These workers produced 2D crystals of LH1-RC cores from Rhodobium marinum with and without the reaction center. Their electron microscopy images showed that removal of the reaction center resulted in a loss of the electron density in the middle of the LH1 ring. Very recently, Karrasch et al. (34) produced 2D crystals of reconstituted LH1 complexes from Rhodospirillum rubrum. These crystals were well enough ordered to be studied by electron diffraction, and a 2D projection map of LH1 at a resolution of 8.5 Å was produced. This map showed a ring structure consisting of 16 αβ pairs, very similar to that for LH2. When the data were processed at lower resolution, it appeared to have sixfold symmetry. However, at higher resolution this pseudo sixfold symmetry broke down to reveal eightfold symmetry. These workers also showed that with the larger ring structure the hole in the middle (diameter, 68 Å) was just big enough to accommodate the reaction center. It is also worth pointing out that the diameter of the LH1 ring seen by Karrasch et al. (34) is very similar to the size of the core complexes seen previously both in Rhodopseudomonas viridis (52, 68) and Rhodobium marinum (50, 51). This new model of the LH1-reaction center core complex as a 16-mer conflicts with the previous measurements of the Bchla/reaction center stochiometry of 24:1 (68, 76). The new model would imply a ratio of 32:1 rather than 24:1. Two more recent attempts to measure this stochiometry have been made (19, 24). Gall (24) measured this ratio for core complexes isolated from seven different species of purple bacteria. The data did show some variability but had an average value of 33 (±4):1. In contrast, Francke and Amesz (19) determined the ratio for core complexes from six different species and found ratios of 24 (±2):1 to 28 (±4):1. Clearly, more work is required to sort out the current ambiguity. It is obvious though that an α12β12 ring is not big enough to enclose the reaction center.
This story is potentially even more complicated. Some species of purple bacteria such as Rhodobacter sphaeroides and Rhodobacter capsulatus contain a gene called pufX (2, 3, 44, 47). This gene encodes a protein which is intimately associated with LH1 (23, 44, 47, 48, 60, 62). In Rhodobacter sphaeroides, deletion of the pufX gene prevents photosynthetic growth but only when LH1 is present (2, 47). A double deletion mutant, PufX− LH1−, still grows photosynthetically (2, 47). This PufX− phenotype appears to result from an inability of the secondary electron acceptor ubiquinone to escape from the reaction center and interact with the cytochrome b-c1 complex. This then blocks photosynthetic electron transport. It has been suggested that the PufX protein resides in the LH1 ring and provides a gate which allows the diffusion of ubiquinone in and out of the reaction center (12). Indeed it has recently been shown in Rhodobacter sphaeroides that deletion of the PufX protein results in an increase in the Bchla/reaction center ratio in the LH1-RC core complex (47, 48, 60).
There have also been some very recent studies which have suggested that in Rhodobacter sphaeroides, the LH1 rings in vivo may not be complete (33). It is now essential, therefore, that a high-resolution structure of an LH1-RC core is determined. We have recently been trying very hard to produce 3D crystals of such complexes (24, 41). We have obtained crystals from four different species of purple bacteria but so far they do not diffract X-rays sufficiently well to allow determination of a high-resolution structure. Watch this space.
Over a number of years Loach and coworkers (45), have developed an in vitro system for reconstituting LH1 complexes from their individual purified components. This approach is now allowing the important structural features required for successful assembly of LH1 complexes to be determined. They are even able to study the effects of reconstituting with mixed apoproteins when the α- and β-apoproteins come from different species.
A MODEL OF THE WHOLE PSU
In order to fully describe the purple bacterial light-harvesting systems, we need to have a complete picture of the organization of the antenna complexes in vivo. This is not yet possible. However, if for the purposes of this minireview, we take the Karrasch et al. (34) structure for LH1 at face value, then it is possible to model a high-resolution generic LH1-RC core complex (27, 56). Two groups have done this, by using the structures of the two LH2 complexes and recognizing the strong structural homology of the LH1 and LH2 apoproteins especially in the transmembrane α-helical regions. The LH1-reaction center core structure can then be put together with the known structure of LH2 to produce a model of the whole PSU (Fig. 5). One striking feature of this model is the way in which the macrocycles of tightly coupled rings of Bchla (the B850 ring in LH2 and the B875 ring in LH1) line up at the same depth in the membrane. This also corresponds very closely to the transmembrane location of the special pair of Bchlas in the reaction center. This minimizes the distance between the rings of Bchl, which in turn maximizes the rate of energy transfer from LH2→LH1 and from LH1 to the reaction center (since distance is one of the major factors that controls the rate of singlet-singlet energy transfer). This model is very useful since it focuses attention on the different types of pigment-pigment interactions which are involved in each of the different energy transfer reactions that occur in the light-harvesting system.
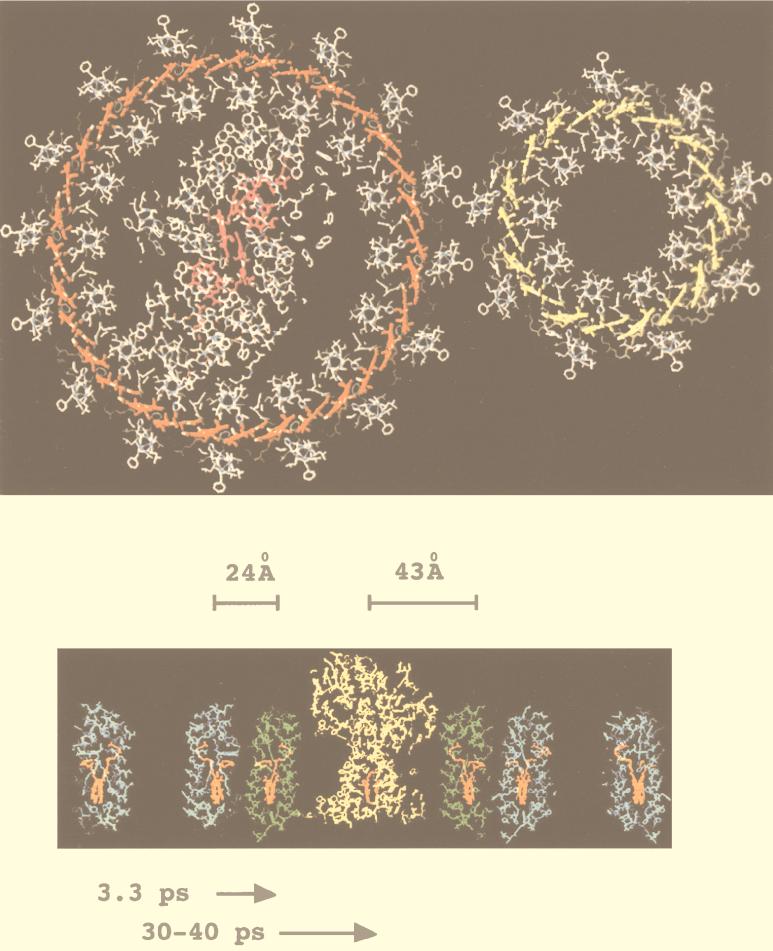
FIG. 5.
Two views of a model of the purple bacterial PSU. (Top) A top view, looking perpendicular to the assumed plane of the membrane. This section is taken at a point in the LH complexes where the tightly coupled rings of Bchlas are located. This figure was adapted from reference 56 with permission from Elsevier Science. The reaction center is located in the center of the LH1 complex. The smaller LH2 sits outside the large LH1 complex. (Bottom) A side view, looking from within the assumed plane of the membrane (blue, LH2; green, LH1; yellow, RC). This section is taken exactly perpendicular to the view shown in the top panel. The distances shown between the different pigment groups (shown in orange) are calculated assuming a space-filling model and the closest possible organization. The times shown are energy transfer times as measured in intact membranes (31, 61, 69, 72).
The situation in vivo, however, is clearly more heterogeneous than these simple models suggest. The composition of the purple bacterial PSU is dynamic and changes depending on the growth conditions as described above. Moreover, previous studies on energy migration in the photosynthetic membranes from a range of different species of purple bacteria have shown that the precise supramolecular organization of the LH2 and LH1-RC cores is also species dependent (14). There is a now an urgent need to examine the architecture of the PSU in intact photosynthetic membranes.
ENERGY TRANSFER WITHIN THE PSU
The purple bacterial PSU has proved to be a very attractive system for physical chemists interested in the study of light-harvesting process. There are two reasons for this: first, there is atomic-resolution, structural information, and second, unlike in plants, there is excellent spectral separation between the different pigment groups in the energy transfer processes. Progress in this area has also been greatly assisted by parallel advances in laser technology which now allow energy transfer events to be probed with femtosecond time resolution (61, 69).
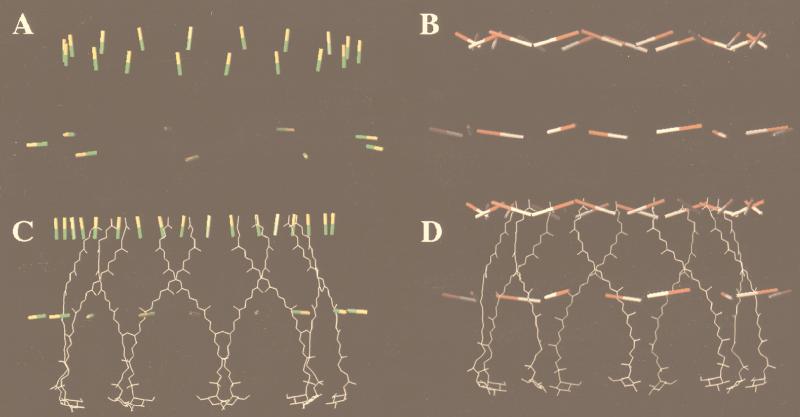
Figure 6 shows the structural context in which the discussion of energy transfer within LH2 should begin (22). The interpigment distances and the relative orientation of the transition dipole moments of the major electronic states are shown.
FIG. 6.
Relative orientations of the Qx and Qy transition dipoles in the B800 and B850 Bchlas and the carotenoid in the LH2 complex from Rhodopseudomonas acidophila. (A) Alignment of the Qx dipoles (yellow-green). (B) Alignment of the Qy dipoles (red-white). (C) Alignment of the carotenoid to the Qx dipoles (the transition dipole movement of the S2 state of the carotenoid runs up and down the long axis of the conjugated double bands). (D) Alignment of the carotenoid with respect to the Qy dipoles. The figure was adapted from reference 22 with permission from Elsevier Science and produced with O (32). The distances between the center of a B800 Bchla and the α-bound and β-bound B850 Bchlas in the same αβ-apoprotein pair are 17.4 and 18.2 Å, respectively. Qx and Qy are labels for the two Bchla absorption bands at ∼590 nm and 800 or 850 nm, respectively. The transition dipoles, which correspond to these absorption bands, lie within the plane of the bacteriochlorin rings, at right angles to each other, diagonally between the nitrogen atoms, which coordinate the central Mg2+. One of the factors which controls the rate of excitation energy transfer between two molecules is the angle between the transition dipoles involved. When the dipoles are parallel, energy transfer is favorable; when they are orthoganal, energy transfer is much less favorable.
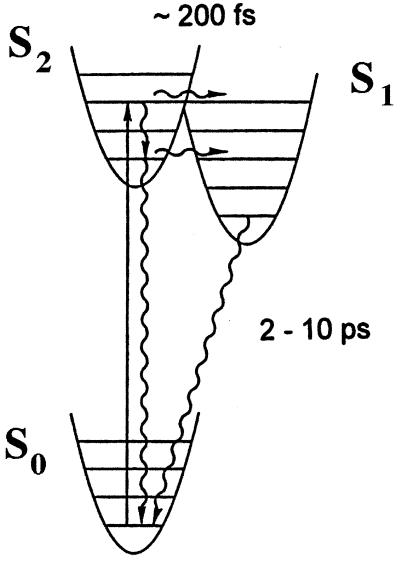
For many years now, it has been well documented that light absorbed by the light-harvesting carotenoids can be used to drive the light reactions in purple bacterial photosynthesis (20, 21, 36). The efficiency of carotenoid→Bchla energy transfer varies from species to species and from LH complex to LH complex, from 30 up to 100% (20). The exact mechanism(s) involved here has (have) not yet been well defined. The basic problem is as follows. For many years, carotenoids were thought to be nonfluorescent (21, 36), implying that their fluorescence lifetimes must very be short indeed. If this were true, then how can there be enough time for such efficient singlet-singlet energy transfer (i.e., light harvesting) to occur before the carotenoid’s excited singlet state is lost by other competing processes? The answer to this question is twofold. First, carotenoids do fluoresce (36), and second, they have a rather unusual photochemistry. In particular, the allowed singlet state transition from the ground state goes to the S2 state rather than the S1 state (29). The direct one-photon-induced transition from the ground state to the S1 state cannot occur because it is symmetry forbidden. Following excitation, the carotenoid S2 state lasts only for a few hundred femtoseconds before it then relaxes into the S1 state. S1 then decays back to the ground state in a few picoseconds (Fig. 7).
FIG. 7.
Schematic representation of the two low-lying excited singlet states of S1 and S2 carotenoids. The approximate positions of the S1 and S2 excited singlet states are shown. The S0→S2 represents the optically allowed (one-photon) transition that gives rise to the carotenoid’s well-known, strong absorption spectrum. The approximate times for the S2→S1 and S1→S0 transitions are also shown.
In the LH2 complex from Rhodopseudomonas acidophila 10050, the efficiency of carotenoid (in this case rhodopin-glucoside) to Bchla singlet-singlet energy transfer is about 55% (21). We can therefore ask two questions: which excited state of the carotenoid is acting as the energy donor and which group of Bchlas (i.e., B800 or B850) receive this energy? If the carotenoid rhodopin-glucoside is excited with a 60-fs excitation pulse, the measured lifetime of the S2 state depends upon where that carotenoid is (45a). In benzylalcohol, its lifetime is ∼130 fs. In LH2, this lifetime is shortened to ∼61 fs. This reduction in lifetime is due to energy transfer to the Bchlas. This can be seen directly by exciting the carotenoid and measuring the kinetics of the arrival of that energy in the B800 or B850 manifolds. Measuring at 851 nm the kinetics of the increase in B850 fluorescence is biphasic. About 70% of the fluorescence rises with a time constant of ∼63 fs. The other 30% rises with a time constant of ∼900 fs. Clearly, the fast phase of this energy transfer event corresponds to direct singlet-singlet energy transfer from the S2 state of rhodopin-glucoside to the B850 manifold. However, what is the slower rise due to? Excitation of LH2 at 800 nm (into B800) has allowed the time course for the B800→B850 singlet-singlet energy transfer to be determined (65, 69). The rate constant for this process is about 900 fs. The slower phase in the carotenoid-to-B850 energy transfer, therefore, reflects that energy which has gone via the B800 Bchls. We have recently been able to confirm this by selectively removing the B800 Bchls from LH2 (45a). In this case, with a B800-less LH2 complex, all of the carotenoid-to-B850 energy transfer is fast. The kinetics of the carotenoid to B800 energy transfer also indicate that the S2 state of rhodopin-glucoside is the major energy donor. On the basis of the energy levels of the carotenoid’s two excited singlet states, it has been suggested that energy transfer from S2 goes by way of the Qx transition (Qx is the Bchla absorption band at ∼590 nm) of Bchla and that from S1 goes by way of the lower-energy Qy transition (69). In the case of LH2 from Rhodopseudomonas acidophila, it appears that nearly all of the carotenoid to Bchla singlet-singlet energy transfer comes from the S2 state (21). In contrast, other antenna complexes, such as LH2 from Rhodobacter sphaeroides (where the efficiency of carotenoid-to-Bchla energy transfer is nearly 100% [20]), are also able to harvest energy from the carotenoid’s S1 state (36).
Once the excited state has reached the B850 manifold, it is very rapidly depolarized (61, 69). This means that it hops very rapidly around the B850 ring. This hopping time has been estimated to be on the order of a few tens of femtoseconds. If there are no other LH complexes nearby which can accept the excitation energy, the excited state will decay in 1 ns. Consequently, the excited state visits each B850 Bchla many times during its lifetime and is, therefore, available for energy transfer out of any site in the ring with equal probability. This is very important because it means that there does not have to be a fixed supramolecular arrangement of LH2 and LH1 complexes in the PSU for efficient energy transfer to occur. As long as the next antenna complex is sufficiently close, energy transfer will occur with equal high efficiency from anywhere in the LH2 ring.
Following fs excitation of LH2, the times of energy transfer for the LH2→LH1 and LH1→reaction center steps can also be measured (61, 69). The kinetics of energy transfer from LH2 to LH1 are somewhat multiexponential. The major and fastest phase takes 3 to 4 ps. The slower phases probably arise from a rather heterogeneous arrangement of the LH complexes in the membrane so that there are several LH2→LH2 steps before an LH1 complex is encountered. The final energy transfer step from LH1 to the reaction center is the slowest and takes 30 to 50 ps. This is clearly due to the larger distance involved compared to LH2→LH1 (Fig. 5). Even though most, if not all, of the times of the energy transfer steps between the absorption of a green photon by an LH2 carotenoid and the arrival of that energy at the reaction center have been resolved, the details of the exact molecular mechanisms involved remain to be precisely defined. A detailed discussion of this and of the extensive range of biophysical techniques currently being employed to tackle this problem are beyond the scope of this minireview. An excellent recent review by Sundström et al. (69), however, can be consulted by those readers who wish to explore this subject in greater detail.
FINAL REMARKS
This is a golden time for those people interested in trying to understand the detailed mechanisms of energy transfer in photosynthetic light-harvesting systems. Apart from the high-resolution structures of the two LH2 complexes described above, detailed structural information is also available for the LHC2 complex from higher plants (39), two water-soluble Bchla-protein complexes (FMO [43, 71]), the water-soluble peridinin-chlorophyll a complex from a dinoflagellate (26), and a whole clutch of phycobiliproteins (e.g., reference 67). As these are subjected to detailed functional studies over the next few years, we can expect the general principles of photosynthetic light harvesting to be established. Current progress in our understanding of purple bacterial light harvesting has been largely led by the acquisition of detailed high-resolution structural information, coupled with a highly multidisciplinary approach to its subsequent exploitation. We expect similar advances will occur in our detailed understanding of the light-harvesting reactions in oxygen-evolving photosynthetic organisms as high-resolution structures of their integral membrane photosystems become available (63, 66). Readers who wish to see more-detailed pictures of LH2 should visit the following two web sites (59a and 71a).
ACKNOWLEDGMENTS
Some of the work described in this minireview was supported by grants from the BBSRC, the Gatsby Charitable Trust, the Human Frontiers of Science Programme, and the EU.
REFERENCES
- 1.Aagaard J, Sistrom W R. Control of the synthesis of reaction centre bacteriochlorophylls in photosynthetic bacteria. Photochem Photobiol. 1972;15:209–225. doi: 10.1111/j.1751-1097.1972.tb06240.x. [DOI] [PubMed] [Google Scholar]
- 2.Barz W P, Francia F, Venturoli G, Melandri B A, Verméglio A, Oesterhelt D. Role of PufX protein in photosynthetic growth of Rhodobacter sphaeroides. 1. PufX is required for efficient light-driven electron transfer and photophosphorylation under anaerobic conditions. Biochemistry. 1995;34:15235–15247. doi: 10.1021/bi00046a032. [DOI] [PubMed] [Google Scholar]
- 3.Barz W P, Verméglio A, Francia F, Verturoli G, Melandri B A, Oesterhelt D. Role of PufX protein in photosynthetic growth of Rhodobacter sphaeroides. 2. PufX is required of efficient ubiquinone/ubiquinol exchange between the reaction centre Q(B) site and the cytochrome b/c1complex. Biochemistry. 1995;34:15248–15258. doi: 10.1021/bi00046a033. [DOI] [PubMed] [Google Scholar]
- 4.Bauer C E, Bird T H. Regulatory circuits controlling photosynthesis gene expression. Cell. 1996;85:5–8. doi: 10.1016/s0092-8674(00)81074-0. [DOI] [PubMed] [Google Scholar]
- 5.Blankenship R E, Madigan M T, Bauer C E. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. [Google Scholar]
- 6.Brunisholz R A, Weimken V, Suter F, Bachofen R, Zuber H. The light-harvesting polypeptides of Rhodospirillum rubrum. II. Localisation of amino terminal regions of light-harvesting polypeptides B870-α and B870-β and the reaction centre subunit at the cytoplasmic side of photosynthetic membrane of Rs. rubrum G-9+ Hoppe-Seyler’s Z Physiol Chem. 1984;365:684–701. doi: 10.1515/bchm2.1984.365.2.689. [DOI] [PubMed] [Google Scholar]
- 7.Brunisholz R A, Zuber H, Valentine J, Lindsay J G, Wooley K J, Cogdell R J. The membrane location of the B890-complex from Rs. rubrumand the effect of carotenoid on the conformation of its two apoproteins at the cytoplasmic surface. Biochim Biophys Acta. 1986;849:295–303. [Google Scholar]
- 8.Burgess J G, Ashby M K, Hunter C N. Chromosomal deletion of genes encoding B800-850 (LH2) polypeptides of Rhodobacter sphaeroides. J Gen Microbiol. 1988;135:1809–1816. doi: 10.1099/00221287-135-7-1809. [DOI] [PubMed] [Google Scholar]
- 9.Bylina E J, Robles S J, Youvan D C. Directed mutations affecting the putative bacteriochlorophyll-binding sites in the light-harvesting 1 antenna of Rhodobacter capsulatus. Isr J Chem. 1988;28:73–78. [Google Scholar]
- 10.Caddick M X, Baumberg S, Hodgson D A, Phillips-Jones M K. Microbial responses to light and time. Symp. Soc. Gen. Microbiol. Vol. 56 1998. [Google Scholar]
- 11.Cogdell R J, Durant I, Valentine J, Lindsay J G, Schmidt K. The isolation and partial characterisation of the light-harvesting pigment-protein complexes of Rhodopseudomonas acidophila. Biochim Biophys Acta. 1983;722:427–435. [Google Scholar]
- 12.Cogdell R J, Fyfe P K, Barrett S J, Prince S M, Freer A A, Isaacs N W, McGlynn P, Hunter C N. The purple bacterial photosynthetic unit. Photosynth Res. 1996;48:55–63. doi: 10.1007/BF00040996. [DOI] [PubMed] [Google Scholar]
- 13.Cogdell R J, Isaacs N W, Freer A A, Arrelano J, Howard T D, Papiz M Z, Hawthornthwaite-Lawless A M, Prince S M. The structure and function of the LH2 (B800-850) complex from the purple photosynthetic bacterium Rhodopseudomonas acidophilastrain 10050. Prog Biophys Mol Biol. 1997;68:1–27. doi: 10.1016/s0079-6107(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 14.Deinum G, Otte S C M, Gardiner A T, Aartsma T J, Cogdell R J, Amesz J. Antenna organisation on Rhodopseudomonas acidophila: a study of the excitation migration. Biochim Biophys Acta. 1991;1060:125–131. [Google Scholar]
- 15.Deisenhofer J, Norris J R. The photosynthetic reaction centre, vol. 1 and 2. San Diego, Calif: Academic Press; 1993. [Google Scholar]
- 16.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Structure of the protein subunits in the photosynthetic reaction centre of Rhodopseudomonas viridisat 3Å resolution. Nature. 1985;318:618–624. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 17.Fowler G J S, Sockalingam G D, Robert B, Grief G G, Hunter C N. Blue shifts in bacteriochlorophyll absorbance correlate with changed hydrogen bonding patterns in light-harvesting LH2 mutants of Rhodobacter sphaeroideswith alterations at α Tyr44 and α Tyr45. Biochem J. 1994;299:695–700. doi: 10.1042/bj2990695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fowler G J S, Visschers R W, Grief G G, van Grondelle R, Hunter C N. Genetically modified photosynthetic antenna complexes with blue-shifted absorbance bands. Nature. 1992;355:848–850. doi: 10.1038/355848a0. [DOI] [PubMed] [Google Scholar]
- 19.Francke C, Amesz J. The size of the photosynthetic unit in purple bacteria. Photosynth Res. 1995;46:347–352. doi: 10.1007/BF00020450. [DOI] [PubMed] [Google Scholar]
- 20.Frank H A, Cogdell R J. Photochemistry of carotenoids. In: Young A, Britton G, editors. Carotenoids in photosynthesis. London, United Kingdom: Chapman and Hall; 1993. pp. 252–326. [Google Scholar]
- 21.Frank H A, Cogdell R J. Carotenoids in photosynthesis. Photochem Photobiol. 1996;63:257–264. doi: 10.1111/j.1751-1097.1996.tb03022.x. [DOI] [PubMed] [Google Scholar]
- 22.Freer A A, Prince S M, Sauer K, Papiz M Z, Hawthornthwaite-Lawless A M, McDermott G, Cogdell R J, Isaacs N W. Pigment-protein interactions and energy transfer in the antenna complex of the photosynthetic bacterium Rhodopseudomonas acidophila. Structure. 1996;4:449–462. doi: 10.1016/s0969-2126(96)00050-0. [DOI] [PubMed] [Google Scholar]
- 23.Fulcher T K, Beatty J T, Jones M R. A demonstration of the key role played by the PufX protein in the functional and structural organization of native and hybrid bacterial photosynthetic core complexes. J Bacteriol. 1998;180:642–646. doi: 10.1128/jb.180.3.642-646.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gall A. Purification, characterisation and crystallisation of a range of Rhodospirillineae pigment-protein complexes. Ph.D. thesis. Glasgow, United Kingdom: University of Glasgow; 1995. [Google Scholar]
- 25.Gudowska-Nowak E, Newton M D, Fajer J. Conformational and environmental effects on bacteriochlorophyll optical spectra: correlations of calculated spectra with structural results. J Phys Chem. 1990;94:5795–5801. [Google Scholar]
- 26.Hoffman E, Wrench P M, Sharples F P, Hiller R G, Welte W, Diederichs K. Structural basis of light-harvesting by carotenoids: peridinin-chlorophyll-protein from Anphidinium carterae. Science. 1996;272:1788–1791. doi: 10.1126/science.272.5269.1788. [DOI] [PubMed] [Google Scholar]
- 27.Hu X, Schulten K. A model for the light harvesting complex 1 (B875) of Rhodobacter sphaeroides. Biophys J. 1998;75:683–694. doi: 10.1016/S0006-3495(98)77558-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu X, Damjanovic T R, Schulten K. Architecture and function of the light-harvesting apparatus of purple bacteria. Proc Natl Acad Sci USA. 1998;95:5935–5941. doi: 10.1073/pnas.95.11.5935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hudson B S, Kohler B E, Schulten K. Linear polyene electronic structure and potential surfaces. In: Lim E C, editor. Excited states. Vol. 6. New York, N.Y: Academic Press; 1982. pp. 22–95. [Google Scholar]
- 30.Hunter C N. Genetic manipulations of the antenna complexes of purple bacteria. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 473–501. [Google Scholar]
- 31.Hunter C N, van Grondelle R, Olsen J D. Photosynthetic antenna proteins: 100 ps before photochemistry starts. Trends Biochem Sci. 1989;14:72–76. doi: 10.1016/0968-0004(89)90047-9. [DOI] [PubMed] [Google Scholar]
- 32.Jones A T, Zou J Y, Cowan S W, Kjeidgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;42:140–149. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 33.Jungas C, Ranck J-L, Rijaud J-L, Joliot P, Verméglio A. Supramolecular organisation of the photosynthetic apparatus of Rhodobacter sphaeroides. EMBO J. 1999;18:534–542. doi: 10.1093/emboj/18.3.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karrasch S, Bullough P A, Ghosh R. The 8.5 Å projection map of the light-harvesting complex 1 from Rhodospirillum rubrumreveals a ring composed of 16 subunits. EMBO J. 1995;14:631–638. doi: 10.1002/j.1460-2075.1995.tb07041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koepke J, Hu X, Muenke C, Schulten K, Michel H. The crystal structure of the light-harvesting complex II (B800-850) from Rhodospirillum molischianum. Structure. 1996;4:581–597. doi: 10.1016/s0969-2126(96)00063-9. [DOI] [PubMed] [Google Scholar]
- 36.Koyama Y, Kuki M, Andersson P O, Gillbro T. Singlet excited states and the light-harvesting function of carotenoids in bacterial photosynthesis. Photochem Photobiol. 1996;63:243–256. [Google Scholar]
- 37.Kramer H J M, van Grondelle R, Hunter C N, Westerhuis W H J, Amesz J. Pigment organisation of the B800-850 antenna complex of Rhodopseudomonas sphaeroides. Biochim Biophys Acta. 1984;765:156–165. [Google Scholar]
- 38.Kraulis P J. Molscript—a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 39.Kühlbrandt W, Wang D N, Fujiyoshi Y. Atomic model of plant light-harvesting complex by electron crystallography. Nature. 1994;367:614–621. doi: 10.1038/367614a0. [DOI] [PubMed] [Google Scholar]
- 40.Lang H P, Hunter C N. The relationship between carotenoid biosynthesis and the assembly of the light-harvesting LH2 complex in Rhodobacter sphaeroides. Biochem J. 1994;298:197–205. doi: 10.1042/bj2980197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law C J. Structure and function studies on the LH1-RC core complex from a range of photosynthetic purple bacteria. Ph.D. thesis. Glasgow, United Kingdom: University of Glasgow; 1999. [Google Scholar]
- 42.Law C J, Cogdell R J, Trissl H-W. Antenna organisation in the purple bacterium Rhodopseudomonas acidophilastudied by fluorescence induction. Photosynth Res. 1997;52:157–165. [Google Scholar]
- 43.Li Y-F, Zhou W, Blankenship R E, Allen J P. Crystal structure of the bacteriochlorophyll a protein from Chlorobium tepidum. J Mol Biol. 1997;271:456–471. doi: 10.1006/jmbi.1997.1189. [DOI] [PubMed] [Google Scholar]
- 44.Lilburn T G, Haith C E, Prince R C, Beatty J T. Pleiotropic effects of puf X gene deletion on the structure and function of the photosynthetic apparatus of Rhodobacter capsulatus. Biochim Biophys Acta. 1992;1100:160–170. doi: 10.1016/0005-2728(92)90077-f. [DOI] [PubMed] [Google Scholar]
- 45.Loach P A, Parkes-Loach P S. Structure-function relationship in core light-harvesting complexes (LH1) as determined by characterisation of the structural subunit and by reconstitution experiments. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 437–471. [Google Scholar]
- 45a.Macpherson, A. N., J. Arellano, N. J. Fraser, R. J. Cogdell, and T. Gillbro. Ultra fast energy transfer for rhodopin glucoside in the LH 2 complex from Rhodopseudomonas acidophila strain 10050, p. 9–14. In G. Garab (ed.), Photosynthesis: mechanisms and effects, vol. 1. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 46.McDermott G, Prince S M, Freer A A, Hawthornthwaite-Lawless A M, Papiz M Z, Cogdell R J, Isaacs N W. Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria. Nature. 1995;374:517–521. [Google Scholar]
- 47.McGlynn P, Hunter C N, Jones M R. The Rhodobacter sphaeroidesPufX protein is not required for photosynthetic competence in the absence of a light-harvesting system. FEBS Lett. 1994;349:349–353. doi: 10.1016/0014-5793(94)00701-2. [DOI] [PubMed] [Google Scholar]
- 48.McGlynn P, Westerhuis W H J, Jones M R, Hunter C N. Consequences for the organisation of reaction centre LH1 core complexes of Rhodobacter sphaeroidesarising from/by deletion of amino acid residues from the C-terminus of the LH1 α-polypeptide. J Biol Chem. 1996;271:3285–3292. doi: 10.1074/jbc.271.6.3285. [DOI] [PubMed] [Google Scholar]
- 49.McLuskey K, Prince S M, Cogdell R J, Isaacs N W. Crystallisation and preliminary X-ray crystallographic analysis of the B800-820 light-harvesting complex from Rhodopseudomonas acidophilastrain 7050. Acta Crystallogr Sect D. 1999;55:885–887. doi: 10.1107/s0907444998016059. [DOI] [PubMed] [Google Scholar]
- 50.Meckenstock R, Krusche K, Brunisholz R A, Zuber H. The light-harvesting core-complex and the B820-subunit from Rhodopseudomonas marina. Part II. Electron microscopic characterisation. FEBS Lett. 1992;311:135–138. doi: 10.1016/0014-5793(92)81384-x. [DOI] [PubMed] [Google Scholar]
- 51.Meckenstuck R, Krusche K, Staehelin L A, Cryklaff M, Brunisholz R A, Zuber H. The six fold symmetry of the B880 light-harvesting membranes of Rhodopseudomonas marina. Biol Chem Hoppe-Seyler. 1994;375:429–438. doi: 10.1515/bchm3.1994.375.7.429. [DOI] [PubMed] [Google Scholar]
- 51a.Michel, H. Personal communication.
- 52.Miller K. Three dimensional structure of a photosynthetic membrane. Nature. 1982;300:53–55. [Google Scholar]
- 53.Montoya G, Cyrkloft M, Sinning I. Two-dimensional crystallisation and preliminary structure analysis of light-harvesting II (B800-850) complex from the purple bacterium Rhodovulum sulphidophilum. J Mol Biol. 1995;250:1–10. doi: 10.1006/jmbi.1995.0353. [DOI] [PubMed] [Google Scholar]
- 54.Olsen J D, Hunter C N. Protein structure modelling of the bacterial light-harvesting complex. Photochem Photobiol. 1994;60:521–535. doi: 10.1111/j.1751-1097.1994.tb05144.x. [DOI] [PubMed] [Google Scholar]
- 55.Otte S C M, Kleinherenbrink F A M, Amesz J. Energy transfer between the reaction centre and the antenna in purple bacteria. Biochim Biophys Acta. 1993;1143:84–90. [Google Scholar]
- 56.Papiz M Z, Prince S M, Hawthornthwaite-Lawless A M, McDermott G, Freer A A, Isaacs W W, Cogdell R J. A model for the photosynthetic apparatus of purple bacteria. Trends Plant Sci. 1996;1:198–206. [Google Scholar]
- 57.Peters J D, Drews G. The transverse membrane orientation of the light-harvesting and reaction centre polypeptides of Rhodopseudomonas capsulata. FEBS Lett. 1998;162:57–60. [Google Scholar]
- 58.Phillips-Jones M K. Light regulation of pigment-protein gene expression in Rhodobacter species. Symp Soc Gen Microbiol. 1998;56:159–184. [Google Scholar]
- 59.Prince S M, Papiz M Z, Freer A A, McDermott G, Hawthornthwaite-Lawless A M, Cogdell R J, Isaacs N W. Apoprotein structure in the LH2 complex from Rhodopseudomonas acidophilastrain 10050: modular assembly and protein pigment interaction. J Mol Biol. 1997;268:412–423. doi: 10.1006/jmbi.1997.0966. [DOI] [PubMed] [Google Scholar]
- 59a.Prince, S. M., et al. Unpublished data.
- 59b.Prince, S. M. February 1999, revision date. [Online.] http://www.chem.gla.ac.uk/protein/LH2/lh2.html. [17 May 1999, last date accessed.]
- 60.Pugh R, McGlynn P, Jones M R, Hunter C N. The LH1-RC complex of Rhodobacter sphaeroidesinteraction between components, time dependent assembly and topology of the PufX. Biochim Biophys Acta. 1998;1366:301–316. doi: 10.1016/s0005-2728(98)00131-5. [DOI] [PubMed] [Google Scholar]
- 61.Pullerits T, Sundström V. Photosynthetic light-harvesting pigment-protein complexes. Toward understanding how and why. Acc Chem Res. 1996;29:381–389. [Google Scholar]
- 62.Recchia R A, Davis C M, Lilburn T G, Beatty J T, Parkes-Loach P A. Isolation of the PufX protein from Rhodobacter capsulatus and Rhodobacter sphaeroidesevidence for its interaction with the α-polypeptide of the core light-harvesting complex. Biochemistry. 1998;37:11055–11063. doi: 10.1021/bi980657l. [DOI] [PubMed] [Google Scholar]
- 63.Rhee K-H, Morris E P, Barber J P, Kühlbrandt W. Three-dimensional structure of the plant photosystem II reaction centre at 8 Å resolution. Nature. 1998;396:283–286. doi: 10.1038/24421. [DOI] [PubMed] [Google Scholar]
- 64.Robert B, Lutz M. Structure of antenna complexes of several Rhodospirillaceae from their resonance Raman spectra. Biochim Biophys Acta. 1985;807:10–23. [Google Scholar]
- 65.Shreve A P, Trautman J K, Frank H A, Owens T G, Albrecht A C. Femtosecond energy transfer processes in the B800-850 light-harvesting complex of Rhodobacter sphaeroides 2.4.1. Biochim Biophys Acta. 1991;1058:280–288. doi: 10.1016/s0005-2728(05)80248-8. [DOI] [PubMed] [Google Scholar]
- 66.Shubert W-D, Klukas O, Krass N, Saenger W, Fromme P, Witt H T. Photosystem I of Synechococcus elongatusat 4Å resolution: comprehensive structure analysis. J Mol Biol. 1997;272:741–769. doi: 10.1006/jmbi.1997.1269. [DOI] [PubMed] [Google Scholar]
- 67.Sidler W. Phycobilisome and phycobiliprotein structures. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Press; 1994. pp. 139–216. [Google Scholar]
- 68.Stark W, Kühlbrandt W, Wildhaber I, Wehrli E, Mühlethaler K. The structure of the photoreceptor unit of Rhodopseudomonas viridis. EMBO J. 1984;3:717–783. doi: 10.1002/j.1460-2075.1984.tb01884.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sundström V, Pullerits T, van Grondelle R. Photosynthetic light-harvesting: reconciling dynamics and structure of purple bacterial LH2 reveals function of photosynthetic unit. J Phys Chem B. 1999;103:2327–2346. [Google Scholar]
- 70.Thornber J R, Trosper T L, Strouse C E. Bacteriochlorophyll in vivo: relationships of spectral forms to specific membrane components. In: Clayton R K, Sistrom W R, editors. The photosynthetic bacteria. London, United Kingdom: Plenum Press; 1978. pp. 133–160. [Google Scholar]
- 71.Tronrud D E, Matthews B W. Structure and X-ray amino acid sequence of the bacteriochlorophyll a protein from Prosthecochloris aestuarrüat 1.9 Å resolution. J Mol Biol. 1986;188:443–454. doi: 10.1016/0022-2836(86)90167-1. [DOI] [PubMed] [Google Scholar]
- 71a.University of Illinois Website. [Online.] http://www.Ks.uiuc.edu/Research/psu/psu.html. [17 May 1999, last date accessed.]
- 72.van Grondelle R, Dekker J P, Gillbro T, Sundström V. Energy transfer and trapping in photosynthesis. Biochim Biophys Acta. 1994;1187:1–65. [Google Scholar]
- 73.Walz T, Jamieson S J, Bowers C M, Bullough P A, Hunter C N. Projection structures of these photosynthetic complexes from Rhodobacter sphaeroides: LH2 at 6Å, LH1 and RC-LH1 at 25Å. J Mol Biol. 1998;282:833–845. doi: 10.1006/jmbi.1998.2050. [DOI] [PubMed] [Google Scholar]
- 74.Youvan D C, Ismail S, Bylina E J. Chromosomal deletion and plasmid complementation of photosynthetic reaction centre and light-harvesting genes from Rhodopseudomonas capsulata. Gene. 1985;38:19–30. doi: 10.1016/0378-1119(85)90199-4. [DOI] [PubMed] [Google Scholar]
- 75.Zuber H, Cogdell R L. Structure and organisation of purple bacterial antenna complexes. In: Blankenship R E, Madigan M T, Bauer C E, editors. Anoxygenic photosynthetic bacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 315–348. [Google Scholar]
- 76.Zuber H, Brunisholz R A. Structure and function of antenna polypeptides and chlorophyll protein complexes: principles and versatility. In: Scheer H, editor. The chlorophylls. Boca Raton, Fla: CRC Press; 1991. pp. 627–703. [Google Scholar]
- 77.Zurdo J, Fernandez C, Ramirez J M. A structural role of the carotenoid in the light-harvesting II protein of Rhodobacter capsulatus. Biochem J. 1993;290:531–537. doi: 10.1042/bj2900531. [DOI] [PMC free article] [PubMed] [Google Scholar]