Abstract
The sigma factor RpoS (ςS) has been described as a general stress response regulator that controls the expression of genes which confer increased resistance to various stresses in some gram-negative bacteria. To elucidate the role of RpoS in Pseudomonas aeruginosa physiology and pathogenesis, we constructed rpoS mutants in several strains of P. aeruginosa, including PAO1. The PAO1 rpoS mutant was subjected to various environmental stresses, and we compared the resistance phenotype of the mutant to that of the parent. The PAO1 rpoS mutant was slightly more sensitive to carbon starvation than the wild-type strain, but this phenotype was obvious only when the cells were grown in a medium supplemented with glucose as the sole carbon source. In addition, the PAO1 rpoS mutant was hypersensitive to heat shock at 50°C, increased osmolarity, and prolonged exposure to high concentrations of H2O2. In accordance with the hypersensitivity to H2O2, catalase production was 60% lower in the rpoS mutant than in the parent strain. We also assessed the role of RpoS in the production of several exoproducts known to be important for virulence of P. aeruginosa. The rpoS mutant produced 50% less exotoxin A, but it produced only slightly smaller amounts of elastase and LasA protease than the parent strain. The levels of phospholipase C and casein-degrading proteases were unaffected by a mutation in rpoS in PAO1. The rpoS mutation resulted in the increased production of the phenazine antibiotic pyocyanin and the siderophore pyoverdine. This increased pyocyanin production may be responsible for the enhanced virulence of the PAO1 rpoS mutant that was observed in a rat chronic-lung-infection model. In addition, the rpoS mutant displayed an altered twitching-motility phenotype, suggesting that the colonization factors, type IV fimbriae, were affected. Finally, in an alginate-overproducing cystic fibrosis (CF) isolate, FRD1, the rpoS101::aacCI mutation almost completely abolished the production of alginate when the bacterium was grown in a liquid medium. On a solid medium, the FRD1 rpoS mutant produced approximately 70% less alginate than did the wild-type strain. Thus, our data indicate that although some of the functions of RpoS in P. aeruginosa physiology are similar to RpoS functions in other gram-negative bacteria, it also has some functions unique to this bacterium.
Microorganisms continuously sense and respond to environmental stimuli, such as starvation, desiccation, osmotic stress, oxidative stress, and changes in temperature. In many instances, an appropriate response to an environmental stimulus requires changes in the levels of specific gene products. One way to accomplish this is by modulating gene expression through the replacement of the main sigma factor of RNA polymerase with an alternative sigma factor that recognizes a specific promoter of the stimulus response gene (19, 39). The sigma factor RpoS (ςS) was originally identified in Escherichia coli and Salmonella typhimurium as an alternative sigma factor that activates the expression of numerous genes required to maintain cell viability during the stationary phase of growth when cells are experiencing nutrient starvation (for reviews, see references 30 and 38). Activation of these genes makes the bacterium more resistant to environmental stresses, such as prolonged starvation, osmotic stress, and oxidative stress. RpoS also plays an important role in exponentially growing cells that are exposed to increased osmolarity (22). In E. coli and S. typhimurium, RpoS induces the expression of more than 30 genes in response to various stresses (33, 42). RpoS also activates the genes necessary for virulence in several bacteria, including E. coli, S. typhimurium, and Shigella flexneri (3, 31, 48, 55). Among pseudomonads, the role of RpoS as a general stress response regulator has been described for Pseudomonas putida (43, 51) and Pseudomonas fluorescens (52). In P. fluorescens, RpoS also modulates the production of several antibiotics and affects the biological control activity of the bacterium.
Pseudomonas aeruginosa is a gram-negative bacterium that is found in various environmental niches, including soil, water, plants, hospital environments, and human infections. As an opportunistic pathogen, this bacterium primarily infects patients who are immunocompromised (11). During infection, P. aeruginosa must survive major environmental changes, including changes in temperature, pH, ionic strength, and the presence of reactive oxygen intermediates and antibodies. With the exception of several genes that have been identified as being involved in the oxidative stress response, such as katA, katB, sodA, sodB, and alginate genes (6, 8, 21, 40, 41, 62), little is known about the stress responses of P. aeruginosa. However, some information regarding the stress responses of this bacterium can be obtained from studies on the regulation of its various exoproducts. P. aeruginosa secretes a battery of extracellular products that are important for virulence: exotoxin A, elastase, LasA protease, phospholipase C, alkaline protease, rhamnolipid, pyocyanin, and alginate. Many of these toxic exoproducts are involved in procuring nutrients and/or protecting the bacterium from the host immune system (14). Furthermore, some of these exoproducts are synthesized in response to the scarcity of specific nutrients (37). For example, exotoxin A and phospholipase C are produced when iron and phosphate are limiting, respectively. Thus, the synthesis of some of these exoproducts may be regulated by various stresses that P. aeruginosa encounters in specific niches.
The P. aeruginosa rpoS gene, which encodes RpoS, or ςS, has been cloned and sequenced by Tanaka and Takahashi (58). The P. aeruginosa rpoS gene, like the E. coli rpoS gene, is growth phase regulated (16). In both E. coli and P. aeruginosa, the level of rpoS mRNA is low during exponential growth but increases severalfold as cells enter the stationary phase. In P. aeruginosa, the induction of rpoS gene expression has been reported to be regulated by the quorum-sensing cascade involving Vfr, the las system, and the rhl system (1, 34, 49). The quorum-sensing systems of P. aeruginosa regulate the synthesis of many virulence factors, including exotoxin A, elastase, LasA protease, and pyocyanin (5, 35). Thus, the synthesis of these virulence factors may be regulated through RpoS.
Because stress responses may play important roles in pathogenesis, we examined the role of RpoS when P. aeruginosa responds to various environmental changes. In this study, we present evidence that RpoS mediates the general stress response and modulates synthesis of several exoproducts, including some secondary metabolites.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
Bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in L broth (LB) or LB supplemented with appropriate antibiotics at 37°C with aeration, unless otherwise indicated. For minimal salts medium, no-carbon–E minimal medium (NCE) (12) supplemented with either glucose (10 mM) or succinate (20 mM) as a carbon source was used. For exotoxin A assays, iron-depleted Trypticase soy broth dialysate was used as previously described (47). For phospholipase C assays and pyocyanin extractions, 1% peptone broth supplemented with 1% glycerol and 1% NaCl was used as the growth medium. For pyoverdine assays, P. aeruginosa was grown in King B medium (28). Following conjugations, pseudomonas isolation agar was used to counterselect against E. coli. Media were solidified with 1.5% Bacto Agar (Difco). The following antibiotic concentrations (per milliliter) were used in this study: ampicillin, 100 μg for E. coli; carbenicillin, 300 μg for P. aeruginosa; gentamicin, 20 μg for E. coli and 300 μg for P. aeruginosa; kanamycin, 50 μg for E. coli. Antibiotics were purchased from Sigma (St. Louis, Mo.).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype or relevant characteristica | Source or reference |
|---|---|---|
| P. aeruginosa | ||
| PAO1 | Prototroph | 23 |
| PA103 | Exotoxin A-overproducing strain | 36 |
| FRD1 | Alginate-overproducing strain | 45 |
| SS6 | PA103 rpoS101::aacCI | This study |
| SS24 | PAO1 rpoS101::aacCI | This study |
| SS138 | FRD1 rpoS101::aacCI | This study |
| E. coli | ||
| DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK l-rpsL nupG | Gibco BRL |
| HB101 | F− Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1 Δ(mcrC-mrr)rpsL20 xyl-5 mtl-1 recA13 | Lab collection |
| Plasmids | ||
| pDB18R | 1.8-kb KpnI-HindIII fragment of P. aeruginosa rpoS in pTZ18R | 58 |
| pRK2013 | Tra1 (RK2), ColE1; Kmr | 15 |
| pSF1 | pMB1 replicon, 1.3-kb oriT from (RK2); Apr | 54 |
| pSS1 | rpoS101::aacCI in pTZ18R; Apr/Cbr Gmr | This study |
| pSS6 | rpoS101::aacCI in pTZ18R plus 1.2-kb EcoRI fragment of oriT from pSF1 | This study |
| pTZ18R | pUC18-derived general cloning vector; Apr/Cbr | Pharmacia |
| pUCGM1 | pUC1918 carrying the aacCI (Gmr) cassette; Apr/Cbr Gmr | 53 |
Kmr, kanamycin resistant; Apr, ampicillin resistant; Cbr carbenicillin resistant; Gmr, gentamicin resistant.
DNA manipulations, transformations, and conjugations.
E. coli DH10B or HB101 was routinely used as a host strain for cloning. Plasmids were purified with QIAprep spin miniprep columns (Qiagen, Santa Clarita, Calif.). DNA fragments were excised from agarose gels and purified with the Qiaex II DNA gel extraction system (Qiagen), according to the manufacturer’s instructions. Restriction enzymes and DNA modification enzymes were purchased from New England Biolabs (Beverly, Mass.), Gibco BRL (Gaithersburg, Md.), or Boehringer Mannheim (Indianapolis, Ind.). For DNA amplifications with a GeneAmp 2400 (Perkin-Elmer, Foster City, Calif.), either Pfu (Stratagene, La Jolla, Calif.) or Amplitaq (Perkin-Elmer) was used. Oligonucleotides were purchased from Operon, Inc. (Alameda, Calif.). DNA was introduced into E. coli or P. aeruginosa either by electroporation or by conjugation. A standard electroporation procedure was used for E. coli with the E. coli gene pulser (Bio-Rad, Hercules, Calif.). For P. aeruginosa, electrocompetent cells were prepared as follows. Cells were grown to mid-log phase (optical density at 600 nm [OD600], 0.5 to 0.7) in 100 ml of LB and harvested by centrifugation. Following two washes with 10 ml of 0.3 M sucrose, the cells were resuspended in 1 ml of 0.3 M sucrose, and 40 μl of this cell suspension was used for electroporation. The conditions used to electroporate P. aeruginosa were 200 Ω, 200 μF, and 1.6 kV. For conjugations, triparental matings were performed with the helper plasmid pRK2013 as previously described (17), with the following modifications. Briefly, 50 to 100 μl of overnight cultures of donor, helper, and recipient strains were mixed in 1 ml of saline, centrifuged, washed with 1 ml of saline, and resuspended in 50 μl of saline. The cell suspension was spotted on an LB plate and incubated overnight at 30°C. Following incubation, the cells were scraped with a sterile cotton swab, resuspended in 2 ml of saline, and plated on selective plates. Introduction of plasmid DNA into P. aeruginosa FRD1 was done exclusively by conjugation.
Construction of P. aeruginosa rpoS mutants.
To generate P. aeruginosa rpoS mutants, the suicide plasmid pSS1, which carries the rpoS101::aacCI null allele, was constructed. Briefly, pDB18R, which carries P. aeruginosa rpoS on a 1.8-kb KpnI-HindIII fragment, was digested with SphI to cut once within rpoS. Following T4 DNA polymerase digestion to blunt the protruding ends, the linearized plasmid was digested with HincII to delete approximately 420 nucleotides within the rpoS gene. The aacCI gene, which encodes gentamicin resistance (Gmr) and which was obtained from pUCGM1 as a SmaI fragment, was inserted to generate the rpoS101 allele. pSS1 was introduced into P. aeruginosa PAO1 and PA103 by electroporation, and the potential rpoS mutants were isolated as having Gmr (encoded by the rpoS101::aacCI allele) but lacking carbenicillin resistance (Cbr) (encoded by the plasmid vector), indicating a double crossover event. Introducing the suicide plasmid pSS6 by conjugation and isolating mutants that had undergone allelic exchange generated an rpoS mutation in FRD1. pSS6 carries a 1.2-kb oriT fragment isolated from pSF1 as an EcoRI fragment in the unique EcoRI site of pSS1. The presence of the rpoS101::aacCI allele in these mutants was verified by two methods, Southern blot hybridization (57) and PCR analysis, to demonstrate the replacement of the wild-type rpoS gene with the rpoS101 allele (data not shown).
For Southern blot analysis, chromosomal DNAs from the putative rpoS mutants and their respective wild-type strains were digested independently with either EcoRI or EcoRV, electrophoresed on agarose gels, and blotted to nitrocellulose. The blots were probed with rpoS, a 420-bp HincII-SphI internal fragment of rpoS, the 820-bp aacCI gene, or the plasmid vector. The rpoS gene and the 820-bp aacCI gene both hybridized to the same-sized EcoRI or EcoRV fragments of chromosomal DNA isolated from putative mutants. Neither the internal rpoS fragment nor the plasmid vector hybridized to chromosomal DNA from the putative mutants. For PCR analysis, oligonucleotides that hybridized to the rpoS flanking sequences were used. In rpoS mutants, we observed the disappearance of the DNA fragment corresponding to the wild-type rpoS gene and the appearance of the larger fragment corresponding to the rpoS101::aacCI allele.
Stress response assays.
The ability of the wild-type cells (PAO1) and the rpoS mutant cells (SS24) to survive during prolonged starvation was determined with cells grown at 37°C with aeration in LB, NCE with 20 mM succinate, or NCE with 10 mM glucose. After cells entered the stationary phase of growth, incubation was continued for several days; cell viability was determined by taking periodic aliquots and measuring the CFU on LB plates. The time at which the cells entered the stationary phase of growth was taken as day 0, and aliquots were taken in periodic increments of 24 h from that initial point. The viability was measured as a percentage of the highest number of CFU for PAO1 and SS24 in each experiment.
To measure cells’ ability to survive heat shock, cells grown overnight in LB at 37°C with aeration were washed twice and diluted in NCE to a density of approximately 7,000 CFU/ml. One milliliter of the diluted culture was placed in a prewarmed tube at 50°C, and the viability was determined by taking periodic aliquots and plating them directly on LB plates to measure the CFU. To measure sensitivity to osmotic stress, cells were grown overnight in LB at 37°C with aeration, washed twice in NCE, and resuspended in NCE, NCE with NaCl, or NCE with sucrose. Resuspended cells were incubated at 37°C with aeration, periodic aliquots were taken, and serial dilutions of the samples were plated on LB plates to determine the CFU. NaCl concentrations of 2, 2.5, and 3 M and sucrose concentrations of 1, 1.5, and 2 M were tested. Sensitivity to hydrogen peroxide (H2O2) was measured on cells grown for 17 h in LB at 37°C as previously described (21). Cells from cultures in mid-log (OD600, 0.3) and stationary phases of growth were plated on LB plates. Sterile Whatman no. 1 filter disks impregnated with 10 μl of 30% H2O2 were placed on top of bacterium-seeded plates and incubated at 37°C for 24 h, and the zones of inhibition were measured.
Enzyme assays.
The catalase assays were performed as described by Beers and Sizer (4) with 19.5 mM H2O2 with cells grown in LB. Cell extracts for the catalase assays were prepared as previously described (6). For the exotoxin A assays, cells were grown in Trypticase soy broth dialysate at 32°C with aeration for 18 h. The cells were harvested by centrifugation, and 10 μl of the supernatant was used to measure the exotoxin A activity, as previously described (47). The values for the ADP-ribosyltransferase activity were measured as counts per minute per 10 μl of supernatant. To measure elastase, LasA protease, and casein-degrading protease activities, cells were grown in LB at 37°C for 14 to 17 h with aeration. Following centrifugation to harvest the cells, the quantity of total proteins in the culture supernatants was determined by the Bradford method (with a kit from Bio-Rad). Elastase activity was determined by the elastin Congo red hydrolysis assay with 20 to 30 μg of supernatant protein, as previously described (46). The elastase activity was calculated as the increase in A495 minute−1 gram of protein−1. The staphylolytic activity of LasA protease was determined with approximately 25 μg of total supernatant protein, as previously described (26). The LasA protease activity was calculated as the rate of decrease in A595 minute−1 milligram of protein−1. To determine the phospholipase C activity, the cells were grown at 30°C for 20 to 24 h in 1% peptone broth supplemented with 1% glycerol and 1% NaCl. The ability of phospholipase C to degrade p-nitrophenylphosphorylcholine was determined as previously described (32) with approximately 10 μg of supernatant protein.
Secondary metabolite assays.
To isolate pyocyanin, cells were grown in peptone broth (1% peptone, 1% NaCl, 1% glycerol) for 17 h at 37°C with aeration. Pyocyanin from 4 ml of supernatant was extracted by repeated extractions (a total of four times) with 1 ml of chloroform (9). The organic layer containing pyocyanin was collected, and the relative amount of pyocyanin isolated was determined by measuring the OD695. The amount of pyoverdine produced and secreted by P. aeruginosa was determined by measuring the fluorescence of pyoverdine when excited at 400 nm, as previously described (50). Briefly, the cells were grown in King B medium for 16 to 17 h at 37°C with aeration. The culture supernatants were serially diluted in 10 mM Tris-HCl (pH 7.5) and excited at 400 nm, and the emission at 460 nm was recorded. The optical density measurements were normalized to the OD600 of the cultures used for the assay.
Alginate assays.
To quantitate the amount of alginate produced when it was grown in liquid medium, cells were grown in LB for 20 to 24 h at 37°C with aeration. Alginate in the culture supernatant was separated from the cells by centrifugation (17,000 × g for 10 min) and dialyzed at 4°C against deionized water for 24 h with three changes of water. Dialysis tubing with a molecular weight cutoff of 10,000 (Spectrum, Houston, Tex.) was used. To quantitate the alginate produced on solid medium, overnight cultures of FRD1 or SS138 were serially diluted in saline (0.85% NaCl), and 0.1 ml of diluted cultures was plated on LB plates. The plates were incubated at 37°C for 25 to 30 h; alginate was collected by scraping the cells and alginate with saline, followed by centrifugation to separate the alginate from the cells. The alginate thus collected was subjected to 24 h of dialysis at 4°C against three changes of deionized water. The amount of alginate present in the samples was quantitated by the carbazole method of Knutson and Jeanes (29), with Macrocystis pyrifera alginate (Sigma) as a standard. The values for alginate production were obtained by normalizing the total amount of alginate produced to the CFU of the culture or the number of colonies on plates.
Twitching-motility assay.
A twitching-motility assay was performed by stabbing a colony of bacteria into the bottom of a petri plate containing 10 ml of L agar (1% agar content). Following incubation at 37°C for 17 h, the volume of agar was reduced with a thick stack of circular paper towels to absorb the moisture. The zone of motility was visualized by staining the agar with Coomassie blue, as described by Alm and Mattick (2).
Chronic-lung-infection assay.
The rat chronic-lung-infection assay was performed as previously described (60). Briefly, agar beads embedded with approximately 3 × 104 cells of either the wild-type bacteria or the rpoS mutant were placed in the left lungs of rats. The lungs of three animals from each group were isolated at days 7 and 14 for bacterial quantification. The lungs of two additional animals from each group were isolated at the same times for histopathological examination as previously described (56). Briefly, sagittal slices of the entire left lobes of the fixed lungs were dehydrated in graded alcohols, embedded in paraffin, and cut into 6-μm-thick sections. Mounted sections were stained with hematoxylin and eosin. Infiltration of the lung by inflammatory cells and exudate was measured microscopically with a Zeiss integrating eyepiece. The number of points overlying the surface area of the infiltrate was divided by the total number of points counted over the entire surface area of the left lobe to measure the percent infiltration. This procedure was repeated with left lobe slices from each animal’s lung.
RESULTS
Construction of P. aeruginosa rpoS mutants.
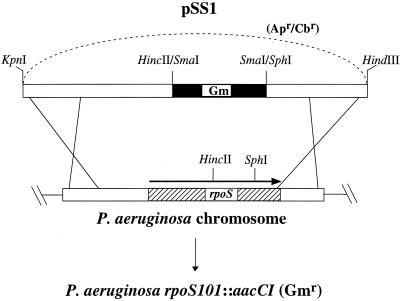
P. aeruginosa rpoS mutants were isolated in order to better understand the potential role of RpoS (ςS) in P. aeruginosa physiology and pathogenesis. Figure 1 shows the schematic diagram for constructing the suicide plasmid, pSS1, which carries the null P. aeruginosa rpoS101::aacCI allele, and the subsequent generation of P. aeruginosa rpoS mutants with pSS1. We generated a null allele of rpoS by deleting the 420-bp HincII-SphI fragment of the rpoS coding sequence and replacing it with an 820-bp SmaI fragment that confers the Gmr encoded by aacCI. We introduced pSS1 into P. aeruginosa and isolated colonies that had undergone the allelic exchange reaction replacing the wild-type rpoS with the rpoS101::aacCI allele. The inheritance of the rpoS101::aacCI allele in P. aeruginosa was confirmed by Southern blot hybridization analysis and/or PCR analysis (data not shown). The rpoS mutants of PAO1, PA103, and FRD1 were designated SS24, SS6, and SS138, respectively (Table 1).
FIG. 1.
Construction of P. aeruginosa rpoS mutants. To generate a null allele of the P. aeruginosa rpoS gene, approximately 420 bp of the rpoS coding sequence on a 1.8-kb fragment was replaced with the aacCI gene, which encodes Gmr, to form pSS1. The wild-type rpoS was replaced with the rpoS101::aacCI allele carried on the suicide plasmid pSS1, as described in Materials and Methods, to generate P. aeruginosa rpoS mutants.
Response of rpoS mutants to environmental stresses.
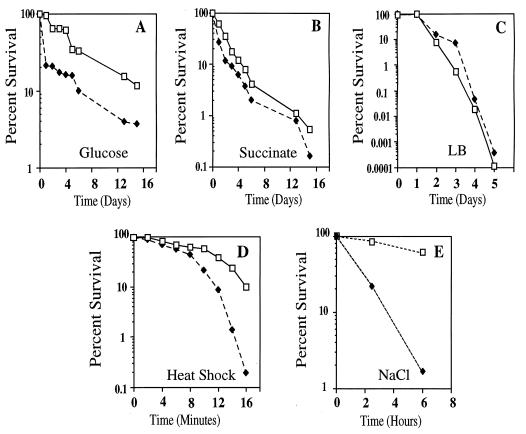
To determine whether RpoS confers cross-protection for P. aeruginosa against various stresses, the resistance of the parent (PAO1) and rpoS mutant (SS24) against prolonged starvation, heat shock, increased osmolarity, and H2O2 was determined. To study its response to prolonged carbon starvation, we grew P. aeruginosa on three different growth media to determine whether growth on a specific carbon source affected the survival of P. aeruginosa. The media used were minimal medium with glucose, minimal medium with succinate, and LB. For P. aeruginosa, glucose is a less preferred carbon source than succinate. There was no difference in growth rate between the parent and rpoS mutant in each of three growth media (data not shown). When bacteria were grown with glucose as the sole carbon source, we observed increased sensitivity by the rpoS mutant to prolonged starvation (Fig. 2A). However, the difference in survival between the parent and the rpoS mutant was not as pronounced as that observed in other bacteria. For example, an E. coli rpoS mutant is almost 50-fold more sensitive to starvation than the parent strain after 9 days of incubation (33). In P. aeruginosa, we consistently observed only a four- to eightfold difference between the parent and rpoS mutant after 12 to 14 days of incubation. In contrast, when bacteria were grown with succinate as a sole carbon source (Fig. 2B) or in LB (Fig. 2C), we observed little difference in survival between the rpoS mutant and the parent. The most striking difference observed among the three growth conditions was the relative resistance of P. aeruginosa to prolonged starvation when the cells were grown with glucose as the carbon source. When cells were grown on glucose as the sole carbon source, approximately 12% of the parent cells and 4% of the rpoS mutant cells were viable after 15 days of starvation. In contrast, when cells were grown on succinate, approximately 1% of the parent cells and 0.8% of the rpoS mutant cells were viable after 13 days of incubation. When cells were grown in LB, no viable cells were recovered after 10 to 16 days of continued incubation (data not shown). Thus, P. aeruginosa is sensitive to prolonged starvation, and RpoS had little effect in protecting the bacterium from prolonged starvation. In addition, the growth of P. aeruginosa on glucose appeared to decrease the sensitivity of the bacterium to prolonged starvation. Although only three substrates were tested, it appeared that survival of P. aeruginosa was enhanced when a poorer substrate was used.
FIG. 2.
Effect of rpoS mutation on stress responses of P. aeruginosa. To assay cell survival after prolonged starvation (A to C), PAO1 (□) and SS24 (⧫) cells were grown in the indicated growth media at 37°C with aeration. Day 0 is the time point at which cells entered stationary phase of growth, and then samples were taken at 24-h intervals. Viability was measured as the number of CFU and was expressed as a percentage of the number of CFU at day 0. Minimal salt medium was supplemented with either 10 mM glucose (A) or 20 mM succinate (B). LB (C) was used as a rich medium. To assay cell survival of heat shock at 50°C (D), stationary-phase cultures of PAO1 (□) and SS24 (⧫) were grown in LB, washed, and transferred to prewarmed tubes. The number of viable cells was measured by plating aliquots on LB plates at each time point and determining the number of CFU. Viability is expressed as a percentage of the number of CFU at time 0. To assay survival of exposure to 2 M NaCl (E), stationary-phase cultures of PAO1 (□) and SS24 (⧫) were grown in LB, washed, resuspended either in minimal salts or in minimal salts supplemented with 2 M NaCl, and incubated at 37°C with aeration. Cell viability was determined at 2 and 6 h. One hundred percent viability corresponds to the CFU present immediately following resuspension in 2 M NaCl. The carbon starvation experiments were performed at least four times, and the other stress response assays were performed at least twice. The data shown are representative of those experiments.
When stationary-phase cultures of the parent (PAO1) and the rpoS mutant (SS24) were exposed to a sudden shift in temperature from 24° to 50°C, the rpoS mutant was more sensitive to heat shock at 50°C. After 8 min of incubation at 50°C, there was a 50-fold difference in survival between the parent and the rpoS mutant (Fig. 2D). When the cells were subjected to an increase in osmotic pressure caused by the addition of a high concentration of salt, the rpoS mutant was more sensitive to a 2 M concentration of sodium chloride (Fig. 2E). After 6 h of exposure, there was approximately a 37-fold difference in relative viability between the parent and the rpoS mutant. Thus, RpoS was essential for resistance against osmotic stress caused by exposure to salt. Against nonionic osmotic stress, the wild type was only slightly more resistant (an approximately threefold difference after 6 h of exposure) to 2 M sucrose than the rpoS mutant (data not shown). This indicated that the role of RpoS in protecting P. aeruginosa from nonionic osmotic stress may be small.
We also assayed whether RpoS mediated protection against H2O2 in P. aeruginosa (Table 2). In exponentially growing cells, there was no difference in sensitivity to H2O2 between the parent and the rpoS mutant. However, when stationary-phase cells were tested, the zone of inhibition caused by H2O2 was approximately 20% larger for the rpoS mutant than for the parent strain. We tested whether this hypersensitivity of the rpoS mutant to H2O2 was due to a decreased production of catalases in the mutant. In stationary-phase cultures, the rpoS mutant produced 60% less catalase activity than the parent strain. Thus, RpoS was required for maximum protection against H2O2 by mediating the production of catalase in P. aeruginosa.
TABLE 2.
Effect of rpoS mutation on oxidative stress in P. aeruginosaa
| Growth phase and phenotype | H2O2 sensitivityb | Catalase activity (U/mg of protein) |
|---|---|---|
| Mid-log | ||
| RpoS+ | 25.8 ± 0.7 | 116 ± 8 |
| RpoS− | 25.5 ± 2.8 | 71 ± 7 |
| Stationary | ||
| RpoS+ | 29.8 ± 0.2 | 1,206 ± 41 |
| RpoS− | 36.3 ± 0.9 | 476 ± 21 |
Values are means ± standard deviations.
Zone of inhibition was measured in millimeters.
Effect of rpoS mutation on production of exoproducts.
P. aeruginosa produces and secretes numerous toxins, proteases, and secondary metabolites that contribute to the pathogenesis of the bacterium. We investigated whether RpoS controls the production and secretion of exotoxin A, elastase, LasA protease, casein-degrading proteases, phospholipase C, pyocyanin, and pyoverdine. The data are shown in Table 3.
TABLE 3.
Effect of rpoS mutation on P. aeruginosa PAO1 exoproductsa
| Exoproduct | Activity in strain:
|
% Relative activity | |
|---|---|---|---|
| PAO1 (RpoS+) | SS24 (RpoS−) | ||
| Exotoxin A | 1,302 ± 46 | 660 ± 20 | 50.7 |
| Elastase | 161 ± 6 | 126 ± 2 | 78.6 |
| LasA protease | 0.587 ± 0.002 | 0.518 ± 0.014 | 88.3 |
| Phospholipase C | 33.45 ± 0.65 | 32.97 ± 0.82 | 98.6 |
| Pyocyanin | 0.347 | 0.716 | 206.3 |
| Pyoverdine | 0.511 | 0.755 | 147.7 |
Exotoxin A activity is measured in counts per minute per 10 μl of supernatant. Elastase activity is measured as the increase in A495 minute−1 gram of protein−1. LasA protease activity is measured as the rate of decrease in A595 minute−1 milligram of protein−1. Phospholipase C activity is measured as the increase in OD405 minute−1 milligram of protein−1. The pyocyanin level was measured as OD695 and the pyoverdine level was measured as OD460 following excitation at OD400 (see Materials and Methods for details). The relative activity was calculated as follows: (activity of the rpoS mutant/activity of the parent) × 100. Values are means ± standard deviations.
Exotoxin A is an ADP-ribosyltransferase that inhibits eukaryotic protein synthesis (36). Exotoxin A activity in the culture supernatants was decreased by 50% in the PAO1 rpoS mutant (SS24). In the exotoxin A-overproducing PA103 and its respective rpoS mutant (SS6), the RpoS− defect resulted in a 70% reduction in exotoxin A activity (18,502 ± 1,276 for the parent versus 5,809 ± 407 for the rpoS mutant). We also determined the effect of the rpoS mutation on the extracellular accumulation of proteases and phospholipase C in the PAO1 background. Culture supernatants of the PAO1 rpoS mutant consistently had 20% less elastase activity and 10% less LasA protease activity. The decreased accumulation of these extracellular proteins in the culture supernatant of rpoS mutant was fully restored to the level of the parental strain when RpoS was provided in trans from a plasmid (data not shown). A measure of casein degradation detects many proteases produced by the bacterium (i.e., elastase and alkaline protease). The difference in the production of casein-degrading protease between the parent strain and the rpoS mutant was negligible (data not shown). Similarly, little difference in the production and secretion of total phospholipase C (Table 3) or hemolytic phospholipase C (data not shown) between the parent and the rpoS mutant was observed. Based on these data, we concluded that RpoS was required for the maximum production of exotoxin A but that it was not required for the maximum production of casein-degrading proteases or phospholipase C.
Pyocyanin and pyoverdine are secondary metabolites produced by P. aeruginosa. Pyocyanin is a blue compound that plays an important role in P. aeruginosa pathogenesis (59). Pyocyanin kills bacteria and inhibits lymphocyte proliferation and ciliary function. The color of P. aeruginosa rpoS mutant culture supernatants was an intense blue relative to parent strains, suggesting that rpoS mutants were producing and secreting more pyocyanin. Measurements showed that culture supernatants from the rpoS mutant contained twice as much pyocyanin as the parent strain PAO1. Conversely, the overproduction of RpoS from a high-copy-number plasmid in PAO1 resulted in a decrease in pyocyanin levels in culture supernatants (data not shown). We also determined the effect of rpoS mutation on the accumulation of pyoverdine, the primary siderophore produced by P. aeruginosa, in culture supernatant (10). Culture supernatant from the rpoS mutant contained 1.5-fold more pyoverdine than the parent strain. These data indicated that RpoS modulates the level of pyocyanin and pyoverdine production in P. aeruginosa.
Effect of rpoS mutation on alginate production.
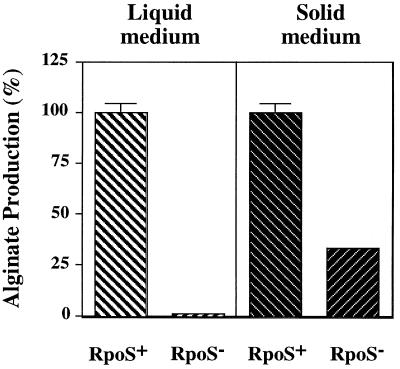
Alginate is an exopolysaccharide that is overproduced by most of the P. aeruginosa strains isolated from CF patients suffering from chronic lung infections (18). The overproduction of alginate contributes to the persistence of the bacterium in the CF lung. To determine whether RpoS is involved in the production of alginate, we examined SS138, the rpoS101::aacCI mutant of FRD1 (an alginate-overproducing CF isolate). As shown in Fig. 3, when the cells were grown in a liquid medium, alginate production was almost completely abolished in the FRD1 rpoS mutant. In contrast, when grown on plates (seeded with 102 CFU/plate), the rpoS mutant produced approximately 35% of the alginate produced by the wild-type strain. Interestingly, when agar plates were seeded with a high cell density (i.e., ≥104 CFU), the rpoS mutant accumulated as much alginate as the wild-type FRD1 (data not shown). Because FRD1 and the rpoS mutant grew at similar rates (data not shown), the difference in alginate production could not be attributed to a growth defect. Thus, RpoS, in combination with cell density, was required for maximum alginate synthesis in P. aeruginosa. Providing RpoS in trans on a multicopy plasmid restored the alginate production of the rpoS mutant to that of the parental strain FRD1 (data not shown).
FIG. 3.
The rpoS mutation affects alginate accumulation in mucoid strain FRD1. To evaluate growth in liquid, FRD1 and its rpoS mutant derivative (SS138) were grown in LB at 37°C with aeration for 20 to 24 h. For growth on plates, approximately 1,000 cells of the parent or the rpoS mutant were spread on L agar plates and incubated at 37°C for 25 to 30 h. Alginate was separated from the cells and quantitated as described in Materials and Methods. The amount of alginate produced was normalized to the number of cells in the culture or to the number of colonies on plates to obtain the amount of alginate produced per CFU. One hundred percent of alginate production corresponds to the level produced by the wild-type FRD1 strain. The values obtained for alginate were as follows: FRD1 (liquid), 1.23 × 10−6 μg CFU−1; SS138 (liquid), 1.39 × 10−8 μg CFU−1; FRD1 (plate), 0.77 μg colony−1; SS138 (plate), 0.27 μg colony−1.
Effect of rpoS mutation on the rat chronic-lung-infection model.
We assessed the role of RpoS in the virulence of P. aeruginosa using the rat chronic-lung-infection model. In this model, the bacteria are embedded in agar beads which protect them from the host immune response (60). The results showed that the rpoS mutant survived as well as the wild-type bacteria when the mutant was embedded in agar beads and placed in rat lungs. Interestingly, the rpoS mutant appeared to cause greater damage to lung tissues than the wild-type strain (Table 4). The increased damage inflicted on the lungs of rats by the rpoS mutant may be due to the overproduction of pyocyanin.
TABLE 4.
Effect of rpoS mutation on chronic lung infection caused by P. aeruginosa
| Parent or mutant | Initial inoculum (CFU/ml) | Quantitative bacteriology (CFU/ml) on daya:
|
Quantitative pathology (% damage) on day:
|
||
|---|---|---|---|---|---|
| 7 | 14 | 7b | 14 | ||
| RpoS+ (PAO1) | 3.5 × 104 | (4.4 ± 4.3) × 105 | (1.3 ± 1.2) × 105 | 18.5 ± 4.9 | 20.9 ± 8.8 |
| RpoS− (SS24) | 2.8 × 104 | (2.3 ± 2.2) × 106 | 1.2 × 106 ± 9.2 × 105 | 32.7 ± 3.8c | 56.2 ± 19.3c |
Values are means ± standard deviations.
Infiltration index.
Significantly different from RpoS+ value (P < 0.05) by Student’s t test.
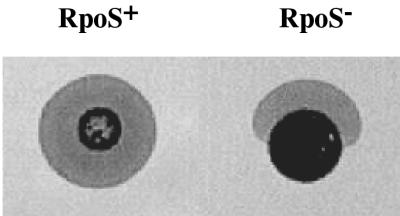
Although the rat chronic-lung-infection model is a good model for assessing the roles of extracellular products in infection, it does not provide information about colonization functions and other initial stages of infection. The colonization of P. aeruginosa is mediated by type IV fimbriae (13), which also mediate the twitching motility. We examined the twitching motility of the rpoS mutant to determine whether RpoS may be involved in P. aeruginosa colonization. As shown in Fig. 4, the zone of motility of the rpoS mutant (SS24) was approximately 60 to 70% of that of the parent (PAO1). In addition, the shape of the zone observed for the rpoS mutant was typically elliptical, rather than the circular shape that was observed for the parent strain. Finally, the rpoS mutant demonstrated altered swarming motility on the top of the agar. These data suggest that RpoS may be involved in the colonization of P. aeruginosa as mediated by the type IV fimbriae.
FIG. 4.
Effect of rpoS mutation on twitching motility. A colony of wild-type (PAO1) or rpoS mutant (SS24) cells was stabbed into the bottom of a petri plate containing 10 ml of L agar (1% agar content). Following incubation at 37°C for 17 h, the volume of agar was reduced by absorbing the moisture with a stack of paper towels, and the zone of motility was visualized by staining the agar with Coomassie brilliant blue.
DISCUSSION
The ability of P. aeruginosa to resist, adapt, and survive in a wide variety of environments makes it a ubiquitous bacterium in nature and likely contributes to its opportunistic pathogenic behavior. As an initial effort to elucidate and understand the stress responses of this bacterium, we isolated and characterized P. aeruginosa rpoS mutants to determine the effect of an RpoS− defect on the stress responses and the synthesis of virulence factors in this bacterium.
Our data demonstrate both similarities and differences between the role of RpoS in P. aeruginosa and its role in other bacteria. Some of the similarities include the RpoS-mediated resistance of P. aeruginosa to heat-shock, H2O2, and osmotic stress. These data indicate that RpoS is a general stress response regulator in P. aeruginosa. However, because the physiology and metabolism of P. aeruginosa are different from those of E. coli, we do not know whether RpoS-mediated resistance to various stresses in P. aeruginosa is similar or even proceeds via analogous genes. For example, the E. coli rpoS mutant is hypersensitive to 15 mM H2O2 (33), while the P. aeruginosa rpoS mutant is resistant to 100 mM H2O2 (data not shown), despite reduced catalase production in the P. aeruginosa rpoS mutant. In E. coli, RpoS regulates the expression of two catalase biosynthetic genes, katE and katG, in order to detoxify H2O2 (25, 44). P. aeruginosa also possesses two catalases, KatA and KatB (6, 40). It was shown that KatA activity is under quorum-sensing control and thus is maximal in stationary phase (20). Because the rpoS gene is also under quorum-sensing control (34), it may be that RpoS directly controls katA expression in stationary phase.
We also found several differences in the role of RpoS between E. coli and P. aeruginosa. One of the differences was the minor role of RpoS in protecting P. aeruginosa from prolonged carbon starvation. P. aeruginosa was quite sensitive to prolonged starvation (Figure 2A to C), despite the fact that P. aeruginosa is almost ubiquitous in nature, including in low-nutrient environments. One possible explanation is that because P. aeruginosa, like other members of the pseudomonads, can metabolize so many compounds as nutrients, it has not developed resistance against prolonged carbon starvation. Alternatively, carbon starvation resistance in P. aeruginosa may be regulated via a mechanism not addressed by our methods and/or by a different regulator than RpoS. In P. putida, protection against carbon starvation has been shown to be regulated by both RpoS (43, 51) and RpoN (27), the sigma factor that regulates nitrogen metabolism in several bacteria.
Another interesting results was the differential effects that various carbon sources had on the subsequent resistance of P. aeruginosa to prolonged carbon starvation. P. aeruginosa grown on glucose was approximately 10- to 15-fold more resistant to prolonged starvation than when grown on succinate and at least 500-fold more resistant than when grown in LB. There are several possibilities for these differences. P. aeruginosa growing in LB or on succinate may accumulate compounds that are toxic to the cells, or growth on glucose may induce the expression of genes under RpoS control that confer enhanced resistance to prolonged starvation. It is intriguing to consider the possibility that growth on a less preferred carbon source (such as glucose) may better prepare the bacterium for the onset of starvation.
RpoS has been demonstrated to be required for production of extracellular toxins or proteases in three other organisms: Yersinia enterocolitica, Vibrio cholerae, and P. fluorescens (24, 52, 61). Of a subset of extracellular products secreted by P. aeruginosa, the accumulation of exotoxin A, pyocyanin, and pyoverdine was most dramatically affected by the RpoS− defect. However, it is unclear whether regulation of these exoproducts by RpoS occurs at the level of synthesis or secretion. The expression of P. aeruginosa xcp operons, which encode the type II secretion apparatus for exotoxin A, elastase, LasA protease, and phospholipase C, among other proteins, is regulated by the quorum-sensing systems (7), which in turn regulate the expression of rpoS (34). This led Chapon-Hervé et al. (7) to propose the possibility that regulation of xcp operons may proceed via RpoS in P. aeruginosa. This proposal was also based on the presence of a potential RpoS consensus promoter upstream of xcp. If the xcp operons are regulated by RpoS, then the rpoS mutant should be deficient for the accumulation of many exoproteins. However, only the accumulation of exotoxin A was significantly affected by the rpoS mutation in our study. Further data are needed to clarify whether RpoS regulates the expression of genes encoding exotoxin A, elastase, and LasA protease directly or regulates the secretion of these proteins via Xcp.
RpoS plays a major role in the production of alginate, and this effect is dependent on whether cells are grown in liquid or on a solid surface. When bacteria are grown on a solid surface, a higher cell density can be reached. Thus, RpoS-mediated alginate production may be cell density dependent. Our data suggest that a high cell density can minimize or even negate the RpoS requirement for the maximum accumulation of alginate and that RpoS is essential for alginate accumulation only when cell density is relatively low. These results pose an interesting question. How can RpoS induce the synthesis or secretion of alginate under low-cell-density conditions when the expression of rpoS itself is induced by high cell density? Perhaps the basal level of rpoS expressed in FRD1 at a low cell density may be sufficient for the induction of alginate production and/or secretion. Alternatively, due to the close proximity of the cells growing on a solid surface, a few cells may be sufficient to induce the quorum-sensing system.
In conclusion, the mutation in rpoS affects a variety of functions in P. aeruginosa. These include the general stress response, the accumulation of virulence factors, and twitching motility. Our data suggest that one of the functions of RpoS in P. aeruginosa is to fine-tune the cell metabolism, which includes modulating the production of several secondary metabolites, in order to maintain the optimal conditions for cells in stationary phase. The two-dimensional gel analysis on PAO1 and SS24 indicated that the rpoS mutation results in an alteration in the production and accumulation of at least 37 proteins (data not shown). Studies to identify the genes regulated by RpoS in P. aeruginosa are in progress.
ACKNOWLEDGMENTS
We thank Kan Tanaka for providing us with pDB18R and pDB19R. We also thank Tricia Maleniak, Robert Kazmierczak, Laura Regassa, Laura Runyen-Janecky, and Jorge Escalante-Semerena for help and encouragement.
This research was supported by Public Health Service grant AI-19146 from the National Institute of Allergy and Infectious Diseases and Veterans Administration Medical Research Funds, awarded to D.E.O. Studies at the University of Calgary were supported by a grant from the Canadian Cystic Fibrosis Foundation, awarded to D.E.W. Studies at the University of Cincinnati were supported by Public Health Service grant AI-40541 from the National Institute of Allergy and Infectious Diseases and Cystic Fibrosis Foundation grant HassetPO97, awarded to D.J.H. Studies at the University of Wisconsin were supported by Public Health Service grant AI-31477 from the National Institute of Allergy and Infectious Diseases, awarded to S.E.H.W. S.-J.S. was supported by Cystic Fibrosis Foundation postdoctoral fellowship SUH96F0.
REFERENCES
- 1.Albus A M, Pesci E C, Runyen-Janecky L J, West S E H, Iglewski B H. Vfr controls quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alm R A, Mattick J S. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol Microbiol. 1995;16:485–496. doi: 10.1111/j.1365-2958.1995.tb02413.x. [DOI] [PubMed] [Google Scholar]
- 3.Arnold K W, Kaspar C W. Starvation- and stationary-phase-induced acid tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 1995;61:2037–2039. doi: 10.1128/aem.61.5.2037-2039.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beers R F, Jr, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 5.Brint J M, Ohman D E. Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J Bacteriol. 1995;177:7155–7163. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown S M, Howell M L, Vasil M L, Anderson A J, Hassett D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapon-Hervé V, Akrim M, Latifi A, Williams P, Lazdunski A, Bally M. Regulation of the xcp secretion pathway by multiple quorum-sensing modulons in Pseudomonas aeruginosa. Mol Microbiol. 1997;24:1169–1178. doi: 10.1046/j.1365-2958.1997.4271794.x. [DOI] [PubMed] [Google Scholar]
- 8.Cochran, W. L., S.-J. Suh, G. A. McFeters, and P. S. Stewart. Unpublished data.
- 9.Cox C D. Role of pyocyanin in the acquisition of iron from transferrin. Infect Immun. 1986;52:263–270. doi: 10.1128/iai.52.1.263-270.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox C D, Graham R. Isolation of an iron-binding compound from Pseudomonas aeruginosa. J Bacteriol. 1979;137:357–364. doi: 10.1128/jb.137.1.357-364.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cross A, Allen J R, Burke J, Ducel G, Harris A, John J, Johnson D, Lew M, MacMillan B, Skalova R, Wenzel R, Tenney J. Nosocomial infections due to Pseudomonas aeruginosa: review of recent trends. Rev Infect Dis. 1983;5(Suppl.):S837–S845. doi: 10.1093/clinids/5.supplement_5.s837. [DOI] [PubMed] [Google Scholar]
- 12.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 13.Doig P, Todd T, Sastry P A, Lee K K, Hodges R S, Paranchych W, Irvin R T. Role of pili in adhesion of Pseudomonas aeruginosa to human respiratory epithelial cells. Infect Immun. 1988;56:1641–1646. doi: 10.1128/iai.56.6.1641-1646.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doring G, Maier M, Muller E, Bibi Z, Tummler B, Kharazmi A. Virulence factors of Pseudomonas aeruginosa. Antibiot Chemother. 1987;39:136–148. doi: 10.1159/000414341. [DOI] [PubMed] [Google Scholar]
- 15.Figurski D, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita M, Tanaka K, Takahashi H, Amemura A. Transcription of the principal sigma-factor genes, rpoD and rpoS, in Pseudomonas aeruginosa is controlled according to the growth phase. Mol Microbiol. 1994;13:1071–1077. doi: 10.1111/j.1365-2958.1994.tb00498.x. [DOI] [PubMed] [Google Scholar]
- 17.Goldberg J B, Ohman D E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Govan J R W, Harris G S. Pseudomonas aeruginosa and cystic fibrosis: unusual bacterial adaptation and pathogenesis. Microbiol Sci. 1986;3:302–308. [PubMed] [Google Scholar]
- 19.Gross C A, Lonetto M, Losick R. Bacterial sigma factors. In: McKnight S, editor. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 129–176. [Google Scholar]
- 20.Hassett, D. J., J.-F. Ma, J. G. Elkins, U. A. Ochsner, S. E. H. West, C. T. Huang, J. Fredericks, S. Burnett, P. S. Stewart, G. McFeters, and T. R. McDermott. Quorum sensing in Pseudomonas aeruginosa controls catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Submitted for publication. [DOI] [PubMed]
- 21.Hassett D J, Schweizer H P, Ohman D E. Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism. J Bacteriol. 1995;177:6330–6337. doi: 10.1128/jb.177.22.6330-6337.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hengge-Aronis R. Back to log phase: ςS as a global regulator in the osmotic control of gene expression in Escherichia coli. Mol Microbiol. 1996;21:887–893. doi: 10.1046/j.1365-2958.1996.511405.x. [DOI] [PubMed] [Google Scholar]
- 23.Holloway B W, Krishnapillai V, Morgan A F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979;43:73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iriarte M, Stainier I, Cornelis G R. The rpoS gene from Yersinia enterocolitica and its influence on expression of virulence factors. Infect Immun. 1995;63:1840–1847. doi: 10.1128/iai.63.5.1840-1847.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanova A, Miller C, Glinsky G, Eisenstark A. Role of rpoS (katF) in oxyR-independent regulation of hydroperoxidase I in Escherichia coli. Mol Microbiol. 1994;12:571–578. doi: 10.1111/j.1365-2958.1994.tb01043.x. [DOI] [PubMed] [Google Scholar]
- 26.Kessler E, Safrin M, Olson J C, Ohman D E. Secreted LasA of Pseudomonas aeruginosa is a staphylolytic protease. J Biol Chem. 1993;268:7503–7508. [PubMed] [Google Scholar]
- 27.Kim Y, Watrud L S, Matin A. A carbon starvation survival gene of Pseudomonas putida is regulated by ς54. J Bacteriol. 1995;177:1850–1859. doi: 10.1128/jb.177.7.1850-1859.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 29.Knutson C A, Jeanes A. A new modification of the carbazole analysis: application to heteropolysaccharides. Anal Biochem. 1968;24:470–481. doi: 10.1016/0003-2697(68)90154-1. [DOI] [PubMed] [Google Scholar]
- 30.Kolter R, Siegele D A, Tormo A. The stationary phase of the bacterial life cycle. Annu Rev Microbiol. 1993;47:855–874. doi: 10.1146/annurev.mi.47.100193.004231. [DOI] [PubMed] [Google Scholar]
- 31.Kowarz L, Coynault C, Robbe-Saule V, Norel F. The Salmonella typhimurium katF (rpoS) gene: cloning, nucleotide sequence, and regulation of spvR and spvABCD virulence plasmid genes. J Bacteriol. 1994;176:6852–6860. doi: 10.1128/jb.176.22.6852-6860.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurioka S, Matsuda M. Phospholipase C activity using p-nitrophenylphosphorylcholine together with sorbitol and its application to studying the metal and detergent requirement of the enzyme. Anal Biochem. 1976;75:281–289. doi: 10.1016/0003-2697(76)90078-6. [DOI] [PubMed] [Google Scholar]
- 33.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 34.Latifi A, Foglino M, Tanaka K, Williams P, Lazdunski A. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhlR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol Microbiol. 1996;21:1137–1146. doi: 10.1046/j.1365-2958.1996.00063.x. [DOI] [PubMed] [Google Scholar]
- 35.Latifi A, Winson M K, Foglino M, Bycroft B W, Stewart G S A B, Lazdunski A, Williams P. Multiple homologues of LuxR and LuxI control the expression of virulence determinants and secondary metabolites through quorum sensing in Pseudomonas aeruginosa PAO1. Mol Microbiol. 1995;17:333–343. doi: 10.1111/j.1365-2958.1995.mmi_17020333.x. [DOI] [PubMed] [Google Scholar]
- 36.Liu P V. Exotoxins of P. aeruginosa. I. Factors that influence the production of exotoxin A. J Infect Dis. 1973;128:506–513. doi: 10.1093/infdis/128.4.506. [DOI] [PubMed] [Google Scholar]
- 37.Liu P V. Extracellular toxins of Pseudomonas aeruginosa. J Infect Dis. 1974;130(Suppl.):S94–S99. doi: 10.1093/infdis/130.supplement.s94. [DOI] [PubMed] [Google Scholar]
- 38.Loewen P C, Hengge-Aronis R. The role of the sigma factor ςS (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi: 10.1146/annurev.mi.48.100194.000413. [DOI] [PubMed] [Google Scholar]
- 39.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 40.Ma, J.-F., U. A. Ochsner, M. G. Klotz, V. K. Nanayakkara, M. L. Howell, Z. Johnson, J. E. Posey, M. L. Vasil, J. J. Monaco, and D. J. Hassett. Bacterioferritin A modulates catalase (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol. 181:3730–3742. [DOI] [PMC free article] [PubMed]
- 41.Mathee K, Ciofu O, Sternberg C K, Lindum P, Campbell J, Jensen P, Jøhnsen A, Givskov M, Ohman D, Molin S, Høiby N, Kharazmi A. Mucoid conversion of Pseudomonas aeruginosa by hydrogen peroxide: a mechanism for virulence activation in the cystic fibrosis lung. Microbiology. 1999;145:1349–1357. doi: 10.1099/13500872-145-6-1349. [DOI] [PubMed] [Google Scholar]
- 42.McCann M P, Kidwell J P, Matin A. The putative ς factor KatF has a central role in development of starvation-mediated general resistance in Escherichia coli. J Bacteriol. 1991;173:4188–4194. doi: 10.1128/jb.173.13.4188-4194.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miura K, Inouye S, Nakazawa A. The rpoS gene regulates OP2, an operon for the lower pathway of xylene catabolism on the TOL plasmid, and the stress response in Pseudomonas putida mt-2. Mol Gen Genet. 1998;259:72–78. doi: 10.1007/s004380050790. [DOI] [PubMed] [Google Scholar]
- 44.Mulvey M R, Switala J, Borys A, Loewen P C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990;172:6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohman D E, Chakrabarty A M. Genetic mapping of chromosomal determinants for the production of the exopolysaccharide alginate in a Pseudomonas aeruginosa cystic fibrosis isolate. Infect Immun. 1981;33:142–148. doi: 10.1128/iai.33.1.142-148.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohman D E, Cryz S J, Iglewski B H. Isolation and characterization of a Pseudomonas aeruginosa PAO mutant that produces altered elastase. J Bacteriol. 1980;142:836–842. doi: 10.1128/jb.142.3.836-842.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohman D E, Sadoff J C, Iglewski B H. Toxin A-deficient mutants of Pseudomonas aeruginosa PA103: isolation and characterization. Infect Immun. 1980;28:899–908. doi: 10.1128/iai.28.3.899-908.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Olsen A, Arnqvist A, Hammar M, Sukupolvi S, Normark S. The RpoS sigma factor relieves H-NS-mediated transcriptional repression of csgA, the subunit gene of fibronectin-binding curli. Mol Microbiol. 1993;7:523–536. doi: 10.1111/j.1365-2958.1993.tb01143.x. [DOI] [PubMed] [Google Scholar]
- 49.Pesci E C, Pearson J P, Seed P C, Iglewski B H. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179:3127–3132. doi: 10.1128/jb.179.10.3127-3132.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prince R W, Cox C D, Vasil M L. Coordinate regulation of siderophore and exotoxin A production: molecular cloning and sequencing of the Pseudomonas aeruginosa fur gene. J Bacteriol. 1993;175:2589–2598. doi: 10.1128/jb.175.9.2589-2598.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramos-González M I, Molin S. Cloning, sequencing, and phenotypic characterization of the rpoS gene from Pseudomonas putida KT2440. J Bacteriol. 1998;180:3421–3431. doi: 10.1128/jb.180.13.3421-3431.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sarniguet A, Kraus J, Henkels M, Muehlchen A M, Loper J E. The sigma factor ςS affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc Natl Acad Sci USA. 1995;92:12255–12259. doi: 10.1073/pnas.92.26.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schweizer H D. Small broad-host-range gentamicin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:831–834. [PubMed] [Google Scholar]
- 54.Selvaraj G, Fong Y C, Iyer V N. A portable DNA sequence carrying the cohesive site (cos) of bacteriophage λ and the mob (mobilization) region of the broad-host-range plasmid RK2: a module for the construction of new cosmids. Gene. 1984;32:235–241. doi: 10.1016/0378-1119(84)90051-9. [DOI] [PubMed] [Google Scholar]
- 55.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sokol P A, Woods D E. Effect of pyochelin on Pseudomonas cepacia respiratory infections. Microb Pathog. 1988;5:197–205. doi: 10.1016/0882-4010(88)90022-8. [DOI] [PubMed] [Google Scholar]
- 57.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 58.Tanaka K, Takahashi H. Cloning, analysis and expression of an rpoS homologue gene from Pseudomonas aeruginosa PAO1. Gene. 1994;150:81–85. doi: 10.1016/0378-1119(94)90862-1. [DOI] [PubMed] [Google Scholar]
- 59.Ulmer A J, Pryjma J, Tarnok Z, Ernst M, Flad H-D. Inhibitory and stimulatory effects of Pseudomonas aeruginosa pyocyanine on human T and B lymphocytes and human monocytes. Infect Immun. 1990;58:808–815. doi: 10.1128/iai.58.3.808-815.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Woods D E, Cryz S J, Friedman R L, Iglewski B H. Contribution of toxin A and elastase to virulence of Pseudomonas aeruginosa in chronic lung infections of rats. Infect Immun. 1982;36:1223–1228. doi: 10.1128/iai.36.3.1223-1228.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yildiz F H, Schoolnik G K. Role of rpoS in stress survival and virulence of Vibrio cholerae. J Bacteriol. 1998;180:773–784. doi: 10.1128/jb.180.4.773-784.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yu H, Schurr M J, Deretic V. Functional equivalence of Escherichia coli ςE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J Bacteriol. 1995;177:3259–3268. doi: 10.1128/jb.177.11.3259-3268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]