Abstract
The final destination of glycosylphosphatidylinositol (GPI)-attached proteins in Saccharomyces cerevisiae is the plasma membrane or the cell wall. Two kinds of signals have been proposed for their cellular localization: (i) the specific amino acid residues V, I, or L at the site 4 or 5 amino acids upstream of the GPI attachment site (the ω site) and Y or N at the site 2 amino acids upstream of the ω site for cell wall localization and (ii) dibasic residues in the region upstream of the ω site (the ω-minus region) for plasma membrane localization. The relationships between these amino acid residues and efficiencies of cell wall incorporation were examined by constructing fusion reporter proteins from open reading frames encoding putative GPI-attached proteins. The levels of incorporation were high in the constructs containing the specific amino acid residues and quite low in those containing two basic amino acid residues in the ω-minus region. With constructs that contained neither specific residues nor two basic residues, levels of incorporation were moderate. These correlations clearly suggest that GPI-attached proteins have two different signals which act positively or negatively in cell wall incorporation for their cellular localization.
Mannoproteins, which are one type of the components of the Saccharomyces cerevisiae cell wall, can be divided into three groups: sodium dodecyl sulfate (SDS)-extractable, reducing-reagent-extractable, and glucanase-extractable mannoproteins (5, 12, 23). Glucanase-extractable mannoproteins are covalently bound to β-1,6-glucan of the cell wall, which forms a large complex with β-1,3-glucan and chitin (9, 10, 11, 13, 17, 19, 20). Many genes encoding glucanase-extractable cell wall mannoproteins have been isolated from Saccharomyces cerevisiae, and so far most of them have been identified as glycosylphosphatidylinositol (GPI)-dependent cell wall proteins (15, 16, 18, 21, 24, 26, 28, 31, 32).
GPI-associated proteins have several common structural characteristics: a signal sequence for secretion in the N terminus and a GPI signal for attachment to GPI in the C terminus. The GPI signal itself has three domains: the region containing the GPI attachment site (the ω site) plus the first and second amino acids downstream of the ω site, a spacer of 5 to 10 amino acids, and a hydrophobic stretch of 10 to 15 amino acids (22, 29). A protein containing the GPI signal is cleaved at the ω site, and the resulting carboxy terminus of the protein is covalently bound to a GPI moiety (23, 30). This reaction occurs in the endoplasmic reticulum. Being associated with membranes by means of the GPI moiety, GPI-attached proteins are then transported to the cell surface and remain on the plasma membrane as GPI-anchored proteins; however, some of them are further processed. They are incorporated into the cell wall by detaching themselves from the GPI moiety and then by linking themselves to β-1,6-glucan of the cell wall (6, 30). These different processes suggest that each GPI-attached protein has a signal for selecting either to be incorporated into the cell wall or to remain on the plasma membrane. Two kinds of amino acid sequences in the region upstream of the ω site (the ω-minus region) have been proposed as being responsible for the selection: dibasic residues for remaining on the plasma membrane (3, 33) and specific amino acid residues at sites 4 or 5 and 2 amino acids upstream of the ω site (ω-4/5 and ω-2 sites, respectively) for incorporation into the cell wall (7). The former residues were determined by sequence analysis, and the latter residues were determined by mutational analysis mainly by using synthetic model sequences. We now need to evaluate these proposals with more authentic sequences of GPI-attached proteins. A number of open reading frames (ORFs) encoding putative GPI-attached proteins have been selected from previous studies (3, 8). In this study, we quantitatively measured the cell wall incorporation of fusion reporter proteins that were constructed from these ORFs and examined the relationship between efficiencies of cell wall incorporation and amino acid residues in their ω-minus regions.
MATERIALS AND METHODS
Nucleotide sequences.
The Saccharomyces Genome Database was used as a source of nucleotide and amino acid sequences of all the ORFs examined, except for YBR078W. Our sequence analysis of YBR078W identified an insertion of C at nucleotide position 1610, resulting in reduction of the ORF size from 468 to 429 amino acids. The C-terminal amino acid sequence used for YBR078W is shown in Table 1.
TABLE 1.
Efficiencies of incorporation of the fusion reporter proteins into cell walls
| ORFa/GPI-attached protein | Amino acid sequence fused to the reporter proteinb | Cell wall incorporationc |
|---|---|---|
| YDR534C | IFTNGKSSTTPQIVNYT G AADSIAAGTGLMGAALAAVIFL* | 0.421 ± 0.045 |
| YIR019C/FLO11 | SGSAVATYSVPSISSTYQ G AANIKVLGNFMWLLLALPVVF* | 0.409 ± 0.122 |
| YNL327W/ETG2 | ITWYSSSTIKPPSISTYS G AAGQLTIRIGSLLLGLISFLL* | 0.380 ± 0.060 |
| YOR214C | PTITPGNITTIGGYE N SSSSLMPSMGILSFLFGLYLLLHP* | 0.338 ± 0.065 |
| YDR134C | TEKPTQQGSSTQTVTSYT G AAVKALPAAGALLAGAAALLL* | 0.325 ± 0.025 |
| YPL130W | TPYSNISSLNEDYD N ASNFLTPTTVALAVLLTILLFIQAY* | 0.315 ± 0.099 |
| YOR009W | TVVSVQSKTTGIVEQTE N GAAKAVIGMGAGALAAVAAMLL* | 0.308 ± 0.074 |
| YER150W | KPTSETSVSSTHDVETNS N AANARAIPGALGLAGAVMMLL* | 0.299 ± 0.022 |
| YDR077W/SED1 | STVVPVSSSASSHSVVINS N GANVVVPGALGLAGVAMLFL* | 0.297 ± 0.062 |
| YOR383C | TSSSATSSSTAELSSYT G AADAITAGTGLMGAALAAVMLL* | 0.291 ± 0.062 |
| YJR151C | SIAHSSASYTVSINT N GAYNFDKDNIFGTAIVAVVALLLL* | 0.280 ± 0.053 |
| YJR004C/Agα1 | IQQNFTSTSLMISTYE G KASIFFSAELGSIIFLLLSYLLF* | 0.265 ± 0.049 |
| YJL078C/PRY3 | TSTTAKLSAYE G AATPLSIFQCNSLAGTIAAFVVAVLFAF* | 0.263 ± 0.053 |
| YLR110C | APKNTTSAAPTHSVTSYT G AAAKALPAAGALLAGAAALLL* | 0.220 ± 0.040 |
| YNL300W | SISSQANTTTHEISTYV G AAVKGSVAGMGAIMGAAAFALL* | 0.214 ± 0.024 |
| YEL040W/UTR2 | SSVSGSASSSTSSMSGN N AGANVAANWRLTVLCVILGYVL* | 0.197 ± 0.060 |
| YLR042C | TKSSQTTNTISSSTSTG G VGSVKPCLYFVLMLETIAYLFS* | 0.179 ± 0.018 |
| YOR382W | TSSSSSSSSSSSSASSS G AAPAAFQGASVGALALGLISYLL* | 0.136 ± 0.026 |
| YLR040C | ALISASNSSSTSRTSQSQ N GAHAKSLYFPMALFGIFAVAL* | 0.123 ± 0.008 |
| YMR006C | SSSTARSSSSTANKA N AAAISYANTNTLMSLLGAITALFGLI* | 0.116 ± 0.038 |
| YJL171C | SATAPSTTSKS N GVALTKMQNGVWYYILAIFTAFTQVVLI* | 0.105 ± 0.028 |
| YIL011W/YIB1 | EKSTNSSSSATSK N AGAAMDMGFFSAGVGAAIAGAAAMLL* | 0.085 ± 0.033 |
| YOL030W/GAS5 | SLLKSAASATSSSQSSSKSK G AAGIIEIPLIFRALAELYNLVL* | 0.081 ± 0.028 |
| YDR055W | SSGASSSSSKSSKG N AAIMAPIGQTTPLVGLLTAIIMSIM* | 0.060 ± 0.028 |
| YBR078w/ECM33 | AQANVSASASSSSSSSKKSK G AAPELVPATSFMGVVAAVGVALL* | 0.059 ± 0.003 |
| YNL190W | GPGEKARKN N AAPGPSNFNSIKLFGVTAGSAAVAGALLLL* | 0.057 ± 0.038 |
| YDR144C/YAP2 | SSTGMLSPTSSSSTRKE N GGHNLNPPFFARFITAIFHHI* | 0.056 ± 0.026 |
| YIR039C/YAP6 | SSFSSSGGSSESTTKKQ N AGYKYRSSFSFSLLSFISYFLL* | 0.055 ± 0.014 |
| YLR194C | YKSTVNGKVASVMSNST N GATAGTHIAYGAGAFAVGALLL* | 0.052 ± 0.055 |
| YLR120C/YAP3 | SGNLTTSTASATSTSSKR N VGDHIVPSLPLTLISLLFAFI* | 0.051 ± 0.029 |
| YDR522C/SPS2 | GKNGAKSQGSSKKME N SAPKNIFIDAFKMSVYAVFTVLFSIIF* | 0.051 ± 0.057 |
| YMR215W/GAS3 | TGSSSASSSSKSK G VGNIVNVSFSQSGYLALFAGLISALL* | 0.046 ± 0.030 |
| YMR008C/PLB1 | ASGSSTHKK N AGNALVNYSNLNTNTFIGVLSVISAVFGLI* | 0.045 ± 0.025 |
| YOL132W/GAS4 | EDADEDKDDLKRKHR N SASISGPLLPLGLCLLFFTFSLFF* | 0.042 ± 0.031 |
Six ORFs, YMR006C, YOL030W, YBR078W, YLR194C, and YDR522C, were originally identified to encode putative GPI-attached proteins through an in silicio sequence analysis by Caro et al. (3). Three ORFs, YIR019C, YOR383C, and YOR382W, were not studied for cell wall localization in our previous study (8).
The potential GPI attachment site (the ω site) of each sequence (3, 8) is indicated in the middle, sandwiched between spacer regions. The amino acid residues I, V, and L at the ω-5 site and Y and N at the ω-2 site are marked in bold, and the basic amino acid residues K and R in the short ω-minus region are marked in bold plus italic. The potential N-glycosylation sites are double underlined.
Cell wall incorporation is represented as ratios of the amounts of laminarinase-extractable cell wall HA-tagged proteins to total amounts of HA-tagged proteins in crude cell lysate, including the cell wall. Values are means with standard deviations from measurements of four different transformants.
Expression of a fusion reporter protein.
A DNA fragment covering the most C-terminal 40 amino acids of a GPI-attached protein and about 200 bp of its 3′ noncoding region was amplified by PCR with primers containing a SalI or BamHI restriction site. The resulting fragment was inserted into the SalI and BamHI sites of a multicopy plasmid, pEαGALHA (8). pEαGALHA contains a reporter gene comprising the signal sequence of yeast invertase, the α-galactosidase from Cyamopsis tetragonoloba, and a hemagglutinin (HA) epitope. The plasmid containing the fusion reporter gene was used to transform the yeast strain YPH499 (27).
Construction of genes encoding GPI proteins tagged with HA.
A fragment containing the 400 bp of the 5′ noncoding region and the N-terminal 25 amino acids of Yer150w was amplified by PCR and inserted into the SacI and BamHI sites of a plasmid, pEHA. pEHA contains the triple-HA epitope sequence between the BamHI and SalI sites of the multicloning site of a multicopy plasmid, YEplac195 (7). Another fragment containing the C-terminal 122 amino acids of Yer150w and the 300 bp of its 3′ noncoding region was amplified by PCR and inserted into the SalI and HindIII sites, generating pEHAYer150w. Similarly, the triple-HA epitope sequence was inserted in frame into the 25th amino acid position of the Yor382w sequence on pEHAYor382w.
Analysis of cell wall-associated proteins.
Cell walls were isolated from yeast transformant cells grown in a selection medium. The yeast cells were broken with glass beads in LY buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 1 mM phenylmethylsulfonyl fluoride) containing 2% SDS. The pellet generated by centrifugation was twice treated with hot 2% SDS in LY buffer by heating it for 10 min at 95°C each time, followed by washing it five times with LM buffer (100 mM sodium acetate [pH 5.5], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride). The washed cell walls were incubated with laminarinase (product no. L5144; Sigma) at a concentration of 0.25 U/ml twice for 1 h each time at 37°C. The treated sample was centrifuged in a microcentrifuge at 10,000 rpm for 2 min at 4°C, and the resulting supernatant was analyzed by SDS-polyacrylamide gel electrophoresis and Western blotting. Protein bands on the blots were detected with the anti-HA monoclonal antibody 12CA5 (Boehringer Mannheim). The band intensities were measured with an Image Master ID (Pharmacia Biotech) and were represented as mean values with standard deviations from measurements of four different transformants.
RESULTS
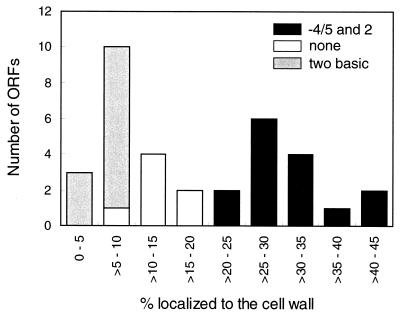
The most C-terminal 40 amino acid sequence of each ORF was fused to a reporter protein comprising the invertase signal sequence, an α-galactosidase from guar, and an HA epitope. The fusion reporter gene is transcribed by the PGK1 promoter (8). A plasmid containing the fusion reporter gene was introduced into the yeast strain YPH499. Cell wall fractions were isolated from each transformant, digested with glucanase, and analyzed on Western blots. The amounts of glucanase-extracted fusion reporter proteins were measured, and the ratios of their amounts to the total amounts of fusion reporter proteins in cells were calculated (Table 1). Table 1 also shows the amino acid sequences of ORFs used to construct the fusion reporter genes. Among the 34 fusion reporter proteins examined, 15 have amino acid residues of I, V, or L at the ω-4/5 site and Y or N at the ω-2 site, 11 have the basic amino acid residues K and R or K alone in the short ω-minus region, and the remaining 8 have none of these distinguishing residues. These fusion reporter proteins were incorporated into the cell wall in various ratios from 0.421 for the fusion reporter protein of Ydr534c (FP-Ydr534c) to 0.042 for FP-Yol132w. Fusion reporter proteins constructed from two well-characterized GPI-dependent cell wall proteins, Sed1p (also called Ydr077w) (25, 32) and Agα1p/Yjr004c (19, 34), showed ratios of 0.297 and 0.265, respectively, while that of a well-characterized GPI-dependent plasma membrane protein, Yap3p/Ylr120c (1, 4, 14), showed a ratio of 0.051. A histogram based on the results in Table 1 clearly indicates the correlation between efficiencies of cell wall incorporation and sequences in the ω-minus region (Fig. 1): cell wall incorporation was higher in the fusion reporter proteins containing the specific amino acid residues I, V, or L at the ω-4/5 site and Y or N at the ω-2 site than in those containing no such specific amino acid residues.
FIG. 1.
Relationship between efficiencies of cell wall incorporation and sequences of the ω-minus region. The data of Table 1 were used for this histogramic representation. Results shown are for sequences with the specific amino acid residue of I, V, or L at the ω-4/5 site and Y or N at the ω-2 site (−4/5 and 2); sequences with two basic amino acid residues R and/or K in the ω-minus region (two basic); and sequences with neither the specific amino acid residues nor the two basic residues (none).
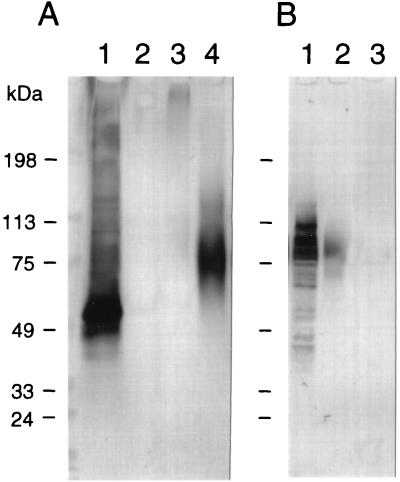
Two proteins, Yer150w, which has V at the ω-5 site and N at the ω-2 site, and Yor382w, which has a stretch of serine residues in the ω-minus region, were examined for their cell wall incorporation. An HA epitope was inserted at the 25th amino acid position from the first methionine of each protein, generating the HA-tagged proteins Yer150w-HA and Yor382w-HA, and their cellular localization was examined. Of the total amount of Yer150w-HA protein, 12.7% was detected as glucanase-extractable cell wall protein (Fig. 2A). This level was much higher than that of Yor382w-HA. The amount of Yor382w-HA released from the cell wall by glucanase was only 2.9% of the total amount of Yor382-HA protein in the cells (Fig. 2B). Yer150w-HA has three potential N-glycosylation sites, while Yor382w-HA has not. Consistent with these characteristics, the Yer150w-HA bands appeared differently on Western blots before and after endoglycosidase H treatment (Fig. 2A, lanes 3 and 4), while the Yor382w-HA bands appeared to be unchanged before and after the treatment (data not shown). These results suggested that N glycosylation occurs at least at one of the three potential sites in Yer150w-HA.
FIG. 2.
Cell wall incorporation of HA-tagged proteins. Cell wall fractions were prepared from transformant cells and analyzed by Western blotting. (A) Ypl130w-HA. Lane 1, crude cell lysate; lane 2, cell wall fraction; lane 3, cell wall fraction treated with laminarinase; lane 4, cell wall fraction treated with laminarinase and endoglycosidase H (7). The sample amounts in lanes 2 to 4 were five times more than that in lane 1. (B) Yor382w-HA. Lane 1, crude cell lysate; lane 2, cell wall fraction treated with laminarinase and endoglycosidase H; lane 3, cell wall fraction. The sample amounts in lanes 2 and 3 were five times more than that in lane 1.
DISCUSSION
We demonstrated in a previous study that the amino acid sequences at the ω-minus region are important for the determination of the final cellular localization of GPI-attached proteins and that the amino acid residues V or I at the ω-4/5 site and V, I, N, or Y at the ω-2 site are required for the efficient incorporation of GPI-attached proteins into the cell wall (7). This requirement, called the ω-4/5 and ω-2 rule, was identified through a mutational analysis of synthetic ω-minus regions. In this study, this rule was verified, as shown in the histogram of Fig. 1. Fusion reporter proteins which have the specific amino acid residues of I, V, or L at the ω-5 site and Y or N at the ω-2 site were incorporated into the cell wall with greater efficiency than those which have no specific amino acid residues in the short ω-minus region. A similar correlation was also observed for intact proteins tagged with HA. The level of cell wall incorporation of Yer150w-HA was much higher than that of Yor382w-HA. Yer150w meets the rule, while Yor382 does not. Our results clearly indicate that these specific residues function as a signal that enhances the incorporation efficiency of GPI-attached proteins into the cell wall. Furthermore, in keeping with the previous findings that replacement of S with Y in a synthetic sequence comprising a serine stretch partially enhances cell wall incorporation (7), efficiencies of the cell wall incorporation of FP-Yir019c and FP-Ypl130w, both of which have only Y at the ω-2 site, were comparable to those of fusion reporter proteins containing the specific amino acid residues in both of the ω-4/5 and ω-2 sites. This suggests that specific amino acid residues only at the ω-2 site in some sequences may be enough for efficient incorporation of GPI proteins into the cell wall. To reach a conclusion, however, further examination will be required.
Levels of glucanase-extractable cell wall proteins varied among the fusion reporter proteins with the specific amino acid residues, suggesting that some factors other than those of the ω-4/5 and ω-2 rule may modify the efficiency of cell wall incorporation. These factors are probably governed by the amino acid sequences themselves, and it is possible that glycosylation of proteins may be one of these factors. However, N glycosylation would be excluded from the possibility. Among the 15 ORFs, YOR214C, YPL130W, YLR110C, and YNL300W have potential N-glycosylation sites within the sequences used for the construction of the fusion reporter proteins (Table 1). In fact, their fusion reporter proteins were detected as broad and smeared bands at considerably higher molecular-mass positions on Western blots (data not shown). Ratios of cell wall incorporation were 0.338, 0.315, 0.220, and 0.214 for FP-Yor214c, FP-Ypl130w, FP-Ylr110c, and FP-Ynl300w, respectively, indicating that there is no correlation between N glycosylation and levels of cell wall incorporation. In our previous study, Ypl130w was not classified as a putative GPI-dependent cell wall protein, because the fusion protein was not detected in the cell wall fraction treated with glucanase (8). In this study, it was clearly detected. This difference in detection may be due to difficulties in detecting N-glycosylated proteins on Western blots. On the other hand, it is possible that O glycosylation of proteins may partly explain the diversity of cell wall incorporation. Considering the S/T richness of the ω-minus regions, it is likely that the fusion reporter proteins are O glycosylated to various extents. Deletion of several genes required for O glycosylation has been known to affect the incorporation efficiency of GPI-attached proteins into the cell wall (2). Further examinations are needed to reach a conclusion.
Vossen et al. have suggested from sequence analysis that a dibasic motif in the ω-minus region may participate in a signal for detaining GPI-attached proteins on the plasma membrane (33). Our results support this proposal. As clearly shown in Fig. 1, incorporation ratios of all the fusion reporter proteins that contain two basic amino acid residues in the ω-minus region into the cell wall were quite low. These ratios were lower than those of the fusion reporter proteins that did not have the two basic amino acid residues. Furthermore, all of the fusion reporter proteins examined in this study were detected in a detergent phase by phase separation with Triton X-114, and the proteins extracted in the detergent phase were phosphatidylinositol-phospholipase C sensitive (data not shown), indicating GPI attachment. Therefore, it can be concluded that with fusion reporter proteins with two basic amino acid residues in the ω-minus region, nearly all the amounts of their proteins remain on the plasma membrane in a GPI-associated form. These two basic residues probably function as a negative factor in the process of cell wall incorporation.
Taken together, these results indicate that GPI-attached proteins have two different signals for determining their cellular localization. These are the amino acid residues V, I, or L at the ω-4/5 site and Y or N at the ω-2 site and the basic residue(s) R and/or K in the short ω-minus region. The former residues act as a positive factor for cell wall incorporation, while the later residues act as a negative factor for incorporation, such that the proteins remain on the plasma membrane as GPI-anchored proteins.
ACKNOWLEDGMENT
We thank S. Bernice Miwa for critical reading of the manuscript.
REFERENCES
- 1.Ash J, Dominguez M, Bergeron J J M, Thomas D Y, Bourbonnais Y. The yeast proprotein convertase encoded by YAP3 is a glycophosphatidylinositol-anchored protein that localizes to the plasma membrane. J Biol Chem. 1995;270:20847–20854. doi: 10.1074/jbc.270.35.20847. [DOI] [PubMed] [Google Scholar]
- 2.Bourdineaud J-P, Van Der Vaart J M, Donzeau M, De Sampaio G, Verrips C T, Lauquin G J-M. Pmt1 monnosyl transferase is involved in cell wall incorporation of several proteins in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:85–98. doi: 10.1046/j.1365-2958.1998.00660.x. [DOI] [PubMed] [Google Scholar]
- 3.Caro L H P, Tettelin H, Vossen J H, Ram A F J, Van Den Ende H, Klis F M. in silicio identification of glycosyl-phosphatidylinositol-anchored plasma membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast. 1997;13:1477–1489. doi: 10.1002/(SICI)1097-0061(199712)13:15<1477::AID-YEA184>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 4.Cawley N X, Wong M, Pu L-P, Tam W, Loh Y P. Secretion of yeast aspartic protease 3 is regulated by its carboxy-terminal tail: characterization of secreted YAP3p. Biochemistry. 1995;34:7430–7437. doi: 10.1021/bi00022a016. [DOI] [PubMed] [Google Scholar]
- 5.Cid V J, Duran A, Del Rey F, Snyder M P, Nombela C, Sanchez M. Molecular basis of cell integrity and morphogenesis in Saccharomyces cerevisiae. Microbiol Rev. 1995;59:345–386. doi: 10.1128/mr.59.3.345-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Nobel H, Lipke P N. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell Biol. 1994;4:42–45. doi: 10.1016/0962-8924(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 7.Hamada K, Terashima H, Arisawa M, Kitada K. Amino acid sequence requirement for efficient incorporation of glycosylphosphatidylinositol-associated proteins into the cell wall of Saccharomyces cerevisiae. J Biol Chem. 1998;273:26946–26953. doi: 10.1074/jbc.273.41.26946. [DOI] [PubMed] [Google Scholar]
- 8.Hamada K, Fukuchi S, Arisawa M, Baba M, Kitada K. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol Gen Genet. 1998;258:53–59. doi: 10.1007/s004380050706. [DOI] [PubMed] [Google Scholar]
- 9.Kapteyn J C, Ram A F J, Groos E M, Kollar R, Montijn R C, Van Den Ende H, Llobell A, Cabib E, Klis F M. Altered extent of cross-linking of β1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall β1,3-glucan content. J Bacteriol. 1997;179:6279–6284. doi: 10.1128/jb.179.20.6279-6284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapteyn J C, Van Den Ende H, Klis F M. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim Biophys Acta. 1999;1426:373–383. doi: 10.1016/s0304-4165(98)00137-8. [DOI] [PubMed] [Google Scholar]
- 11.Kapteyn J C, Montijn R C, Vink E, De La Cruz J, Llobell A, Douwes J E, Shimoi H, Lipke P N, Klis F M. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked β-1,3-/β-1,6-glucan heteropolymer. Glycobiology. 1996;6:337–345. doi: 10.1093/glycob/6.3.337. [DOI] [PubMed] [Google Scholar]
- 12.Klis F M. Cell wall assembly in yeast. Yeast. 1996;10:851–869. doi: 10.1002/yea.320100702. [DOI] [PubMed] [Google Scholar]
- 13.Kollar R, Reinhold B B, Petrakova E, Yeh H J, Ashwell G, Drgonova J, Kapteyn J C, Klis F M, Cabib E. Architecture of the yeast cell wall β(1→6)-glucan interconnects mannoprotein, β(1→3)-glucan, and chitin. J Biol Chem. 1997;272:17762–17775. doi: 10.1074/jbc.272.28.17762. [DOI] [PubMed] [Google Scholar]
- 14.Komano H, Fuller R S. Shared functions in vivo of a glycosyl-phosphatidylinositol-linked aspartyl protease, Mkc7, and the proprotein processing protease Kex2 in yeast. Proc Natl Acad Sci USA. 1995;92:10752–10756. doi: 10.1073/pnas.92.23.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo K, Inouye M. TIP1, a cold-shock-inducible gene of Saccharomyces cerevisiae. J Biol Chem. 1991;266:17537–17544. [PubMed] [Google Scholar]
- 16.Kowalski L R Z, Kondo K, Inouye M. Cold-shock induction of a family of TIP1-related proteins associated with the membrane in Saccharomyces cerevisiae. Mol Microbiol. 1995;15:341–353. doi: 10.1111/j.1365-2958.1995.tb02248.x. [DOI] [PubMed] [Google Scholar]
- 17.Lipke P N, Ovalle R. Cell wall architecture in yeast: new structure and new challenges. J Bacteriol. 1998;180:3735–3740. doi: 10.1128/jb.180.15.3735-3740.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lipke P N, Wojciechowicz D, Kurjan J. Agα1 is the structural gene for the Saccharomyces cerevisiae α-agglutinin, a cell surface glycoprotein involved in cell-cell interactions during mating. Mol Cell Biol. 1989;9:3155–3165. doi: 10.1128/mcb.9.8.3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C-F, Montijn R C, Brown J L, Klis F, Kurjan J, Bussey H, Lipke P N. Glycosyl phosphatidylinositol-dependent cross-linking of α agglutinin and β1,6-glucan in the S. cerevisiae cell wall. J Cell Biol. 1995;128:333–340. doi: 10.1083/jcb.128.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Montijn R C, Van Rinsum J, Van Schagen F A, Klis F M. Glucomannoproteins in the cell wall of Saccharomyces cerevisiae contain a novel type of carbohydrate side-chain. J Biol Chem. 1994;269:19338–19342. [PubMed] [Google Scholar]
- 21.Moukadiri I, Armero J, Abad A, Sentandreu R, Zueco J. Identification of a mannoprotein present in the inner layer of the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1997;179:2154–2162. doi: 10.1128/jb.179.7.2154-2162.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nuoffer C, Horvath A, Riezman H. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J Biol Chem. 1993;268:10558–10563. [PubMed] [Google Scholar]
- 23.Orlean P. Biogenesis of yeast wall and surface components. In: Pringle J R, Broach J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. 3. Cell cycle and cell biology. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 229–362. [Google Scholar]
- 24.Roy A, Lu C F, Marykwas D L, Lipke P N, Kurjan J. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol Cell Biol. 1991;11:4196–4206. doi: 10.1128/mcb.11.8.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimoi H, Kitagaki H, Ohmori H, Iimura Y, Ito K. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J Bacteriol. 1998;180:3381–3387. doi: 10.1128/jb.180.13.3381-3387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimoi H, Iimura Y, Obata T. Molecular cloning of CWP1: a gene encoding a Saccharomyces cerevisiae cell wall protein solubilized with Rarobacter faecitabidus protease I. J Biochem. 1995;118:302–311. doi: 10.1093/oxfordjournals.jbchem.a124907. [DOI] [PubMed] [Google Scholar]
- 27.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Teunissen A W R H, Steensma H Y. The dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast. 1995;11:1001–1013. doi: 10.1002/yea.320111102. [DOI] [PubMed] [Google Scholar]
- 29.Udenfriend S, Kodukula K. How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu Rev Biochem. 1995;64:563–591. doi: 10.1146/annurev.bi.64.070195.003023. [DOI] [PubMed] [Google Scholar]
- 30.Van Der Vaart J M, Verrips C T. Cell wall proteins of Saccharomyces cerevisiae. Biotechnol Genet Eng Rev. 1998;15:387–411. doi: 10.1080/02648725.1998.10647963. [DOI] [PubMed] [Google Scholar]
- 31.Van Der Vaart J M, Caro L H P, Chapman J W, Klis F M, Verrips C T. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3104–3110. doi: 10.1128/jb.177.11.3104-3110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Der Vaart J M, Biesebeke R, Chapman J W, Toshka H Y, Klis F M, Verrips C T. Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl Environ Microbiol. 1997;63:615–620. doi: 10.1128/aem.63.2.615-620.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vossen J H, Muller W H, Lipke P N, Klis F M. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J Bacteriol. 1997;179:2202–2209. doi: 10.1128/jb.179.7.2202-2209.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wojciechowicz D, Lu C-F, Kurjan J, Lipke P N. Cell surface anchorage and ligand-binding domains of the Saccharomyces cerevisiae cell adhesion protein α-agglutinin, a member of the immunoglobulin superfamily. Mol Cell Biol. 1993;13:2554–2563. doi: 10.1128/mcb.13.4.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]