Abstract
Oral microbiota play a prominent role in canine periodontal disease and wet foods are often blamed for poor oral health, but canine oral microbial communities have been poorly studied. We aimed to determine differences in oral health measures, breath odor, and oral microbiota populations of dogs fed wet or dry food. Twelve adult dogs fed either a commercial dry (extruded) or commercial wet (canned) food for 6 wk were studied. Breath samples were measured for sulfur compounds, teeth were scored for plaque, calculus, and gingivitis by a blinded veterinary dentist, salivary pH was measured, and supragingival (SUP) and subgingival (SUB) plaque samples were collected for microbiota analysis. Plaque DNA was extracted and Illumina sequencing was conducted. Phylogenetic data were analyzed using the CosmosID bioinformatics platform and SAS 9.4, with P <0.05 being significant and P <0.10 being trends. Plaque coverage tended to be higher (P < 0.10) in dogs fed wet vs. dry food, but other oral health scores were not different. Dogs fed dry food had higher (P < 0.05) salivary pH and lower (P < 0.05) breath sulfur concentrations than those consuming wet food. Bacterial alpha diversity was higher in SUP than SUB samples, and a clear separation in beta diversity was observed between sample sites on principal coordinates analysis (PCoA) plots. In SUP samples, dogs fed wet food had a higher alpha diversity than dogs fed dry food, with PCoA plots showing a separation between wet and dry food. Relative abundances of Firmicutes, Synergistetes, and 10 bacterial genera were different (P < 0.05) in SUB samples of dogs fed wet vs. dry food. Relative abundances of Fusobacteria and over 20 bacterial genera were different (P < 0.05) in SUP samples of dogs fed wet vs. dry food. In general, oral health-associated bacterial taxa (Pasteurella, Capnocytophaga, Corynebacterium) were higher, while bacteria associated with poor oral health (Fretibacterium fastidiosum, Filifactor alocis, Treponema medium, Tannerella forsythia, Porphyromonas canoris, Porphyromonas gingivalis) were lower in dogs fed dry food. Such shifts in the oral microbiota may impact periodontal disease risk, but longer dietary intervention studies are required to confirm their role in the disease process. Our results suggest that dogs fed dry extruded foods have lower breath odor and tooth plaque buildup and an oral microbiota population more closely associated with oral health than dogs fed wet canned foods.
Keywords: canine health, next-generation sequencing, oral microbiome, periodontal disease
Twelve healthy adult dogs were used to determine differences in oral health measures, breath odor, and oral bacteria populations of dogs consuming wet or dry foods. Our results suggest that dogs consuming dry foods have lower breath odor, less tooth plaque buildup, and an oral microbiota population more closely associated with health than dogs consuming wet foods.
Introduction
Periodontal disease is the most frequently diagnosed oral disease of dogs and is initiated by the buildup of plaque on the tooth surface (Niemiec, 2012). In research on beagles, it was identified that there was tooth attachment loss (≥1 mm) in 20% of dogs aged 1 yr and 84% in dogs aged more than 3 yr (Kortegaard et al., 2008). In the United States, oral disease was reported in 18.2% of dogs visiting primary veterinary practices (Wallis and Holcombe, 2020). While periodontal disease is initiated by the buildup of plaque on the tooth surface, the plaque formation occurs in stages. Initially, salivary glycoproteins adhere to the tooth surface to form the pellicle, which is followed by bacterial adhesion and plaque maturation. After that, plaque can become mineralized to form calculus, which provides a porous surface to which new plaque can adhere. Finally, the inflammation (gingivitis) and resulting tissue damage occurs because of an improperly regulated immune response to bacterial infection (Niemiec, 2012). Therefore, minimizing plaque formation is key to preventing the progression of periodontal disease.
The diet offered to pets may impact periodontal disease development. Because most dog owners feed a complete and balanced commercial diet, any nutrient deficiencies that could impact host immune response and consequent periodontal disease are rare. The primary means by which a diet may impact the initiation and progression of periodontal disease is by its texture (abrasive action), affecting the accumulation of plaque (Gorrel, 1998). For decades, researchers have studied the effects of dietary texture as a mechanical method to prevent plaque accumulation in dogs. Dogs fed hard/solid foods have been reported to retain essentially normal teeth and gums compared with animals fed the same food that was ground or minced (Burwasser and Hill, 1939; Krasse, 1960; Saxe et al., 1967). Most importantly, one study reported that even the minimal chewing required for minced food had some cleansing or protective effects compared to not chewing at all (i.e., dogs fed by gastric intubation) (Egelberg, 1965a).
Although it is known that diet texture and physical abrasion can reduce the accumulation of plaque on teeth, its impact on the oral microbiota community is unknown in dogs. In cats, it was shown that animals fed dry foods exclusively had a higher bacterial diversity in their oral microbiome than those fed wet foods exclusively, as well as a higher abundance of Porphyromonas spp. and Treponema spp. (Adler et al., 2016). To our knowledge, similar studies have not been conducted in dogs. There has been a limited number of studies characterizing the canine oral microbiota of healthy vs. diseased dogs (Davis et al., 2013; Wallis et al., 2021; Niemiec et al., 2022), how oral habitats differ (Ruparell et al., 2020a; Oba et al., 2021b), and how treat consumption may affect the oral microbiota (Ruparell et al., 2020b; Oba et al., 2021a), but impact of diet type has not been evaluated to our knowledge.
Given the lack of knowledge regarding the effects of diet type on the oral microbiota in dogs, the current study aimed to determine the differences in oral microbiota and health outcomes of dogs consuming wet or dry foods. Compared to dogs consuming a dry diet, we hypothesized that consumption of a wet diet would increase (worsen) oral health scores, increase halitosis, reduce salivary pH, and increase the relative abundance of bacteria associated with poor oral health.
Materials and Methods
Animals
Twelve adult female beagle dogs (mean age = 6.0 ± 1.12 yr; mean body weight = 10.78 ± 1.23 kg; mean body condition score = 6.1 ± 0.64) were used in this study. All procedures were approved by the University of Illinois Institutional Animal Care and Use Committee prior to experimentation (IACUC #19247). Dogs were housed individually in pens (1.0 m wide by 1.8 m long) in a humidity- and temperature-controlled animal facility. Dogs had access to fresh water at all times and were fed once daily to maintain body weight and a healthy body condition score (Laflamme, 1997). Dogs were weighed once weekly prior to feeding, with a body weight of all dogs remaining constant throughout the study. Dogs were fed at 0800 each morning and were given 1 h to consume their food. Leftover food was weighed each day to calculate intake.
Prior to the start of the study, clinical indices, including a serum chemistry panel and hematology, and a physical examination were performed to confirm health of all animals. For 3 mo prior to the dietary intervention, all animals consumed the same diet (Purina Dog Chow Complete Adult with Real Chicken, Nestle Purina PetCare Company, St. Louis, MO) and were fed to maintain body weight. Also, a dental evaluation was performed by a veterinary dentist to confirm the presence and integrity of all teeth to be scored to confirm trial eligibility.
Dogs were allotted to one of two treatments and fed for 6 wk, which has been shown to be enough time for oral health outcomes and microbiota to change (Carroll et al., 2020; Engel et al., 2020; Oba et al., 2021a). Dietary treatments included 1) dry diet (Purina Dog Chow Complete Adult with Real Chicken, Nestle Purina PetCare Company) and 2) wet diet (Pedigree Choice Cuts in Gravy with Beef, Mars Petcare US, Franklin, TN) (Supplementary Table 1). At the end of the experiment, a blinded veterinary dentist scored teeth, breath samples were measured for malodor, salivary pH was analyzed, and supragingival (SUP) and subgingival (SUB) plaque samples were collected. No treats, chew toys, or other dental interventions were permitted for the duration of the study.
Anesthesia methods
All dogs had their food withheld for at least 12 h prior to anesthesia, but were allowed water until transportation. The hair over the left or right cephalic vein was clipped, the site was aseptically prepared, and a 20-gauge intravenous catheter was placed in the cephalic vein for the administration of sedative and anesthetic agents, and intravenous fluids. Following catheterization, butorphanol (0.3 mg/kg) was administered intravenously and dogs were preoxygenated. Anesthesia was then induced with etomidate with or without a coinduction of midazolam (0.3 mg/kg) or lidocaine (2 mg/kg). Dogs were orotracheally intubated and transferred to isoflurane delivered in oxygen to maintain anesthesia. Intravenous fluids (Lactated ringer’s solution) were run at 5 mL/kg/h throughout anesthesia and active heating with a forced-air warmer was provided to maintain normothermia. Cardiovascular and respiratory function was monitored continuously using an anesthetic multiparameter monitor. Supplementary anesthetic agents and cardiovascular support were administered as needed based on the decision of the attending veterinary anesthesiologist.
Salivary pH
Salivary pH was measured using pH strips (Fisherbrand Plastic pH Strips; pH range = 6.5 to 13) on the same day and time of dental scoring. All dogs had their food withheld for at least 12 h prior to salivary pH measurement, using two strips on each side per dog (four, in total). The salivary pH reported was the mean of the four strips. Saliva samples were collected where it naturally pools (in the cheek pouch and under the tongue) for 30 s.
Halitosis measurement
Breath samples were analyzed for total volatile sulfur compound concentrations using a halimeter (Interscan Corp, Simi Valley, CA). Halimeter measurements were conducted 7 h after feeding. Halitosis measurements were obtained for each dog using a clean plastic straw (i.e., a clean straw was used for each measurement) as an extension of the halimeter air drawing hose. The highest reading of volatile sulfur compounds over a period of approximately 30 s was displayed by the halimeter and recorded. The machine was allowed to return to 0 (about 60 to 120 s) before the next measurement was taken. Each dog was measured three times and a mean score was calculated.
Dental scoring
Gingivitis, plaque, and calculus scoring were conducted by a board-certified veterinary dentist according to a modified version of previous scoring systems (Mühlemann and Son, 1971; Gorrel et al., 1999). For each measurement, the I3, C, P3, P4, and M1 teeth on the upper jaw (maxilla) and the C, P3, P4, and M1 teeth on the lower jaw (mandible) were scored. These two teeth were chosen because they are the teeth from which plaque samples for microbiota analysis were collected. Thus, the dental score data would be more compatible with the sample collection site.
To assess gingivitis, after an initial visual evaluation of the gingiva, a periodontal probe (Williams model, Cislak Manufacturing, Inc., Niles, IL) was placed subgingivally on the buccal side of each tooth, and values were assigned via visual assessment of inflammation and bleeding, if present, upon probing. Each tooth was graded by the average of the three scores obtained per tooth. The score for each dog is the mean score for all teeth scored. Plaque levels were evaluated by using Trace Disclosing Solution (Young Dental, Earth City, MO) to cover the teeth, followed by a gentle rinse of water to remove the excess. Plaque was hence revealed and subsequently scored for coverage and thickness according to Gorrel et al. (1999) using the anatomical landmarks described in Hennet et al. (2006) to divide the teeth into gingival and occlusal portions. The gingival and occlusal values for each tooth were averaged to obtain a tooth total score. The average plaque coverage was multiplied by the average of plaque thickness to obtain a whole mouth mean calculus score for each animal. Calculus scores were based on the visual assessment of coverage and thickness on the mesial, buccal, and distal portions of the tooth. The tooth score is the average of the scores for each of the three-tooth surfaces. The average of calculus coverage was multiplied by the average of calculus thickness to obtain a whole mouth mean calculus score for each animal.
Pocket depth in millimeters was based on height from bottom of pocket to gingival margin, <2 mm = normal sulcus; >2 and <3 mm = slight; >3 and <5 mm = moderate; >5 mm = severe. The tooth score is the average of pocket depth for each tooth. The average of pocket depth of all teeth was used to obtain a whole mouth mean pocket score for each animal.
Plaque sample collection
Once scored, plaque samples were collected for microbiota analysis and the teeth surfaces were cleaned. Teeth were assessed using a sterile periodontal probe on the gingival margin and sweeping along the base of the crown. SUB and SUP plaque samples were collected from the fourth premolar and first molar mandibular teeth and the fourth premolar and first molar maxillary teeth. Plaque samples were placed into sterile 2.0 mL cryovials (CryoELITE, Wheaton, Millville, NJ) and immediately placed on dry ice until storage at −80 °C, where they were stored until analysis.
Microbiota analysis
Total DNA from plaque samples were extracted using Mo-Bio PowerSoil Kits (MO BIO Laboratories, Inc., Carlsbad, CA), followed by quantification of extracted DNA using a Qubit 3.0 Fluorometer (Life Technologies, Grand Island, NY). Quality of extracted DNA was assessed by electrophoresis using agarose gels (E-Gel EX Gel 1%; Invitrogen, Carlsbad, CA). DNA samples were sent to CosmosID (Rockville, MD) for library creation and sequencing. DNA libraries were prepared using the Illumina Nextera XT library preparation kit (Illumina, San Diego, CA), with a modified protocol. Libraries were then sequenced on an Illumina HiSeq 183 platform (2 × 150 bp). Unassembled sequencing reads were directly analyzed by CosmosID bioinformatics platform (CosmosID Inc.) described elsewhere (Hasan et al., 2014; Lax et al., 2014; Ottesen et al., 2016; Ponnusamy et al., 2016) for microbiome analysis and profiling of organism relative abundance. CosmosID uses their proprietary curated microbial genomics database, which contains nearly 170,000 phylogenetically organized genomes and gene sequences. The database enables multikingdom identification of bacteria, viruses, phages, fungi, and protists, with bacteria being the focus of this study. Alpha diversity was estimated using Chao1, the Shannon Index, and the Simpson Index. Beta diversity was calculated using Jaccard distance measures and presented using a principal coordinates analysis plot.
Statistical analysis
All data were analyzed using SAS (version 9.4, SAS Institute, Cary, NC) using the Mixed Models procedure, with dog being considered a random effect and diet was considered a fixed effect. Data normality was checked using the univariate procedure and Shapiro–Wilk statistic, with log transformation being used when normal distribution was lacking. If after the logarithmic transformation of the data, the data did not reach normality, the data were analyzed using the npar1way procedure and Wilcoxon statistic. Data were reported as means, with P < 0.05 considered significant and P < 0.10 considered a trend. Linear discriminant analysis effect size (LEfSe) (Segata et al., 2011) was used to evaluate the genetic sequences and to identify genera that were enriched at the various habitats and dietary groups.
Results
Dental scoring and salivary pH
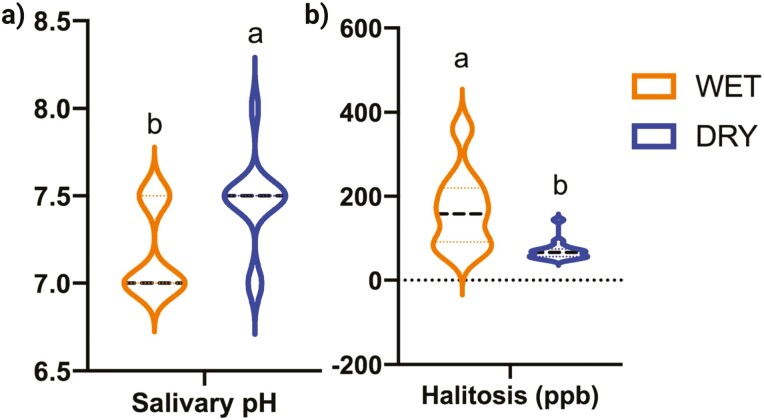
All dental scores are presented in Table 1, while volatile sulfur compound concentrations and salivary pH are presented in Figure 1. None of the dental scores were affected by diet type, except for plaque coverage that tended to be higher (P = 0.0738) in dogs consuming wet food than those consuming dry food. Dogs consuming the dry food had a higher (P = 0.0156) salivary pH and lower (P = 0.0293) breath odor than dogs consuming the wet food.
Table 1.
Plaque, gingivitis, and calculus scores and pocket depths of healthy adult dogs consuming commercially available wet or dry food
| Dry food | Wet food | SEM | P-value | |
|---|---|---|---|---|
| Plaque coverage | 3.28 | 3.55 | 0.094 | 0.0738 |
| Plaque thickness | 2.55 | 2.65 | 0.114 | 0.9361 |
| Plaque score1 | 8.44 | 9.37 | 0.545 | 0.2623 |
| Gingivitis score | 1.23 | 1.25 | 0.080 | 0.8414 |
| Calculus coverage | 2.56 | 2.86 | 0.238 | 0.3977 |
| Calculus thickness | 1.69 | 1.64 | 0.154 | 0.8057 |
| Calculus score1 | 4.61 | 4.72 | 0.701 | 0.9100 |
| Pocket depth (mm) | 2.52 | 2.25 | 0.135 | 0.1813 |
Plaque and calculus scores range from 0 (low) to 12 (maximum).
Figure 1.
Salivary pH (a) and volatile sulfur concentrations (b) from healthy adult dogs consuming commercially available wet or dry food. Groups with different superscripts differ (P < 0.05).
Oral microbiota
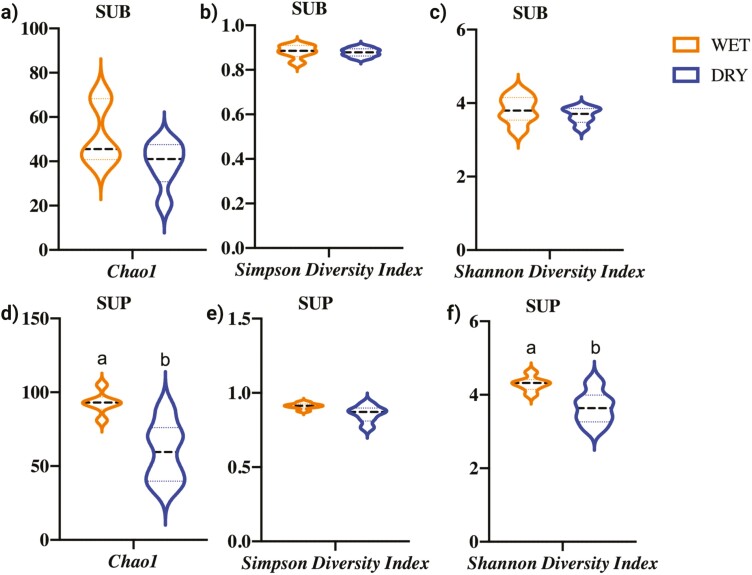
Illumina sequencing produced a total of 595.2 million sequences, with an average of 24.80 ± 13.2 million sequences per sample. Analyses were conducted with all samples being rarified to a level of 5.48 million sequences. Oral bacterial alpha diversity appeared to be different between sample types (SUB vs. SUP) and diet fed (wet food vs. dry food), but depended on the measure used. The Chao1 index, an abundance-based estimator of species richness if all bacterial species were identified in samples, was higher (P < 0.05) in SUP than SUB samples (Supplementary Figure 1) and higher (P < 0.05) in SUP samples of dogs consuming a wet food than those consuming a dry food (Figure 2). Chao1 gives more weight to low-abundance species and is particularly useful for data sets skewed toward low-abundance species (Kim et al., 2017). The Simpson index, an estimator of species richness and species evenness, was not affected by sample type or diet. The Shannon diversity index, another estimator of species richness and species evenness, was affected by diet, with SUP samples of dogs consuming a wet food having a higher (P < 0.05) index than SUP of dogs consuming a dry food (Figure 2). The Shannon diversity index was not affected by sample type or diet in SUB samples.
Figure 2.
Bacterial alpha diversity indices of dogs consuming a commercially available wet or dry food as assessed by the Chao1 index (a and d), Simpson diversity index (b and e), and Shannon diversity index (c and f) in canine plaque samples (SUB, subgingival; SUP, supragingival). Groups with different superscripts differ (P < 0.05).
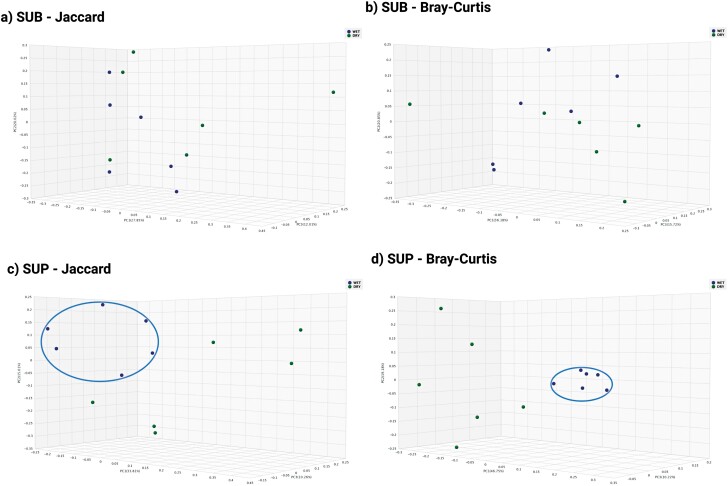
Principal coordinates analysis plots based on the Jaccard and Bray–Curtis indexes were created to represent the beta diversity of plaque samples. The Jaccard index, which is based on the presence or absence of bacterial taxa (Schroeder and Jenkins, 2018), showed that sample types (SUB, SUP) clustered separately from one another (Supplementary Figure 2). The Jaccard index also showed that dogs consuming wet vs. dry foods clustered separately in SUP samples, but not in SUB samples (Figure 3). The Bray–Curtis index, which takes into account the abundance of each bacterial taxa present and estimates the dissimilarity of samples (Mbareche et al., 2020), showed that sample types (SUB, SUP) clustered separately from one another (Supplementary Figure 2). The Bray–Curtis index also showed that dogs consuming wet vs. dry foods clustered separately in SUP samples, but not in SUB samples (Figure 3).
Figure 3.
Bacterial beta diversity indices of plaque samples (SUB, subgingival; SUP, supragingival) of dogs consuming a commercially available wet or dry food as assessed by the Jaccard index (a and c) or Bray–Curtis index (b and d).
The most abundant bacterial phyla in SUB and SUP samples were Bacteroidetes (77% and 83%, respectively), Proteobacteria (12% and 10%, respectively), and Firmicutes (6% and 3%, respectively). The most abundant genera in SUB samples were Porphyromonas (65%), unclassified Porphyromonadaceae (5%), Campylobacter (4%), and Bergeyella (3%), while Porphyromonas (69%), Bergeyella (6%), Neisseria (5%), and Capnocytophaga (5%) were the most abundant in SUP samples. The relative abundance of Bacteroidetes was lower (P < 0.05), while the relative abundances of Actinobacteria, Firmicutes, and Spirochaetes were higher (P < 0.05) in SUB samples than SUP samples (Supplementary Figure 3). The relative abundances of Bergeyella, Capnocytophaga, Fretibacterium, Moraxella, and Neisseria were lower (P < 0.05), while the relative abundances of Aerobacter, Campylobacter, Conchiformibius, Corynebacterium, Desulfobulbus, Filifactor, Lactobacillus, Parvimonas, unclassified Peptostreptococcaceae, unclassified Porphyromonadaceae, Prevotella, Streptococcus, and Treponema were higher (P < 0.05) in SUB samples than in SUP samples (Supplementary Figure 4).
In SUB samples, the relative abundances of Capnocytophaga cynodegmi and Conchiformibius were lower (P < 0.05) in dogs consuming wet food than those consuming dry food (Table 2). In contrast, the relative abundances of Firmicutes, Synergistetes, Fretibacterium, Tannerella, unclassified Peptostreptococcaceae, Streptococcus, Tannnerella forsythia, Peptostreptococcaceae bacterium oral taxon 113, and Fretibacterium fastidiosum were higher (P < 0.05) in dogs consuming wet food than those consuming dry food (Table 2).
Table 2.
Oral bacteria (relative abundance, %) that were significantly different between treatments present in subgingival plaque samples from healthy adult dogs consuming a commercially available wet or dry food
| Oral bacteria | Dry | Wet | SEM | P-value | ||
|---|---|---|---|---|---|---|
| Subgingival plaque samples | ||||||
| Phyla | Genus | Species | ||||
| Actinobacteria | 1.90 | 3.01 | 0.80 | 0.32 | ||
| Bacteroidetes | 81.04 | 73.64 | 2.65 | 0.08 | ||
| Capnocytophaga | 3.27 | 1.24 | 0.84 | 0.11 | ||
| Capnocytophaga cynodegmi | 0.79a | 0.33b | 0.13 | 0.03 | ||
| Porphyromonas | 67.60 | 62.08 | 4.01 | 0.35 | ||
| Tannerella | 0.27b | 0.64a | 0.07 | 0.002 | ||
| Tannerella forsythia | 0.27b | 0.64a | 0.07 | 0.002 | ||
| Euryarchaeota | 0.06 | 0.24 | 0.10 | 0.66 | ||
| Firmicutes | 3.97b | 7.81a | 0.82 | 0.01 | ||
| Filifactor | 0.97 | 2.01 | 0.42 | 0.11 | ||
| Unclassified Peptostreptococcaceae | 0.56b | 1.94a | 0.36 | 0.04 | ||
| Peptostreptococcaceae bacterium oral taxon 113 | 0.56b | 1.94a | 0.36 | 0.04 | ||
| Streptococcus | 0.53b | 1.42a | 0.21 | 0.01 | ||
| Streptococcus agalactiae | 0.33b | 0.99a | 0.14 | 0.02 | ||
| Fusobacteria | 0.11 | 0.39 | 0.11 | 0.11 | ||
| Proteobacteria | 11.27 | 12.07 | 1.44 | 0.70 | ||
| Neisseria | 2.32 | 2.24 | 0.62 | 0.93 | ||
| Arcobacter | 1.29 | 1.47 | 0.48 | 0.85 | ||
| Conchiformibius | 1.32a | 0.20b | 0.22 | 0.004 | ||
| Spirochaetes | 1.61 | 2.60 | 0.48 | 0.18 | ||
| Treponema | 1.61 | 2.60 | 0.48 | 0.18 | ||
| Synergistetes | 0.04b | 0.21a | 0.05 | 0.03 | ||
| Fretibacterium | 0.04b | 0.21a | 0.05 | 0.03 | ||
| Fretibacterium fastidiosum | 0.04b | 0.21a | 0.05 | 0.03 | ||
Groups with different superscripts differ (P < 0.05).
In SUP samples, the relative abundances of Arcobacter, Capnocytophaga, Pasteurella, Conchiformibius, Actinomyces timonensis, Capnocytophaga canimorsus, Conchiformibius steedae, Neisseria weaveri, Pasteurella dagmatis, Porphyromonas cangingivalis, and unclassified Treponema were lower (P < 0.05) in dogs consuming wet food than those consuming dry food (Table 3). In contrast, the relative abundances of unclassified Porphyromonadaceae, unclassified Bacteroidales, unclassified Peptostreptococcaceae, Tannerella, Filifactor, Treponema medium, Porphyromonas gingivalis, Porphyromonas canoris, Tannnerella forsythia, Streptococcus agalactiae, Peptostreptococcaceae bacterium oral taxon 113, Porphyromonadaceae bacterium COT-184 OH4590, and Filifactor alocis were higher (P < 0.05) in dogs consuming wet food than those consuming dry food (Table 3).
Table 3.
Oral bacteria (relative abundance, %) that were significantly different between treatments present in supragingival plaque samples from healthy adult dogs consuming a commercially available wet or dry food
| Oral bacteria | Dry | Wet | SEM | P-value | ||
|---|---|---|---|---|---|---|
| Supragingival plaque samples | ||||||
| Phyla | Genus | Species | ||||
| Actinobacteria | 1.72 | 1.79 | 1.01 | 0.18 | ||
| Actinomyces | 1.13 | 0.10 | 0.78 | 0.37 | ||
| Actinomyces timonensis | 1.09a | 0.04b | 0.77 | 0.03 | ||
| Bacteroidetes | 82.43 | 83.35 | 1.99 | 0.75 | ||
| Unclassified Bacteroidales | 0.01b | 0.04a | 0.01 | 0.003 | ||
| Capnocytophaga | 6.65a | 2.86b | 1.13 | 0.04 | ||
| Capnocytophaga canimorsus | 1.96a | 0.58b | 0.34 | 0.01 | ||
| Unclassified Porphyromonadaceae | 0.46b | 1.99a | 0.43 | 0.03 | ||
| Porphyromonas | 67.46 | 72.46 | 4.19 | 0.42 | ||
| Porphyromonadaceae bacterium COT-184 OH4590 | 0.46b | 1.99a | 0.43 | 0.03 | ||
| Porphyromonas cangingivalis | 16.8a | 9.44b | 2.05 | 0.03 | ||
| Porphyromonas canoris | 5.07b | 8.34a | 0.91 | 0.03 | ||
| Porphyromonas gingivalis | 0.28b | 0.67a | 0.07 | 0.003 | ||
| Tannerella | 0.15b | 0.59a | 0.07 | 0.001 | ||
| Tannerella forsythia | 0.15b | 0.59a | 0.07 | 0.001 | ||
| Candidatus Gracilibacteria | 0.06 | 0.01 | 0.02 | 0.19 | ||
| Euryarchaeota | 0.05 | 0.36 | 0.12 | 0.09 | ||
| Firmicutes | 2.50 | 4.14 | 0.57 | 0.07 | ||
| Filifactor | 0.42b | 1.05a | 0.16 | 0.02 | ||
| Filifactor alocis | 0.42b | 1.05a | 0.16 | 0.02 | ||
| Unclassified Peptostreptococcaceae | 0.13b | 0.95a | 0.10 | 0.0001 | ||
| Peptostreptococcaceae bacterium oral taxon 113 | 0.13b | 0.95a | 0.10 | 0.0001 | ||
| Streptococcus agalactiae | 0.16b | 0.56a | 0.11 | 0.03 | ||
| Fusobacteria | 0.04b | 0.32a | 0.06 | 0.01 | ||
| Proteobacteria | 12.05 | 8.21 | 1.41 | 0.08 | ||
| Neisseria | 5.38 | 4.37 | 1.00 | 0.49 | ||
| Neisseria weaveri | 2.37a | 0.33b | 0.42 | 0.0002 | ||
| Arcobacter | 1.77a | 0.09b | 0.34 | 0.01 | ||
| Conchiformibius | 0.29a | 0.09b | 0.04 | 0.003 | ||
| Conchiformibius steedae | 0.29a | 0.05b | 0.04 | 0.002 | ||
| Pasteurella | 2.77a | 0.18b | 0.86 | 0.01 | ||
| Pasteurella dagmatis | 2.59a | 0.11b | 0.84 | 0.001 | ||
| Spirochaetes | 0.96 | 1.27 | 0.26 | 0.43 | ||
| Unclassified Treponema | 0.34a | 0.02b | 0.12 | 0.01 | ||
| Treponema medium | 0.05b | 0.17a | 0.03 | 0.01 | ||
Groups with different superscripts differ (P < 0.05).
LEfSe identified four phyla and 15 genera that were affected by sample type [linear discriminant analysis (LDA) ≥ 3]. Two phyla were enriched in SUP plaque (Chloroflexi and Gracilibacteria) and seven genera were at least three-fold higher in SUP plaque (Bergeyella, Capnocytophaga, Neosseroa, Pasteurella, unclassified Chloroflexi, Melaminivora, Haemophilys). Two phyla were at least three-fold higher in SUB plaque (Spirochaetes and Firmicutes), and eight genera were at least three-fold higher in SUB plaque (Rothia, Conchiformibius, Filifactor, Cardiobacterium, Treponema, Desulfobulbus, Campylobacter, Porphyromonadaceae) (Supplementary Figure 5).
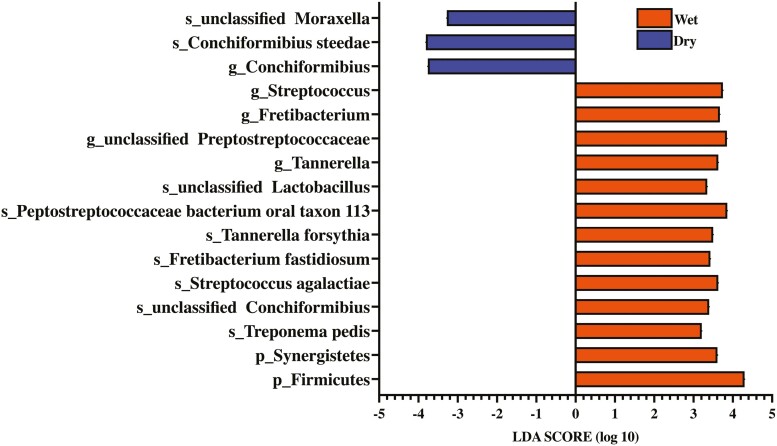
LEfSe identified two bacterial phyla, five bacterial genera, and nine bacterial species that were enriched in dogs consuming wet or dry foods in SUB samples (LDA ≥ 3) (Figure 4). Firmicutes, Synergistetes, Streptococcus, Fretibacterium, unclassified Peptostreptococcaceae, Tannerella, Treponema pedis, unclassified Chonciformibius, Fretibacterium fastidiosum, Tannerella forsythia, Peptostreptococcaceae bacterium oral taxon 113, and unclassified Lactobacillus were enriched in dogs consuming wet food, while Conchiformibius, unclassified Moraxella, and Conchiformibius steedae were enriched in dogs consuming dry food.
Figure 4.
Linear discriminant analysis effect size (LEfSe) identified bacteria enriched in subgingival plaque samples of healthy adult dogs consuming commercially available wet or dry food [linear discriminant analysis (LDA) score ≥ 3].
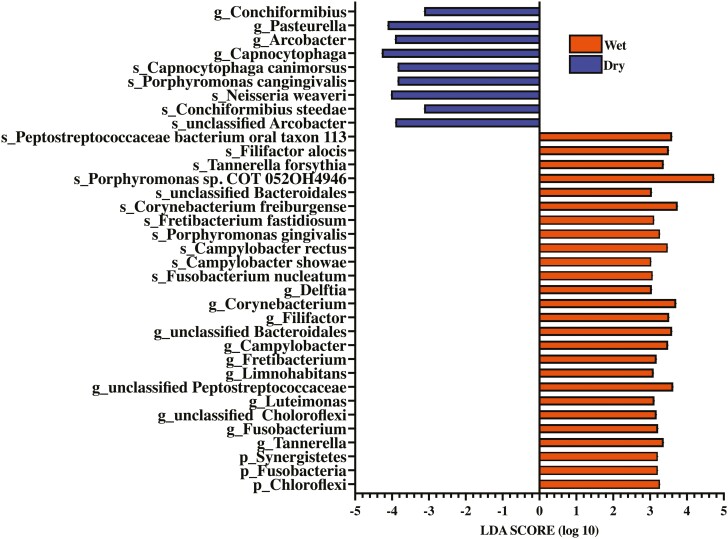
LEfSe also identified three bacterial phyla, 16 bacterial genera, and 16 bacterial species that were enriched in dogs consuming wet or dry foods in SUP samples (LDA ≥ 3) (Figure 5).
Figure 5.
Linear discriminant analysis effect size (LEfSe) identified bacteria enriched in supragingival plaque samples of healthy adult dogs consuming commercially available wet or dry food [linear discriminant analysis (LDA) score ≥ 3].
Chloroflexi, Fusobacteria, Synergistetes, Tannerella, Fusobacterium, unclassified Chloroflexi, Luteimonas, unclassified Peptostreptococcaceae, Limnohabitans, Fretibacterium, Campylobacter, unclassified Bacteroidales, Filifactor, Corynebacterium, Delftia, Fusobacterium nucleatum, Campylobacter showae, Camplylobacter rectus, Porphyromonas gingivalis, Fretibacterium fastidiosum, Corynebacterium freiburgense, unclassified Bacteroidales, Propyromonas sp. COT 052OH4946, Tannerella forsythia, Filifactor alocis, and Peptostreptococcaceae bacterium oral taxon 113 were enriched in dogs consuming wet food, while Conchiformibius, Pasteurella, Arcobacter, Capnocytophaga, unclassified Arcobacter, Conchiformibius steedae, Neisseria weaver, Porphyromonas cangingivalis, and Capnocytophaga canimorsus were enriched in dogs consuming dry food.
Discussion
The effects of diet on oral health is well known, with the main effect on the initiation and progression of periodontal disease being due to texture and its ability to impact the accumulation of plaque (Gorrel, 1998). In the past, it was shown that diet texture is a mechanical method to prevent plaque accumulation (Burwasser and Hill, 1939; Krasse, 1960; Egelberg, 1965a; Saxe et al., 1967). Dogs fed hard foods retained essentially normal teeth and gums, while those fed soft foods (same food, but ground and mixed with water) developed gingivitis, plaque, and calculus (Burwasser and Hill, 1939). Additionally, with solid food, gums appear normal and most gingival crevices remain free of bacteria, whereas with soft food (same food, but minced and mixed into a mush) gingivitis occurred, more crevices gave positive bacterial cultures, and the resulting microbiota resembled that associated with periodontal disease (Krasse, 1960).
In a study comparing a commercial dry dog food and a wet food form, results showed that dogs fed the dry diet acquired less “soft debris” on their teeth (Saxe et al., 1967). Another study proved the importance of chewing in the prevention of plaque accumulation by feeding dogs by gastric intubation or oral consumption of minced food. In that study, gingival exudation tended to increase to a greater extent during tube feeding, suggesting that even the minimal chewing required to consume minced food had some cleansing or protective effects (Egelberg, 1965b). Data from the present study agrees with those in previous studies, with dogs consuming wet food having greater plaque buildup than those consuming dry food. Additionally, dogs consuming dry food had lower breath odor and a higher salivary pH than those consuming wet food.
The main odor components in the mouth are small, volatile compounds that can be produced through bacterial metabolism. The main bacterial-derived odorants include hydrogen sulfide, methanethiol, volatile sulfur compounds, trimethylamine, indole, skatole, putrescine, cadaverine, acetone, pyridine, and ammonia (Solis and Volpe, 1973; Tonzetich, 1977; Persson et al., 1990; Yaegaki and Sanada, 1992; Tangerman and Winkel, 2007; Tangerman, 2009; Takeshita et al., 2012; Gulsahi et al., 2014; Nani et al., 2017; Mogilnicka et al., 2020). Most of these molecules are present under normal physiological conditions, but excessive production results in an unpleasant smell that is noticed by pet owners. Oral malodor can be affected by diet, which may contain odorants or substances that may be fermented into odorants by bacteria (Mogilnicka et al., 2020). Gram-negative bacteria (e.g., Bacteroides forsythus, Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans, and Prevotella intermedia) have been implicated in the production of volatile sulfur compounds originating from sulfur-containing substrates in food (Awano et al., 2002). In the present study, animals consuming wet food had higher relative abundance of Porphyromonas gingivali, which may be linked with the higher total volatile sulfur compound concentrations, but more research would be needed to provide a causal relationship.
The higher salivary pH of dogs consuming dry food (dry: 7.46 ± 0.07; wet: 7.17 ± 0.07) in the current study may be viewed as being beneficial to oral health. The consumption of acidic food as well as bacterial metabolism that reduces salivary pH leads to dental demineralization (Fejerskov and Kidd, 2008). In contrast, alkaline salivary pH can reduce demineralization and enhance the remineralization process (Edgar and O’Mullane, 1990). Even though periodontal disease is one of the most common oral diseases in dogs, a small dataset exists on saliva composition and oral health biomarkers in dogs. Traditionally, the mean salivary pH of medium-sized dogs has been reported to range between 7.2 and 8.1 (Larmas and Scheinin, 1971; Smeets-Peeters et al., 1998). In a more recent study, mean salivary pH of dogs was reported to be 8.53 ± 0.34, and not different among the three dog breeds (Dachshund, Labrador Retriever, Jack Russel) tested (Lavy et al., 2012). In another recent study reported a salivary pH of 7.93 ± 0.46 in dogs without oral disease, with neutered and intact male and female dogs not being different (Iacopetti et al., 2017).
In addition to plaque accumulation, dietary composition, physical properties, etc. may influence the oral microbiota populations, including the diversity of bacteria. For example, cats consuming a high-protein, low-carbohydrate wet food (~70 % moisture) had a more diverse oral microbiome, with enrichment of bacteria associated with both gingival health and periodontal disease than cats consuming a dry extruded kibble (highly refined, cereal-based, dehydrated rations) diet (Adler et al., 2016). While greater bacterial diversity has been associated with oral disease, it is not observed consistently. In one dog study, plaque sample operational taxonomic unit (OTU) counts from dogs with mild periodontitis and gingivitis were higher than those with healthy gingiva (Davis et al., 2013). In other studies, however, differences in the number of OTU or the Shannon diversity index were not observed among healthy and diseased groups (Wallis et al., 2015; Niemiec et al., 2022). In the present study, SUP samples of dogs consuming the wet food had a more diverse microbiota than those consuming the dry food. Additionally, beta diversity analysis clearly showed a distinct separation between SUP sample microbiota populations of dogs consuming the wet and dry foods. Alpha and beta diversity indices were not different between dietary groups in SUB samples, however.
The current understanding of the canine oral microbiome and the impact that diet has on its composition is limited. Overall, the bacterial profile present in the current study is similar to those published previously. In a previous study, Porphyromonas was the most abundant bacterial genus in SUB plaque samples of healthy dogs, with Moraxella and Bergeyella also being highly abundant (Davis et al., 2013). In another study that measured oral bacteria 28 d after dental cleaning and polishing by a veterinary dentist, Porphyromonas was the most abundant bacterial genus in all samples type (SUP and SUB), followed by Moraxella, Fusobacterium, and Enhydrobacter in SUB samples, and Moraxella, Enhydrobacter, and Neisseria in SUP samples (Oba et al., 2021a). A third study agreed, with Porphyromonas being the most abundant bacterial genus present in all samples (SUP and SUB), followed by Fusobacterium, Treponema, Moraxella, and Fusibacter in SUB samples, and Moraxella, Enhydrobacter, Fusobacterium, and Corynebacterium in SUP samples (Oba et al., 2021b).
Similar to those previous publications, which used 16S rRNA-based sequencing, Porphyromonas was the most abundant bacterial genus in all sample type (SUP and SUB) in the present study, along with unclassified Porphyromonadaceae, Campylobacter, and Bergeyella in SUB samples, and Bergeyella, Neisseria, and Capnocytophaga in SUP samples. Moraxella (~0.2%), Fusobacterium (~0.2%), Treponema (~1.6%), and Corynebacterium (~1.6%) were present at a low abundance, while Enhydrobacter and Fusibacter were not detected in the present study, however. These differences among studies may be due to the sequencing techniques and databases used. It has been previously shown that 16S rRNA amplicon-based sequencing yields quantitatively and qualitatively different results than those coming from shotgun metagenomics. Compared with the 16S rRNA-based methods, shotgun metagenomics allows for much deeper characterization of the microbiome complexity, allowing identification of a larger number of species for each sample (Laudadio et al., 2018).
In the current study, we aimed to examine two of the main factors thought to influence the composition of the oral microbiome—dietary moisture and texture. While differences were observed in bacterial diversity and abundance among dogs consuming wet vs. dry foods, the implications for oral health is still unclear. In SUB plaque samples of humans with generalized aggressive periodontitis and generalized chronic periodontitis, higher levels of Bacteroidales, Filifactor alocis, Fretibacterium fastidiosum, Fretibacterium sp. HOT 360, Porphyromonas gingivalis, Selenomonas sputigena, Tannerella forsythia, and TM7 sp. HOT 356 are present when compared with subjects in a healthy control group (Oliveira et al., 2016). Filifactor alocis has emerged as an important periodontal pathogen in humans (Vashishta et al., 2019; Schlafer et al., 2010). Tannerella forsythia, Porphyromonas gingivalis, and Treponema denticola also constitute the “red complex” in humans—a group of bacteria associated with periodontal disease and classified as highly virulent (Hajishengallis, 2014; Trindade et al., 2014; Hajishengallis, 2015; Lamont and Hajishengallis, 2015). Furthermore, Treponema medium has been isolated from SUB samples of patients with adult periodontitis (Umemoto et al., 1997; Nakazawa et al., 2003), with the number of Treponema medium being increased in SUB samples from deep periodontal pockets (Asai et al., 2002). In the present study, Fretibacterium fastidiosum and Tannerella forsythia were higher in SUB samples of dogs consuming wet food than those consuming dry food. Additionally, Treponema medium, Filifactor alocis, Porphyromonas gingivalis, and Tannerella forsythia were higher in SUP samples of dogs consuming wet food than those consuming dry food.
Porphyromonas spp. are well-known bacteria associated with the development of periodontal diseases, with Porphyromonas cangingivalis being a highly abundant bacterial group in the oral cavity of healthy dogs and animals with inflamed periodontal tissues (Dewhirst et al., 2012; Davis et al., 2013; Wallis et al., 2015; do Nascimento Silva et al., 2017). Porphyromonas gingivalis is one of the principal bacteria thought to contribute to the development of human and animal periodontal disease (Genco et al., 1998; Norris and Love, 2000; Özavci et al., 2019). Porphyromonas in general are black-pigmented species from the gingival sulcus of dogs with naturally occurring periodontal disease, with counts of P. intermedia being positively correlated with the amount of plaque and the degree of gingivitis (Allaker et al., 1997). Porphyromonas canoris, P. salivosa, P. cangingivalis, P. cansulci, P. crevioricanis, and Prevotella denticola have been isolated from healthy dogs, but at a low level (3% to 9% of dogs) (Allaker et al., 1997). In the present study, dogs consuming the wet food had a higher relative abundance of Porphyromonas canoris and Porphyromonas gingivalis, with dogs consuming the dry food having a higher relative abundance of Porphyromonas cangingivalis. This shift in the oral microbiota due to diet type may indicate that the consumption of a high-moisture, soft food can lead to an undesirable change in the oral microbiota.
This study had some limitations, including the lack of using a clean mouth model, a low number of dogs studied, and the study of a single dog breed. In future studies, it would be interesting to evaluate other dietary formats available on the market (dehydrated, freeze-dried, raw, semi-moist, homemade) to evaluate how different textures and densities impact oral health outcomes and microbiota. It would also be interesting to test different inclusion levels of protein, carbohydrate, or fat to determine how dietary macronutrients impact oral microbiota populations. Finally, because dog breeds differ in jaw structure, tooth spacing and orientation, and risk of periodontal disease, it is suggested that various dog breeds be studied in future studies.
In the current study, plaque, calculus, and gingivitis scores were not different between dogs consuming wet food or dry foods, but plaque buildup tended to be lower, breath sulfur concentrations were lower, and salivary pH was higher in dogs consuming the dry food. Oral bacterial diversity was higher in dogs consuming the wet food. Also, oral health-associated bacterial genera (Pasteurella, Capnocytophaga, and Corynebacterium) were higher, while bacteria associated with poor oral health (Fretibacterium fastidiosum, Filifactor alocis, Treponema medium, Tannerella forsythia, Porphyromonas canoris, and Porphyromonas gingivalis) were lower in dogs consuming the dry food. Such shifts in the oral microbiota may impact periodontal disease risk, but longer dietary intervention studies are required to confirm or reject this hypothesis. In addition to conducting longer intervention studies, more research is needed to determine whether other dietary components may influence the microbial populations.
Supplementary Material
Acknowledgment
Funding was provided by USDA Hatch Grant ILLU-538-937.
Glossary
Abbreviations
- LDA
linear discriminant analysis
- LEfSe
linear discriminant analysis effect size
- OTU
operational taxonomic units
- SUB
subgingival source
- SUP
supragingival source.
Contributor Information
Patrícia M Oba, Department of Animal Sciences, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Kelly M Sieja, Department of Animal Sciences, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Stephanie C J Keating, Department of Veterinary Clinical Medicine, College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Teodora Hristova, Department of Veterinary Clinical Medicine, College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Amy J Somrak, Department of Veterinary Clinical Medicine, College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Kelly S Swanson, Department of Animal Sciences, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA; Department of Veterinary Clinical Medicine, College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA; Division of Nutritional Sciences, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Conflict of Interest Statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Adler, C. J., Malik R., Browne G. V., and Norris J. M.. . 2016. Diet may influence the oral microbiome composition in cats. Microbiome 4:1–9. doi: 10.1186/s40168-016-0169-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allaker, R. P., de Rosayro R., Young K. A., and Hardie J. M.. . 1997. Prevalence of Porphyromonas and Prevotella species in the dental plaque of dogs. Vet. Rec. 140:147–148. doi: 10.1136/vr.140.6.147. [DOI] [PubMed] [Google Scholar]
- Asai, Y., Jinno T., Igarashi H., Ohyama Y., and Ogawa T.. . 2002. Detection and quantification of oral treponemes in subgingival plaque by real-time PCR. J. Clin. Microbiol. 40:3334–3340. doi: 10.1128/JCM.40.9.3334-3340.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano, S., Gohara K., Kurihara E., Ansai T., and Takehara T.. . 2002. The relationship between the presence of periodontopathogenic bacteria in saliva and halitosis. Int. Dent. J. 52:212–216. doi: 10.1002/j.1875-595x.2002.tb00927.x. [DOI] [PubMed] [Google Scholar]
- Burwasser, P., and Hill T. J.. . 1939. The effect of hard and soft diets on the gingival tissues of dogs. J. Dent. Res. 18:389–393. doi: 10.1177/00220345390180040801. [DOI] [Google Scholar]
- Carroll, M. Q., Oba P. M., Sieja K. M., Alexander C., Lye L., de Godoy M. R. C., He F., Somrak A. J., Keating S. C. J., Sage A. M., . et al. 2020. Effects of novel dental chews on oral health outcomes and halitosis in adult dogs. J. Anim. Sci. 98:1–7. doi: 10.1093/jas/skaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, I. J., Wallis C., Deusch O., Colyer A., Milella L., Loman N., and Harris S.. . 2013. A cross-sectional survey of bacterial species in plaque from client owned dogs with healthy gingiva, gingivitis. PLoS One 8:1–12. doi: 10.1371/journal.pone.0083158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst, F. E., Klein E. A., Thompson E. C., Blanton J. M., Chen T., Milella L., Buckley C. M. F., Davis I. J., Bennett M.-L., and Marshall-Jones Z. V.. . 2012. The canine oral microbiome. PLoS One 7:e36067. doi: 10.1371/journal.pone.0036067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, W. M., and O’Mullane D. M.. . 1990. The role of saliva in demineralization and remineralization of teeth. In: Saliva and dental health. 1st ed. British Dental Association,London, United Kingdom. p. 19–24. [Google Scholar]
- Egelberg, J. 1965a. Local effects of diet on plaque formation and gingivitis development in dogs. I. Effect of hard and soft diets. Odontol. Rev. 16:31–41. [PubMed] [Google Scholar]
- Egelberg, J. 1965b. Local effect of diet on plaque formation and development of gingivitis in dogs. III. Effect of frequency of meals and tube feeding. Odontol. Rev. 16:50–60. [PubMed] [Google Scholar]
- Engel, A. S., Kranz H. T., Schneider M., Tietze J. P., Piwowarcyk A., Kuzius T., Arnold W., and Naumova E. A.. . 2020. Biofilm formation on different dental restorative materials in the oral cavity. BMC Oral Health 20:1–10. doi: 10.1186/S12903-020-01147-X/TABLES/3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejerskov, O., and Kidd E.. . 2008. The role of saliva. Dent. Caries 2:189–208. [Google Scholar]
- Genco, C. A., Van Dyke T., and Amar S.. . 1998. Animal models for Porphyromonas gingivalis-mediated periodontal disease. Trends Microbiol. 6:444–449. doi: 10.1016/s0966-842x(98)01363-8. [DOI] [PubMed] [Google Scholar]
- Gorrel, C. 1998. Periodontal disease and diet in domestic pets. J. Nutr. 128:2712S–2714S. doi: 10.1093/jn/128.12.2712S. [DOI] [PubMed] [Google Scholar]
- Gorrel, C., Warrick J., and Bierer T. L.. . 1999. Effect of a new dental hygiene chew on periodontal health in dogs. J. Vet. Dent. 16:77–81. doi: 10.1177/089875649901600203. [DOI] [PubMed] [Google Scholar]
- Gulsahi, A., Evirgen Ş., Öztaş B., Genç Y., and Çetinel Y.. . 2014. Volatile sulphur compound levels and related factors in patients with chronic renal failure. J. Clin. Periodontol. 41:814–819. doi: 10.1111/jcpe.12280. [DOI] [PubMed] [Google Scholar]
- Hajishengallis, G. 2014. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35:3–11. doi: 10.1016/j.it.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis, G. 2015. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan, N. A., Young B. A., Minard-Smith A. T., Saeed K., Li H., Heizer E. M., McMillan N. J., Isom R., Abdullah A. S., Bornman D. M., . et al. 2014. Microbial community profiling of human saliva using shotgun metagenomic sequencing. PLoS One 9:e97699. doi: 10.1371/journal.pone.0097699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennet, P., Servet E., Salesse H., and Soulard Y.. . 2006. Evaluation of the Logan & Boyce plaque index for the study of dental plaque accumulation in dogs. Res. Vet. Sci. 80:175–180. doi: 10.1016/j.rvsc.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Iacopetti, I., Perazzi A., Badon T., Bedin S., Contiero B., and Ricci R.. . 2017. Salivary pH, calcium, phosphorus and selected enzymes in healthy dogs: a pilot study. BMC Vet. Res. 13:1–7. doi: 10.1186/s12917-017-1256-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B.-R., Shin J., Guevarra R. B., Lee J. H., Kim D. W., Seol K.-H., Lee J.-H., Kim H. B., and Isaacson R. E.. . 2017. Deciphering diversity indices for a better understanding of microbial communities. J. Microbiol. Biotechnol. 27:2089–2093. doi: 10.4014/jmb.1709.09027. [DOI] [PubMed] [Google Scholar]
- Kortegaard, H. E., Eriksen T., and Baelum V.. . 2008. Periodontal disease in research beagle dogs—an epidemiological study. J. Small Anim. Pract. 49:610–616. doi: 10.1111/j.1748-5827.2008.00609.x. [DOI] [PubMed] [Google Scholar]
- Krasse, B. 1960. Effect of consistency of diet on bacteria in gingival pockets in dogs. Odont. Rev. 11:152–165. [Google Scholar]
- Laflamme, D. 1997. Development and validation of a body condition score system for dogs. Canine Pract. 22:10–15. [Google Scholar]
- Lamont, R. J., and Hajishengallis G.. . 2015. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21:172–183. doi: 10.1016/j.molmed.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larmas, M., and Scheinin A.. . 1971. Studies on dog saliva: I. Some physico-chemical characteristics. Acta Odontol. Scand. 29:205–214. doi: 10.3109/00016357109026516. [DOI] [PubMed] [Google Scholar]
- Laudadio, I., Fulci V., Palone F., Stronati L., Cucchiara S., and Carissimi C.. . 2018. Quantitative assessment of shotgun metagenomics and 16S rDNA amplicon sequencing in the study of human gut microbiome. OMICS 22:248–254. doi: 10.1089/omi.2018.0013. [DOI] [PubMed] [Google Scholar]
- Lavy, E., Goldberger D., Friedman M., and Steinberg D.. . 2012. pH values and mineral content of saliva in different breeds of dogs. Isr. J. Vet. Med. 67:244–248. [Google Scholar]
- Lax, S., Smith D. P., Hampton-Marcell J., Owens S. M., Handley K. M., Scott N. M., Gibbons S. M., Larsen P., Shogan B. D., Weiss S., . et al. 2014. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science 345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbareche, H., Dumont-Leblond N., Bilodeau G. J., and Duchaine C.. . 2020. An overview of bioinformatics tools for DNA meta-barcoding analysis of microbial communities of bioaerosols: digest for microbiologists. Life 10:185. doi: 10.3390/life10090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogilnicka, I., Bogucki P., and Ufnal M.. . 2020. Microbiota and malodor—etiology and management. Int. J. Mol. Sci. 21:2886. doi: 10.3390/ijms21082886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlemann, H. R., and Son S.. . 1971. Gingival sulcus bleeding—a leading symptom in initial gingivitis. Helv. Odontol. Acta 15:107–113. [PubMed] [Google Scholar]
- Nakazawa, F., Hoshino E., Fukunaga M., Jinno T., Asai Y., Yamamoto H., and Ogawa T.. . 2003. Amended biochemical characteristics and phylogenetic position of Treponema medium. Oral Microbiol. Immunol. 18:127–130. doi: 10.1034/j.1399-302x.2003.00015.x. [DOI] [PubMed] [Google Scholar]
- Nani, B. D., de Lima P. O., Marcondes F. K., Groppo F. C., Rolim G. S., de Moraes A. B. A., Cogo-Müller K., and Franz-Montan M.. . 2017. Changes in salivary microbiota increase volatile sulfur compounds production in healthy male subjects with academic-related chronic stress. PLoS One 12:e0173686. doi: 10.1371/journal.pone.0173686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- do Nascimento Silva, A., de Avila E. D., Nakano V., and Avila-Campos M. J.. . 2017. Pathogenicity and genetic profile of oral Porphyromonas species from canine periodontitis. Arch. Oral Biol. 83:20–24. doi: 10.1016/j.archoralbio.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Niemiec, B. A. 2012. Veterinary periodontology. In: Niemiec B. A., editor. Etiology and pathogenesis of periodontal disease. John Wiley & Sons, Hoboken, NJ; p.18–34. [Google Scholar]
- Niemiec, B. A., Gawor J., Tang S., Prem A., and Krumbeck J. A.. . 2022. The bacteriome of the oral cavity in healthy dogs and dogs with periodontal disease. Am. J. Vet. Res. 83:50–58. doi: 10.2460/ajvr.21.02.0027. [DOI] [PubMed] [Google Scholar]
- Norris, J. M., and Love D. N.. . 2000. The association of two recombinant proteinases of a feline strain of Porphyromonas gingivalis with periodontal disease in cats. Vet. Microbiol. 71:69–80. doi: 10.1016/s0378-1135(99)00154-6. [DOI] [PubMed] [Google Scholar]
- Oba, P. M., Carroll M. Q., Alexander C., Somrak A. J., Keating S. C. J., Sage A. M., and Swanson K. S.. . 2021a. Dental chews positively shift the oral microbiota of adult dogs. J. Anim. Sci. 99:1–18. doi: 10.1093/jas/skab100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oba, P. M., Carroll M. Q., Alexander C., Valentine H., Somrak A. J., Keating S. C. J., Sage A. M., and Swanson K. S.. . 2021b. Microbiota populations in supragingival plaque, subgingival plaque, and saliva habitats of adult dogs. Anim. Microbiome 3:38. doi: 10.1186/s42523-021-00100-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, R. R. D. S., Fermiano D., Feres M., Figueiredo L. C., Teles F. R. F., Soares G. M. S., and Faveri M.. . 2016. Levels of candidate periodontal pathogens in subgingival biofilm. J. Dent. Res. 95:711–718. doi: 10.1177/0022034516634619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen, A., Ramachandran P., Reed E., White J. R., Hasan N., Subramanian P., Ryan G., Jarvis K., Grim C., Daquiqan N., . et al. 2016. Enrichment dynamics of Listeria monocytogenes and the associated microbiome from naturally contaminated ice cream linked to a listeriosis outbreak. BMC Microbiol. 16:275. doi: 10.1186/s12866-016-0894-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Özavci, V., Erbas G., Parin U., Yüksel H. T., and Kirkan S.. . 2019. Molecular detection of feline and canine periodontal pathogens. Vet. Anim. Sci. 8:100069. doi: 10.1016/j.vas.2019.100069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson, S., Edlund M.-B., Claesson R., and Carlsson J.. . 1990. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol. Immunol. 5:195–201. doi: 10.1111/j.1399-302x.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- Ponnusamy, D., Kozlova E. V., Sha J., Erova T. E., Azar S. R., Fitts E. C., Kirtley M. L., Tiner B. L., Andersson J. A., Grim C. J., . et al. 2016. Cross-talk among flesh-eating Aeromonas hydrophila strains in mixed infection leading to necrotizing fasciitis. Proc. Natl Acad. Sci. USA 113:722–727. doi: 10.1073/pnas.1523817113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruparell, A., Inui T., Staunton R., Wallis C., Deusch, O. and Holcombe L. J.. . 2020a. The canine oral microbiome: variation in bacterial populations across different niches. BMC Microbiol. 20:42 doi: 10.1186/s12866-020-1704-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruparell, A., Warren M., Staunton R., Deusch O., Dobenecker B., Wallis C., O'Flynn C., McGenity P., and Holcombe L. J.. . 2020b. Effect of feeding a daily oral care chew on the composition of plaque microbiota in dogs. Res. Vet. Sci. 132:133–141. doi: 10.1016/j.rvsc.2020.05.001. [DOI] [PubMed] [Google Scholar]
- Saxe, S. R., Greene J. C., Bohannan H. M., and Vermillion J. R.. . 1967. Oral debris, calculus, and periodontal disease in the beagle dog. Periodontics 5:217–225. [PubMed] [Google Scholar]
- Schlafer, S., Riep B., Griffen A. L., Petrich A., Hübner J., Berning M., Friedmann A., Göbel U. B., and Moter A.. . 2010. Filifactor alocis—involvement in periodontal biofilms. BMC Microbiol. 10:1–13. doi: 10.1186/1471-2180-10-66/FIGURES/6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, P. J., and Jenkins D. G.. . 2018. How robust are popular beta diversity indices to sampling error? Ecosphere 9:e02100. doi: 10.1002/ecs2.2100. [DOI] [Google Scholar]
- Segata, N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., and Huttenhower C.. . 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeets-Peeters, M., Watson T., Minekus M., and Havenaar R.. . 1998. A review of the physiology of the canine digestive tract related to the development of in vitro systems. Nutr. Res. Rev. 11:45–69. doi: 10.1079/NRR19980005. [DOI] [PubMed] [Google Scholar]
- Solis, M. C., and Volpe A. R.. . 1973. Determination of sulfur volatiles in putrefied saliva by a gas chromatography-microcoulometric titrating system. J. Periodontol. 44:775–778. doi: 10.1902/jop.1973.44.12.775. [DOI] [PubMed] [Google Scholar]
- Takeshita, T., Suzuki N., Nakano Y., Yasui M., Yoneda M., Shimazaki Y., Hirofuji T., and Yamashita Y.. . 2012. Discrimination of the oral microbiota associated with high hydrogen sulfide and methyl mercaptan production. Sci. Rep. 2:215. doi: 10.1038/srep00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangerman, A. 2009. Measurement and biological significance of the volatile sulfur compounds hydrogen sulfide, methanethiol and dimethyl sulfide in various biological matrices. J. Chromatogr. B 877:3366–3377. doi: 10.1016/j.jchromb.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Tangerman, A., and Winkel E. G.. . 2007. Intra- and extra-oral halitosis: finding of a new form of extra-oral blood-borne halitosis caused by dimethyl sulphide. J. Clin. Periodontol. 34:748–755. doi: 10.1111/j.1600-051X.2007.01116.x. [DOI] [PubMed] [Google Scholar]
- Tonzetich, J. 1977. Production and origin of oral malodor: a review of mechanisms and methods of analysis. J. Periodontol. 48:13–20. doi: 10.1902/jop.1977.48.1.13. [DOI] [PubMed] [Google Scholar]
- Trindade, F., Oppenheim F. G., Helmerhorst E. J., Amado F., Gomes P. S., and Vitorino R.. . 2014. Uncovering the molecular networks in periodontitis. Proteomics Clin. Appl. 8:748–761. doi: 10.1002/prca.201400028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umemoto, T., Nakazawa F., Hoshino E., Okada K., Fukunaga M., and Namikawa I.. . 1997. Treponema medium sp. nov., isolated from human subgingival dental plaque. Int. J. Syst. Bacteriol. 47:67–72. doi: 10.1099/00207713-47-1-67/CITE/REFWORKS. [DOI] [PubMed] [Google Scholar]
- Vashishta, A., Jimenez-Flores E., Klaes C., Tian S., Miralda I., Lamont R., and Uriarte S.. . 2019. Putative periodontal pathogens, Filifactor alocis and Peptoanaerobacter stomatis, induce differential cytokine and chemokine production by human neutrophils. Pathogens 8:59. doi: 10.3390/pathogens8020059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis, C., and Holcombe L. J.. . 2020. A review of the frequency and impact of periodontal disease in dogs. J. Small Anim. Pract. 61:529–540. doi: 10.1111/jsap.13218. [DOI] [PubMed] [Google Scholar]
- Wallis, C., Marshall M., Colyer A., O’Flynn C., Deusch O., and Harris S.. . 2015. A longitudinal assessment of changes in bacterial community composition associated with the development of periodontal disease in dogs. Vet. Microbiol. 181:271–282. doi: 10.1016/j.vetmic.2015.09.003. [DOI] [PubMed] [Google Scholar]
- Wallis, C., Saito E. K., Salt C., Holcombe L. J., and Desforges N. G.. . 2021. Association of periodontal disease with breed size, breed, weight, and age in pure-bred client-owned dogs in the United States. Vet. J. 275:105717. doi: 10.1016/j.tvjl.2021.105717. [DOI] [PubMed] [Google Scholar]
- Yaegaki, K., and Sanada K.. . 1992. Volatile sulfur compounds in mouth air from clinically healthy subjects and patients with periodontal disease. J. Periodontal Res. 27:233–238. doi: 10.1111/j.1600-0765.1992.tb01673.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.