Abstract
Sialic acids are used as a receptor by several viruses and variations in the linkage type or C-5 modifications affect the binding properties. A species barrier for multiple viruses is present due to α2,3- or α2,6-linked sialic acids. The C-5 position of the sialic acid can be modified to form N-acetylneuraminic acid (Neu5Ac) or N-glycolylneuraminic acid (Neu5Gc), which acts as a determinant for host susceptibility for pathogens such as influenza A virus, rotavirus, and transmissible gastroenteritis coronavirus. Neu5Gc is present in most mammals such as pigs and horses but is absent in humans, ferrets, and dogs. However, little is known about C-5 content in wildlife species or how many C-5 modified sialic acids are present on N-linked glycans or glycolipids. Using our previously developed tissue microarray system, we investigated how 2 different lectins specific for Neu5Gc can result in varying detection levels of Neu5Gc glycans. We used these lectins to map Neu5Gc content in wild Suidae, Cervidae, tigers, and European hedgehogs. We show that Neu5Gc content is highly variable among different species. Furthermore, the removal of N-linked glycans reduces the binding of both Neu5Gc lectins while retention of glycolipids by omitting methanol treatment of tissues increases lectin binding. These findings highlight the importance of using multiple Neu5Gc lectins as the rich variety in which Neu5Gc is displayed can hardly be detected by a single lectin.
Keywords: host tropism, influenza A, sialic acid, tissue microarray, wildlife
Introduction
It has been widely accepted that for many pathogens the crucial determinant in host species susceptibility is the sialic acid type and its linkage on target tissues. Sialic acids are variably present in tissues and secretions of different species and can contain a plethora of additional modifications (Matrosovich et al. 2000; Tumpey et al. 2007; Paulson and de Vries 2013; de Graaf and Fouchier 2014). These modifications influence host-related functions and many viruses, in turn, have adapted to specific sialic acid modifications (Rogers et al. 1986; Varki and Gagneux 2012; Song et al. 2016; Barnard et al. 2019; Broszeit et al. 2019; Tortorici et al. 2019; Morniroli et al. 2020).
One such modification is at the C-5 carbon, which can contain an N-acetyl or N-glycolyl structure (Varki et al. 2015), forming N-acetylneuraminic acid (Neu5Ac) and N-glycolylneuraminic acid (Neu5Gc), respectively . In mammals, the Neu5Gc modification is created via hydroxylation of Neu5Ac by the cytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMAH) enzyme (Ito et al. 2000). This enzyme is expressed through the CMAH gene, which is present in many mammalian species but is non-functional in certain species such as dogs, ferrets, seals, hedgehogs, over a 100 different monkey species, and humans (Suzuki 2006; Ng et al. 2014; Springer et al. 2014; Peri et al. 2018). Furthermore, CMAH is also dysfunctional in white-tailed deer and 2 different bat lineages, 2 species that are important for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) zoonosis (Chandler et al. 2021; Alkhovsky et al. 2022), which may be an interesting detail since sialic acids are suggested to be used as attachment mediating co-receptors by SARS-CoV-2 (Sun 2021; Nguyen et al. 2022). In contrast, mammals such as horses and pigs have a functional CMAH gene and display high levels of Neu5Gc (Suzuki et al. 1997, 2000). Previous research has shown that avian, human, and equine influenza A virus hemagglutinins (HAs) bind in an exclusive manner to either Neu5Ac or Neu5Gc (Connor et al. 1994; Broszeit et al. 2019; Spruit et al. 2022). Specificity is also seen for other pathogens such as rotavirus and transmissible gastroenteritis virus, where strains alter between Neu5Gc or Neu5Ac preference based on their host species (Schultze et al. 1996; Yu et al. 2012; Burzynska et al. 2021). Therefore, it is important to elucidate the biochemical display of Neu5Gc and Neu5Ac on the epithelial surface of domestic and wild animal tissues as glycobiology appears to play a key role in driving host-pathogen interactions.
We aimed to determine the Neu5Gc content on epithelial cells in wildlife with 2 different lectins, which are defined as glycoproteins capable of binding glycan structures found on glycoproteins and glycolipids. Therefore, both the antibody and recombinant viral attachment protein used in this study will be classified as lectins (Taylor et al. 2015). The anti-Neu5Gc IgY antibody (αNeu5Gc) was used previously and is available commercially (Paul et al. 2020), and an H5/Vietnam/1203/04 influenza A HA protein specific for Neu5Gc (H5VNY161A HA protein), which we produced and used frequently in previous studies (de Vries et al. 2014; Broszeit et al. 2019; Spruit et al. 2021) has been used as well. We determined the dependency of these 2 lectins on the glycan classes such as N-linked glycans or glycolipids. We then determined the Neu5Ac and Neu5Gc content on the respiratory tissues of wild animals including Suidae and Cervidae species, tiger (Panthera tigris), and European hedgehog (Erinaceus europaeus) and showed how 2 different lectins specific for Neu5Gc have varying specificities in animal tissues depending on the glycan classes.
Using lectin histochemistry, we observed that the αNeu5Gc and H5VNY161A HA protein can differentiate between the glycan classes. Wild Suidae has fluctuating levels of Neu5Gc and Neu5Ac content between the upper and lower respiratory tissues. In Cervidae, the receptor recognition of the αNeu5Gc antibody and H5VNY161A HA protein differed, with αNeu5Gc recognizing a Neu5Gc glycan that is not bound by H5VNY161A HA, highlighting why it is essential to use several lectins to analyze glycan contents on tissues. Overall, we show that Neu5Gc display is highly variable in the respiratory system of different species.
Results
The αNeu5Gc antibody displays batch variability in Neu5Gc binding
We started by comparing the 2 Neu5Gc specific lectins, αNeu5Gc antibody and H5VNY161A HA protein, on the nasal tissue of horse, an established positive control animal for Neu5Gc detection (Broszeit et al. 2019; Spruit et al. 2022). The results showed that there was a difference in the binding consistency of the αNeu5Gc antibody between different vials (Fig. 1A). Histochemical staining with various αNeu5Gc antibody batches and H5VNY161A HA protein indicated that, while the H5VNY161A HA showed consistent binding over all nasal tissues in the horse tissue microarray (TMA), the αNeu5Gc antibody displayed varying binding properties, with vial 1 and 3 showing roughly similar binding patterns, whereas vial 2 shows no binding beyond the first horse (Fig. 1A). Histochemical stains with αNeu5Gc (vial 3) and H5VNY161A over the entire respiratory tract of the horse showed that the αNeu5Gc, while able to detect Neu5Gc presence in the epithelial layers, produced a much lower signal compared with H5VNY161A, whereas the H5VNWT HA shows that Neu5Ac is also present in the nasal epithelia of horse (Fig. 1B). These results show that the polyclonal nature of the αNeu5Gc antibody can be subject to vial variation. The αNeu5Gc antibody may also detect a narrower range of Neu5Gc receptors than the H5VNY161A stained the tissues deeper in the epithelial layer into the goblet cells in submucosal glands, suggesting there is Neu5Gc presence in goblet cells that the αNeu5Gc does not recognize. The difference in Neu5Gc range between the 2 lectins prompted us to analyze whether these lectins have a variable specificity to the different glycan classes.
Fig. 1.

Comparison of αNeu5Gc antibodies and Neu5Gc & Neu5Ac specific HA proteins. A) Lectin histochemical staining of various equine nasal tissues with H5VNY161A and various batches of the αNeu5Gc IgY antibody to showcase batch variation in Neu5Gc detection. B) Staining of equine respiratory tissues with the H5VNWT for α2,3-linked Neu5Ac presence, H5VNY161A & αNeu5Gc IgY (vial 3) for Neu5Gc presence and Maackia amurensis (MALII) lectin as a control for sialic acid presence.
Removal of N-linked Neu5Gc glycans reduces binding of both the αNeu5Gc antibody and the H5VNY161A HA protein on equine respiratory tissues
To test whether Neu5Gc is presented on N-glycans, we removed these N-glycans with PNGase F. Any residual binding by the lectins indicated O-linked glycans or glycolipid structures containing Neu5Gc. For both αNeu5Gc and H5VNY161A, a slight reduction in binding affinity can be observed after PNGase F treatment of the horse respiratory tissues (Fig. 2A), however, neither of the Neu5Gc specific lectins lost binding completely, to any of the tissues. Two controls were performed to confirm these findings. The H5VNWT HA (Fig. 2B), which is α2,3-linked Neu5Ac specific, showed that removal of N-linked glycans did not affect the presence of Neu5Ac sialic acids. The LCA (Lens culinaris agglutinin) lectin (Fig. 2B) specific for N-linked glycans showed that PNGase F did almost completely remove N-linked glycans. This confirms that although both αNeu5Gc and H5VNY161A can bind Neu5Gc containing N-linked glycans, the N-linkage is not an absolute requirement. Furthermore, the results suggest that a vast majority of binding comes from other glycan classes, potentially O-linked glycans or glycolipids. Indeed, the αNeu5Gc antibody was raised against a glycolipid (Diaz et al. 2009), whereas the H5VNY161A was serendipitously discovered after an alanine screen of conserved amino acid residues (Wang et al. 2012).
Fig. 2.
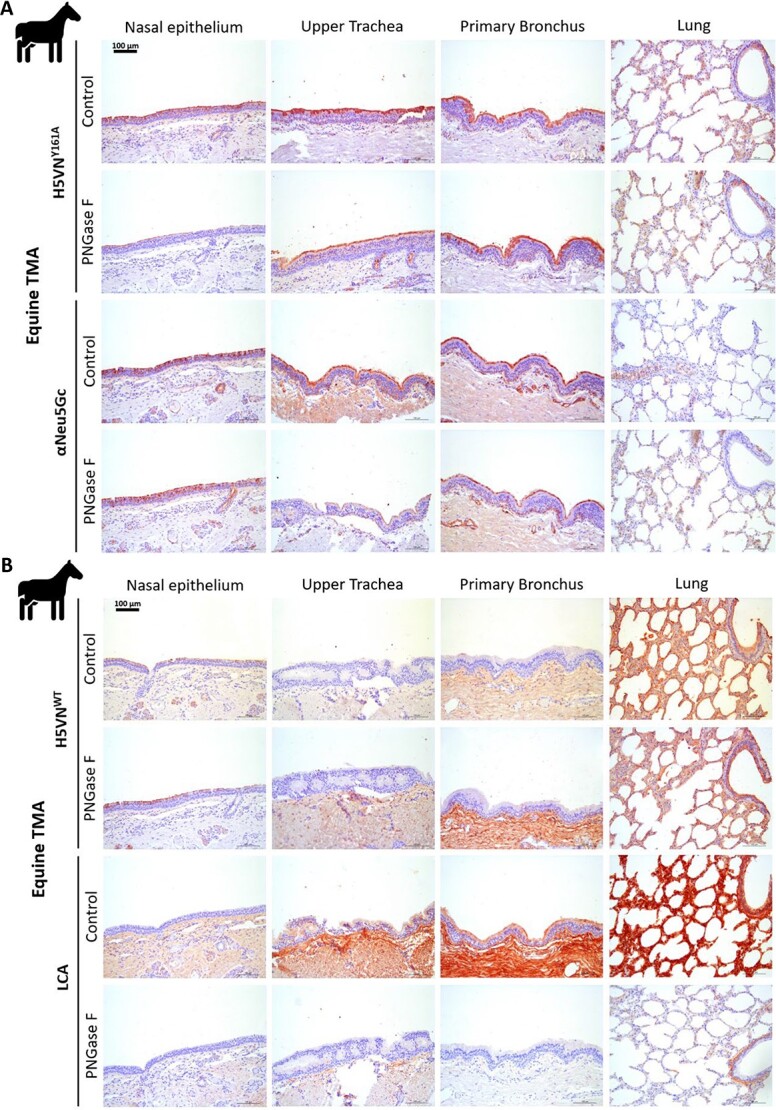
Lectin histochemistry of PNGase F treated tissues to remove N-linked glycans in equine. A) Lectin histochemical staining of H5VNY161A and αNeu5Gc IgY on PNGase F treated and untreated equine respiratory tissues to visualize dependency on N-linked glycans. B) Staining of PNGase F treated equine respiratory tissues with H5VNWT for α2,3-linked Neu5Ac presence and Lens culinaris agglutinin (LCA) lectin as a control for the presence of N-linked glycans.
Retention of glycolipids by omitting methanol treatment increases the binding of the H5VNY161A HA protein to equine respiratory tissues
It is a common practice in tissue histochemical assays to treat tissues with methanol to remove any potential innate peroxidase activity (Bussolati and Radulescu 2011; Magaki et al. 2019). However, these treatments, along with the paraffin-embedding procedure, potentially remove glycolipids that are embedded in the epithelial membrane (Table 1). We, therefore, conducted histochemistry experiments in which we did not treat horse respiratory tissue with methanol to compare whether retention of glycolipids affected the binding properties of αNeu5Gc and H5VNY161A to Neu5Gc containing glycans. A comparison of H5VNY161A binding on methanol-treated and untreated tissue shows that untreated tissue gave a higher binding signal compared with methanol-treated tissues (Fig. 3A). Striking however is that untreated tissue gave a lower signal for the αNeu5Gc lectin (Fig. 3A). Cholera toxin subunit B (CTX) protein, specific for glycolipids, showed that treatment with methanol removes all glycolipids on the cell surface consistently and that there are almost no glycolipids present in nasal tissue (Fig. 3B). Meanwhile, the precomplexed antibody control without any HA protein confirmed that there is no nonspecific binding of antibodies to glycolipids. These results show that glycolipid binding by H5VNY161A is possible but not an absolute requirement for binding to horse respiratory tissues. Now with an established protocol that no longer uses methanol to detect Neu5Gc structures, we aimed to study its presence in understudied wildlife species.
Table 1.
Tissue preparation, embedding, and processing steps with indications of potential loss of glycolipids and mucins.
| Process | Potential loss of glycolipids | Potential loss of mucins |
|---|---|---|
| Tissue fixation | ||
| Paraformaldehyde | ✔ | ✔ |
| Ethanol 70% | ||
| Dehydration & embedding | ||
| Ethanol 70% | ||
| Ethanol 96% | ||
| Xylene | ✔ | ✔ |
| Paraffin | ||
| Rehydration sectioned tissue | ||
| Xylene | ✔ | ✔ |
| Ethanol 96% | ||
| Ethanol 70% | ||
| Water | ||
| Histochemical staining treatments | ||
| Methanol treatment | ✔ |
Fig. 3.

Lectin histochemistry of methanol-treated and untreated equine tissues to retain glycolipids. A) Lectin histochemical staining of H5VNY161A and αNeu5Gc IgY proteins on methanol untreated equine respiratory tissues to visualize dependency of glycolipids. B) Staining of methanol untreated equine respiratory tissues with Cholera Toxin subunit B lectin (CTX) as a glycolipid control and a precomplexed antibody-only control of α-strep-tag mouse-HRP and goat-α-mouse-HRP.
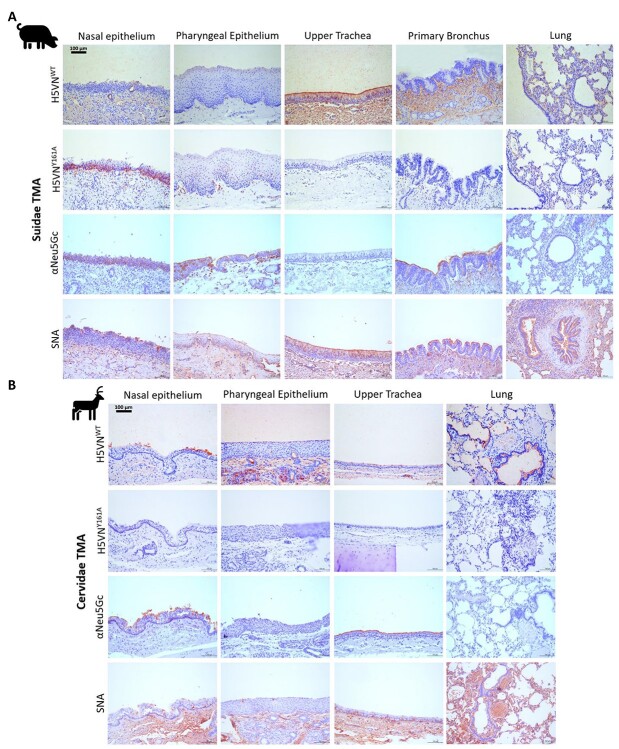
Both Suidae and Cervidae show a variance in Neu5Gc display
We then moved on to testing the Neu5Gc content in wild animals that can come into proximity to domestic species. For this, wild Suidae and Cervidae species were selected as their Neu5Gc content has not been mapped. Lectin histochemical stains showed that for wild Suidae (Fig. 4A, Sus scrofa shown) the content of Neu5Gc varied over the respiratory tract. Higher Neu5Gc concentrations were present in the nasal epithelium than further down the respiratory tract. The α2,6-linked specific SNA lectin showed binding over the entire respiratory tract. On the other hand the α2,3-linked Neu5Ac specific H5VNWT protein gains binding lower into the respiratory tract. This suggests that there is a gradient of Neu5Gc to Neu5Ac in the respiratory tract of wild Suidae. For the Cervidae however a consistent lack of binding could be seen for H5VNY161A (Fig. 4B, Capreolus capreolus shown) as well as only sporadic binding of the α2,6-linked specific SNA lectin, but curiously the αNeu5Gc did display binding over the entire respiratory tract except for the lung. This indicates that some Neu5Gc containing structures are present in Cervidae, which are recognized by αNeu5Gc, but not by H5VNY161A HA. Lastly, the H5VNWT did show binding on the Cervidae respiratory tract confirming that there is also α2,3-linked Neu5Ac present on the tissues in the epithelial layer alongside the Neu5Gc structures that the αNeu5Gc bound.
Fig. 4.
Lectin histochemistry for Neu5Ac and Neu5Gc detection on wild Suidae and Cervidae. A) Lectin histochemical staining of Suidae (Sus scrofa shown) respiratory tissues with H5VNWT for α2,3-linked Neu5Ac presence, H5VNY161A & αNeu5Gc IgY for Neu5Gc presence and Sambucus nigra (SNA) lectin as a control for α2,6-linked sialic acid presence. B) Staining of Cervidae (Capreolus capreolus shown) respiratory tissues with the lectins indicated in A.
Tiger is a Neu5Gc species while European hedgehog is a Neu5Ac species
Lastly, 2 wildlife species were tested from which in literature the presence of an intact CMAH gene was studied. The tiger was marked as a species containing Neu5Gc possibly over the entire respiratory system, whereas the European hedgehog was expected to not contain any Neu5Gc (Peri et al. 2018). For the tiger, only the Neu5Gc specific H5VNY161A mutant HA and α2,6-linked specific SNA lectin showed binding over the entire respiratory tract (Fig. 5A), whereas in the European hedgehog (Fig. 5B) only the Neu5Ac specific H5VNWT HA and α2,6-linked specific SNA lectin showed binding. These results in terms of Neu5Gc contents matched expectations found in the literature (Table 2; Peri et al. 2018). However, while H5VNY161A did show binding in tiger respiratory tissues αNeu5Gc did not, demonstrating again that there are Neu5Gc structures that only 1 of the 2 Neu5Gc lectins recognizes.
Fig. 5.
Lectin histochemistry for Neu5Ac and Neu5Gc detection on CMAH active and deficient species. A) Tiger respiratory tissues with H5VNWT for α2,3-linked Neu5Ac presence, H5VNY161A & αNeu5Gc IgY for Neu5Gc presence and Sambucus nigra (SNA) lectin as a control for α2,6-linked sialic acid presence. The black arrows highlight locations of binding for H5VNY161A. B) Staining of European hedgehog respiratory tissues with the lectins indicated in A.
Table 2.
Overview CMAH gene status in all tested domestic and wild animal species from literature (Peri et al. 2018).

|
Evaluation of the presence of intact CMAH genes
For the formation of Neu5Gc, an intact CMAH enzyme is required, but the presence of a seemingly intact CMAH gene does not automatically lead to (high) expression of the CMAH enzyme, and therefore presence of Neu5Gc (Jahan et al. 2021). Previously, the presence of intact CMAH genes was investigated for species of which a reference genome is available (Peri et al. 2018). It was shown that horses, wild boar, roe deer, and tigers have intact CMAH genes, whereas there is an incomplete CMAH gene in European hedgehogs. For some species (peccary and red river hog), no genome is available and thus the presence of an intact CMAH gene is unclear. We investigated whether intact CMAH genes were present in the other animals investigated in this article (Table 2), of which the genomes were present in the whole-genome shotgun database (Supplementary Table S1), which are the red deer, hog deer, and reindeer. In the red deer and hog deer, complete CMAH genes were observed. In reindeer, one exon could not be found and therefore we cannot be certain whether the CMAH gene yields an active enzyme. However histochemical staining results for reindeer matched the results found in Cervidae (Fig. 4A and B) suggesting that the complete CMAH gene is present and does yield active enzymes.
Discussion
In this study, we used a Neu5Gc specific HA protein of H5VN and an αNeu5Gc antibody to explore the Neu5Gc content in the respiratory tract of domestic and wild animals. Lectin histochemistry on horse respiratory tissue showed that the H5VNY161A and αNeu5Gc antibody can bind both N-linked glycans and glycolipids containing Neu5Gc, however, signals from the HA proteins were more intense. Importantly, we found a considerable vial variation between the αNeu5Gc antibodies. We, therefore, as with many glycan determinants, recommend using multiple lectins for a similar target.
Testing of wild animals such as wild Suidae and Cervidae shows that, just as with domesticated animals such as horses, wild animals can contain both Neu5Ac and Neu5Gc (Suzuki et al. 1985; Barnard et al. 2020). With the wild Suidae, the display of Neu5Gc and Neu5Ac changes when descending from the upper respiratory tract into the lower parts, seeing a gradual switch from Neu5Gc to Neu5Ac. This contrasts with what is seen in domestic Suidae which are entirely Neu5Gc, save for the lungs (Suzuki et al. 1997; Burlak et al. 2013; Byrd-Leotis et al. 2014). These results highlight the differences between domesticated and wild species and how pathogen susceptibility might therefore vary between the 2. Interestingly, in Cervidae we observed the first difference between the 2 Neu5Gc lectins with H5VNY161A displaying no binding while the αNeu5Gc did. This suggests that the CMAH enzyme in Cervidae does not convert a specific subset of Neu5Ac to Neu5Gc, one that is not recognized by the HA protein. Although it remains unknown which specific Neu5Gc structures are present on the epithelial surface, these findings do again highlight why it is essential to use several different lectins to analyze Neu5Gc glycan display on tissues.
The results found with the methanol treatments also show the importance of taking glycolipids into account as they can contain Neu5Gc presenting structures. When it comes to glycolipids it is important to note that the process of creating paraffin-embedded tissues comes with a loss of secreted mucins and surface glycolipids (Table 1), 2 structures that can not only contain Neu5Gc but are also factors that play a key role in pathogen evasion and tropism via (decoy) receptors. For mucins, it is known that paraffin-embedding does not lead to a total loss as mucin histochemical stainings are still possible using Periodic acid–Schiff (PAS) and Alcian Blue stain methods (Ali et al. 2013). Loss of glycolipids is also a known occurrence from paraffin-embedding procedures (Alroy et al. 1986), however, to but is not complete as we found that some glycolipids are still present using cholera toxin when excluding the methanol treatment from histochemistry. To which extent this paraffin-embedding influences glycolipid Neu5Ac and Neu5Gc display on respiratory tissues is unknown as Neu5Gc glycolipid presence can vary per species, and is still poorly understood. Histology using frozen tissue methods however demonstrates that mucin and glycolipid retention is far superior when using cryo-embedded tissues methods embedding compared to paraffin-embedding (Cohen et al. 2012). The downside however is that cryo-embedding requires fresh tissue samples collected quickly after the animal has died, which is not always possible when working with exotic animals and was beyond the possible scope of this study.
Lastly, in tiger and European hedgehog, we see that tiger is a Neu5Gc species, whereas European hedgehog is a Neu5Ac species. These findings show that wildlife species can also have an entire respiratory tract containing only Neu5Gc or Neu5Ac in terms of receptor presentation. Curiously for tiger, we see again a difference between the 2 Neu5Gc lectins with the H5VNY161A HA protein being able to bind, whereas the αNeu5Gc antibody cannot. This is most likely caused by a subclass of Neu5Gc glycans that the polyclonal αNeu5Gc does not recognize. This could be tied to the blood group as Neu5Gc is a blood group determinant in domesticated cats (Andrews et al. 1992; Bighignoli et al. 2007). Finally, we demonstrate that European hedgehogs do not contain Neu5Gc as suspected due to an incomplete CMAH gene.
Overall, we show that Neu5Gc content is highly variable in different species, highlighting the need of mapping the loss of CMAH functionalities in different mammals. Mapping these highly variable contents is important as it will shed light on potential pathogen evasion strategies and potential animals with a low species barrier for zoonosis. We also highlight the importance of using multiple Neu5Gc lectins for mapping as the rich variety in which Neu5Gc is displayed can lead to certain lectins being unable to bind a specific glycan class of Neu5Gc.
Material and methods
Expression vectors and protein production
Human codon-optimized HA sequences from A/Vietnam/1203/04 (H5N1) (GenBank: EF541403)—for both the H5VNWT and H5VNY161A mutant—were cloned into the pCD5 vector as previously described (de Vries et al. 2010; Nemanichvili et al. 2021). Both the plasmids have been submitted to Addgene as pCD5-H5_Vietnam_1203/04-sfGFP_ST (H5VNWT, catalog #182546) and pCD5-H5_Vietnam_1203/04-Y161A-sfGFP_ST (H5VNY161A, catalog #182547). The resulting pCD5-HA-GCN4-sfGFP-Strep encodes for the hemagglutinin of the HA protein in frame with a trimerization domain, superfolder green fluorescent protein (sfGFP), and Strep-tag located C-terminally. The H5VNY161A mutant was generated by site-directed mutagenesis (Q5, New England Biolabs, Ipswich, MA, United States) of the HA using primers introducing a single point mutation in the DNA codon changing tyrosine at position 161 into alanine (Y161A; Broszeit et al. 2019). Both HA proteins were produced using a mammalian cell culture system as previously described (de Vries et al. 2010; Nemanichvili et al. 2021).
Tissue collection and TMAs creation using 3D-printed array plates
Tissues originating from the respiratory tract (nasal epithelium, pharyngeal epithelium, upper trachea, lower trachea, primary bronchus, and lung) of selected species were collected from and assembled in 5 TMAs (Table 3). Tissues were obtained from dead animals, which were sent for diagnostic and educational purposes to the Veterinary Pathologic Diagnostic Center, Division of Pathology, Faculty of Veterinary Medicine, Utrecht University. No animals were killed for this study. After collection, tissues were fixed in buffered pH 6.9 formaldehyde solution 4% m/v for 24–48 h at room temperature and subsequently transferred into a 70% ethanol solution for storage. Tissues were processed according to standard paraffin-embedding procedures (Fischer et al. 2008), in brief, the fixed tissues were dehydrated in steps through 70% ethanol, 96% ethanol, and xylene followed by embedding into paraffin blocks, these procedures are known to likely remove some glycolipids and secreted mucins during the process, 2 factors that play key roles in viral tropism (Table I). Sections cut from these tissue blocks onto slides were then deparaffinized and rehydrated in steps through xylene, 96% alcohol, 70% alcohol, and water. Microscopic evaluation of hematoxylin and eosin-stained slides was performed to determine tissue quality before selection and punching of tissue cores for incorporation into microarrays using a 2-mm biopsy punch pen (Miltex). These tissue cores were then incorporated into an empty 3D-printed array plate.
Table 3.
The species composition of the 5 tissue microarrays.
| TMA | English name |
|---|---|
| TMA 1 (Equine) | |
| (5x) Equus ferus caballus | Horse |
| TMA 2 (Suidae) | |
| (4x) Sus scrofa | Wild boar |
| (1x) Pecari tajacu | Peccary |
| (1x) Potamochoerus porcus | Red river hog |
| TMA 3 (Cervidae) | |
| (2x) Capreolus capreolus | Roe deer |
| (2x) Cervus elaphus | Red deer |
| (1x) Hyelaphus porcinus | Hog deer |
| (1x) Rangifer tarandus | Reindeer |
| TMA 4 (exotic wildlife) | |
| (1x) Panthera tigris | Tiger |
| TMA 5 (Erinaceidae) | |
| (5x) Erinaceus europaeus | European hedgehog |
The 3D-printed array plates were designed as previously described (Nemanichvili et al. 2021). Once the plate was filled with the required amount of tissue cores it was embedded into paraffin and left to cool. These TMA blocks were then cut into thin 0.4-μm slices onto microscopy slides for immunohistochemical staining.
Lectin histochemical staining
Tissue slides were prepared for lectin histochemical staining as previously described (Wickramasinghe and Verheije 2015; Nemanichvili et al. 2018). Tissues were treated for 30 min in methanol with 1% v/v peroxidase to inhibit endogenous peroxidase activity. For tissue staining 5 μg/mL of the HA proteins were precomplexed with α-strep-tag mouse-horseradish peroxidase (HRP; IBA Lifesciences, Göttingen, Lower Saxony, Germany) primary antibody and goat-α-mouse-HRP (Thermofisher, Waltham, MA, United States) secondary antibody at 4:2:1 molar ratio as previously described (Nemanichvili et al. 2021). Hematoxylin stains were performed to visualize tissue structure and morphology before covering the stained slides with coverslips using AquaTex (Merck). For stains with αNeu5Gc, tissues were stained with a 1:300 dilution of αNeu5Gc IgY (Biolegend, San Diego, CA, United States) followed by 1:100 dilution of goat anti-chicken IgY-HRP (Thermofisher, Waltham, MA, United States). For initial testing 3 different vials of αNeu5Gc were used to test batch variance. Vial 1 was from an earlier batch while vials 2 and 3 were from the same batch. After initial testing shown in Fig. 1A, it was vial 3 that was used for subsequent stains. For removal of N-linked glycans, tissues were treated overnight with 50 μL/mL PNGase F (New England Biolabs, Ipswich, MA, United States) in a humidity chamber at 37°C before being stained. The lectin L. culinaris Agglutinin (LCA; Vector Laboratories, Burlingame, CA, United States) was used at 2 μg/mL as a control for N-linked glycan presence. For retention of glycolipids, the 30-min treatment of tissues in methanol with 1% v/v peroxidase was omitted. The recombinant HRP conjugated Cholera Toxin Subunit B (CTX) (Thermofisher, Waltham, MA, United States) of 1 μg/μL was used at a 1:200 dilution as a control for glycolipid presence. The lectins Sambucus nigra agglutinin (SNA) and Maackia amurensis (MALII; Vector Laboratories, Burlingame, CA, United States) were used at 2.5 and 5 μg/mL, respectively to confirm the presence of sialic acids on the tissue. All lectin histochemical stains and controls were performed 3 times in independent assays (n = 3).
Analysis of the CMAH genes
The CMAH genes of the horse (Equus caballus), roe deer (C. capreolus), wild boar/domestic pig (S. scrofa), tiger (P. tigris), and European hedgehog (E. europaeus) were already investigated previously (Peri et al. 2018). No genome was available for the red river hog (Potamochoerus porcus) and peccary (Pecari tajacu). We analyzed the CMAH genes of the red deer (Cervus elaphus), hog deer (Axis porcinus), and reindeer (Rangifer tarandus). The genomes were present in the whole-genome shotgun contigs (WGS) collection. Using blastn, we searched for the 15 exons of the chimpanzee (Pan troglodytes) CMAH gene in the respective genome, with a search-optimized for more dissimilar sequences (discontinuous megablast). The first exon was too small to find significant similarity and the 15th exon always gave >100 hits. Therefore, these 2 exons were not analyzed. The presence, orientation, and order of the exons was investigated (Supplementary Table S1 and Supplementary File S1). The exons were aligned with the chimpanzee exons and additional bases were added when the blastn search did not indicate the complete exon. Finally, the presence of stop codons in the exons was investigated. A stop codon was detected in all analyzed animals in exon 14 and therefore this was considered an artifact, possibly still yielding an active CMAH enzyme.
Supplementary Material
Acknowledgments
We would like to thank the Centre for Cell Imaging at the Faculty of Veterinary Medicine, Utrecht University for use of the microscopy facilities and 3D-printing services provided. We would also like to thank the Veterinary Pathologic Diagnostic Centre at the Faculty of Veterinary Medicine, Utrecht University for their assistance in acquiring and processing all the required animal tissues. We also thank Lisanne van der Voort of the Utrecht University, Faculty of Veterinary Medicine, Communications & Marketing Department for her graphical designs of the pictograms. We would finally like to thank Mariette F. Ducatez of the French National Research Institute for Agriculture, Food and the Environment for her guidance and input.
Contributor Information
Nikoloz Nemanichvili, Division of Pathology, Department of Biomolecular Health Sciences, Faculty of Veterinary Medicine, Utrecht University, Yalelaan 1, 3584 CL Utrecht, The Netherlands.
Cindy M Spruit, Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Utrecht University, Universiteitsweg 99, 3584 CG Utrecht, The Netherlands.
Alinda J Berends, Division of Pathology, Department of Biomolecular Health Sciences, Faculty of Veterinary Medicine, Utrecht University, Yalelaan 1, 3584 CL Utrecht, The Netherlands.
Andrea Gröne, Division of Pathology, Department of Biomolecular Health Sciences, Faculty of Veterinary Medicine, Utrecht University, Yalelaan 1, 3584 CL Utrecht, The Netherlands.
Jolianne M Rijks, Dutch Wildlife Health Centre, Department of Biomolecular Health Sciences, Faculty of Veterinary Medicine, Utrecht University, Yalelaan 1, 3584 CL Utrecht, The Netherlands.
Monique H Verheije, Division of Pathology, Department of Biomolecular Health Sciences, Faculty of Veterinary Medicine, Utrecht University, Yalelaan 1, 3584 CL Utrecht, The Netherlands.
Robert P de Vries, Department of Chemical Biology and Drug Discovery, Utrecht Institute for Pharmaceutical Sciences, Utrecht University, Universiteitsweg 99, 3584 CG Utrecht, The Netherlands.
Funding
This research was supported by an ERC Starting Grant from the European Commission (802780) and a Beijerinck Premium of the Royal Dutch Academy of Sciences awarded to Robert P. de Vries.
Conflict of interest statement None declared.
Abbreviations
CMAH, cytidine monophospho-N-acetylneuraminic acid hydroxylase; CTX, cholera toxin subunit B;, HA, hemagglutinin; HRP, horseradish peroxidase; H5VN, H5 HA from A/Vietnam/1203/2004; LCA, Lens culinaris agglutinin; MALII, Maackia amurensis lectin II; Neu5Ac, N-acetylneuraminic acid; Neu5Gc, N-glycolylneuraminic acid; sfGFP, superfolder green fluorescent protein; SNA, Sambucus nigra lectin; TMA, tissue microarray; WGS, whole-genome shotgun
References
- Ali U, Nagi A, Naseem N, Ullah E. Mucin histochemistry in tumours of colon, ovaries and lung. Arch Histol Cytol. 2013:7:163. [Google Scholar]
- Alkhovsky S, Lenshin S, Romashin A, Vishnevskaya T, Vyshemirsky O, Bulycheva Y, Lvov D, Gitelman A. SARS-like coronaviruses in horseshoe bats (Rhinolophus spp.) in Russia, 2020. Viruses. 2022:14:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alroy J, Ucci AA, Goyal V, Woods W. Lectin histochemistry of glycolipid storage diseases on frozen and paraffin-embedded tissue sections. J histochem Cytochem. 1986:34:501–505. [DOI] [PubMed] [Google Scholar]
- Andrews GA, Chavey PS, Smith JE, Rich L. N-Glycolylneuraminic acid and N-Acetylneuraminic acid define feline blood group A and B antigens. Blood. 1992:79:2485–2491. [PubMed] [Google Scholar]
- Barnard KN, Wasik BR, LaClair JR, Buchholz DW, Weichert WS, Alford-Lawrence BK, Aguilar HC, Parrish CR. Expression of 9-O- and 7,9-O-acetyl modified sialic acid in cells and their effects on influenza viruses. MBio. 2019:10:e02490–e02419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard KN, Alford-Lawrence BK, Buchholz DW, Wasik BR, LaClair JR, Yu H, Honce R, Ruhl S, Pajic P, Daugherity EK, et al. Modified sialic acids on mucus and erythrocytes inhibit influenza A virus hemagglutinin and neuraminidase functions. J Virol. 2020:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bighignoli B, Niini T, Grahn RA, Pedersen NC, Millon LV, Polli M, Longeri M, Lyons LA. Cytidine monophospho-N-acetylneuraminic acid hydroxylase (CMAH) mutations associated with the domestic cat AB blood group. BMC Genet. 2007:8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broszeit F, Tzarum N, Zhu X, Nemanichvili N, Eggink D, Leenders T, Li Z, Liu L, Wolfert MA, Papanikolaou A, et al. N-Glycolylneuraminic acid as a receptor for influenza A viruses. Cell Rep. 2019:27:3284, e3286–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlak C, Bern M, Brito AE, Isailovic D, Wang Z-Y, Estrada JL, Li P, Tector AJ. N-linked glycan profiling of GGTA1/CMAH knockout pigs identifies new potential carbohydrate xenoantigens. Xenotransplantation. 2013:20:277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burzynska P, Sobala LF, Mikolajczyk K, Jodlowska M, Jaskiewicz E. Sialic acids as receptors for pathogens. Biomol Ther. 2021:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussolati G, Radulescu RT. Blocking endogenous peroxidases in immunohistochemistry: a mandatory, yet also subtle measure. Appl Immunohistochem Mol Morphol. 2011:19. [DOI] [PubMed] [Google Scholar]
- Byrd-Leotis L, Liu R, Bradley KC, Lasanajak Y, Cummings SF, Song X, Heimburg-Molinaro J, Galloway SE, Culhane MR, Smith DF, et al. Shotgun glycomics of pig lung identifies natural endogenous receptors for influenza viruses. Proc Natl Acad Sci U S A. 2014:111:E2241–E2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JC, Bevins SN, Ellis JW, Linder TJ, Tell RM, Jenkins-Moore M, Root JJ, Lenoch JB, Robbe-Austerman S, DeLiberto TJ, et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc Natl Acad Sci U S A. 2021:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Varki NM, Jankowski MD, Gagneux P. Using unfixed, frozen tissues to study natural mucin distribution. J Vis Exp. 2012:e3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994:205:17–23. [DOI] [PubMed] [Google Scholar]
- Diaz SL, Padler-Karavani V, Ghaderi D, Hurtado-Ziola N, Yu H, Chen X, Brinkman-Van der Linden EC, Varki A, Varki NM. Sensitive and specific detection of the non-human sialic acid N-glycolylneuraminic acid in human tissues and biotherapeutic products. PLoS One. 2009:4:e4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer AH, Jacobson KA, Rose J, Zeller R. Paraffin embedding tissue samples for sectioning. CSH protocols. 2008, 2008:pdb.prot4989. [DOI] [PubMed] [Google Scholar]
- de Graaf M, Fouchier RA. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J. 2014:33:823–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Suzuki Y, Suzuki T, Takada A, Horimoto T, Wells K, Kida H, Otsuki K, Kiso M, Ishida H, et al. Recognition of N-glycolylneuraminic acid linked to galactose by the α2,3 linkage is associated with intestinal replication of influenza A virus in ducks. J Virol. 2000:74:9300–9305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahan M, Thomson PC, Wynn PC, Wang B. The non-human glycan, N-glycolylneuraminic acid (Neu5Gc), is not expressed in all organs and skeletal muscles of nine animal species. Food Chem. 2021:343:128439. [DOI] [PubMed] [Google Scholar]
- Magaki S, Hojat SA, Wei B, So A, Yong WH. An introduction to the performance of immunohistochemistry. Methods Mol Biol. 2019:1897:289–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci Maria R, Donatelli I, Kawaoka Y. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000:74:8502–8512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morniroli D, Gianni ML, Consales A, Pietrasanta C, Mosca F. Human sialome and coronavirus disease-2019 (COVID-19) pandemic: an understated correlation? Front Immunol. 2020:11:1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemanichvili N, Tomris I, Turner HL, McBride R, Grant OC, van der Woude R, Aldosari MH, Pieters RJ, Woods RJ, Paulson JC, et al. Fluorescent trimeric hemagglutinins reveal multivalent receptor binding properties. J Mol Biol. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemanichvili N, Berends A, Wubbolts R, Gröne A, Rijks J, de Vries R, Verheije M. Tissue microarrays to visualize influenza D attachment to host receptors in the respiratory tract of farm animals. Viruses. 2021:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PSK, Böhm R, Hartley-Tassell LE, Steen JA, Wang H, Lukowski SW, Hawthorne PL, Trezise AEO, Coloe PJ, Grimmond SM, et al. Ferrets exclusively synthesize Neu5Ac and express naturally humanized influenza A virus receptors. Nat Commun. 2014:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L, McCord KA, Bui DT, Bouwman KM, Kitova EN, Elaish M, Kumawat D, Daskhan GC, Tomris I, Han L, et al. Sialic acid-containing glycolipids mediate binding and viral entry of SARS-CoV-2. Nat Chem Biol. 2022:18:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Bachar Abramovitch S, Padler-Karavani V. Specific detection of Neu5Gc in animal tissues by immunohistochemistry. In: Costa C, editors. Xenotransplantation: methods and protocols. New York, NY: Springer US; 2020. pp. 59–72 [DOI] [PubMed] [Google Scholar]
- Paulson JC, de Vries RP. H5N1 receptor specificity as a factor in pandemic risk. Virus Res. 2013:178:99–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri S, Kulkarni A, Feyertag F, Berninsone PM, Alvarez-Ponce D. Phylogenetic distribution of CMP-Neu5Ac hydroxylase (CMAH), the enzyme synthetizing the proinflammatory human xenoantigen Neu5Gc. Genome Biol Evol. 2018:10:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GN, Herrler G, Paulson JC, Klenk HD. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J Biol Chem. 1986:261:5947–5951. [PubMed] [Google Scholar]
- Schultze B, Krempl C, Ballesteros ML, Shaw L, Schauer R, Enjuanes L, Herrler G. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J Virol. 1996:70:5634–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Qi J, Khedri Z, Diaz S, Yu H, Chen X, Varki A, Shi Y, Gao GF. An open receptor-binding cavity of hemagglutinin-esterase-fusion glycoprotein from newly-identified influenza D virus: basis for its broad cell tropism. PLoS Pathog. 2016:12:e1005411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer SA, Diaz SL, Gagneux P. Parallel evolution of a self-signal: humans and new world monkeys independently lost the cell surface sugar Neu5Gc. Immunogenetics. 2014:66:671–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruit CM, Nemanichvili N, Okamatsu M, Takematsu H, Boons GJ, de Vries RP. N-glycolylneuraminic acid in animal models for human influenza A virus. Viruses. 2021:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruit CM, Zhu X, Tomris I, Carrasco MR, Han AX, Broszeit F, van der Woude R, Bouwman KM, Luu MMT, Matsuno K, et al. N-glycolylneuraminic acid binding of avian and equine H7 influenza A viruses. J Virol. 2022:jvi0212021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun XL. The role of cell surface sialic acids for SARS-CoV-2 infection. Glycobiology. 2021:31:1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A. Genetic basis for the lack of N-glycolylneuraminic acid expression in human tissues and its implication to human evolution. Proc Jpn Acad Ser B Phys Biol Sci. 2006:82:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Matsunaga M, Matsumoto M. N-Acetylneuraminyllactosylceramide, GM3-NeuAc, a new influenza A virus receptor which mediates the adsorption-fusion process of viral infection. Binding specificity of influenza virus A/Aichi/2/68 (H3N2) to membrane-associated GM3 with different molecular species of sialic acid. J Biol Chem. 1985:260:1362–1365. [PubMed] [Google Scholar]
- Suzuki T, Horiike G, Yamazaki Y, Kawabe K, Masuda H, Miyamoto D, Matsuda M, Nishimura SI, Yamagata T, Ito T, et al. Swine influenza virus strains recognize sialylsugar chains containing the molecular species of sialic acid predominantly present in the swine tracheal epithelium. FEBS Lett. 1997:404:192–196. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Ito T, Suzuki T, Holland Robert E, Chambers Thomas M, Kiso M, Ishida H, Kawaoka Y. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol. 2000:74:11825–11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor ME, Drickamer K, Schnaar RL, Etzler ME, Varki A. Discovery and classification of glycan-binding proteins. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, et al., editors. Essentials of Glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2015. Copyright 2015–2017 by The Consortium of Glycobiology. In: Jolla L, editors. California All rights reserved. pp. 361–372. [Google Scholar]
- Tortorici MA, Walls AC, Lang Y, Wang C, Li Z, Koerhuis D, Boons GJ, Bosch BJ, Rey FA, de Groot RJ, et al. Structural basis for human coronavirus attachment to sialic acid receptors. Nat Struct Mol Biol. 2019:26:481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solórzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, et al. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007:315:655–659. [DOI] [PubMed] [Google Scholar]
- Varki A, Gagneux P. Multifarious roles of sialic acids in immunity. Ann N Y Acad Sci. 2012:1253:16–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Schnaar RL, Schauer R. Sialic acids and other nonulosonic acids. Essentials of Glycobiology. 2015:179–195. [Google Scholar]
- de Vries RP, de Vries E, Bosch BJ, de Groot RJ, Rottier PJM, de Haan CAM. The influenza A virus hemagglutinin glycosylation state affects receptor-binding specificity. Virology. 2010:403:17–25. [DOI] [PubMed] [Google Scholar]
- de Vries RP, Zhu X, McBride R, Rigter A, Hanson A, Zhong G, Hatta M, Xu R, Yu W, Kawaoka Y, et al. Hemagglutinin receptor specificity and structural analyses of respiratory droplet-transmissible H5N1 viruses. J Virol. 2014:88:768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Tscherne DM, McCullough C, Caffrey M, Garcia-Sastre A, Rong L. Residue Y161 of influenza virus hemagglutinin is involved in viral recognition of sialylated complexes from different hosts. J Virol. 2012:86:4455–4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickramasinghe INA, Verheije MH. Protein histochemistry using coronaviral spike proteins: studying binding profiles and sialic acid requirements for attachment to tissues. In: Maier HJ, Bickerton E, Britton P, editors. Coronaviruses: methods and protocols. Springer New York: New York, NY; 2015. pp. 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Dang VT, Fleming FE, von Itzstein M, Coulson BS, Blanchard H. Structural basis of rotavirus strain preference toward N-acetyl- or N-glycolylneuraminic acid-containing receptors. J Virol. 2012:86:13456–13466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.