Abstract
Context
Chronic kidney disease (CKD) causes multiple interrelated disturbances in mineral metabolism. Genetic studies in the general population have identified common genetic variants associated with circulating phosphate, calcium, parathyroid hormone (PTH), and fibroblast growth factor 23 (FGF23).
Objective
In this study we aimed to discover genetic variants associated with circulating mineral markers in CKD.
Methods
We conducted candidate single-nucleotide variation (SNV) analysis in 3027 participants in the multiethnic Chronic Renal Insufficiency Cohort (CRIC) to determine the associations between SNVs and circulating levels of mineral markers.
Results
SNVs adjacent to or within genes encoding the regulator of G protein–coupled signaling 14 (RGS14) and the calcium-sensing receptor (CASR) were associated with levels of mineral metabolites. The strongest associations (P < .001) were at rs4074995 (RGS14) for phosphate (0.09 mg/dL lower per minor allele) and FGF23 (8.6% lower), and at rs1801725 (CASR) for calcium (0.12 mg/dL higher). In addition, the prevalence of hyperparathyroidism differed by rs4074995 (RGS14) genotype (chi-square P < .0001). Differential inheritance by race was noted for the minor allele of RGS14. Expression quantitative loci (eQTL) analysis showed that rs4074995 was associated with lower RGS14 gene expression in glomeruli (P = 1.03 × 10–11) and tubules (P = 4.0 × 10–4).
Conclusion
We evaluated genetic variants associated with mineral metabolism markers in a CKD population. Participants with CKD and the minor allele of rs4074995 (RGS14) had lower phosphorus, lower plasma FGF23, and lower prevalence of hyperparathyroidism. The minor allele of RGS14 was also associated with lower gene expression in the kidney. Further studies are needed to elucidate the effect of rs4074995 on the pathogenesis of disordered mineral metabolism in CKD.
Keywords: CKD-MBD, genetics, mineral metabolism, chronic kidney disease
Mineral metabolism disturbances develop in nearly all individuals with chronic kidney disease (CKD) and are associated with fracture risk, soft-tissue and vascular calcification, and cardiovascular-related mortality (1). Chronic kidney disease–mineral bone disorder (CKD-MBD) is monitored closely with the goal of maintaining optimal circulating levels of intact parathyroid hormone (PTH), calcium, phosphate, and 25-hydroxyvitamin D. Fibroblast growth factor 23 (FGF23), a more recently recognized marker of mineral metabolism, is not typically monitored in clinical practice, but is widely considered an integral component of CKD-MBD (2).
Genome-wide association studies (GWAS) of circulating mineral metabolism markers in the general population have identified common variants associated with circulating PTH, calcium, phosphate, and FGF23. Many of these variants reside within or near genes encoding regulators of vitamin D, calcium, and phosphate homeostasis. For example, variants have been discovered within CYP24A1 (cytochrome P450 family 24 subfamily A member 1), CASR (the calcium-sensing receptor), and RGS14/SLC34A1 (regulator of G protein signaling 14/Solute carrier family 34 member 1) (3-6). CYP24A1 encodes 24-hydroxylase, an enzyme responsible for the catabolism of both 25-hydrodroxyvitamin D and 1,25-dihydroxyvitamin D. Its actions are induced by the high FGF23 environment of CKD and contribute to decreased vitamin D levels and secondary hyperparathyroidism (7). CASR is found on the parathyroid glands and alters PTH secretion in response to circulating calcium levels. It is downregulated in CKD and serves as a target for therapeutic intervention (8, 9). Although not previously implicated in CKD, the protein product of RGS14 and the protein product of SLC34A1, NaPi2a, are in close proximity within the renal tubule and both function in regulating urinary phosphate excretion (10-12). The effect of these variants among individuals with CKD, who have the most pronounced disturbances in mineral metabolism, is unknown. On the one hand, metabolic drivers arising from the loss of kidney function, including phosphate retention and impaired calcitriol synthesis, may overwhelm relatively modest genetic effects. On the other hand, the unique environment of CKD may augment specific genetic associations or identify previously unrecognized genetic effects. Moreover, it is possible that genetic associations may differ according to racial background, given known racial-ethnic differences in PTH and vitamin D metabolism in patients with CKD (13-17). Elucidating these associations, specifically within a CKD population, may provide further insight into the mineral metabolism differences noted by race. In addition, understanding the contribution of genetics to mineral marker levels is critically important given that mineral metabolism abnormalities are systematically evaluated and treated among patients with CKD. As such, a better understanding of these genetic associations will assist in the recognition of those at risk for abnormalities in mineral metabolism and allow prediction of the treatment response according to genotype in the future, thereby applying principles of precision medicine to the treatment of CKD-MBD.
We conducted a candidate single-nucleotide variation (SNV, formerly single-nucleotide polymorphism [SNP]) analysis of mineral metabolism markers in a CKD cohort to evaluate known genetic variants and to test for potential differences by kidney function. In addition, we characterized racial differences in the inheritance pattern of genetic variants and their impacts on circulating levels of mineral metabolism markers in CKD.
Materials and Methods
Study Population
The Chronic Renal Insufficiency Cohort Study (CRIC) is an observational study of adults aged 21 to 74 years that seeks to determine risk factors for cardiovascular disease (CVD) and CKD progression (18, 19). Since 2003, participants with an estimated glomerular filtration rate (eGFR) of 20 to 70 mL/min/1.73 m2 have been recruited across 13 sites spread throughout the United States. Participants underwent extensive clinical evaluation at 6-month intervals until death or withdrawal of informed consent. Key exclusions included individuals with polycystic kidney disease, renal cancer, organ transplantation, recent immunosuppressive therapy or chemotherapy, New York Heart Association class III to IV heart failure, dialysis treatment lasting longer than 1 month, cirrhosis, myeloma, HIV, institutionalization, pregnancy, inability to consent, and enrollment in other studies.
Genotyping and Imputation
For the present study, we used publicly available genotype data obtained from the Database of Genotypes and Phenotypes (dbGap; study accession phs000524.v1.p1). Among all 3939 CRIC participants, 3635 participants were previously genotyped using the Illumina Human Omni 1-Quad Array Platform, and 3527 samples met quality control standards, as previously described (20). After exclusion of self-reported race other than Black or White, 3027 participants who met quality control standards remained. When a candidate SNV was not among the directly genotyped SNVs, imputation with reference to the 1000 Genomes Phase 3 genotypes was performed using the University of Michigan Imputation Server (21). Only SNVs with imputation R2 greater than 0.80 were retained for subsequent analysis.
Measurement of Mineral Markers
Levels of serum calcium, phosphate, PTH, and plasma FGF23 were measured in previous studies of the CRIC population, and these values were obtained from dbGap (22). FGF23 levels were measured using a second-generation c-terminal assay (Immutopics) at a central CRIC laboratory. Plasma PTH levels were measured using the intact assay (Scantibodies) (22). Levels of 25-hydroxyvitamin D, 1,25-dihydroxyvitamin D, and 24,25-dihydroxycholecalciferol were unavailable.
Statistical Analysis
The associations between genotypes and serum calcium, phosphate, PTH, and plasma FGF23 were performed using sequentially adjusted linear regression models. Owing to skewed distributions, PTH and FGF23 were natural log-transformed. The predictor variable of interest for all models was SNV genotype under an additive genetic model. Covariates for model 1 included age, sex, and the first 10 principal components of ancestry. Model 2 was additionally adjusted for body mass index (BMI), eGFR, and, to account for potential nonlinear effects on mineral metabolites, eGFR squared. Model 3 additionally included the potential intermediate effects of calcium, phosphate, PTH, and FGF23, with each model excluding the outcome variable as appropriate. All analyses were stratified according to self-reported race, and subsequently combined into transethnic meta-analysis results using inverse-variance weighting implemented in the software package METAL (23). SNVs were excluded based on imputation quality score less than 0.3, transethnic minor allele frequency (MAF) less than 0.05, call rate less than 95% and Hardy-Weinberg equilibrium P value less than 10–5. Data processing and statistical analyses were performed with PLINK v1.9 and R (version 4.1.0) (24).
Expression Quantitative Loci Analysis
To further investigate the influence of SNVs on gene expression in the kidney, we performed expression quantitative loci (eQTL) analysis using renal compartment–specific tissue data sets (25). In a genomic data set of 151 human kidneys with 121 tubule and 119 glomerulus samples (~70% from White individuals and 18% from Black individuals), we performed compartment-based eQTL analysis separately for the 2 compartments by using a linear model and a ± 1-Mb window around the transcription start site. In addition, to estimate the posterior probabilities that mineral metabolism phenotypes and gene expression share common causal variants in the gene region, we conducted colocalization analyses using coloc (v.3.2.1).
Selection of Single-Nucleotide Variations for Testing in Chronic Kidney Disease
Candidate SNVs were chosen based on previously reported genome-wide associations with mineral metabolism markers in the general population (3-6, 26). The first population-based study by Kestenbaum et al (6) detected SNVs associated with serum phosphate in a population of 16 264 participants of European Ancestry with an eGFR greater than 45 mL/min/1.73 m2. The second population-based study by Robinson-Cohen et al (4) performed a GWAS of FGF23 in 16 624 participants of European ancestry with an eGFR greater than or equal to 30 mL/min/1.73 m2. A third GWAS of circulating PTH by Robinson-Cohen and colleagues (3) was performed in 29 155 participants of European Ancestry with an eGFR greater than or equal to 30 mL/min/1.73 m2. Finally, O’Seaghdha et al (5) performed a genome-wide association meta-analysis of calcium in a discovery cohort of 39 400 European individuals. Kidney function was not specified in this cohort. When SNVs from the general population were unavailable in the CRIC cohort, a proxy SNV with the strongest correlation (minimum R2 = 0.7) was chosen. When a proxy SNV with adequate linkage disequilibrium was unavailable, the SNV of interest was excluded from the analysis. Phenotyping and genotyping protocols of each study have been previously described (3-6, 26). Bonferroni correction was applied to each marker based on the number of SNVs tested for a given marker. P values less than the Bonferroni-corrected P value for each marker were considered statistically significant for regression analyses.
Results
Population Characteristics
The study population consisted of 3027 participants from the Chronic Renal Insufficiency Cohort (CRIC) with available genotype and phenotype data (Table 1). The mean (SD) age of the cohort was 58.5 (10.9) years, 45% were female and 46% self-identified with Black race. Nearly half had a diagnosis of diabetes and 34% had prevalent CVD at baseline. The mean (SD) eGFR was 44.8 (14.8) mL/min/1.73 m2 with a range of 13 to 104 mL/min/1.73 m2. The albumin-to-creatinine ratio and PTH levels were higher in Black individuals, which also demonstrated a slightly higher prevalence of diabetes. A total of 39% of the cohort had a serum PTH concentration at or above the upper limit of normal (65 pg/mL).
Table 1.
Baseline characteristics of genotyped participants in the Chronic Renal Insufficiency Cohort Study
| Characteristic | Overall | White participants | Black participants | P |
|---|---|---|---|---|
| No. | 3027 | 1635 | 1392 | |
| Age, y | 58.5 (10.9) | 58.9 (11.0) | 58.1 (10.7) | .25 |
| Female, n (%) | 1358 (44.8) | 649 (39.7) | 709 (50.9) | < .01 |
| Body mass index | 32.3 (7.9) | 31.2 (7.3) | 33.6 (8.4) | < .01 |
| Prevalent diabetes, n (%) | 1427 (47.1) | 709 (43.4) | 718 (51.6) | < .01 |
| Prevalent CVD, n (%) | 1038 (34.3) | 503 (30.8) | 535 (38.4) | < .01 |
| eGFR, mL/min/1.73 m2 | 44.8 (14.8) | 45.4 (14.7) | 44.1 (14.9) | .12 |
| Systolic blood pressure, mm Hg | 127.8 (21.9) | 123.2 (19.4) | 133.1 (23.4) | < .01 |
| Diastolic blood pressure, mm Hg | 71.4 (12.9) | 69.3 (11.5) | 73.9 (14.1) | < .01 |
| Albumin-to-creatinine ratio, mg/g | 44.3 (7.9-391.4) | 29.6 (6.4-308.6) | 75.0 (11.3-513.7) | < .01 |
| Calcium, mg/dL | 9.2 (0.5) | 9.2 (0.5) | 9.2 (0.5) | .24 |
| Phosphate, mg/dL | 3.7 (0.7) | 3.6 (0.7) | 3.8 (0.7) | < .01 |
| Parathyroid hormone, pg/mL | 53 (34-87) | 45 (31-71) | 66 (40-111) | < .01 |
| FGF23, RU/mL | 144 (95-237) | 141 (94- 222) | 149 (96-253) | .02 |
Mean (SD), median (IQR), or n (%), as appropriate.
Abbreviations: CVD, cardiovascular disease; eGFR, estimated glomerular filtration rate; FGF23, fibroblast growth factor 23.
Bolded values are those reaching significance.
Association of Genetic Single-Nucleotide Variations With Mineral Metabolism Markers
Phosphate
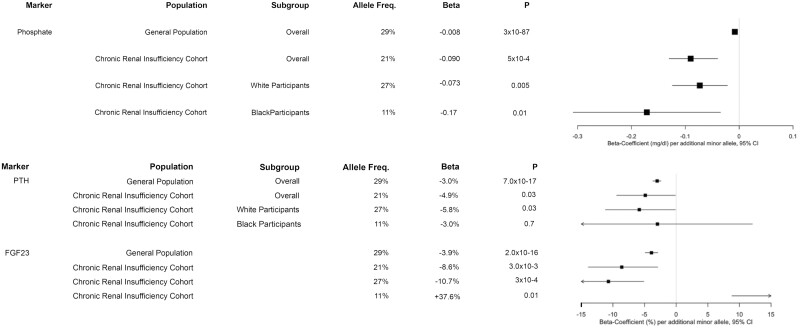
Ten SNVs found to be significantly associated with serum phosphate in the general population underwent replication in this CKD population (Table 2) (6). In analyses including all participants regardless of race, rs4074995 (located within the intronic region of the protein-coding gene for the regulator of G protein signaling 14 [RGS14]) was significantly associated with phosphate. In model 2, each minor allele was associated with 0.09 mg/dL lower phosphate (95% CI, –0.13 to –0.04; P = 5 × 10–4) (Fig. 1). This association persisted in White participants after stratification by race, where each minor allele was associated with 0.09 mg/dL lower phosphate (95% CI, –0.14 to –0.04; P = 9 × 10–4) in model 1 (Supplementary Tables S1 and S2) (27). After further adjustment for GFR, BMI, calcium, PTH, and FGF23, this association was no longer significant. The direction of effect was consistent with the general population, but the magnitude of the association was greater in CKD (see Fig. 1). Mean and median values of mineral markers by genotype demonstrate lower serum phosphate as well as higher 24-hour urine phosphate and fractional excretion of phosphate for the minor allele of RGS14 (Table 3).
Table 2.
Additive model association results for phosphate among all individuals (n = 3027)
| General White population (6, 26) | CRIC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SNV | Host/Adjacent gene | Minor allele | MAF | Model 1c | Model 2c | Model 3c | |||||
| β (SE)a | P | β (95% CI) | P | β (95% CI) | P | β (95% CI) | P | ||||
| rs1697421 |
ALPL
NBPF3 |
A | 0.35 | 0.044 | 3 × 10–16 | 0.01 (–0.03 to 0.05) |
.67 | 0.001 (–0.03 to 0.03) |
.99 | –0.001 (–0.035 to 0.033) |
.94 |
| rs1801725 | CASR | A | 0.14 | –0.01 (0.0006) |
1 × 10–81 | –0.06 (–0.11 to –0.01) |
.04 | –0.04 (–0.09 to 0.01) |
.15 | –0.033 (–0.084 to 0.017) |
.19 |
| rs4074995 | RGS14 | A | 0.21 |
–0.008
(0.0004) |
3 × 10 –87 |
–0.09
(–0.14 to –0.04) |
3 × 10 -4 |
–0.09
(–0.13 to –0.04) |
5 × 10 -4 |
–0.048
(–0.086 to –0.010) |
.01 |
| rs9469578 | IHPK3 | T | 0.13 | –0.064 | 5 × 10–10 | –0.03 (–0.11 to 0.05) |
.50 | –0.05 (–0.12 to 0.03) |
.23 | –0.04 (–0.10 to 0.02) |
.16 |
| rs453639 | ENPP3 | C | 0.25 | 0.036 | 4 × 10–7 | 0.01 (–0.03 to 0.05) |
.58 | 0.02 (–0.02 to 0.06) |
.33 | 0.02 (–0.02 to 0.06) |
.36 |
| rs947583 | PDE7B | C | 0.28 | 0.035 | 2 × 10–9 | –0.01 (–0.05 to 0.03) |
.63 | –0.01 (–0.05 to 0.03) |
.64 | 0.01 (–0.03 to 0.04) |
.64 |
| rs11063205b | C12orf4 | A | 0.09 | 0.052 | 4 × 10–8 | –0.04 (–0.16 to 0.08) |
.52 | –0.06 (–0.17 to 0.05) |
.29 | –0.003 (–0.06 to 0.06) |
.92 |
| rs10799702 | ALPL | G | 0.39 | –0.01 (0.0004) |
5 × 10–239 | 0.0002 (–0.03 to 0.03) |
.99 | 0.01 (–0.02 to 0.04) |
.54 | 0.01 (–0.02 to 0.04) |
.45 |
| rs17145704 | LINC00709 | G | 0.12 | –0.01 (0.0007) |
4 × 10–22 | 0.01 (–0.06 to 0.09) |
.73 | 0.03 (–0.04 to 0.10) |
.43 | 0.05 (0.003 to 0.09) |
.04 |
| rs209957 | CYP24A1 | G | 0.23 | 0.003(0.0004) | 1 × 10–14 | 0.004 (–0.04 to 0.05) | .86 | 0.02 (–0.03 to 0.06) | .43 | 0.05 (0.01 to 0.08) | .01 |
Abbreviations: BMI, body mass index; CRIC, Chronic Renal Insufficiency Cohort; eGFR, estimated glomerular filtration rate; FGF23, fibroblast growth factor 23; MAF, minor frequency allele; PTH, parathyroid hormone; SNV, single-nucleotide variation.
a SEs unavailable in original genome-wide association studies (Kestenbaum et al) (6); SEs provided for SNVs from the UK Biobank.
b rs11063205 is proxy for rs2970818 (R2 = 0.7).
c Bonferroni-corrected significance threshold = 0.005.
Model 1 includes age, sex, and first 10 principal components of ancestry.
Model 2 includes model 1 + eGFR and eGFR-squared + BMI.
Model 3 includes model 2 + calcium, PTH, and FGF23.
Bolded values are those reaching significance.
Figure 1.
Mineral marker associations with rs4074995 within RGS14. Comparison of the associations between RGS14 single-nucleotide polymorphisms between the general population, the Chronic Renal Insufficiency Cohort (CRIC) population, and the CRIC population stratified by White and Black race. Results of model 2 are presented. Phosphate and calcium are presented as mg/dL difference per additional minor allele. Parathyroid hormone (PTH) and fibroblast growth factor 23 (FGF23) are presented as percentage difference per additional minor allele.
Table 3.
Mineral metabolism markers by rs4074995 genotype in the Chronic Renal Insufficiency Cohort population
| Variant | Host/Adjacent gene |
Genotype | No. | PTH, pg/mL median (IQR) |
Calcium, mg/dL, mean (SD) | Phosphate, mg/dL, mean (SD) | FGF23, RU/mL, median (IQR) | FEP, %, median (IQR) | 24-h urine phosphate, mg/d, mean (SD) | eGFR, mL/min/1.73 m2, mean (SD) |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4074995 | RGS14 | G_G | 1940 | 57.2 (37-91) | 9.2 (0.5) | 3.7 (0.7) | 150 (96-247) | 26 (19-36) | 736 (352) | 44 (15) |
| A_G | 942 | 48 (33-84) | 9.2 (0.4) | 3.7 (0.6) | 141 (96-222) | 28 (20-39) | 807 (348) | 45 (15) | ||
| A_A | 138 | 40 (28-64) | 9.2 (0.5) | 3.5 (0.6) | 126 (92-185) | 28 (22-37) | 836 (337) | 46 (15) |
FEP = [(urine phosphate × serum creatinine)/(serum phosphate × urine creatinine)] × 100.
Abbreviations: eGFR, estimated glomerular filtration rate; FEP, fractional excretion phosphate; FGF23, fibroblast growth factor 23; IQR, interquartile range; PTH, parathyroid hormone.
Bolded values are those reaching significance.
Fibroblast growth factor 23
Within the general population, 5 SNVs were significantly associated with plasma FGF23 (Table 4) (4). Of these SNVs, rs4074995 was also associated with FGF23 when all races were evaluated together, with 8.6% lower FGF23 per additional minor allele (95% CI, –13.9 to –2.9; P = 3 × 10–3) in model 2 and attenuation of this association after adjustment for phosphate (–6.4% [–11.8 to –0.7]; P = .03) (see Fig. 1). In race-stratified analyses, White participants demonstrated an association consistent with the general population, whereas the direction of effect was reversed and the magnitude of association greater in Black participants: rs4074995 (RGS14) was associated with 10.7% lower FGF23 in White participants (95% CI, –16, –5.1; P = 3 × 10–4) but 37.6% higher FGF23 in Black participants in model 2 (95% CI 6.7-77.4), P = .01) (Supplementary Tables S3 and S4) (27). These associations persisted both in White and Black participants after adjustment for calcium, phosphate, and PTH.
Table 4.
Additive model association results for fibroblast growth factor 23 among all individuals (n = 3027)
| SNV | Host/Adjacent gene |
Minor allele | MAF | General White population (4) | CRIC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 1d | Model 2d | Model 3d | |||||||||
| % Diffζ (95% CI) |
P | % Diffe (95% CI) |
P | % Diffe (95% CI) |
P | % Diffe (95% CI) |
P | ||||
| rs17216707 | CYP24A1 | C | 0.15 | –5.4 (–6.4 to –4.4) | 3 × 10–24 | 3.2 (–2.0 to 8.7) | .23 | 2.7 (–2.0 to 7.5) | .24 | 3.1 (–1.9 to 7.7) | .18 |
| rs687289a | ABO | G | 0.38 | 3.7 (2.7 to 4.7) | 6 × 10–17 | 0.8 (–3.1 to 4.8) | .71 | –0.5 (–3.9 to 3.1) | .79 | –0.9 (–4.3 to 2.6) | .61 |
| rs4074995 b | RGS14 | A | 0.21 | –3.9 (–4.9 to –2.9) | 2 × 10 –16 | –10.0 (–15.7 to –3.8) | 2 × 10 –3 | –8.6 (–13.9 to –2.9) | 3 × 10 –3 | –6.4 (–11.8 to –0.7) | .03 |
| rs17479566 | LINC01506 | T | 0.18 | 3.1 (2.1 to 4.1) | 2 × 10–9 | 0.3 (–6.5 to 7.6) | .94 | 1.2 (–5.0 to 7.8) | .71 | 0.8 (–5.4 to 7.3) | .82 |
| rs13330604c | LINC01229 | T | 0.20 | 3.5(2.3 to 4.7) | 5 × 10–9 | 0.7 (–6.9 to 8.9) | .86 | 3.1 (–4.0 to 10.6) | .40 | 3.0 (–4.0 to 10.5) | .42 |
Abbreviations: CRIC, Chronic Renal Insufficiency Cohort; MAF, minor allele frequency; SNV, single-nucleotide variation.
a rs687289 is a proxy for rs2769071 (R2 = 0.98).
b rs4074995 is a proxy for rs11741640 (R2 = 0.98); this proxy was used for simplification, in concordance with the results from other markers.
c rs13330604 is a proxy for rs9925837 (R2 = 0.9).
d Bonferroni-corrected significance threshold = 0.01.
e Represents the relative difference in serum markers per minor allele obtained by exponentiation of the β coefficients.
Model 1 for FGF23 includes age, sex, and first 10 principal components of ancestry.
Model 2 for FGF23 includes model 1 + eGFR, and eGFR-squared + BMI.
Model 3 for FGF23 includes model 2 + calcium, phosphate, and PTH.
Bolded values are those reaching significance.
Parathyroid hormone
Within the general population, 5 SNVs were significantly associated with serum PTH levels (Table 5) (3). None of these SNVs demonstrated significant associations with PTH within the entire CKD population. However, although narrowly missing significance thresholds, rs4074995 (RGS14) was associated with a 5.8% lower PTH for each additional minor allele (95% CI, –10.2 to –1.2; P = .01) in the fully adjusted model. Consistent with this, the prevalence of secondary hyperparathyroidism was lowest in the homozygous minor allele genotype (Fig. 2) (28). The association between the minor allele and lower PTH levels was consistent with the association seen in the general population (3). In addition, a similar association was seen among White and Black participants (–5.8% [95% CI, –11.2 to –0.1] and –5.8% [95% CI, –17.8 to 7.9], respectively) (Supplementary Tables S5 and S6) (27). Also narrowly missing significance thresholds was rs1801725 (CASR), which was associated with 6.1% higher PTH per additional minor allele (95% CI, 1.1-11.3; P = .02] (see Table 5). The direction of this association was consistent with that reported in the general population although the magnitude was approximately 2-fold greater in the CKD population (6.1% vs 3.0%; see Table 5). This finding persisted in the White population, who demonstrated 7.9% higher PTH per additional minor allele (95% CI, 2.5-13.4; P = 3 × 10–3) (see Supplementary Table S5) (27). There was no significant association among Black participants (see Supplementary Table S6) (27).
Table 5.
Additive model association results for parathyroid among all individuals (n = 3027)
| SNV | Host/Adjacent gene | Minor allele | MAF | General White population (3) | CRIC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 1b | Model 2b | Model 3b | |||||||||
| % Diffc (95% CI) |
P | % Diffc (95% CI) |
P | % Diffc (95% CI) |
P | % Diffc (95% CI) |
P | ||||
| rs6127099 | CYP24A1 | T | 0.28 | 7.3 (6.6 to 7.9) | 4 × 10–53 | –0.52 (–4.3 to 3.4) | .79 | 0.21 (–3.04 to 3.6) | .90 | –0.11 (–3.3 to 3.2) | .95 |
| rs4074995 | RGS14 | A | 0.21 | –3.0 (–2.4 to –3.7) | 7 × 10–17 | –5.8 (–10.9 to –0.43) | .04 | –4.9 (–9.4 to –0.12) | .03 | –5.8 (–10.2 to –1.2) | .01 |
| rs219779 | CLDN14 | A | 0.27 | –4.1 (–3.5 to –4.7) | 4 × 10–16 | –3.9 (–8.3 to 0.61) | .07 | –3.0 (–6.7 to 0.98) | .20 | –2.0 (–5.7 to 1.9) | .3 |
| rs4443100 | RTDR1 | G | 0.35 | 2.02 (1.4 to 2.6) | 9 × 10–9 | –1.5 (–4.9 to 2.1) | .41 | –1.1 (–4.1 to 2.02) | .49 | –1.2 (–4.1 to 1.8) | .42 |
| rs1801725a | CASR | A | 0.14 | 3.0 (2.4 to 3.7) | 4.8 × 10–8 | –0.04 (–5.6 to 5.9) | .99 | 2.9 (–2.1 to 8.1) | .26 | 6.1 (1.1 to 11.3) | .02 |
Abbreviations: BMI, body mass index; CRIC, Chronic Renal Insufficiency Cohort; eGFR, estimated glomerular filtration rate; FGF23, fibroblast growth factor 23; MAF, minor allele frequency; PTH, parathyroid hormone; SNV, single-nucleotide variation.
a rs1801725 is proxy for rs73186030 (R2 = 0.96).
b Bonferroni-corrected significance threshold = 0.01.
c Represents the relative difference in serum markers per minor allele obtained by exponentiation of the β coefficients.
Model 1 for PTH includes age, sex, and first 10 principal components of ancestry.
Model 2 for PTH includes model 1 + eGFR, and eGFR-squared + BMI.
Model 3 for PTH includes model 2 + calcium, phosphate, and FGF23.
Bolded values are those reaching significance.
Figure 2.
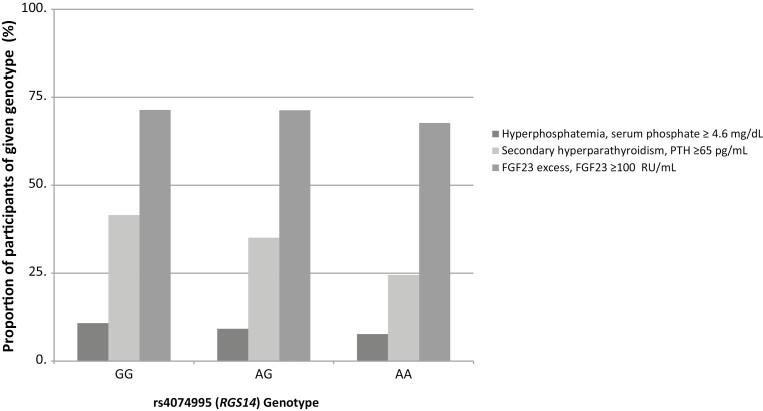
The prevalence of abnormal mineral markers by rs4074995 (RGS14) genotype (28). The prevalence of hyperparathyroidism differs by rs4074995 (RGS14) genotype (chi-square P < .0001). The minor allele is A. The AA genotype had the lowest prevalence of hyperparathyroidism. The prevalence of hyperphosphatemia and fibroblast growth factor 23 (FGF23) excess were not significantly different between the genotypes: hyperphosphatemia (chi-square P = .08) and FGF23 excess (chi-square P = .8).
Calcium
We examined 9 SNVs with significant associations with serum calcium within the general population, including 7 from a previously published GWAS and 2 novel SNVs from the recent GWAS of participants from the UK Biobank (Table 6) (5, 26). Of these SNVs, rs1801725, a missense variant located within the coding region of CASR was significantly associated with calcium (0.12 mg/dL higher per additional minor allele [95% CI 0.08-0.16; P = 4.1 × 10–5]) (see Table 6). The direction and magnitude of the association between rs1801725 and calcium were similar to that reported in the general population (5). This finding was consistent among White participants in race-stratified analyses, in whom rs1801725 was associated with 0.12 mg/dL higher calcium (95% CI, 0.08-0.17; P = 2 × 10–8) (Supplementary Table S7) (27). This SNV was not associated with serum calcium among Black participants (Supplementary Table S8) (27).
Table 6.
Additive model association results for calcium among all individuals (n = 3012)
| SNV | Host/Adjacent gene | Minor allele | MAF | General White population (5, 26) | CRIC | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model 1e | Model 2e | Model 3e | |||||||||
| β (SE) | P | β (95% CI) | P | β (95% CI) | P | β (95% CI) | P | ||||
| rs1801725 | CASR | A | 0.14 | 0.071 (0.004) | 9 × 10 –86 | 0.11 (0.07 to 0.15) | 8.6 × 10-7 | 0.10 (0.06 to 0.15) | 1.3 × 10-5 | 0.12 (0.08 to 0.16) | 4.1 × 10 -5 |
| rs838709a | DGKD | C | 0.23 | 0.018 (0.003) | 8 × 10–11 | 0.03 (–0.006 to 0.07) | .10 | 0.03 (–0.004 to 0.07) | .08 | 0.02 (–0.01 to 0.05) | .26 |
| rs1260333b | GCKR | T | 0.40 | 0.017 (0.003) | 1 × 10–10 | –0.001 (–0.03 to 0.03) | .94 | 0.0006 (–0.03 to 0.03) | .96 | 0.02 (–0.01 to 0.04) | .23 |
| rs17145704c | LINC00709 | G | 0.12 | 0.027 (0.005) | 5 × 10–9 | –0.006 (–0.07 to 0.06) | .86 | –0.01 (–0.074 to 0.05) | .74 | –0.01 (–0.08 to 0.06) | .86 |
| rs3814964d | NAP1L4 | A | 0.28 | –0.018 (0.003) | 1 × 10–10 | 0.03 (–0.001 to 0.06) | .06 | 0.027 (–0.002 to 0.06) | .06 | 0.03 (–0.01 to 0.07) | .08 |
| rs7336933 | VWA8 | A | 0.18 | –0.022 (0.004) | 9 × 10–10 | –0.03 (–0.07 to 0.02) | .21 | –0.03 (–0.07 to 0.02) | .26 | –0.01 (–0.04 to 0.02) | .44 |
| rs1570669 | CYP24A1 | G | 0.44 | –0.018 (0.003) | 9 × 10–12 | –0.003 (–0.03 to 0.02) | .82 | –0.004 (–0.03 to 0.02) | .78 | –0.03 (–0.06 to –0.01) | .01 |
| rs11063205 | C12orf4 | A | 0.06 | 0.0021 (0.0036) | 6 × 10–9 | –0.07 (–0.16 to 0.03) | .19 | –0.06 (–0.16 to 0.04) | .23 | 0.02 (–0.02 to 0.08) | .33 |
| rs6556307 | NSD1/RGS14 | A | 0.14 | –0.0016 (0.0003) | 1 × 10–8 | 0.005 (–0.03 to 0.04) | .77 | 0.004 (–0.03 to 0.04) | .84 | –0.01 (–0.04 to 0.03) | .64 |
Abbreviations: BMI, body mass index; CRIC, Chronic Renal Insufficiency Cohort; eGFR, estimated glomerular filtration rate; FGF23, fibroblast growth factor 23; MAF, minor allele frequency; PTH, parathyroid hormone; SNV, single-nucleotide variation.
a rs838709 is proxy for rs1550532 (R2 = 0.96).
b rs1260333 is proxy for rs780094 (R2 = 0.8).
c rs17145704 is proxy for rs10491003 (R2 = 1).
d rs3814964 is proxy for rs7481584 (R2 = 1).
e Bonferroni-corrected significance threshold = 0.006.
Model 1 includes age, sex, and first 10 principal components of ancestry.
Model 2 includes model 1 + eGFR, and eGFR-squared + BMI.
Model 3 includes model 2 + phosphate, PTH, and FGF23.
Expression quantitative loci analysis of rs4074995 in the kidney
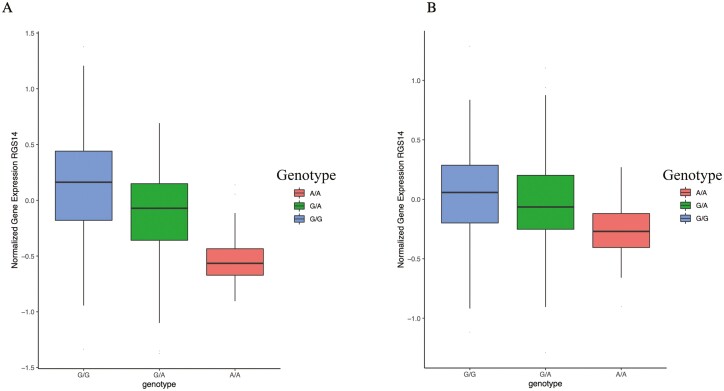
Our candidate SNV analysis in the CKD population showed that rs4074995 (RGS14) was significantly associated with multiple critical biomarkers of CKD-MBD. Using eQTL, we found that RGS14 gene expression was significantly associated with the rs4074995 variant genotype; the minor allele of rs4074995 was associated with lower RGS14 expression both in the glomerulus (P = 1.0 × 10–11) and renal tubule (P = 4.0 × 10–4) (Fig. 3A and 3B). In addition, colocalization analysis revealed a strong correlation between the tubule and glomerular eQTLs and GWAS effect sizes of SNVs located within ± 100 kb of RGS14 for FGF23 (posterior probability = 0.97) and PTH (posterior probability = 0.99).
Figure 3.
Expression quantitative loci analysis of RGS14 in renal tissue. A, The minor allele (A) of rs4074995 is associated with decreased gene expression of RGS14 in glomeruli (P = 1.03 × 10–11). B, The minor allele (A) of rs4074995 is associated with decreased gene expression of RGS14 in renal tubules (P = 3.99 × 10–4).
Discussion
Major advances in computational technology and large-scale genome sequencing have shed light on the genetic architecture of the CKD population and led to the discovery of causal genes and genetic variants linked to disease states (29). However, the genetic architecture related to CKD-MBD is poorly characterized. In this study, we evaluated the associations between genetic variants and mineral metabolism markers within a CKD population in comparison to the general population. We replicated SNVs that were within the RGS14 and CASR genes, finding that these SNVs were also associated with differences in circulating mineral metabolism markers in people with CKD. Significant associations were observed at rs4074995 (RGS14) for phosphate and FGF23 with a nearly significant association with PTH. SNP rs1801725 (CASR) was significantly associated with calcium and PTH. This study also highlights differences in MAFs between the White and Black population.
The minor allele of rs4074995 was associated with lower circulating phosphate and FGF23. This SNV is intronic to the RGS14 gene and located upstream of several genes that regulate tubular phosphate reabsorption including SLC34A1, which encodes for renal type 2a sodium phosphate cotransporter (NaPi-IIa). We confirmed our findings by eQTL analysis to show a significant association of rs4074995 minor allele with lower RGS14 gene expression both in glomerular and tubular compartments, and with colocalization studies demonstrating strong correlations between RGS14 gene expression and circulating FGF23 and PTH. These findings align with recent preclinical studies that provide evidence for RGS14 to play a key role in the convergence of FGF23- and PTH-dependent signaling pathways in the proximal tubule to inhibit phosphate reabsorption via NaPi-IIa (10-12). Potentially related to the presence of RGS14 in the kidney, rs4074995 has also been associated with estimated glomerular filtration rate (30). Still, how this SNV is associated with decline in kidney function in CKD is yet to be determined by future GWAS.
Our SNV and eQTL analyses provide insights into potential mechanisms and biologic implications of the rs4074995 variant inheritance pattern on the regulation of phosphate homeostasis. The direction of our association was consistent with the general population but the magnitude of association was greater in CKD and larger than the association with circulating phosphate levels achieved by dietary phosphate restriction in clinical trials conducted in nondialysis CKD participants (31). Of note, published phenome-wide association studies have shown associations of an SNV in close linkage disequilibrium with rs4074995 (rs12654812, R2 = 0.77) with an increased risk of nephrolithiasis in the presence of the minor allele (32-34). In addition, the Finnish Biobank (FinnGen) and the UK Biobank both demonstrate a positive association of rs4074995 with nephrolithiasis and urolithiasis (odds ratio = 1.14, P = 5.4 × 10–7 and odds ratio = 1.14 P = 1.1 × 10–10, respectively). Together, these studies provide evidence for rs4074995 as a genetic determinant of mineral metabolism in disease states such as CKD and nephrolithiasis. Moreover, the consistency of this finding in the general population, CKD, and with nephrolithiasis provides compelling justification for additional studies to determine the biological role of this gene in regulating mineral metabolism.
Whether the contribution of RGS14 SNVs to circulating FGF23 levels is indirect via regulation of renal phosphate reabsorption is unknown. Prior gene expression analysis suggests a direct mechanism and demonstrated that increased RSG14 expression is associated with increased FGF23 expression in heart, arteries, and skeletal muscle (4). Interestingly, rs4074995 (RGS14) demonstrated a substantially differential association with FGF23 levels between White and Black participants, where the association was negative in White participants and positive in Black participants. While this finding could suggest a biological difference, it could also represent a false discovery and should be subjected to replication. On closer investigation, the number of Black participants homozygous for the minor allele (n = 18) is rather small; yet this group demonstrates the highest FGF23 level. Given the small size of this group, it may be subject to the effect of outliers in FGF23 thereby driving a false association. Whether the minor allele of rs4074995 (RGS14) protects an individual from complications related to CKD-MBD (fractures, CVD, vascular calcification, and mortality) remains to be determined. However, the strong contribution of RGS14 to circulating levels of mineral metabolism markers underscores the need for further investigation of its role in CKD-MBD.
CASR signal transduction tightly regulates PTH secretion in response to acute changes in circulating ionized calcium (8). In CKD-MBD, CASR expression is decreased within the hyperplastic parathyroid gland and this decrease is associated with increased PTH secretion (9). In the CRIC CKD cohort and in the general population, serum calcium levels were strongly associated with rs1801725 (CASR), a missense variant located within the coding region of CASR. rs1801725 was also associated with higher PTH, a relationship seen only in White participants. Previous studies in a large cohort of hemodialysis patients showed similar associations between the minor allele of rs1801725 and higher PTH levels in participants of European ancestry (35, 36). The lack of association in Black individuals may reflect an underpowered analysis due to differences in MAFs (MAF for rs1801725 of 0.03 in Black participants as compared to 0.17 in White participants). Of note, the strong associations between SNVs within CYP24A1 and mineral markers found in the general population are notably absent in CKD. It is possible that in the context of CKD, acquired changes in vitamin D metabolism (eg, decreased 1-alpha hydroxylase activity and increased 24-hydroxylase activity resulting in reduced active vitamin D) have larger relative roles compared with genetic determinants of vitamin D metabolism. This merits further investigation (7, 37).
One strength of this study is the extension of the findings in the general population to identify potential determinants of mineral metabolism markers within the CKD population for whom the biologic implications and potential treatment options are most relevant. The candidate SNV approach allows for the testing of important SNVs such as rs4074995, which may have important implications in CKD but, owing to small sample sizes, might not reach the stringent significance thresholds of GWAS. In addition, the racial makeup of the CRIC cohort allowed for the investigation of racial differences in inheritance patterns and genetic associations. Finally, the availability of multiple markers of mineral metabolism allowed for concurrent adjustment of each marker to determine independent associations. This study also has important limitations. First, despite evaluating a relatively large CKD population, the sample size likely limited our ability to detect some of the genetic associations previously seen within the general population. Second, the sample size possibly contributed to the lack of associations within Black participants, in whom the MAFs were much lower. Third, given the CRIC population did not contain all SNVs discovered in the UK Biobank, it is possible that missed associations remain as these SNVs were not able to be investigated in this study. Finally, further studies are needed to investigate the phenotype associations of these SNVs particularly as they pertain to CKD-MBD–related outcomes, including cardiovascular mortality, fracture, and cardiovascular-related death.
In conclusion, genetic associations with mineral metabolism markers previously revealed in the general population are evident within the CKD population. In particular, SNVs within RGS14 highlight the need for further studies to elucidate the effect of this gene in CKD-MBD.
Acknowledgments
The CRIC Study was conducted by the CRIC Study Investigators and supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). The CRIC investigators supplied the data from the CRIC Study reported here. This manuscript was not prepared in collaboration with the Investigators of the CRIC Study and does not necessarily reflect the opinions or views of the CRIC Study or the NIDDK.
Glossary
Abbreviations
- BMI
body mass index
- CASR
calcium-sensing receptor
- CKD
chronic kidney disease
- CKD-MBD
chronic kidney disease–mineral bone disorder
- CRIC
Chronic Renal Insufficiency Cohort
- CVD
cardiovascular disease
- dbGap
Database of Genotypes and Phenotypes
- eGFR
estimated glomerular filtration rate
- eQTL
expression quantitative loci
- FGF23
fibroblast growth factor 23
- GWAS
genome-wide association studies
- MAF
minor allele frequency
- PTH
parathyroid hormone
- SNV
single-nucleotide variation
Contributor Information
Marciana L Laster, Department of Pediatrics, University of California, Los Angeles, Los Angeles, California 90095-1752, USA.
Bryce Rowan, Department of Biostatistics, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA.
Hua-Chang Chen, Department of Biostatistics, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA.
Tae-Hwi Schwantes-An, Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, Indiana 46202, USA.
Xin Sheng, Department of Medicine and Genetics, University of Pennsylvania, Philadelphia, Pennsylvania 19104, USA.
Peter A Friedman, Department of Pharmacology and Chemical Biology, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania 15213, USA.
T Alp Ikizler, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA; Vanderbilt O’Brien Center for Kidney Disease, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA.
Janet S Sinshiemer, Department of Human Genetics and Computational Medicine, David Geffen School of Medicine at UCLA, Los Angeles, California 90095-1752, USA; Department of Biostatistics, UCLA Fielding School of Public Health, Los Angeles, California 90095-1752, USA.
Joachim H Ix, Department of Medicine, University of California, San Diego, San Diego, California 92161, USA.
Katalin Susztak, Department of Medicine and Genetics, University of Pennsylvania, Philadelphia, Pennsylvania 19104, USA.
Ian H de Boer, Department of Medicine, University of Washington, Seattle, Washington 98195-6420, USA.
Bryan Kestenbaum, Kidney Research Institute, University of Washington, Seattle, Washington 98195-6420, USA.
Adriana Hung, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA.
Sharon M Moe, Clinical Translational Sciences Institute, Indiana University School of Medicine, Indianapolis, Indiana 46202, USA; Department of Medicine, Indiana University School of Medicine, Indianapolis, Indiana 46202, USA.
Farzana Perwad, Department of Pediatrics, University of California, San Francisco, San Francisco, California 94143, USA.
Cassianne Robinson-Cohen, Department of Medicine, Vanderbilt University Medical Center, Nashville, Tennessee, USA; Vanderbilt O’Brien Center for Kidney Disease, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA.
Financial Support
The authors report the following National Institutes of Health (NIH) grants outside the submitted work: National Institute of Diabetes and Digestive and Kidney Diseases K23DK123378 (M.L.L.), NIH R01DKK105811 (P.A.F), NIH R01GM140632 (P.A.F), National Human Genome Research Institute (NHGRI; No. U01HG007703 to J.S.S.), NHGRI R01HG009120 (J.S.S.), NHGRI R01HG006139 (J.S.S.), National Institute of Allergy and Infectious Diseases (NIAID; No. R01AI153044 to J.S.S), National Institute of General Medical Sciences (NIGMS; No. 1R35GM141798 to J.S.S.), and R01DK122075 (C.R.C).
Disclosures
T.S.A. is a paid consultant for Target RWE. S.M.M. is a scientific advisor for Sanifit, Ardeylx, and Amgen. J.H.I. is a DSMB member of Sanifit International and Ad Board member for AstraZeneca, Jnana Pharmaceuticals, and Ardelyx, Inc. The other authors have nothing to disclose.
Data Availability
All data analyzed during this study are included in the dbGaP data repository, study accession phs000524.v1.p1.
References
- 1. Kidney Disease: Improving Global Outcomes (KDIGO) CKD-MBD Work Group. KDIGO clinical practice guideline for the diagnosis, evaluation, prevention, and treatment of chronic kidney disease-mineral and bone disorder (CKD-MBD). Kidney Int Suppl. 2009;76(113):S1-130. [DOI] [PubMed] [Google Scholar]
- 2. Kidney Disease: Improving Global Outcomes (KDIGO) CKD-MBD Work Group. KDIGO 2017 clinical practice guideline update for the diagnosis, evaluation, prevention, and treatment of chronic kidney disease-mineral and bone disorder (CKD-MBD). Kidney Int Suppl (2011). 2017;7(1):1-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Robinson-Cohen C, Lutsey P, Kleber M, et al. Genetic variants associated with circulating parathyroid hormone. J Am Soc Nephrol. 2017;28(5):1553-1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Robinson-Cohen C, Bartz T, Lai D, et al. Genetic variants associated with circulating fibroblast growth factor 23. J Am Soc Nephrol. 2018;29(10):2583-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. O’Seaghdha C, Wu H, Yang Q, et al. Meta-analysis of genome-wide association studies identifies six new loci for serum calcium concentrations. PLoS Genet. 2013;9(9):e1003796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kestenbaum B, Glazer N, Köttgen A, et al. Common genetic variants associate with serum phosphorus concentration. J Am Soc Nephrol. 2010;21(7):1223-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones G, Prosser D. Kaufmann M. 25-Hydroxyvitamin D-24-hydroxylase (CYP24A1): its important role in the degradation of vitamin D. Arch Biochem Biophys. 2012;523(1):9-18. [DOI] [PubMed] [Google Scholar]
- 8. Conigrave AD, Ward DT. Calcium-sensing receptor (CaSR): pharmacological properties and signaling pathways. Best Pract Res Clin Endocrinol Metab. 2013;27(3):315-331. [DOI] [PubMed] [Google Scholar]
- 9. Cañadillas S, Canalejo A, Santamaría R, et al. Calcium-sensing receptor expression and parathyroid hormone secretion in hyperplastic parathyroid glands from humans. J Am Soc Nephrol. 2005;16(7):2190-2197. [DOI] [PubMed] [Google Scholar]
- 10. Biber J, Hernando N, Forster I. Phosphate transporters and their function. Annu Rev Physiol. 2013;75:535-550. [DOI] [PubMed] [Google Scholar]
- 11. Sneddon WB, Ruiz GW, Gallo LI, et al. Convergent signaling pathways regulate parathyroid hormone and fibroblast growth factor-23 action on NPT2A-mediated phosphate transport. J Biol Chem. 2016;291(36):18632-18642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Friedman P, Sneddon W, Mamonova T, et al. RGS14 regulates PTH- and FGF23-sensitive NPT2A-mediated renal phosphate uptake via binding to the NHERF1 scaffolding protein. J Biol Chem. 2022;289(5):101836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jovanovich A, Chonchol M, Cheung AK, et al. Racial differences in markers of mineral metabolism in advanced chronic kidney disease. Clin J Am Soc Nephrol. 2012;7(4):640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gutiérrez OM, Parsa A, Isakova T, et al. Genetic African ancestry and markers of mineral metabolism in CKD. Clin J Am Soc Nephrol. 2016;11(4):653-662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsu S, Zelnick LR, Lin YS, et al. Differences in 25-hydroxyvitamin D clearance by eGFR and race: a pharmacokinetic study. J Am Soc Nephrol. 2021;32(1):188-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hsu S, Hoofnagle AN, Gupta DK, et al. Race, ancestry, and vitamin d metabolism: the Multi-Ethnic Study of Atherosclerosis. J Clin Endocrinol Metab. 2020;105(12):e4337-e4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robinson-Cohen C, Hoofnagle A, Ix J, et al. Racial differences in the association of serum 25-hydroxyvitamin D concentration with coronary heart disease events. JAMA 2013;310(2): 179-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Denker M, Boyle S, Anderson AH, et al. Chronic Renal Insufficiency Cohort Study Investigators. Chronic Renal Insufficiency Cohort Study (CRIC): overview and summary of selected findings. Clin J Am Soc Nephrol. 2015;10(11):2073-2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feldman HI, Appel LJ, Chertow GM, et al. Chronic Renal Insufficiency Cohort (CRIC) Study Investigators. The Chronic Renal Insufficiency Cohort (CRIC) Study: design and methods. J Am Soc Nephrol. 2003;14(7 Suppl 2):S148-S153. [DOI] [PubMed] [Google Scholar]
- 20. Parsa A, Kanetsky PA, Xiao R, et al. Chronic Renal Insufficiency Cohort (CRIC) Study Investigators. Genome-wide association of CKD progression: the Chronic Renal Insufficiency Cohort Study. J Am Soc Nephrol. 2017;28(3):923-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Das S, Forer L, Schönherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Isakova T, Cai X, Lee J, et al. CRIC Study Investigators. Longitudinal evolution of markers of mineral metabolism in patients with CKD: the Chronic Renal Insufficiency Cohort (CRIC) Study. Am J Kidney Dis. 2020;75(2):235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qiu C, Huang S, Park J, et al. Renal compartment-specific genetic variation analyses identify new pathways in chronic kidney disease. Nat Med. 2018;24(11):1721-1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pan-UK Biobank. Pan-ancestry genetic analysis of the UK Biobank. 2020. September 11, 2020. https://pan.ukbb.broadinstitute.org
- 27. Laster ML, Rowan B, Chen HC,. et al. Supplemental data for “Genetic variants associated with mineral metabolism traits in chronic kidney disease.” Harvard Dataverse, V1. 2022.. Accessed on July 3, 2022. doi: 10.7910/DVN/XCE7VC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Isakova T, Wahl P, Vargas GS, et al. Fibroblast growth factor 23 is elevated before parathyroid hormone and phosphate in chronic kidney disease. Kidney Int. 2011;79(12):1370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tin A, Köttgen A. Genome-wide association studies of CKD and related traits. Clin J Am Soc Nephrol. 2020;15(11):1643-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. The University of Cambridge. Pheno Scanner. 2018. Accessed on April 27, 2021. http://www.phenoscanner.medschl.cam.ac.uk [Google Scholar]
- 31. Isakova T, Gutiérrez OM, Smith K, et al. Pilot study of dietary phosphorus restriction and phosphorus binders to target fibroblast growth factor 23 in patients with chronic kidney disease. Nephrol Dial Transplant. 2011;26(2):584-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen W, Chou W, Chu H, et al. The rs1256328 (ALPL) and rs12654812 (RGS14) polymorphisms are associated with susceptibility to calcium nephrolithiasis in a Taiwanese population. Sci Rep. 2019;9(1):17296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Long J, Chen Y, Lin H, et al. Significant association between RGS14 rs12654812 and nephrolithiasis risk among Guangxi population in China. J Clin Lab Anal. 2018;32(6):e22435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang L, Feng C, Ding G, et al. Association study of reported significant loci at 5q35.3, 7p14.3, 13q14.1 and 16p12.3 with urolithiasis in Chinese Han ethnicity. Sci Rep. 2017;7:45766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Grzegorzewska AE, Bednarski D, Świderska M, Mostowska A, Jagodziński PP. The Calcium-sensing receptor gene polymorphism rs1801725 and calcium-related phenotypes in hemodialysis patients. Kidney Blood Press Res. 2018;43(3):719-734. [DOI] [PubMed] [Google Scholar]
- 36. Moe SM, Wetherill L, Decker BS, et al. Calcium-sensing receptor genotype and response to cinacalcet in patients undergoing hemodialysis. Clin J Am Soc Nephrol. 2017;12(7):1128-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang W, Ma F, Wang L, et al. The association analysis between CYP24A1 genetic polymorphisms and the risk of ischemic stroke in Chinese Han population. Brain Behav. 2020;10(2):e01503. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data analyzed during this study are included in the dbGaP data repository, study accession phs000524.v1.p1.