Abstract
Context
The adrenal cortex consists of zona glomerulosa (ZG), fasciculata (ZF), and reticularis. Aldosterone-producing cell clusters (APCCs) that strongly express aldosterone synthase (CYP11B2) are frequently found in adult adrenals and harbor somatic mutations that are also detected in aldosterone-producing adenomas (APAs). Primary aldosteronism is mainly caused by APAs or idiopathic hyperaldosteronism (IHA). We presume that APCCs are causing IHA and are precursors of APAs. However, the gene expression characteristics and especially the development of APCCs are not well understood.
Objective
This study aimed to analyze the transcriptome of APCCs at single-cell resolution and infer the developmental trajectory.
Methods
Single-cell RNA sequencing (scRNA-seq) of 2 adult adrenals was performed.
Results
Immunohistochemical analyses confirmed the 2 adrenals had APCCs. scRNA-seq data of 2928 adrenal cells were obtained and 1765 adrenocortical cells were identified based on unsupervised clustering and the marker gene expression. The adrenocortical cells were divided into 6 clusters, of which 3 clusters (923 cells) were composed of APCC/ZG cells. By further subclustering, the APCC/ZG cells were divided into 3 clusters (clusters C1, C2, and C3), we finally identified APCC cluster (C3) and ZG cluster (C1). Cluster C2 seemed to be ZG-to-ZF transitional cells. RNA velocity analysis inferred the developmental direction from cluster ZG-cluster-C1 to APCC-cluster-C3. The scRNA-seq additionally revealed that many CYP11B2-positive cells were positive for CYP11B1 and/or CYP17A1, which were essential for cortisol but not for aldosterone production.
Conclusions
Our results revealed the gene expression characteristics of APCC at single-cell resolution and show that some ZG cells remodel to APCC.
Keywords: single-cell RNA sequencing, transcriptomic study, adrenal gland, aldosterone-producing cell cluster
The adrenal cortex is composed of 3 histologically and functionally distinct layers: zona glomerulosa (ZG), zona fasciculata (ZF), and zona reticularis (ZR) (1). ZG is the outermost layer and produces aldosterone, which is involved in the regulation of Na+/K+ balance and blood pressure. ZF is located beneath ZG and produces glucocorticoids, which play an important role in regulating glucose metabolism and immune responses. ZR is the innermost layer of the adrenal cortex, which produces adrenal androgens in primates. Aldosterone production in ZG is mainly regulated by angiotensin II (renin-angiotensin system), potassium, and to a lesser extent ACTH (2).
Primary aldosteronism (PA) is the most common form of secondary hypertension, and its prevalence is between 5% and 20% of hypertensive patients (3). PA is caused by autonomous production of aldosterone and significantly increases cardiovascular complications not only by the sodium/water retention but also by direct aldosterone action on vessels, heart, and brain (4). Thus, the control of aldosterone is essential for PA patients. PA is primarily classified into aldosterone-producing adenoma (APA, generally unilateral) and bilateral PA (also called idiopathic hyperaldosteronism [IHA]). The former is often curable by unilateral adrenalectomy, whereas the latter is mostly treated by lifelong mineralocorticoid receptor antagonists. The principle of these treatments has not changed for decades. To develop next-generation therapies, it is necessary to clarify the pathophysiology of PA, specifically how APAs develop and what adrenal lesions are responsible for IHA.
We previously established immunohistochemical protocols that distinguish the localization of cytochrome P450 family 11 subfamily B member 2 (aldosterone synthase, CYP11B2) and cytochrome P450 family 11 subfamily B member 1 (cortisol synthase, CYP11B1) in human adrenal tissue sections and found the presence of aldosterone-producing cell clusters (APCCs) that strongly express CYP11B2 in many normal adults (5). APCCs produce high concentrations of aldosterone (6, 7) and frequently harbor somatic mutations of ion pump/channel genes (8), which are similarly found in APA (9-12). Intriguingly, APCCs are also found in the adrenal cortex adjacent to APAs (1), and APCCs appear to produce aldosterone autonomously (13, 14). Moreover, we reported possible APCC-to-APA transitional lesions (6, 14-16). These pieces of evidence may support the hypotheses that some APCCs are the precursors of APA and are the lesions responsible for IHA when they are large or exist in a large number. In mice, adrenocortical cells develop from Gli1-positive stem cells in the capsule and/or Shh-positive progenitor cells in subcapsular/ZG region, and ZG cells convert into ZF cells centripetally (17, 18). Adrenocortical cell renewal is sexually dimorphic and relies on different progenitor populations before and after puberty (19, 20). However, how adrenocortical cells, including APCCs, differentiate in humans has not been elucidated.
Recently, advances in single-cell RNA sequencing (scRNA-seq) technology have made it possible to analyze the transcriptomes of complex and heterogeneous tissues and estimate cellular developmental trajectory (21). scRNA-seq has been applied to various normal tissues, cancers, and chronic diseases to identify novel cells, elucidate the etiology of diseases, and promote clinical applications for diagnosis and treatment. We previously reported a tissue transcriptome study comparing laser-captured tissues of ZG, ZF, ZR, and APCC (8). In this study, we aimed to identify transcriptome characteristics of ZG and APCC cells from human adrenals at the single-cell resolution using scRNA-seq while referencing the laser-captured tissue transcriptome study, thereby determining the developmental trajectory of adrenocortical cells while focusing on ZG and APCC cells.
Materials and Methods
Institutional Review Boards
This study was performed under the approval of the institutional review boards of Saitama Medical University International Medical Center (approval #: 18-185), Keio University School of Medicine (20090018), and Kyushu University (889-00). Written informed consent was obtained from all cases.
Human Samples
Adult human adrenal samples for scRNA-seq and immunostaining were collected from 2 cases in the Saitama Medical University International Medical Center. Samples 1 (SIMC-Uro #12142, a unique nonsequential patient control number in the Department of Uro-oncology, Saitama Medical University International Medical Center; Supplementary Figure 1) and 2 (SIMC-Uro #12936; Supplementary Figure 2) were obtained from a 62-year-old male at radical nephrectomy for renal cell carcinoma and a 29-year-old male at adrenalectomy for pheochromocytoma, respectively (22). The diagnoses were made based on clinical data, imaging features, and postoperative histopathological data.
Immunohistochemistry
Serial sections (4 μm) from formalin-fixed paraffin-embedded specimens of the adrenal glands were deparaffinized and treated with Target retrieval solutions (Dako). Immunohistochemical staining was performed using a Dako autostainer universal system according to the manufacturer’s protocol. Staining for CYP11B2, CYP11B1, CYP17A1, HSD3B2, and CD56 was performed using primary and secondary antibodies described in Table S1 (22).
Dispersing and Freezing Adrenal Cells
The adrenal glands were placed in ice-cold DMEM (Nacalai Tesque, Inc., #08458-45) immediately after removal. The adrenal medulla was dissected and removed from the adrenal gland under microscopy while on ice, and the remaining adrenocortical tissue-enriched sample was cut into small pieces with a scalpel. Fifty milliliters of cosmic calf serum (CCS; HyClone, #SH30087.03) was added to 450 mL of DMEM to prepare 10% DMEM/CCS. Approximately one-third to two-thirds of removed normal adrenal glands were minced (yellow arrowheads in Figures S1 and S2) (22) and shaken at 37 °C for 1 hour at 90 rpm in 100 mL collagenase solution (1 mg/mL collagenase [Merck, C5138], and 4 U/mL DNase [Promega, M6101] in 10% DMEM/CCS). The digested tissue solution was passed through a cell strainer (Falcon Cell Strainer 100 µm Nylon, #352360), centrifuged at 300g for 5 minutes, and the supernatant was discarded. Two milliliters of Red Blood Cell Lysis Buffer (Roche, #11814389001) was added to the pellet and mixed by inverting gently for 10 minutes and centrifuged at 300g for 5 minutes, and the supernatant was discarded. The cell pellet was resuspended in 10 mL of 10% DMEM/CCS and underwent cell counting. The cells were pelleted again, resuspended in Bambanker cell freezing media (Japan Genetics, #CS-02-001), and frozen at -80 °C.
Fluorescence-activated Cell Sorting
Fluorescence-activated cell sorting (FACS) was performed to enrich adrenocortical cells. Cells were resuspended in PBS at a concentration of 1 × 106/mL and incubated with phycoerythrin/Cyanine7-conjugated mouse anti-human CD31 antibody (BioLegend Cat# 303117, RRID:AB_2114314), fluorescein isothiocyanate (FITC)-conjugated mouse anti-human CD24 antibody (BioLegend Cat# 311104, RRID:AB_314853), FITC-conjugated mouse anti-human CD45 antibody (BioLegend Cat# 368507, RRID:AB_2566367), allophycocyanin (APC)-labeled mouse monoclonal anti-human CD56 antibody (BioLegend Cat# 304610, RRID:AB_314452), and eBioscience Fixable Viability Dye eFluor 520 (ThermoFisher Scientific, #65-0867). CD31, CD24, and CD45 antibodies and Fixable Viability Dye were used to label endothelial cells, adrenal medulla cells (23), lymphocytes, and dead cells, respectively. CD56 antibody was used to label subcapsular adrenocortical cells including APCCs (24) (also see our previous observation: Figure S3) (22). The stained cells were sorted using BD FACSAria II (Becton-Dickinson). Doublets and debris were excluded on 2 following gating steps (forward scatter-height vs forward scatter-area, forward scatter-area vs side scatter-area). Adrenal medulla cells, endothelial cells, lymphocytes, and dead cells were excluded in FITC vs phycoerythrin/Cyanine7 gating step (Figure S4) (22). The specific and nonspecific bindings of APC-labeled anti-CD56 antibodies were determined by comparison with the fluorescence intensity of APC-conjugated isotype IgG (BioLegend Cat# 400221, RRID:AB_2891178), and the cellular regions of specific binding to CD56 antibody and nonspecific binding to CD56 antibody were set as gate A and gate B, respectively, in the FITC vs APC gating step (Figure S4) (22). Sample 1 contained enough cells in gate A (about 6300 cells), and cells for scRNA-seq were collected from gate A only. Sample 2 contained only a few cells in gate A (about 1200 cells); therefore, cells for scRNA-seq were collected from gates A and B. All data were analyzed with FlowJo software (version 9.9.6).
scRNA-seq
The sorted cell suspensions were concentrated by centrifugation. The libraries of scRNA-seq were prepared from the cell suspensions using Chromium controller (10X Genomics) and Chromium Single Cell 3′ Reagent Kits v3.1 according to the manufacturer’s instructions. Pair-end sequencing of the libraries was conducted using NovaSeq6000 (Illumina).
scRNA-seq Data Processing and Analysis
Raw sequencing data processing, quality control, doublets removal, batch correction, and data normalization
Sequencing data were analyzed using Cell Ranger 3.1.0 (10X Genomics). The “mkfastq” command was used to convert the data to the FASTQ format, and the “count” command was used to generate raw gene-barcode matrices aligned to 10X Genomics GRCh38 Ensembl 93 genome (version 3.0.0) (25). The gene-barcode matrices were imported into the R package Seurat (version 4.0.1) (26), an R package for scRNA-seq analysis, and subsequent scRNA-seq downstream analysis was performed. To filter out low-quality cells, the criteria for high-quality cells were set as cells with a gene number ≥ 500 and a percentage of mitochondrial genes < 10%, and nonqualifying cells were excluded. The R package DoubletFinder (Ver 2.0.3) (27) was used with default settings to detect doublet cells, and cells that were determined to be doublets were excluded. The “SCTransform” function of Seurat was used to normalize the filtered gene expression data using regularized negative binomial regression method, and the normalized data were used for dimensionality reduction and unsupervised clustering. The percentages of mitochondrial genes were entered into the “vars.to.regress” option and regressed as unwanted sources of variation. The “SelectIntegrationFeatures,” “PrepSCTIntegration,” “FindIntegrationAnchors,” and “IntegrateData” functions were used to correct for batch effects. The “NormalizeData” function was used to create log-normalized gene expression data for use in differential expression analysis and visualization of gene expression levels. The “expression level” in figures and tables refers to the log-normalized counts per gene.
Dimensionality reduction and clustering
Principal component analysis of the gene expression data was performed using the “RunPCA” function of Seurat, and the dimensionality of the data was reduced to the top 30 principal components. The “RunUMAP” function was used to reduce the dimensionality of the principal component analysis data using Uniform Manifold Approximation and Projection (UMAP) method, The UMAP data were used to visualize the gene expression data on 2 dimensions. Unsupervised clustering of the cells based on the Louvain algorithm was performed with the “FindNeighbors” and “FindClusters” functions. The “resolution” parameters of the “FindClusters” function were adjusted between 0.4 and 1.2 according to the developer’s instructions (26).
Differential expression analysis, gene ontology enrichment analysis
Differentially expressed genes (DEGs) were detected by Wilcoxon rank-sum test using the “FindAllMarkers” or “FindMarkers” function of Seurat. The Bonferroni method was used to correct for multiple comparisons. Genes with an adjusted P value < 0.01 and a log2 processed average fold change greater than 0.25 were defined as DEGs.
Gene ontology (GO) enrichment analysis for DEGs of each cluster was performed using the R package clusterProfiler (Ver 4.2.1) (28). GO terms in the biological processes subontology were tested using Fisher exact test, and terms with an adjusted P value corrected for the Benjamini-Hochberg method less than 0.05 were defined as significant.
Trajectory analysis with Monocle 3
Trajectory analysis was performed using the R package Monocle 3 (Ver 1.0.0) (29). In the trajectory analysis with Monocle 3, the lineage of development is identified based on the similarity of gene expression profile between cells, and the pseudotime of cells is calculated along the identified lineage. The gene expression data preprocessed with Seurat were imported in Monocle 3, and the developmental trajectories and pseudotime were inferred following the developer’s instructions. The “resolution” parameter of the “cluster_cells” function was set to 0.01.
RNA velocity analysis
RNA velocity represents the direction and velocity of cells in transcriptome space and is calculated from the expression states of unspliced and spliced mRNAs. The CellRanger output BAM files were processed by the python package velocyto (Ver 0.17.17) (30) to generate loom files with spliced and unspliced gene counts. These loom files were merged and the RNA velocity was calculated using the R package velocyto.R (version 0.6) (30). The parameters of the “RunVelocity” function were set to kCells = 25, fit.quantile = 0.02, and deltaT = 1. The RNA velocities were projected on UMAP plot with the “show.velocity.on.embedding.cor” function and the visualization parameters were set to n = 100, grid.n = 20, arrow.scale = 0.6, and scale = log.
Results
scRNA-seq Analysis of the Human Adult Adrenal Glands
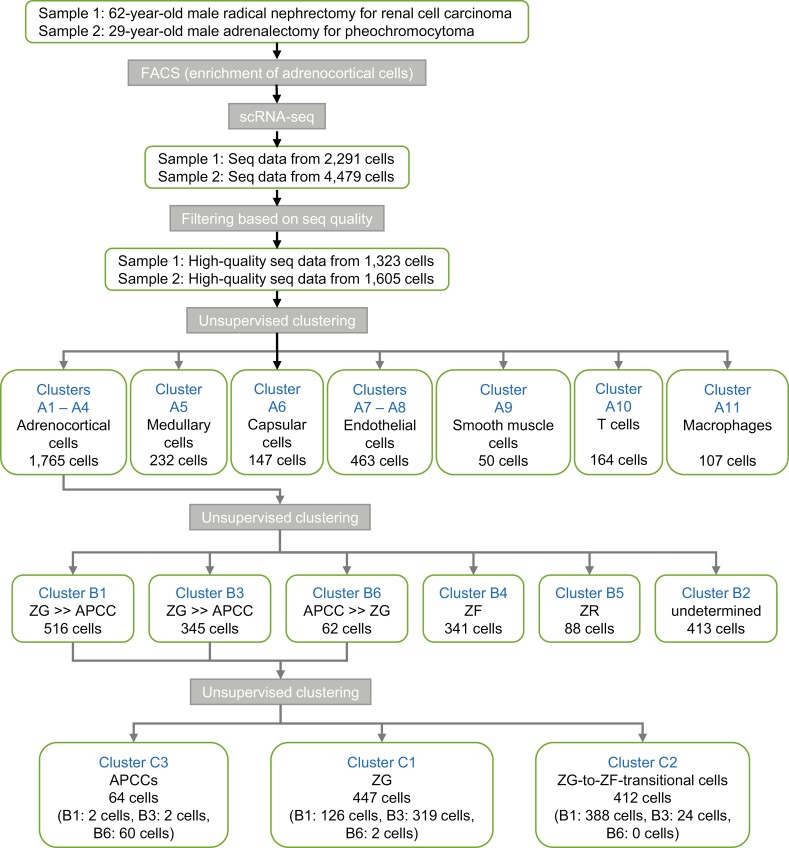
We performed droplet-based scRNA-seq on human adrenocortical cells collected from 2 adult males, with no apparent abnormalities in adrenocortical tissues, using the 10X Chromium system. A total of 6770 cells (sample 1: 2291; sample 2: 4479) were obtained, of which 407 cells (sample 1: 186; sample 2: 221) had positive CYP11B2 counts (defined as CYP11B2+ cells). Filtering was performed and 2928 high-quality cells were obtained (Fig. 1). An average of 10829 ± SD transcripts transcribed from 2565 ± SD genes were detected in each cell.
Figure 1.
The flow of the study and unsupervised clustering for single-cell RNA sequencing of 2 adrenal glands.
Aldosterone-producing Portions in Samples 1 and 2
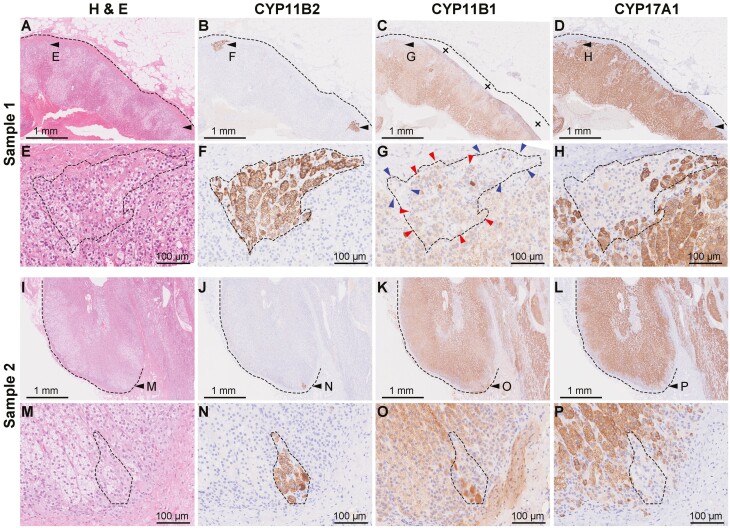
To confirm the presence of aldosterone-producing portions in the sample tissues histologically, histochemical and immunohistochemical staining was performed using the portion possessed for pathological diagnoses. Sample 1 had normal histology that consisted of ZG, ZF, and ZR (Fig. 2A). The adrenal harbored apparent APCCs (black arrowheads in Fig. 2A-B). We previously reported that APCCs are negative for CYP11B1 (5). However, we noticed that the APCC seemed to have weakly positive cells for CYP11B1 (Fig. 2C), whereas CYP17A1 seemed negative in APCCs as previously reported (Fig. 2D). Detailed observation with dotted line surrounding an APCC (Fig. 2E-2F), we noticed that the APCC consisted of CYP11B1-positive cells (red arrowheads in Fig. 2G) and CYP11B1-negative cells (blue arrowheads). The APCCs also harbor CYP17A1-positive cells (Fig. 2H). Sample 2 also had APCCs (Fig. 2I-2P) and were also contain CYP11B1-positive cells and CYP17A1-positive cells. CD56 was positive for ZG and the outer part of ZF as previously reported, and APCCs were present in the CD56-positive region in both samples (Figure S5) (22). Overall, the samples had APCCs, and detailed observation showed that APCCs contain some CYP11B1-positive and CYP17A1-positive cells.
Figure 2.
Histochemical and immunohistochemical staining for samples 1 and 2. (A-D) Low-magnification findings of histochemical and immunohistochemical staining for sample 1 (A, hematoxylin-eosin; B, CYP11B2; C, CYP11B1; D, CYP17A1). Dotted line indicates the outline of the adrenal gland and black arrows indicate APCCs. In panel C, the crosses indicate tissue folding. (E-H) High-magnification findings of histochemical and immunohistochemical staining for APCC of sample 1 (E, hematoxylin-eosin; F, CYP11B2; G, CYP11B1; H, CYP17A1). Dotted line indicates the outline of the APCC. In panel G, red arrowheads indicate CYP11B1-positive cells and blue arrowheads indicate CYP11B1-negative cells. (I-L) Low-magnification findings of histochemical and immunohistochemical staining for sample 2 (I, hematoxylin-eosin; J, CYP11B2; K, CYP11B1; L, CYP17A1). Dotted line indicates the outline of the adrenal gland and black arrowhead indicates APCC. (M-P) High-magnification findings of histochemical and immunohistochemical staining for APCC of sample 2 (M, hematoxylin-eosin; N, CYP11B2; O, CYP11B1; P, CYP17A1). Dotted line indicates the outline of the APCC.
Identification of Adrenocortical Cells
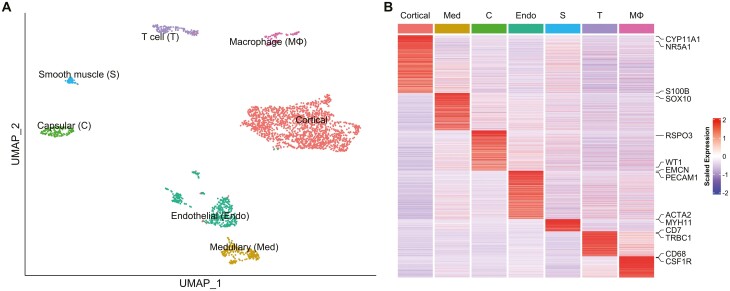
We performed UMAP visualization using Seurat (see Materials and Methods) on 2928 high-quality cells. The distribution of cells in each sample showed that the batch effect between samples 1 and 2 was properly corrected (Figure S6A) (22). Unsupervised clustering divided these cells into 11 clusters (Figure S6B) (22). Based on the marker gene expression patterns in each cluster, we were able to classify these clusters into adrenocortical cells (n = 1765; clusters A1-A4; marked by upregulation of CYP11A1 and NR5A1) (21), medullary cells (n = 232; cluster A5; SOX10, S100B) (31, 32), capsular cells (n = 147; cluster A6; WT1, RSPO3) (33, 34), endothelial cells (n = 463; clusters A7 and A8; PECAM1, EMCN) (21), smooth muscle cells (n = 50; cluster A9; MYH11, ACTA2) (21), T cells (n = 164; cluster A10; CD7, TRBC1) (21), and macrophages (n = 107; cluster A11; CD68, CSF1R) (21) (Fig. 3 and Fig. S6C) (22). Thus, adrenocortical cells were the most abundant (accounting for 60.3%), suggesting that the enrichment of adrenocortical cells using FACS probably worked to some extent (see Materials and Methods).
Figure 3.
Clustering of adrenal cells. (A) UMAP plot showing the adrenal cell types. Dots: single cells. Colors: cell types. (B) Heatmap showing the average expression levels of DEGs in each adrenal cell type. Columns are each cell type; rows are individual genes. Gene names shown are the genes that were used to identify the cell types. The expression level is the log-normalized counts per gene. Average expression levels are z-scored.
In adrenocortical cells (clusters A1-A4), when compared with the whole high-quality cell population analyzed, 1256 genes were upregulated more than 1.19-fold (>logFC 0.25) with an adjusted P value < 0.01 (Table S2) (22). In these genes, steroidogenic genes such as CYP17A1, HSD3B2, and CYP21A2 were also highly upregulated (12.4-, 11.8-, and 10.0-fold; Table S2), supporting that the clusters include adrenocortical cells (1, 22).
Clustering of the Adrenocortical Cells
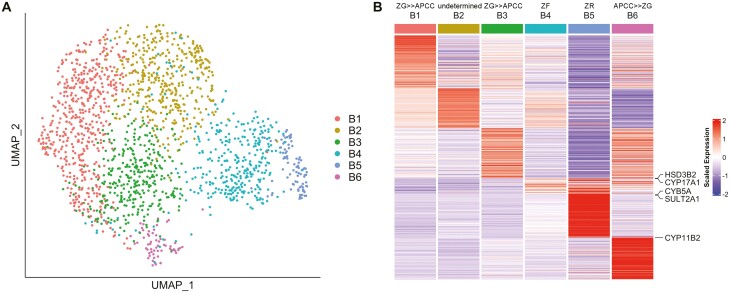
To further characterize adrenocortical cells, we subsequently performed unsupervised clustering of 1765 cells in clusters A1 through A4. The cells were equally distributed between the 2 samples (Figure S7A) (22). The adrenocortical cells, which were originally clustered into 4 clusters, were reclassified into 6 clusters in this round of unsupervised clustering (clusters B1-B6; Fig. 4A, Figure S7B) (22), and up- or down-regulated genes in each cluster were identified (Table S3) (22). Black lines in Figure S7B indicate developmental trajectory, which is discussed in the following section. CYP11B2 was significantly upregulated (11.6-fold) in cluster B6 (Figure S7C-D) (22). Although CYP11B1 expression was detected throughout all 6 clusters (FigureS7E-F) (22), the low expression of CYP17A1 in cluster B6, along with the high expression of CYP11B2 (Figures S7G-H) (22) indicated that these cells have the characteristics of aldosterone-producing cells. In our previous DNA microarray study using laser-micro-dissected samples (Gene Expression Omnibus, accession no. GSE68889), we reported differentially expressed genes between APCC and ZG (APCC-vs-ZG genes), between APCC and ZF (APCC-vs-ZF genes), as well as between ZG and ZF (ZG-vs-ZF genes) (8). Here, we counted the genes commonly up- or down-regulated in the laser-micro-dissection study among the differentially expressed genes of clusters B1 through B6 in the current study as shown in Table 1 (common genes are marked by *, †, ‡ in Table S3) (22). Among 283 genes in cluster B6 (up- and down-regulated genes: 178 and 105), 10 genes (3.53%) were commonly up- or down-regulated in APCC-vs-ZG genes, which was higher than those in other clusters (0.00%-0.83%). This result supports that cluster B6 contained many APCC cells. Clusters B1, B3, and B6, contained commonly up- or down-regulated genes with APCC-vs-ZG genes (8.37%, 6.83%, and 13.43%) and with ZG-vs-ZF genes (10.84%, 7.55%, and 10.25%), whereas clusters B2, B4, and B5 had much fewer common genes (0.71%-2.12%). Thus, we deduced that the clusters B1, B3, and B6 contained ZG cells and in particular the cluster B6 contained APCC cells and possibly induced ZG cells.
Figure 4.
Clustering of adrenocortical cells. (A) UMAP plot showing the adrenocortical cell clusters. Dots: single cells. Colors: clusters. (B) Heatmap showing the average expression levels of DEGs in each adrenocortical cell cluster. Columns are each cluster; rows are individual genes. Gene names shown are the genes that were used to identify the cell types. The expression level is the log-normalized counts per gene. Average expression levels are z-scored.
Table 1.
The number of commonly up- or down-regulated differentially expressed genes between our previous studya and clusters B1-B6
| Clusters | CYP11B2 + cells | No. of genes in clusters | APCC vs ZG | APCC vs ZF | ZG vs ZF | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Up | Down | Total | Up | Down | Total | Up | Down | Total | Up | Down | Total | ||
| B1 | 46 | 169 | 34 | 203 | 0 | 1 | 1 (0.49%) | 15 | 2 | 17 (8.37%) | 19 | 3 | 22 (10.84%) |
| B2 | 14 | 135 | 148 | 283 | 0 | 0 | 0 (0%) | 2 | 0 | 2 (0.71%) | 6 | 0 | 6 (2.12%) |
| B3 | 50 | 167 | 111 | 278 | 0 | 1 | 1 (0.36%) | 17 | 2 | 19 (6.83%) | 17 | 4 | 21 (7.55%) |
| B4 | 39 | 56 | 292 | 348 | 2 | 1 | 3 (0.86%) | 1 | 2 | 3 (0.86%) | 1 | 2 | 3 (0.86%) |
| B5 | 1 | 165 | 921 | 1086 | 4 | 5 | 9 (0.83%) | 5 | 3 | 8 (0.74%) | 4 | 7 | 11 (1.01%) |
| B6 | 43 | 178 | 105 | 283 | 10 | 0 | 10 (3.53%) | 38 | 0 | 38 (13.43%) | 27 | 2 | 29 (10.25%) |
a Nishimoto K et al. (8).
In contrast, cells expressing CYB5A (13.6-fold) and SULT2A1 (8.5-fold), marker genes of the ZR (35, 36), were highly enriched in the cluster B5 (Fig. 4B, Figure S7I, Table S3) (22), suggesting that this cluster consisted mainly of ZR cells. The trajectory analysis showed that the clusters B1/B3 may differentiate into 3 directions: to (1) the cluster B2 (upward direction on Figure S7B) (22), (2) the clusters B4/B5 (right direction), and (3) the cluster B6 (downward direction). In mice adrenal cortex, ZG cells were shown to migrate centripetally and differentiate into ZF cells (17, 18). In the adrenal cortex of Prkar1a knockout mice, it was also shown that ZF cells differentiate into ZR-like cells by constitutive PKA activation (20). The clusters B1/B3 contained CYP11B2+ cells (Figure S7D) (22); therefore, we hypothesized that noninduced or weakly induced ZG cells, which were mostly contained in the clusters B1/B3, were differentiated into the cluster B6 (APCC/induced ZG cluster) and to ZR (cluster B5) via ZF (cluster B4). The highest upregulated genes in the cluster B4 were HSD3B2 and CYP17A1 (3.41- and 2.93-fold; marker genes of the ZF (5); Fig. 4B; Table S3) (22) and GO enrichment analyses showed upregulated genes in the cluster B4 were associated with glucocorticoid production named “steroid hormone biosynthetic process” and “glucocorticoid metabolic process” (Table S4) (22), supporting the hypothesis that the cluster B4 is enriched with cortisol-producing cells. Based on the GO enrichment analyses, cells in cluster B2 seemed to be associated with cellular functions including cellular responses and translation rather than steroid biosynthesis, and thus we could not definitively classify the identity of the cluster B2 cells. Consequently, we identified the presumable cortisol-producing cluster B4 and ZR-enriched cluster B5 with their lists of DEGs (Table S3) (22). These were likely to differentiate continuously from ZG clusters B1/B3, as shown in mice (18), and differ from the direction of differentiation from ZG to APCC/induced ZG cluster (Figure S7B) (22).
Identification of APCC and ZG-to-ZF Transitional Cells
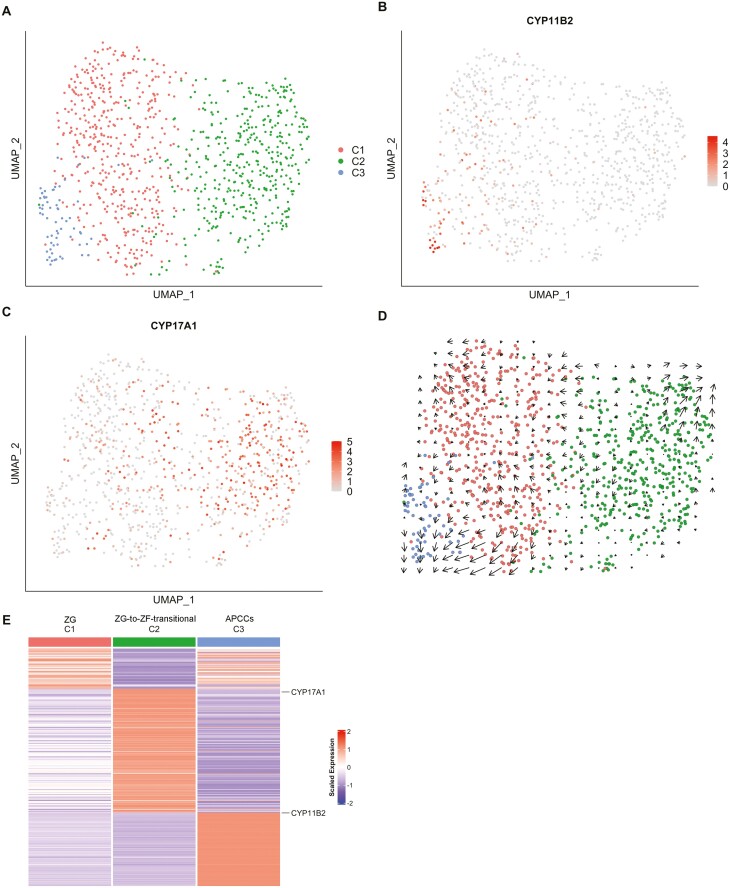
As mentioned previously, we deduced that the clusters B1, B3, and B6 contained ZG cells and APCC cells. We also assumed that they may contain transitional cells from ZG to ZF because clusters B1/B3 exist between clusters 4 and 6 on UMAP (Figure S7B) (22). To further characterize the clusters B1, B3, and B6 and appraise the transcriptional characteristics of cells in these zones (ie, APCC, ZG, and ZG-to-ZF transitional zone), we performed unsupervised clustering of 923 cells in the clusters B1, B3, and B6 (516, 345, and 62 cells) (Fig. 5), which reclassified the cells into 3 clusters: cluster C1, C2, and C3 (447, 412, and 64 cells) (Figs. 1 and 5). The percentage of CYP11B2+ cells was lowest in the C1 (18 cells, 4.4%, red dots in Fig. 5A) and higher in the cluster C2 (98 cells, 21.9%, green dots) and cluster C3 (82.8%, 53 cells, blue dots) (Fig. 5B). Whereas numbers of CYP17A1 expressing cells in clusters C1, C2, and C3 were 107 (23.9%), 241 (58.5%), and 9 (14.1%) (Fig. 5C). The expression levels of CYP11B2 and CYP17A1 (Fig. 5B and 5C) suggested that clusters C3, C1, and C2 represent APCC (APCC-cluster C3), ZG cells including noninduced ZG cells (ZG-cluster C1), and ZG-to-ZF transitional cells (ZG-to-ZF-cluster C2). In addition, we found triple-positive cells expressing CYP11B2, CYP11B1, and CYP17A1 and double-positive cells expressing CYP11B2 and CYP11B1 in APCC-cluster C3 (Table S5) (22). This was consistent with the findings of the immunohistochemical staining described previously (Fig. 2).
Figure 5.
Clustering of ZG cells. (A) UMAP plot showing the ZG cell clusters. Dots: single cells. Colors: clusters. (B) Scatterplots showing the expression level of CYP11B2 projected onto the UMAP plot. Dots: single cells. Dot color: expression level. (C) Scatterplots showing the expression level of CYP17A1 projected onto the UMAP plot. Dots: single cells. Dot color: expression level. (D) The result of RNA velocity analysis of ZG cells projected onto the UMAP plot, showing the developmental direction from ZG cells (cluster C1) to APCC cells (cluster C3). Dots: single cells. Colors: cell types. Arrows: RNA velocity estimates (divided into 20 grids horizontally and vertically, the RNA velocity observed in each grid is aggregated and displayed). (E) Heatmap showing the average expression levels of DEGs in each ZG cell cluster. Columns are each cluster; rows are individual genes. Gene names shown are the genes that were used to identify the cell types. The expression level is the log-normalized counts per gene. Average expression levels are z-scored.
To investigate the differentiation relationship between ZG cells and APCC cells, RNA velocity analysis was performed. The arrows in Fig. 5D indicate the direction and rate of cell differentiation. Interestingly, the arrows between ZG-cluster C1 and APCC-cluster C3 directed from C1 to C3, suggesting that some ZG cells remodel to APCCs. The arrows between ZG-cluster C1 and ZG-to-ZF-cluster C2 were small and inconsistent in direction, therefore the direction of differentiation was not well estimated in this analysis.
Gene expression change from ZG (cluster C1) to APCC (cluster C3) and ZG (cluster C1) to ZG-to-ZF transitional cells (cluster C2)
We compared the gene expression between ZG-cluster C1 and APCC-cluster C3. Steroidogenic genes for aldosterone synthesis, including CYP11B2, HSD3B2, and CYP21A2, were highly upregulated in APCC-cluster C3 (9.07-, 7.05-, and 2.81-fold) (Table S6) (22). MC2R, TSPAN12, and CLRN1, which we previously reported to be upregulated in APCC, were also upregulated in APCC-cluster C3 (2.42-, 1.99-, 1.52-fold), supporting that this cluster consisted of APCC cells. In comparison between ZG-cluster C1 and ZG-to-ZF-cluster C2, CYP17A1 was upregulated and CYP11B2 was downregulated in ZG-to-ZF-cluster C2 (2.72- and 0.65-fold), supporting that this cluster consisted of ZG-to-ZF-transitional cells (Table S6) (22).
GO enrichment analyses showed that the upregulated genes in APCC-cluster C3 enriched in terms related to steroidogenesis (Table S7) (22). Similarly, the upregulated genes in ZG-to-ZF-cluster C2 enriched in terms related to steroidogenesis. These results suggested that the differentiation of ZG into APCC involves significant changes in the expression of steroidogenic genes.
Discussion
In this study, we performed scRNA-seq of human adult adrenals and, successfully differentiated plausible cell clusters corresponding to adrenocortical cells/zones of APCC, ZG, ZF, ZR, and novel ZG-to-ZF transitional zone based on their gene expression profiles. Moreover, we herein deduced that APCC cells develop from ZG cells by trajectory/RNA velocity analyses (29, 30), which matches our hypothesis that ZG cells remodel to APCCs with ion channel or pump somatic mutations (5, 15, 37).
Recently, scRNA-seq studies on adrenocortical cells were reported from 2 laboratories. Han et al constructed a huge database called human cell landscape based on scRNA-seq from many organs including adrenals (21). The same laboratory used the scRNA-seq data of the adrenals and reported a single-cell comparative analysis of human and mouse adult adrenals to reveal the conserved genetic networks in these mammalian systems (38). A second group performed scRNA-seq of adult human adrenals and revealed that there were 5 types of immune cells in the adrenals (39). Thus, these groups have successfully clustered the adrenal cortex by scRNA-seq. In addition to these classical zones, we describe here novel clusters of APCC and ZG-to-ZF transitional zone.
The genes upregulated more than 4-fold except steroidogenic genes in APCC-cluster-C3 were HOPX (HOP homeobox), VPREB3 (v-set pre-B cell surrogate light chain 3), and COL15A1 (Collagen Type XV 1 Chain) (Table S6) (22). These genes except COL15A1 were reported to be upregulated in APA as well (40, 41). APCC and APA had several upregulated genes in common, which was consistent with our hypothesis that APCC is a precursor of some APAs. The DEGs between APCC-cluster C3 and ZG-cluster C1 may be of use for future adrenal studies in humans, because the current in vitro (eg, H295R cells) and in vivo models (ie, CYP11B2-cre mice (18)) are limited to use as they are derived from adrenocortical cancer cells and mice, respectively.
In this study, FACS was performed to enrich adrenocortical cells by excluding CD24/CD31/CD45-positive cells (adrenal medulla cells, endothelial cells, lymphocytes) and collecting CD56-positive cells (ZG and the outer part of ZF). For sample 2, CD56-negative cells were also collected because of the small number of CD56-positive cells. In the scRNA-seq analysis of Huang et al without FACS, adrenocortical cells accounted for 17% of the total, whereas in this study they accounted for 60%, suggesting that FACS enriched the adrenocortical cells (39). Sample 2, in which CD56 sorting was not performed, also had the highest percentage of adrenocortical cells (45%), suggesting that exclusion of CD24/CD31/CD45-positive cells was effective in enriching adrenocortical cells and collection of CD56-positive cells may not contribute much to enrichment.
In conclusion, our study is the first report of APCC cells and ZG-to-ZF transitional cells analyzed by scRNA-seq. Our results suggested that APCC has a similar mechanism of aldosterone synthesis with ZG and develops from ZG. This fits in our hypothesis that the APCC develops from ZG and becomes a precursor of APA.
Acknowledgments
We thank Ms. Noriko Akiyama and Ritsuko Toda in Saitama Medical University International Medical Center for great assistance on ‘Dispersing and Freezing Adrenal Cells’; Ms. Kiyomi Imamura in Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo for ‘scRNA-seq’; and Morphotechnology Co., Ltd in Sapporo, Japan for ‘Immunohistochemistry’. And we thank Drs. Miki Inoue, Takashi Baba and Ken-Ichirou Morohashi in Kyushu University for excellent assistances on data analyses.
Glossary
Abbreviations
- APA
aldosterone-producing adenoma
- APC
allophycocyanin
- APCC
aldosterone-producing cell cluster
- CCS
cosmic calf serum
- CYP11B1
cytochrome P450 family 11 subfamily B member 1
- CY11B2
cytochrome P450 family 11 subfamily B member 2
- DEG
differentially expressed gene
- FACS
fluorescence-activated cell sorting
- FITC
fluorescein isothiocyanate
- GO
gene ontology
- IHA
idiopathic hyperaldosteronism
- PA
primary aldosteronism
- scRNA-seq
single-cell RNA sequencing
- UMAP
Uniform Manifold Approximation and Projection
- ZF
zona fasciculata
- ZG
;zona glomerulosa
- ZR
zona reticularis
Contributor Information
Norifusa Iwahashi, Department of Medicine and Bioregulatory Science, Graduate School of Medical Sciences, Kyushu University, Fukuoka 812-8582, Japan.
Hironobu Umakoshi, Department of Medicine and Bioregulatory Science, Graduate School of Medical Sciences, Kyushu University, Fukuoka 812-8582, Japan.
Tsugio Seki, Department of Medical Education, School of Medicine, California University of Science and Medicine, Colton, CA 92324, USA.
Celso E Gomez-Sanchez, Department of Pharmacology and Toxicology and Medicine, University of Mississippi Medical Center, Jackson, MS 39216, USA.
Kuniaki Mukai, Medical Education Center, Keio University School of Medicine, Tokyo 160-8582, Japan.
Makoto Suematsu, Department of Biochemistry, Keio University School of Medicine, Tokyo 160-8582, Japan.
Yuta Umezawa, Department of Uro-Oncology, Saitama Medical University International Medical Center, Saitama 350-1298, Japan.
Mototsugu Oya, Department of Urology, Keio University School of Medicine Tokyo 160-8582, Japan.
Takeo Kosaka, Department of Urology, Keio University School of Medicine Tokyo 160-8582, Japan.
Masahide Seki, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Chiba 277-0882, Japan.
Yutaka Suzuki, Department of Computational Biology and Medical Sciences, Graduate School of Frontier Sciences, The University of Tokyo, Chiba 277-0882, Japan.
Yutaka Horiuchi, Department of Microbiology, Faculty of Medicine, Saitama Medical University, Saitama 350-0495, Japan.
Yoshihiro Ogawa, Department of Medicine and Bioregulatory Science, Graduate School of Medical Sciences, Kyushu University, Fukuoka 812-8582, Japan.
Koshiro Nishimoto, Department of Uro-Oncology, Saitama Medical University International Medical Center, Saitama 350-1298, Japan.
Funding
This study was supported by Shimadzu Corporation (to M.S.); the Mitsubishi Foundation (to Y.O.); SECOM Science and Technology Foundation (to Y.O.); Grant-in-Aid for Exploratory Research (20K21604 to Y.O.); Grant-in-Aid for Scientific Research (S) (22H04993 to Y.O.), Grant-in-Aid for Scientific Research (C) (18K09205 to K.N., 17K09890 to K.M.), and Grant-in-Aid for Early-Career Scientists (20K17493 to H.U.) from Japan Society for the Promotion of Science; the Uehara Memorial Foundation (to H.U.); Kaibara Morikazu Medical Science Promotion Foundation (to H.U.); Takeda Science Foundation (to H.U.); National Heart, Lung and Blood Institute grant (R01HL144847) and the National Institute of General Medical Sciences grant (U54GM115428) from National Institutes of Health in the United States (to C.E.G.-S.); the Department of Veteran Affairs (BX00468 to C.E.G.-S.); Hidaka Project from Saitama Medical University International Medical Center (to Y. U); and Grant-in-Aid from Saitama Medical University (21-B-1-15 to K.N).
Conflict of Interest
The authors have nothing to disclose.
Data Availability
Some or all of the data generated or analyzed in this study are contained in the data repository described in this publication or in the bibliography. Supplemental materials are available on GitHub (https://github.com/NorifusaIwahashi/Supplemental-data-for-Characterization-of-APCC-at-single-cell-resolution). Sequence data are deposited to ArrayExpress (accession number E-MTAB-11837).
References
- 1. Melmed S, Koenig R, Rosen C, Auchus R, Goldfine A.. Williams Textbook of Endocrinology E-book. 14th ed. Elsevier; 2019. [Google Scholar]
- 2. Hattangady NG, Olala LO, Bollag WB, Rainey, WE. Acute and chronic regulation of aldosterone production. Mol Cell Endocrinol. 2012;350(2):151-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Funder JW, Carey RM, Mantero F, et al. The management of primary aldosteronism: case detection, diagnosis, and treatment: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2016;101(5):1889-1916. [DOI] [PubMed] [Google Scholar]
- 4. Ohno Y, Sone M, Inagaki N, et al. ; Nagahama Study, JPAS Study Group.. Prevalence of cardiovascular disease and its risk factors in primary aldosteronism: a multicenter study in Japan. Hypertension 2018;71(3):530-537. [DOI] [PubMed] [Google Scholar]
- 5. Nishimoto K, Nakagawa K, Li D, et al. Adrenocortical zonation in humans under normal and pathological conditions. J Clin Endocrinol Metab. 2010;95(5):2296-2305. [DOI] [PubMed] [Google Scholar]
- 6. Sugiura Y, Takeo E, Shimma S, et al. Aldosterone and 18-oxocortisol coaccumulation in aldosterone-producing lesions. Hypertension 2018;72(6):1345-1354. [DOI] [PubMed] [Google Scholar]
- 7. Zhang Z, Sugiura Y, Mune T, et al. Immunohistochemistry for aldosterone synthase CYP11B2 and matrix-assisted laser desorption ionization imaging mass spectrometry for in-situ aldosterone detection. Curr Opin Nephrol Hypertens. 2019;28(2):105-112. [DOI] [PubMed] [Google Scholar]
- 8. Nishimoto K, Tomlins SA, Kuick R, et al. Aldosterone-stimulating somatic gene mutations are common in normal adrenal glands. Proc Natl Acad Sci USA. 2015;112(33):E4591-E4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choi M, Scholl UI, Yue P, et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science 2011;331(6018):768-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scholl UI, Goh G, Stölting G, et al. Somatic and germline CACNA1D calcium channel mutations in aldosterone-producing adenomas and primary aldosteronism. Nat Genet. 2013;45(9):1050-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Azizan EAB, Poulsen H, Tuluc P, et al. Somatic mutations in ATP1A1 and CACNA1D underlie a common subtype of adrenal hypertension. Nat Genet. 2013;45(9):1055-1060. [DOI] [PubMed] [Google Scholar]
- 12. Beuschlein F, Boulkroun S, Osswald A, et al. Somatic mutations in ATP1A1 and ATP2B3 lead to aldosterone-producing adenomas and secondary hypertension. Nat Genet. 2013;45(4):440-4, 444e1-2. [DOI] [PubMed] [Google Scholar]
- 13. Nanba K, Tsuiki M, Sawai K, et al. Histopathological diagnosis of primary aldosteronism using CYP11B2 immunohistochemistry. J Clin Endocrinol Metab. 2013;98(4):1567-1574. [DOI] [PubMed] [Google Scholar]
- 14. Nishimoto K, Umakoshi H, Seki T, et al. ; JRAS Study Group. Diverse pathological lesions of primary aldosteronism and their clinical significance. Hypertens Res. 2021;44(5):498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishimoto K, Seki T, Kurihara I, et al. Case report: nodule development from subcapsular aldosterone-producing cell clusters causes hyperaldosteronism. J Clin Endocrinol Metab. 2016;101(1):6-9. [DOI] [PubMed] [Google Scholar]
- 16. Nishimoto K, Koga M, Seki T, et al. Immunohistochemistry of aldosterone synthase leads the way to the pathogenesis of primary aldosteronism. Mol Cell Endocrinol. 2017;441:124-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. King P, Paul A, Laufer E. Shh signaling regulates adrenocortical development and identifies progenitors of steroidogenic lineages. Proc Natl Acad Sci USA. 2009;106(50):21185-21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Freedman BD, Kempna PB, Carlone DL, et al. Adrenocortical zonation results from lineage conversion of differentiated zona glomerulosa cells. Dev Cell. 2013;26(6):666-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grabek A, Dolfi B, Klein B, et al. The adult adrenal cortex undergoes rapid tissue renewal in a sex-specific manner. Cell Stem Cell 2019;25(2):290-296.e2. [DOI] [PubMed] [Google Scholar]
- 20. Dumontet T, Sahut-Barnola I, Septier A, et al. PKA signaling drives reticularis differentiation and sexually dimorphic adrenal cortex renewal. JCI Insight 2018;3(2):e98394. doi 10.1172/jci.insight.98394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Han X, Zhou Z, Fei L, et al. Construction of a human cell landscape at single-cell level. Nature 2020;581(7808):303-309. [DOI] [PubMed] [Google Scholar]
- 22.Supplemental-data-for-Characterization-of-APCC-at-single-cell-resolution. The purpose of this repository is to publish supplemental data for the article “Characterization of aldosterone-producing cell cluster (APCC) at single-cell resolution.” Github Available at: https://github.com/NorifusaIwahashi/Supplemental-data-for-Characterization-of-APCC-at-single-cell-resolution. Accessed March 6, 2022.
- 23. Akashi T, Shirasawa T, Hirokawa, K. Gene expression of CD24 core polypeptide molecule in normal rat tissues and human tumor cell lines. Virchows Arch. 1994;425(4):399-406. [DOI] [PubMed] [Google Scholar]
- 24. Caroccia B, Fassina A, Seccia TM, et al. Isolation of human adrenocortical aldosterone-producing cells by a novel immunomagnetic beads method. Endocrinology 2010;151(3):1375-1380. [DOI] [PubMed] [Google Scholar]
- 25. Zheng GXY, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hao Y, Hao S, Andersen-Nissen E, et al. Integrated analysis of multimodal single-cell data. Cell 2021;184(13):3573-3587.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McGinnis CS, Murrow LM, Gartner, ZJ. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst 2019;8(4):329-337.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu G, Wang L-G, Yan G-R, He Q-Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics 2015;31(4):608-609. [DOI] [PubMed] [Google Scholar]
- 29. Cao J, Spielmann M, Qiu X, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019;566(7745):496-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. La Manno G, Soldatov R, Zeisel A, et al. RNA velocity of single cells. Nature 2018;560(7719):494-498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lloyd RV, Blaivas M, Wilson, BS. Distribution of chromogranin and S100 protein in normal and abnormal adrenal medullary tissues. Arch Pathol Lab Med. 1985;109(7):633-635. [PubMed] [Google Scholar]
- 32. Reiprich S, Stolt CC, Schreiner S, Parlato R, Wegner, M. SoxE proteins are differentially required in mouse adrenal gland development. Mol Biol Cell. 2008;19(4):1575-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bandiera R, Vidal VPI, Motamedi FJ, et al. WT1 maintains adrenal-gonadal primordium identity and marks a population of AGP-like progenitors within the adrenal gland. Dev Cell. 2013;27(1):5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vidal V, Sacco S, Rocha AS, et al. The adrenal capsule is a signaling center controlling cell renewal and zonation through Rspo3. Genes Dev. 2016;30(12):1389-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Suzuki T, Sasano H, Takeyama J, et al. Developmental changes in steroidogenic enzymes in human postnatal adrenal cortex: immunohistochemical studies. Clin. Endocrinol. 2000;53(6):739-747. [DOI] [PubMed] [Google Scholar]
- 36. Rege J, Nakamura Y, Wang T, et al. Transcriptome profiling reveals differentially expressed transcripts between the human adrenal zona fasciculata and zona reticularis. J Clin Endocrinol Metab. 2014;99(3):E518-E527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nishimoto K, Seki T, Hayashi Y, et al. Human adrenocortical remodeling leading to aldosterone-producing cell cluster generation. Int. J. Endocrinol 2016;2016:7834356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lai S, Ma L, E W, et al. Mapping a mammalian adult adrenal gland hierarchy across species by microwell-seq. Cell Regen 2020;9(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang L, Liao J, Chen Y, et al. Single-cell transcriptomes reveal characteristic features of cell types within the human adrenal microenvironment. J Cell Physiol. 2021;236(11):7308-7321. doi: 10.1002/jcp.30398. [DOI] [PubMed] [Google Scholar]
- 40. Wang T, Satoh F, Morimoto R, et al. Gene expression profiles in aldosterone-producing adenomas and adjacent adrenal glands. Eur J Endocrinol. 2011;164(4):613-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Felizola SJA, Katsu K, Ise K, et al. Pre-B lymphocyte protein 3 (VPREB3) expression in the adrenal cortex: precedent for non-immunological roles in normal and neoplastic human tissues. Endocr Pathol. 2015;26(2):119-128. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Some or all of the data generated or analyzed in this study are contained in the data repository described in this publication or in the bibliography. Supplemental materials are available on GitHub (https://github.com/NorifusaIwahashi/Supplemental-data-for-Characterization-of-APCC-at-single-cell-resolution). Sequence data are deposited to ArrayExpress (accession number E-MTAB-11837).