Abstract
A Staphylococcus aureus mutant (SPW1) which is unable to survive long-term starvation was shown to have a transposon insertion within a gene homologous to the sodA family of manganese-dependent superoxide dismutases (SOD). Whole-cell lysates of the parental 8325-4 strain demonstrated three zones of SOD activity by nondenaturing gel electrophoresis. The activities of two of these zones were dependent on manganese for activity and were absent in SPW1. The levels of SOD activity and sodA expression were growth-phase dependent, occurring most during postexponential phase. This response was also dependent on the level of aeration of the culture, with highest activity and expression occurring only under high aeration. Expression of sodA and, consequently, SOD activity could be induced by methyl viologen but only during the transition from exponential- to postexponential-phase growth. SPW1 was less able to survive amino acid limitation and acid stress but showed no alteration in pathogenicity in a mouse abscess model of infection compared to the parental strain 8325-4.
The aerobic environment is potentially toxic to life due to the high reactivity of oxygen. When oxygen becomes partially reduced, reactive oxygen species such as superoxide anion (O2.), hydrogen peroxide (H2O2), and hydroxyl radical (.OH) are often formed (17). Internal generation of these reactive species results in damage to DNA, proteins, and lipids (23). As a defense against these toxic effects, organisms have evolved superoxide dismutases (SODs) which detoxify O2. to hydrogen peroxide (H2O2), which in turn is generally broken down to water by catalase. SODs are classified according to the metal ion cofactor required for activity; the copper-zinc type (Cu/Zn-SOD), the manganese type (Mn-SOD), the iron type (Fe-SOD), and the recently identified nickel type (Ni-SOD) (18, 26).
Escherichia coli has three SODs, which differ in their location and temporal expression. Both Fe- and Mn-SOD are present in the cytoplasm, where they protect DNA and proteins from oxidation. Expression of the Fe-SOD, encoded by sodB, is constitutive and therefore is thought to play a housekeeping role (21), while the levels of Mn-SOD, encoded by sodA, fluctuate, increasing in response to elevated levels of internal O2. and upon changes in growth phase (13). The Cu/Zn-SOD, encoded by sodC, is located in the periplasm and protects the periplasmic and membrane constituents from exogenous superoxide (4, 24). To date, bacterial Cu/Zn-SOD has been identified only in gram-negative pathogens, where it is thought to protect against host defense mechanisms (4, 29, 38, 45). After engulfment of bacteria by professional phagocytes, the induction of highly microbiocidal reactive oxygen metabolites during the oxidative burst occurs, resulting in killing (2, 37).
Staphylococcus aureus is medically important as the cause of many nosocomial infections (42). The ability of S. aureus to survive in nutrient-limiting and stressful conditions contributes to its transmissibility in the hospital environment, which occurs primarily via direct contact (e.g., hand to wound), airborne carriage, and contact with surfaces such as indwelling devices (e.g., catheters). Recently we have characterized the starvation survival and stress responses of S. aureus and have identified several loci involved in these processes (7, 10, 11, 43, 44). During a screen for starvation survival mutants, a gene showing homology to the sodA family of SOD was identified. This study describes the characterization of the SOD and the demonstration of its role in stress resistance but not pathogenicity in a mouse abscess model of infection.
MATERIALS AND METHODS
Cloning and sequencing of sodA.
A plasmid, pSPW1, which contained chromosomal DNA flanking the lacZ proximal region of the Tn insertion within S. aureus SPW1, was generated in a previous study (43). The insert in pSPW1 (approximately 5 kbp) was excised, DIG labelled according to the protocol of Boehringer Mannheim GmbH (Germany), and used to probe a λ ZAP Express library of partial Sau3A digest (2 to 10 kb) of S. aureus 8325-4 genomic DNA (15). A clone containing a 3.3-kbp genomic DNA fragment spanning the SPW1 transposon insertion site was identified, and a stable phagemid (pSPW100) was excised from λ ZAP Express in E. coli XLOLR (Stratagene). A primer walking-based approach was used to sequence a 1,120-bp region (GenBank accession no. AF121672).
Strains and growth conditions.
Strains and plasmids used in this study are listed in Table 1. S. aureus 8325-4 (31) and SPW1 (43) were grown in a chemically defined medium (CDM) containing 0.1% glucose (22, 44) or in brain heart infusion (BHI) broth (Oxoid). Agar plates were prepared by the addition of 1% agar (wt/vol) to the above-described culture. Liquid cultures were grown with high aeration (10 ml of medium in a 250-ml flask, 250 rpm orbital shaking) or low aeration (100 ml of medium in a 250-ml flask, 125 rpm linear shaking). Amino acid- and glucose-limited starvation cultures were prepared as described in Watson et al. (44), and starved cells were recovered as described by Clements and Foster (10). Viable counts were determined by serial dilution of cultures with phosphate-buffered saline (PBS) and plating on CDM agar (1% wt/vol). Results shown are representative of at least three independent experiments showing less than 10% variation, unless otherwise stated.
TABLE 1.
Bacterial strains
| Strain | Genotype or relevant characteristics | Origin (reference) |
|---|---|---|
| 8325-4 | Wild-type strain cured of known prophages | R. Novick (31) |
| SPW1 | sodA::Tn917-LTV1 Eryr | Laboratory stock (43) |
| PC6911 | agrΔ::tetM Tcr | Laboratory stock (6) |
| MC51 | sodA::Tn917-LTV1 agrΔ::tetM Tcr Eryr | This study |
| PC1839 | sarA::km Kmr | Laboratory stock (6) |
| MC52 | sodA:Tn917-LTV1 sarA::km Kmr Eryr | This study |
| PC400 | sigB::tet Tcr | Laboratory stock (6) |
| MC53 | sodA::Tn917-LTV1 sigB::tet Tcr Eryr | This study |
Construction of reporter fusion strains.
The sodA::Tn917-LTV1 locus was transduced from SPW1 into PC6911 (agr::tet), PC1839 (sar::km) and PC400 (sigB::tet) by phage transduction, by using Φ11 as carrier (32), selecting for transductants on erythromycin (5 μg ml−1) containing BHI agar, giving rise to strains MC51, MC52, and MC53, respectively.
β-Galactosidase and luciferase assays.
β-Galactosidase assays with cell lysates, with 4-methylumbelliferyl-β-d-galactoside as substrate, and luciferase assays were performed as previously described (6).
Pathogenicity study.
A mouse abscess model of infection was used as described in Chan et al. (7). Results from six mice were recorded, and their significance was determined by the Mann-Whitney test.
Oxygen free-radical resistance assay.
Cells during different phases of growth in CDM were pelleted by centrifugation (5,000 rpm, 10 min), resuspended to a cell density of 5 × 106 CFU ml−1, and incubated at 37°C in either an equal volume of PBS containing 1 mM xanthine plus 0.1 U of xanthine oxidase (external free-radical generation [16]) or 10 mM methyl viologen (internal free-radical generation [20]). Viability was determined by CFU ml−1 on CDM agar.
SOD activity assay.
Cell lysates were prepared by resuspension of cells in lysis buffer (10 mM Tris-HCl [pH 8], 1 mM EDTA, 25 μg of lysostaphin [Sigma]/ml−1), followed by repeated freeze thawing until cell lysis was observed by microscopic examination. One hundred micrograms of soluble protein was loaded on a 12.5% (wt/vol) nondenaturing polyacrylamide gel, and electrophoresis was carried out according to standard procedures (28) but without sodium dodecyl sulphate in the gel or in the electrophoresis buffer. Enzyme activity was visualized by negative staining by the nitroblue tetrazolium method (3). Quantification of SOD activity in cell lysates was determined by the inhibition of the auto-oxidation of pyrogallol as described by Marklund and Marklund (30).
Metal depletion and reconstitution of crude cell extracts.
Metal depletion was performed by dialyzing cell extracts against metal depletion buffer (20 mM 8-hydroxyquinoline, 2.5 M guanidium chloride, 5 mM Tris-HCl [pH 3.8], 0.1 mM EDTA) as described by Kirby et al. (27). Reconstitution of metal-depleted cell extracts with either manganese chloride or ferric and ferrous ammonium sulphate was performed as described by Vasconcelos and Deneer (41).
Acid tolerance and the adaptive response.
Acid tolerance and the acid-adaptive response were determined as described in Chan et al. (7). Briefly, for acid tolerance, cells were resuspended in CDM acidified to pH 2, and viability was determined by CFU ml−1. For the acid-adaptive response, cells were resuspended in CDM (pH 4) for 1 h, prior to determination of acid tolerance (7).
RESULTS AND DISCUSSION
The transposon insertion of SPW1 is within a sodA homologue.
The Tn917-LTV1 insertion mutant, SPW1, was isolated in a previous study, screening for starvation survival mutants of S. aureus (43). Chromosomal DNA flanking the transposon insertion was cloned and sequenced as described in Materials and Methods. Sequence analysis of this region (1,120 bp) by using the Staden and GCG packages (SEQNET; Daresbury Laboratory, Warrington, United Kingdom), revealed that the Tn917 had inserted into an ORF of 226 amino acids which encodes a putative protein of 23 kDa. The Tn917 insertion occurred after residue 54. Database searches revealed that the ORF has sequence similarity to the SOD family of proteins (Fig. 1). Comparison of the amino acid sequence to other bacterial SOD of the Mn2+ type (sodA; E. coli [P00448], Salmonella typhimurium [P43019], Listeria monocytogenes [P28764], Bacillus subtilis [P54375]) and the Fe2+ type (sodB; E. coli [P09157], Pseudomonas putida [P77928], Legionella pneumophila [P31108]) showed that the sequence had greater similarity to the sodA family of Mn-SOD. Thus, the S. aureus gene characterized in this study was designated sodA. The four Mn2+ metal ion binding residues characteristic of Mn-SOD were conserved (34). The greatest identities are to SodA from B. subtilis and L. monocytogenes, with which the S. aureus SodA shows 80% identity over 226 and 203 amino acids, respectively. Additionally the alignment revealed the lack of a 7-amino-acid deletion (position 59 to 66; Fig. 1) conserved amongst the Fe-SOD and the presence of a 1-amino-acid deletion conserved amongst the Mn-SOD. Poyart et al. (36) identified a putative SOD gene from S. aureus RN4220 (a derivative of 8325), using PCR with degenerate primers based on the sequence of several gram-positive Mn-SODs. The alignment of this sequence (Z49245) with the sodA identified in this study demonstrates only 71.3% identity, and therefore they most probably represent different enzymes. Both sequences have residues which are conserved among the Mn-SOD family.
FIG. 1.
Alignment of the S. aureus SodA amino acid sequence with those of E. coli [P00448, P09157], S. typhimurium [P43019], L. pneumophila [P31108], B. subtilis [P54375], P. putida [P77928], L. monocytogenes [P28764], and the putative S. aureus enzyme [S54793] (only partially sequenced). Numbers represent the position of the amino acid sequence. Fe and Mn denote metal iron cofactor of enzyme. Residues in bold are predicted to bind metal iron cofactor. Boxed residues have been predicted to bind Mn2+. Asterisks pinpoint deletions, which discriminate Mn- from Fe-SOD.
The sodA gene is likely to be monocistronic, as putative Rho-independent terminators are present up- and downstream of the gene (ΔG−6.2 and −14.4 kJ mol−1, respectively, calculated according to Tinoco et al. [39]).
S. aureus SodA is a manganese-requiring enzyme.
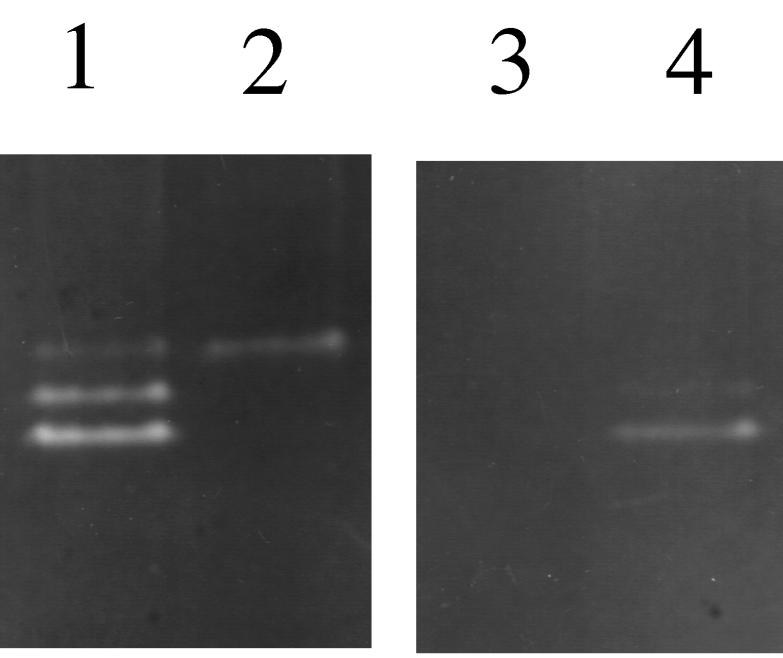
The SOD activity of S. aureus 8325-4 was analyzed by nondenaturing polyacrylamide gel electrophoresis staining for SOD activity (Fig. 2, lane 1). Three bands of SOD activity were identified, with the upper band demonstrating the least activity. Comparison of whole-cell lysates of SPW1 with S. aureus 8325-4 showed that the lower two bands of activity were absent in SPW1 (Fig. 2, lanes 1 and 2). To determine the metal ion requirement of the superoxide dismutases, whole-cell lysates of S. aureus 8325-4 were treated to remove all metal ions. As can be seen in Fig. 2, lane 3, this abolished all SOD activity from the lysate. Replacement of the metal ions with Mn2+ restored activity of the lower two bands, but not the upper band of activity, establishing that they require Mn2+ for activity (Fig. 2, lane 4). The activity of the upper band was sensitive to H2O2 (data not shown), which is typical of Fe-SODs, although the addition of Fe2+ did not restore this activity to metal ion-depleted cell lysates of S. aureus 8325-4.
FIG. 2.
SOD activity of whole-cell extracts of S. aureus 8325-4 and SPW1. Extracts were separated on a 12.5% (wt/vol) nondenaturing polyacrylamide gel, and stained for SOD activity. Lane 1, 8325-4; lane 2, SPW1; lane 3, 8325-4 metal ion depleted; lane 4, 8325-4 metal ion depleted, reconstituted with Mn2+.
Since SODs are dimeric proteins, it may be that the two activities associated with SodA are due to a homodimer and a heterodimer with the proposed low-level, constitutive, Fe-SOD. This type of activity profile has been demonstrated in E. coli (9) and would explain why inactivation of sodA results in the loss of two bands of activity.
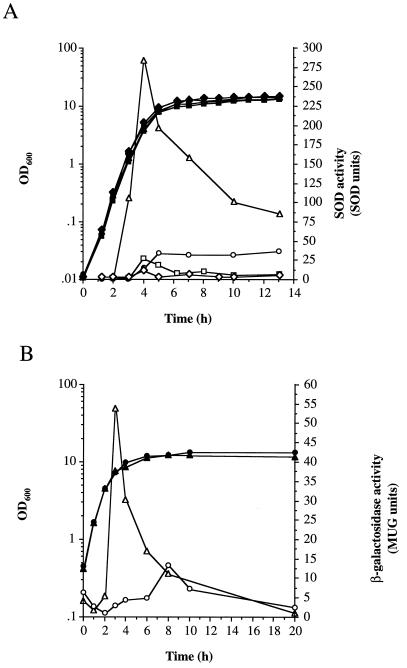
SOD activity and sodA expression during growth of S. aureus 8325-4.
SOD activity of bacterial cell lysates during different phases of growth (Fig. 3A) was determined by the inhibition of the auto-oxidation of pyrogallol (30). Activity during the exponential phase of growth was low (2 U) but increased 18-fold (36 U) upon entry into postexponential phase (5 h), remaining high throughout stationary phase (13 h). This increase was observed only when cultures were incubated with high aeration; low aeration resulted in a 10-fold increase in SOD activity after 4 h, which rapidly fell to 8 U after 6 h incubation. The stationary-phase increase in SOD activity was dependent on sodA, since SPW1 demonstrated only a constitutive basal level of SOD activity throughout growth (Fig. 3A). The basal level of SOD observed in the sodA mutant is probably due to the activity of the putative Fe-dependent superoxide dismutase, which would protect the cell from low-level O2..
FIG. 3.
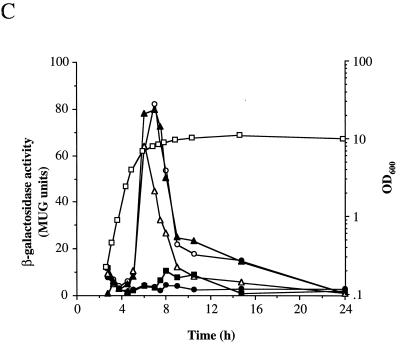
SOD activity and expression of sodA during growth and the effect of methyl viologen. (A) Total SOD activity. 8325-4 (○,●,▵,▴,□,■) and SPW1 (⧫,◊) were grown under high (▵,▴,○,●,⧫,◊) or low (□,■) aeration in BHI or in BHI with 10 μM methyl viologen (▵,▴). Closed symbols represent OD600, and open symbols represent SOD activity as described in Materials and Methods. (B) Expression of sodA. SPW1 was grown in BHI with high aeration with (▵,▴) or without (○,●) 10 μM methyl viologen. Growth was measured by OD600 (closed symbols) and β-galactosidase activity (open symbols) as described in Materials and Methods. (C) Effect of growth phase on methyl viologen induction of sodA. Expression of sodA was measured during growth of SPW1 in BHI (●) and in BHI with 10 μM methyl viologen added 0 h (○), 2.75 h (▴), 4.5 h (▵), and 7 h (■) after inoculation. Cells were incubated with high aeration, and expression was measured as described in Materials and Methods. Growth as measured by OD600 is shown for one representative culture (□).
The SPW1 Tn917-LTV1 insertion generated a lacZ transcriptional fusion with sodA, enabling its expression to be monitored throughout growth, by measuring the levels of β-galactosidase activity. Expression of sodA was low during exponential-phase growth (1 U at 2 h) but increased 13-fold (13 U at 8 h) during the stationary phase, when cultures were incubated with high aeration, correlating with the SOD activity results. The fact that SPW1 is a sodA mutant may affect the finite levels of expression. Culture under conditions of high aeration may result in increased internal superoxide levels, which leads to oxidative stress for the cell. Expression of sodA is induced to deal with this potentially lethal assault.
Increased levels of SOD during stationary-phase growth have been observed in many bacteria, including E. coli, B. subtilis, L. monocytogenes, and Legionella (5, 25, 38, 41). The increase in SOD in cells during stationary-phase growth may not result from an increase in O2. generation but may be a mechanism to prevent the accumulation of O2. damage to proteins, since damaged proteins will accumulate due to the low rate of de novo protein synthesis and, hence, low protein turnover (33). Since SPW1 (sodA) is less able to survive in the stationary phase of growth, as are SOD-deficient mutants of E. coli (5) and Legionella (38), the protective role of SOD during starvation survival is supported.
Induction of SOD activity and sodA expression by methyl viologen.
Methyl viologen, when accumulated within bacterial cells, is a potent generator of O2. via redox cycling (20). When methyl viologen was incubated with S. aureus 8325-4, an approximately ninefold maximal increase in SOD activity over the untreated control was observed (Fig. 3A). This correlated with an approximately fourfold increase in sodA expression compared to the untreated culture (Fig. 3B). Interestingly, the methyl viologen induction of SOD activity and sodA expression was growth-phase dependent, occurring specifically during the post-exponential phase, irrespective of the time of addition of methyl viologen to the culture (Fig. 3C and data not shown). If, however, methyl viologen was added after stationary phase was reached, there was no induction of sodA expression.
In E. coli, addition of methyl viologen leads to an immediate induction of sodA expression (40), whereas in B. subtilis the addition of methyl viologen does not induce sodA at all (25). The range of methyl viologen effects could be due to differences in permeability.
Role of sodA in oxidative stress resistance.
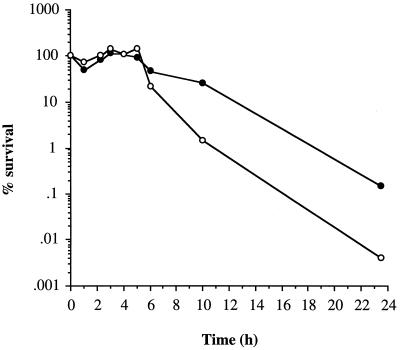
Oxidative stress can be induced internally, within bacterial cells, by the addition of methyl viologen, while external free radicals can be generated by the incubation of cells with xanthine and xanthine oxidase (16). Surprisingly, the addition of methyl viologen to exponential-phase cells had no significant effect on the growth rate but did lead to a slight reduction in yield upon entry to stationary phase of S. aureus 8325-4 and SPW1 (data not shown). The addition of methyl viologen to stationary-phase cells (8 h postexponential phase; glucose limited) of 8325-4 had no effect on the rate of loss of viability (data not shown) but caused a 100-fold reduction in CFU of SPW1 after 24 h compared to the untreated control (Fig. 4). Untreated SPW1 showed the same death kinetics as 8325-4 over the duration of the experiment. Incubation of S. aureus 8325-4 or SPW1 in PBS containing xanthine and xanthine oxidase had no effect on the rate of growth, yield, or loss of cell viability irrespective of the growth phase of the cells (data not shown). Thus, lack of SodA results in increased sensitivity to internally generated oxidative stress, but this is apparent only in the stationary phase, which coincides with maximal sodA expression.
FIG. 4.
Effect of methyl viologen on cell survival. Cells (8 h postexponential phase) of SPW1 were resuspended in PBS (●) or in PBS containing 10 μM methyl viologen (○). Cell viability was determined by plating on CDM agar.
Role of sodA in starvation survival and recovery from starvation.
SPW1 was initially isolated from a screen of mutants that demonstrated a decreased ability to survive amino acid starvation (43). Further analysis of the starvation defect demonstrated that the increased rate of loss of viability was dependent on the aeration of the culture during starvation. SPW1 demonstrated identical starvation kinetics to 8325-4 in amino acid-limiting CDM when the cultures were incubated statically, whereas SPW1 lost viability at a faster rate when the cultures were incubated with shaking (43; data not shown). The inactivation of sodA in SPW1 had no effect on the ability of the starved cells to recover and resume growth after starvation (data not shown), by standard recovery protocols (10).
SodA is involved in acid tolerance and the acid-adaptive response and is induced by acid.
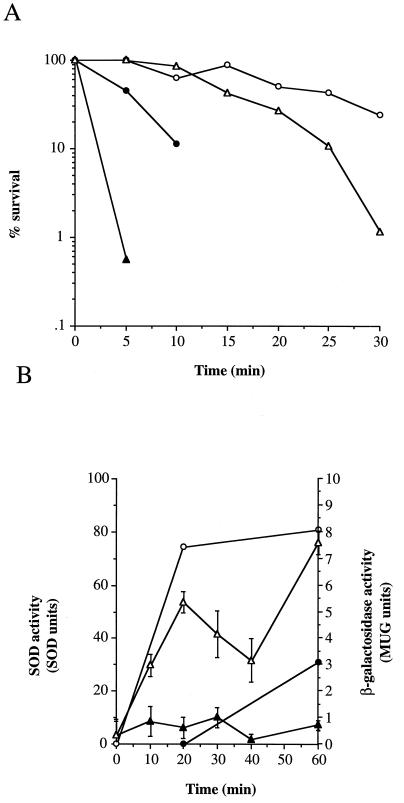
We have previously shown that S. aureus develops increased resistance to acid stress upon starvation or by incubation of cells at a lower but nonlethal pH as part of the stress response (7). Exponential-phase cells of S. aureus 8325-4 are sensitive to CDM acidified to pH 2 but become tolerant when adapted for 1 h at pH 4 prior to challenge at pH 2 (Fig. 5A; 7). SodA is involved in acid resistance, since exponential-phase cells of SPW1 were less tolerant to CDM (pH 2) than S. aureus 8325-4 (>103-log loss of viability after 10 min compared to 1-log loss, respectively; Fig. 5A). SodA also has a role in the acid-adaptive response, as sodA showed a >2-log drop in viability compared to a 0.6-log drop by 8325-4 (30 min, pH 2 treatment after pH 4 adaptation [Fig. 5A]).
FIG. 5.
Role of SodA in acid tolerance and adaptation. (A) S. aureus 8325-4 (●,○) and SPW1 (▴,▵) were grown and treated with acid as described in Materials and Methods. Exponential-phase cells were either incubated at pH 4 (37°C) for 1 h (○,▵) or untreated (●,▴) prior to pH 2 assault. Viability was determined by plating on CDM agar. Results are representative of three independent experiments showing less than fivefold variability. (B) Induction of sodA expression and SOD activity during acid adaptation. Exponential-phase cells (OD600 = 0.8) were resuspended in CDM pH 7 (●,▴) or pH 4 (○,▵). Expression of sodA (SPW1, ▴,▵) and total SOD activity (8325-4, ●,○) were determined as described in Materials and Methods. Error bars represent the standard deviations of three independent samples.
Exponential-phase cells (optical density at 600 nm [OD600] = 0.8) of S. aureus 8325-4 develop increased acid resistance when incubated at pH 4 prior to challenge at pH 2 (Fig. 5A). The expression of sodA was compared between cells resuspended at pH 4 (adaptive) and pH 7 (nonadaptive) and is shown in Fig. 5B. The level of sodA expression remained relatively constant at pH 7 (0.2 to 0.8 MUG U), while at pH 4, the level increased almost 10-fold to 7.6 MUG U after 60 min. When total levels of SOD activity were measured in S. aureus 8325-4, a similar rise in SOD activity was observed (Fig. 5B).
How sodA is involved in acid resistance is unknown, but acid stress may lead to uncoupling of the respiratory chain, resulting in the increased internal synthesis of O2.. Alternatively, or additionally, inactivation of sodA may result in increased cellular damage from the effects of O2., rendering the cell more sensitive to acid stress.
Regulation of sodA expression.
In order to identify potential regulators of sodA in S. aureus, we examined sodA expression in strains defective in several global gene regulators. SigB is an alternative sigma factor (46), which is preferentially expressed in the stationary phase and has a role in acid and oxidative stress resistance (7). The global regulators of toxin production, agr and sarA (8, 35), are also preferentially expressed during stationary phase. We have recently shown that sigB expression is partially controlled by SarA (7). The sodA::lacZ fusion was introduced into a set of strains bearing defined mutations in sigB, agr, or sarA. No alteration in the pattern of sodA::lacZ expression was observed in any of the backgrounds (data not shown), during growth or induction by methyl viologen. In E. coli, superoxide stress induces the soxRS regulon, resulting in the synthesis of SodA and DNA repair mechanisms (13). To date, there is no evidence for a comparable mechanism in S. aureus.
Role of sodA in pathogenicity.
Cu/Zn-SODs have been shown to be an important virulence factors in several pathogens, including E. coli, Neisseria meningitidis, and Salmonella typhimurium (1, 14, 45). It has been suggested that SOD is involved in protecting S. aureus from neutrophils, since it was shown that the attachment of SOD to the surface of S. aureus decreased the rate of killing (19). A mouse abscess model of S. aureus infection (7) was therefore used to determine the contribution of sodA to pathogenicity. No significant difference between S. aureus 8325-4 and SPW1 was observed in regard to the size of the lesion formed or the number of bacteria that were recovered from the lesion (data not shown).
Interestingly, SodA of Haemophilus influenzae has been shown to play a role in nasopharyngeal colonization (12), which is also a preferred site of S. aureus colonization. It is therefore possible that SodA may be important in the successful colonization of specific niches.
The resistance to oxidative stress involves the interplay between several resistance mechanisms and components. In order to understand how S. aureus responds and adapts to oxidative stress inside and outside the host, it is important to identify all the components of the resistance mechanism and to determine how they are regulated in response to potentially lethal stresses. It is by such exquisite mechanisms of adaptation that S. aureus has become such a versatile and successful pathogen.
ACKNOWLEDGMENTS
This work was supported by the Royal Society (S.J.F.), BBSRC (S.P.W.), and the BBSRC/Celsis Connect Program (M.O.C.).
REFERENCES
- 1.Battistoni A, Donnarumma G, Greco R, Valenti P, Rotilio G. Overexpression of a hydrogen peroxide-resistant periplasmic Cu, Zn superoxide dismutase protects Escherichia coli from macrophage killing. Biochem Biophys Res Commun. 1998;243:804–807. doi: 10.1006/bbrc.1998.8182. [DOI] [PubMed] [Google Scholar]
- 2.Beaman L, Beaman B L. The role of oxygen and its derivatives in microbial pathogenesis. Annu Rev Microbiol. 1984;38:27–48. doi: 10.1146/annurev.mi.38.100184.000331. [DOI] [PubMed] [Google Scholar]
- 3.Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. doi: 10.1016/0003-2697(71)90370-8. [DOI] [PubMed] [Google Scholar]
- 4.Benov L T, Fridovich I. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J Biol Chem. 1994;269:25310–25314. [PubMed] [Google Scholar]
- 5.Benov L, Fridovich I. A superoxide dismutase mimic protects sodA sodB Escherichia coli against aerobic heating and stationary-phase death. Arch Biochem Biophys. 1995;332:291–294. doi: 10.1006/abbi.1995.1465. [DOI] [PubMed] [Google Scholar]
- 6.Chan P F, Foster S J. The role of environmental factors in the regulation of virulence determinant expression in Staphylococcus aureus 8325-4. Microbiology. 1998;144:2469–2479. doi: 10.1099/00221287-144-9-2469. [DOI] [PubMed] [Google Scholar]
- 7.Chan P F, Foster S J, Ingham E, Clements M O. The alternative sigma factor, ςB controls the environmental stress response but not starvation survival or pathogenicity in a mouse abscess model, in Staphylococcus aureus. J Bacteriol. 1998;180:6082–6089. doi: 10.1128/jb.180.23.6082-6089.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clare D A, Blum J, Fridovich I. A hybrid superoxide dismutase containing both functional iron and manganese. J Biol Chem. 1984;259:5932–5936. [PubMed] [Google Scholar]
- 10.Clements M O, Foster S J. Starvation-recovery of Staphylococcus aureus 8325-4. Microbiology. 1998;144:1755–1763. doi: 10.1099/00221287-144-7-1755. [DOI] [PubMed] [Google Scholar]
- 11.Clements M O, Watson S P, Poole R K, Foster S J. CtaA of Staphylococcus aureus is required for starvation survival, recovery, and cytochrome biosynthesis. J Bacteriol. 1999;181:501–507. doi: 10.1128/jb.181.2.501-507.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Mello R A, Langford P R, Kroll J S. Role of bacterial Mn-cofactored superoxide dismutase in oxidative stress response, nasopharyngeal colonisation, and sustained bacteremia caused by Haemophilus influenzae type b. Infect Immun. 1997;65:2700–2706. doi: 10.1128/iai.65.7.2700-2706.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demple B. Regulation of bacterial oxidative stress genes. Annu Rev Genet. 1991;25:315–337. doi: 10.1146/annurev.ge.25.120191.001531. [DOI] [PubMed] [Google Scholar]
- 14.Farrant J L, Sansone A, Canvin J R, Pallen M J, Langford P R, Wallis T S, Dougan G, Kroll J S. Bacterial copper and zinc-cofactored superoxide dismutase contributes to the pathogenesis of systemic salmonellosis. Mol Microbiol. 1997;25:785–796. doi: 10.1046/j.1365-2958.1997.5151877.x. [DOI] [PubMed] [Google Scholar]
- 15.Foster S J. Molecular characterisation and functional analysis of the major autolysin of Staphylococcus aureus 8325-4. J Bacteriol. 1995;177:5723–5725. doi: 10.1128/jb.177.19.5723-5725.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fridovich I. Quantitative aspects of the production of superoxide anion radical by milk xanthine oxidase. J Biol Chem. 1970;245:4053–4057. [PubMed] [Google Scholar]
- 17.Fridovich I. The biology of oxygen radicals. Science. 1978;201:875–880. doi: 10.1126/science.210504. [DOI] [PubMed] [Google Scholar]
- 18.Fridovich I. Superoxide radical and superoxide dismutases. Annu Rev Biochem. 1995;64:97–112. doi: 10.1146/annurev.bi.64.070195.000525. [DOI] [PubMed] [Google Scholar]
- 19.Hampton M B, Kettle A J, Winterbourn C C. Involvement of superoxide and myeloperoxidase in oxygen-dependent killing of Staphylococcus aureus by neutrophils. Infect Immun. 1996;64:3512–3517. doi: 10.1128/iai.64.9.3512-3517.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassan H M, Fridovich I. Superoxide radicals and the oxygen enhancement of the toxicity of paraquat in Escherichia coli. J Bacteriol. 1978;253:8143–8148. [PubMed] [Google Scholar]
- 21.Hopkin K A, Papazian M A, Steinman H M. Functional differences between manganese and iron superoxide dismutases in Escherichia coli K-12. J Bacteriol. 1992;267:24253–24258. [PubMed] [Google Scholar]
- 22.Hussain M, Hastings J G M, White P J. A chemically defined medium for slime production by coagulase-negative staphylococci. J Med Microbiol. 1991;34:143–147. doi: 10.1099/00222615-34-3-143. [DOI] [PubMed] [Google Scholar]
- 23.Imlay J A, Linn S. DNA damage and oxygen radical toxicity. Science. 1988;240:1302–1309. doi: 10.1126/science.3287616. [DOI] [PubMed] [Google Scholar]
- 24.Imlay K R, Imlay J A. Cloning and analysis of sodC, encoding the copper-zinc superoxide dismutase of Escherichia coli. J Bacteriol. 1996;178:2564–2571. doi: 10.1128/jb.178.9.2564-2571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inaoka T, Matsumura Y, Tsuchido T. Molecular cloning and nucleotide sequence of the superoxide dismutase gene and characterization of its product from Bacillus subtilis. J Bacteriol. 1998;180:3697–3703. doi: 10.1128/jb.180.14.3697-3703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim E-J, Chung H-J, Suh B, Hah Y C, Roe J-H. Transcriptional and post-transcriptional regulation by nickel of sodN gene encoding nickel-containing superoxide dismutase from Streptomyces coelicolor Muller. Mol Microbiol. 1998;27:187–195. doi: 10.1046/j.1365-2958.1998.00674.x. [DOI] [PubMed] [Google Scholar]
- 27.Kirby T, Blum J, Kahane I, Fridovich I. Distinguishing between Mn-containing and Fe-containing superoxide dismutases in crude extracts of cells. Arch Biochem Biophys. 1980;201:551–555. doi: 10.1016/0003-9861(80)90544-5. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Langford P R, Loynds B M, Kroll J S. Copper-zinc superoxide dismutase in Haemophilus species. J Gen Microbiol. 1992;138:517–522. doi: 10.1099/00221287-138-3-517. [DOI] [PubMed] [Google Scholar]
- 30.Marklund S, Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem. 1974;47:469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- 31.Novick R P. Properties of a high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–156. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 32.Novick R P. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 33.Nystrom T, Gustavsson N. Maintenance energy requirement: what is required for stasis survival of Escherichia coli? Biochem Biophys Acta. 1998;1365:225–231. doi: 10.1016/s0005-2728(98)00072-3. [DOI] [PubMed] [Google Scholar]
- 34.Parker M W, Blake C C F. Iron- and manganese-containing superoxide dismutases can be distinguished by analysis of their primary structures. FEBS Lett. 1988;229:377–382. doi: 10.1016/0014-5793(88)81160-8. [DOI] [PubMed] [Google Scholar]
- 35.Peng H-L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterisation, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poyart C, Berche P, Trieu-Cuot P. Characterisation of superoxide dismutase genes from Gram-positive bacteria by polymerase chain reaction using degenerate primers. FEMS Microbiol Lett. 1995;131:41–45. doi: 10.1016/0378-1097(95)00232-t. [DOI] [PubMed] [Google Scholar]
- 37.Segal A W. The electron transport chain of the microbiocidal oxidase of phagocytic cells and its involvement in the molecular pathology of chronic granulomatous disease. J Clin Investig. 1989;83:1785–1793. doi: 10.1172/JCI114083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St. John G, Steinman H M. Periplasmic copper-zinc superoxide dismutase of Legionella pneumophilia: role in stationary-phase survival. J Bacteriol. 1996;178:1578–1584. doi: 10.1128/jb.178.6.1578-1584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tinoco I, Borer P N, Dengler B, Irvine M, Uhlenbeck M C, Crothers D M, Gralla J. Improved estimation of secondary structure of ribonucleic acids. Nature. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 40.Touati D. Transcriptional and posttranscriptional regulation of manganese superoxide dismutase biosynthesis in Escherichia coli, studied with operon and protein fusions. J Bacteriol. 1988;170:2511–2520. doi: 10.1128/jb.170.6.2511-2520.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vasconcelos J A P, Deneer H G. Expression of superoxide dismutase in Listeria monocytogenes. Appl Environ Microbiol. 1994;60:2360–2366. doi: 10.1128/aem.60.7.2360-2366.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waldvogel F A. Staphylococcus aureus (including toxic shock syndrome) In: Mandell G L, Douglas R G, Bennet J E, editors. Principles and practices of infectious diseases. New York, N.Y: Churchill Livingstone; 1995. pp. 1489–1510. [Google Scholar]
- 43.Watson S P, Antonio M, Foster S J. Isolation and characterisation of Staphylococcus aureus starvation-induced, stationary-phase mutants defective in survival or recovery. Microbiology. 1998;144:3159–3169. doi: 10.1099/00221287-144-11-3159. [DOI] [PubMed] [Google Scholar]
- 44.Watson S P, Clements M O, Foster S J. Characterisation of the starvation-survival response of Staphylococcus aureus. J Bacteriol. 1998;180:1750–1758. doi: 10.1128/jb.180.7.1750-1758.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilks K E, Dunn K L R, Farrant J L, Reddin K M, Gorringe A R, Langford P R, Kroll J S. Periplasmic superoxide dismutase in meningococcal pathogenicity. Infect Immun. 1998;66:213–217. doi: 10.1128/iai.66.1.213-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu S, De Lencastre H, Tomasz A. Sigma-B, putative operon encoding alternative sigma factor of Staphylococcus aureus RNA polymerase: molecular cloning and DNA sequencing. J Bacteriol. 1996;178:6036–6042. doi: 10.1128/jb.178.20.6036-6042.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]