Abstract
Context
Polymorphisms in the gene encoding the glucagon-like peptide-1 receptor (GLP1R) are associated with type 2 diabetes but their effects on incretin levels remain unclear.
Objective
We evaluated the physiologic and hormonal effects of GLP1R genotypes before and after interventions that influence glucose physiology.
Design
Pharmacogenetic study conducted at 3 academic centers in Boston, Massachusetts.
Participants
A total of 868 antidiabetic drug-naïve participants with type 2 diabetes or at risk for developing diabetes.
Interventions
We analyzed 5 variants within GLP1R (rs761387, rs10305423, rs10305441, rs742762, and rs10305492) and recorded biochemical data during a 5-mg glipizide challenge and a 75-g oral glucose tolerance test (OGTT) following 4 doses of metformin 500 mg over 2 days.
Main Outcomes
We used an additive mixed-effects model to evaluate the association of these variants with glucose, insulin, and incretin levels over multiple timepoints during the OGTT.
Results
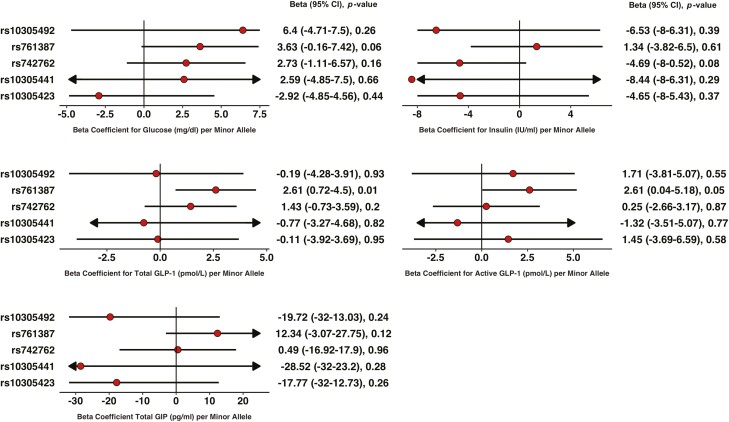
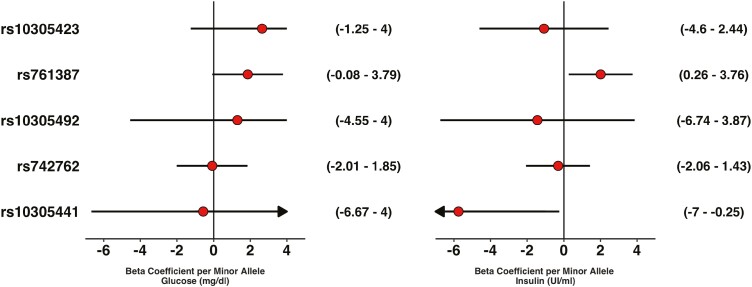
During the OGTT, the G-risk allele at rs761387 was associated with higher total GLP-1 (2.61 pmol/L; 95% CI, 1.0.72-4.50), active GLP-1 (2.61 pmol/L; 95% CI, 0.04-5.18), and a trend toward higher glucose (3.63; 95% CI, -0.16 to 7.42 mg/dL) per allele but was not associated with insulin. During the glipizide challenge, the G allele was associated with higher insulin levels per allele (2.01 IU/mL; 95% CI, 0.26-3.76). The other variants were not associated with any of the outcomes tested.
Conclusions
GLP1R variation is associated with differences in GLP-1 levels following an OGTT load despite no differences in insulin levels, highlighting altered incretin signaling as a potential mechanism by which GLP1R variation affects T2D risk.
Keywords: genetics, GLP1R, incretin response, pharmacogenetics, type 2 diabetes
Clinical trials have shown interindividual differences in glycemic response to antidiabetic medications (1); yet clinicians continue to make therapeutic decisions based on the population average parameters of efficacy. Given the difficulty in predicting which patient with type 2 diabetes (T2D) would respond better to 1 drug over another, antidiabetic drugs are often prescribed sequentially or in combination. Frequent glucose monitoring is used to infer drug response. This “trial and error” approach could contribute to polypharmacy or delay the time it takes for a patient to achieve glycemic control. Although clinical practice guidelines recommend tailoring treatment for T2D according to patient characteristics, genetic factors that modulate pharmacologic effects remain elusive and ignored (2, 3).
Genome-wide association studies (GWAS) have identified hundreds of genetic variants that modify T2D risk and related glycemic traits, including variants at GLP1R, which encodes the glucagon-like peptide-1 (GLP-1) receptor (4, 5). An analysis of genetic coding variants in GLP1R identified an association of the minor (A) allele at a low-frequency nonsynonymous coding variant (A316T, rs10305492) with lower fasting glucose (FG) and T2D risk but higher 2-hour glucose (6). Activation of the receptor by GLP-1 stimulates adenylyl cyclase pathways, which enhance pancreatic beta-cell insulin secretion through the incretin pathway (7). In addition to incretin mimetics, sulfonylurea and metformin, 2 commonly prescribed glucose-lowering drugs, have been shown to interact with the incretin pathway or modulate insulin secretion (8, 9). Previous studies have suggested that metformin may induce GLP-1 secretion by enteroendocrine L cells in both fasting and postprandial states, contributing to the drug’s glucose-lowering effect (8-11). Metformin has also been described to interact the with the incretin axis by enhancing the expression of GLP1R and related insulinotropic islet receptors (12). With respect to sulfonylureas, studies have shown that incretins potentiate sulfonylurea-induced insulin secretion and sulfonylureas augment the incretin effect, incretins and sulfonylureas may work synergistically to lower glycemia (13, 14). Because the GLP-1 receptor is central to incretin action and insulin secretion, genetic variation at GLP1R may partly explain interindividual differences in glycemic response to these drugs.
To improve our understanding of the role of genetic variation at GLP1R in the incretin response in human participants free of disease, we leveraged the Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans (SUGAR-MGH) (15, 16). For our primary objective, we sought to determine whether polymorphisms in GLP1R previously associated with T2D risk or glycemic traits were associated with differences in the incretin response. As a secondary objective, we evaluated the physiologic and hormonal effects of these polymorphisms before and after glipizide and metformin administration, interventions that are known to influence glucose physiology.
Methods
Study Design and Participants
The study design and baseline participant characteristics of SUGAR-MGH (clinical trial registration no. NCT01762046, www.clinicaltrials.gov) have been described in detail previously (15, 16). The protocol was approved by the Partners Human Research Committee, and written informed consent was obtained from all participants. In brief, SUGAR-MGH is a National Institutes of Health-funded clinical research study conducted at 3 academic medical centers within Boston, MA. Between 2008 and 2015, 1000 eligible participants were preferentially enrolled if they were at risk of developing T2D (metabolic syndrome, obesity, history of gestational diabetes and/or polycystic ovary syndrome, family history of T2D) or had T2D managed only on lifestyle interventions and not antidiabetic agents. Of the 1000 participants, 890 were successfully genotyped.
Study Procedures and Measurements
The SUGAR-MGH protocol required 2 visits to the clinical research center. In visit 1, following an overnight fast of at least 8 hours, plasma glucose and insulin were measured at regular intervals over 240 minutes after a single, open-label, oral dose of 5 mg of glipizide. After a washout period of 5 days, patients were instructed to take 4 doses of 500 mg of metformin over 48 hours to define the acute effects of metformin after approaching steady-state concentration. Participants took 1 dose of metformin day 6, and 2 doses of metformin on day 7. On day 8, following an overnight fasting of 8 hours, participants received the fourth dose of metformin before undergoing an oral glucose tolerance test (OGTT). Plasma glucose and insulin were measured at regular intervals over 120 minutes. In a subset of participants (n = 143), GLP-1 and glucose-dependent insulinotropic polypeptide (GIP) were measured at selected time points (0, 5, 10, 15, 30, 60, and 120 minutes). A pictorial description of the study workflow can be found in Supplementary Figure 1 (17).
Laboratory Measurements
Plasma glucose was measured by a hexokinase assay (Roche, Indianapolis, IN). Insulin levels were determined by radioimmunoassay (Beckman Coulter, Fullerton, CA). The intra-assay coefficient of variability (CV) for the insulin assay was 2.2% to 4.4% and the interassay CV was 2.9% to 6%. C-peptide and glucagon were measured by radioimmunoassay (KKPED1; Siemens, Erlangen, Germany, and LINOplex Kit, HENDO-65K-Rev; Linco Research, St. Charles, MO, respectively). The CV of the glucagon assay was 10.9% to 13.3%. In prechilled EDTA tubes containing DPP-IV inhibitor (Millipore, Billerica, MA), the incretin hormones (GLP-1 and GIP) were measured from blood samples. Active GLP-1 (7-36,7-37) was measured using the GLP-1 (active) ELISA Kit (Millipore); total GLP-1 was measured using the GLP-1 (7-36,9-36) ELISA kit from Alpco Immunoassays (Salem, NH); and GIP was measured by using the Human GIP Total ELISA Kit from Millipore.
Genotyping and Imputation
Of the 1000 participants, 890 were successfully genotyped and underwent quality control (QC) analyses. Samples were genotyped with the Illumina Multi-Ethnic Genotyping Array, including 1.7 million markers enriched for exome content with > 400,000 markers missense, nonsense, indels, and synonymous variants. A 3-step QC protocol was applied using PLINK (18, 19), including 2 stages of variant removal and an intermediate stage of sample exclusion. We excluded genetic markers with proportion of missingness ≥ 0.05, Hardy-Weinberg equilibrium P ≤ 1 × 10-20 for all the cohort or minor allele frequency < 0.01 and repeated the protocol after sample exclusion. We excluded individuals with sex discordance, sample call rates ≥ 0.02, or in-sample relatedness (pairs with PI-HAT ≥ 0.125; the individual with the higher proportion of missingness was removed). After QC, 868 subjects remained for the analysis. Genotypes were phased with SHAPEIT2 (20). We then performed genotype imputation with IMPUTE2 with 1000G phase 3 (21), HRC (22), and TOPMed (23) reference panels using the Michigan and the TOPMed Imputation servers (24). We retained variants with r2 > 0.8 and a minor allele frequency > 0.005 for further analyses.
Selection of Polymorphisms Within GLP1R
We selected genetic variants within or near GLP1R (Build 37 Chromosome 6:38,966,574-39,105,519) with evidence for strong association with T2D, random glucose, FG, 2-hour glucose, or hemoglobin A1c at genome-wide significance (P < 5 × 10-8) by mining the Type 2 Diabetes Knowledge Portal (25). We pruned the list of genetic variants for linkage disequilibrium (r2 > 0.5) using LDlink European Panel and excluded variants with minor allele frequency < 1% in our dataset (26). The following genetic variants were included in the study: rs761387 (P = 5 × 10-8 for T2D), rs10305423 (P = 1.4 × 10-8 for random glucose), rs10305441 (P = 1.2 × 10-15 for T2D), rs742762 (P = 5.8 × 10-10 for T2D), and rs10305492 (P = 3.7 × 10-21 for random glucose). GWAS effect estimate of these variants and their association with T2D and glycemic traits are provided in Supplemental Table 1 and Supplemental Table 2 (17).
Outcomes
Our primary outcome was the incretin response, including glucose, insulin, and GLP-1 levels during an OGTT. Our secondary outcomes included prediabetes/diabetes status defined by FG ≥ 100 mg/dL or 2-hour glucose ≥ 140 mg/dL, predefined pharmacological endpoints for the glipizide and metformin challenges (glucose trough adjusted for baseline, time to glucose trough, time to glucose trough adjusted for baseline, insulin peak adjusted for baseline, and change in glucose from the trough to 240 minutes divided by 240 minus the time at trough, and difference in FG before and after 3 doses of metformin 500 mg) (16), and changes in glucose and insulin levels from baseline during the glipizide challenge ([ glucose at baseline to trough], [glucose at baseline to 90 minutes], [glucose at baseline to 120 minutes], area over the glucose curve from baseline to 120 minutes, [glucose from baseline to trough]/time to trough, [insulin at baseline to insulin peak], [insulin at baseline to 60 minutes], area under the insulin curve from baseline to 240 minutes, and [insulin from baseline to peak])/time to insulin peak.)
Statistical Analysis
We defined major homozygous as 2 copies of the major allele, minor homozygous as 2 copies of the minor allele, and heterozygous as 1 copy of the minor allele. Laboratory values during the OGTT were plotted against time by number of alleles. Using a trapezoidal method with and without adjusting for baseline values, the areas under the curves (AUC) for insulin, glucose, and incretins were calculated during the OGTT.
To estimate additive genetic effects on the incretin response (glucose, insulin, and incretin hormones) during the OGTT with multiple timepoints, we used repeated measures mixed-effect models with a random covariate that accounted for intra-individual correlation. We used linear regression models to estimate genetic effects on the predefined pharmacological endpoints. All models were adjusted for body mass index (BMI), age, gender, FG, self-reported ethnicity, and ancestry principal components. As the underlying genetic susceptibility for T2D may contribute to inter-individual variability on the outcome, in a sensitivity analysis, we performed mixed-effect models of the incretin response with adjustment for a T2D polygenic risk score constructed using the PRS-CS method (27) and summary statistics from a largescale T2D GWAS (28).
To evaluate the association between GLP1R genetic variants and prediabetes/diabetes status in this study, we performed a logistic regression model with impaired fasting glucose/diabetes at baseline (FG ≥ 100 mg/dL) and impaired glucose tolerance/diabetes (2-hour glucose ≥ 200 mg/dL) during the OGTT. To evaluate for effect modification by age, we included an interaction term between single-nucleotide polymorphisms and age in the mixed-effect models. To evaluate whether GLP1R variation influences the activity of dipeptidyl peptidase-4, an enzyme that deactivates incretins, we performed mixed-effect models for the ratio between active and total GLP-1 levels during the OGTT on the GLP1R polymorphisms. To evaluate the influence of genetic polymorphisms at GLP1R on insulin and glucose response during the glipizide challenge, we performed an additive linear model on the changes in insulin and glucose levels from baseline.
Effect estimates were reported with 95% CI. As we examined the effects of 5 GLP1R variants, we used a P value of 0.05/5 (ie, 0.01) to declare statistical significance. All statistical analyses were performed using R version 4.0.2 (R Foundation for Statistical Computing, Vienna, Austria) and using the lme4 and the lmerTest packages (29, 30).
Results
The 868 participants were balanced across both sexes and had a wide age range (32-59 years) with a mean age of 47 years old. A total of 656 participants (75.67%) were overweight or obese and 684 (78.80%) had an FG below 100 mg/dL (Table 1). We note that there was a larger proportion of self-reported white participants in those with incretin measurements (N = 143) compared with those without incretin measurements (N = 725), and allele frequencies differed slightly between groups (Supplemental Table 3) (17). We also note that their average glucose levels were slightly higher. We recognize that the incretin effect may differ by diabetes status (31). However, the limited sample size of participants with incretin measurements prevented us from performing stratified analyses by fasting glucose categories.
Table 1.
Baseline demographics of the study participants
| Characteristic | n = 868 |
|---|---|
| Male, n (%) | 407 (46.9) |
| Age, y | 47.19 (16.12) |
| Race, n (%) | |
| White | 599 (69.0) |
| Black | 185 (21.3) |
| Asian | 45 (5.2) |
| Other | 39 (4.5) |
| Height, m | 169.23 (9.61) |
| Weight, kg | 86.69 (22.28) |
| BMI, kg/m2 | 30.19 (7.21) |
| Systolic blood pressure, mm Hg | 125.41 (17.68) |
| Diastolic blood pressure, mm Hg | 76.12 (10.31) |
| Resting pulse, per minute | 69.00 (12.56) |
| Respiratory rate, per minute | 17.49 (2.01) |
| Fasting glucose, mg/dL | 92.62 (17.06) |
| Glucose levels, n (%) | |
| Normal | 684 (78.8) |
| Impaired | 144 (16.6) |
| Diabetes | 40 (4.6%) |
| rs10305441 (%) | |
| GG | 852 (98.2) |
| AG | 14 (1.6) |
| AA | 2 (0.2) |
| rs761387 (%) | |
| AA | 674 (77.6) |
| GA | 181 (20.9) |
| GG | 13 (1.5) |
| rs10305423 (%) | |
| CC | 811 (94.5) |
| TC | 46 (5.4) |
| TT | 1 (0.1) |
| rs10305492 (%) | |
| GG | 846 (97.6) |
| AG | 21 (2.4) |
| AA | 0 (0) |
| rs742762 (%) | |
| AA | 664 (76.6) |
| CA | 190 (21.9) |
| CC | 13 (1.5) |
Data for categorical variables is presented as frequencies (%) and for numerical variables mean (SD).
BMI, body mass index.
None of the variants were significantly associated with prediabetes/diabetes (P > 0.05), though there was a suggestive association between rs761387 and impaired glucose tolerance/diabetes that did not reach statistical significance but was consistent with reported results in larger sample sizes (odds ratio, 2.45; 95% CI, 0.89-3.39, per minor allele, P value = 0.08) (32). Given that only 25 participants had either 2-hour glucose level ≥ 200 mg/dL or FG > 126 mg/dL, we were unable to assess the association of the polymorphisms with glucose levels in the diabetes range (Supplementary Table 4) (17).
From repeated measures mixed-effect models using all timepoints during the OGTT, the G allele (minor allele) at rs761387 was associated with higher levels of total and active GLP-1 over 120 minutes (2.61 pmol/L per allele; 95% CI, 0.72-4.50, P = 0.01 and 2.61 pmol/L per allele, 95% CI, 0.04-5.18, P = 0.04, respectively) but was not associated with insulin levels (P = 0.61; Fig. 1C and 1D). Carriers of the G allele had higher glucose levels, but this association did not reach statistical significance (3.63 mg/dL per allele; 95% CI, -0.16 to 7.42, P = 0.06).
Figure 1.
Association of polymorphisms in GLP1R variants with glucose, insulin, and incretin levels during an oral glucose tolerance test. (A) Glucose, (B) insulin, (C) total GLP-1, (D) active GLP-1, and (E) active GIP. Beta coefficients were obtained from additive linear mixed models for measurements at all timepoints between 0 and 120 minutes. *Statistically significant.
From linear regression models, the G allele at rs761387 was associated with higher levels of active GLP-1 at baseline (0.91 pmol/L; 956% CI, 0.006-1.83 per minor allele, P = 0.04) and higher AUC for total and active GLP-1 without adjustments for baseline levels at time 0 (405.09 pmol/L per minor allele, 145.96-664.22, P = 0.002 and 451.22 pmol/L per minor allele; 95% CI, 115.96-786.48, P = 0.01) (Supplemental Table 5) (17). We found no association between rs761387 and AUC glucose, AUC insulin, or AUC GLP-1 after adjusting for baseline levels (P > 0.1). Adjusting our mixed models with the T2D polygenic score did not alter any of the genetic effects on the incretin response (Supplemental Table 6) (17).
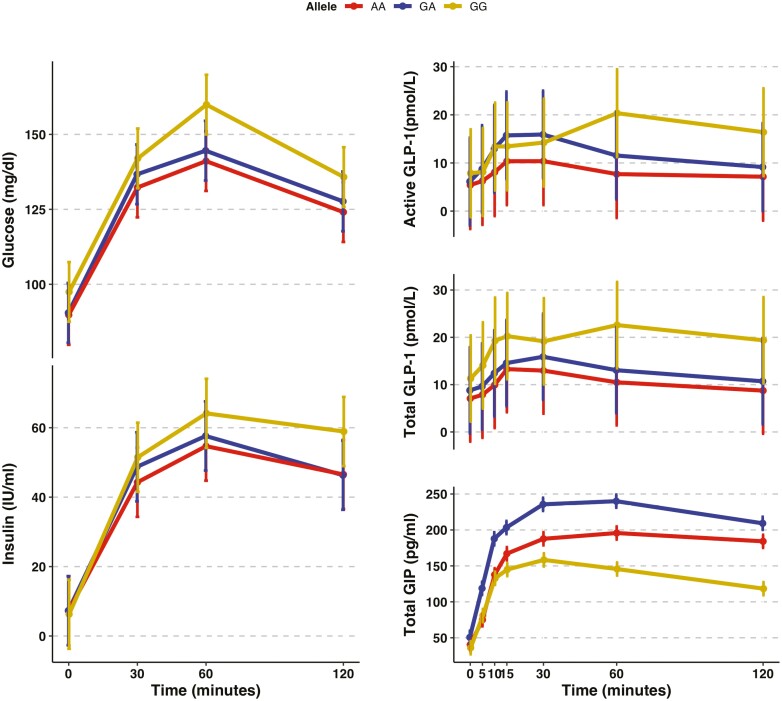
The largest difference in total GLP-1 levels between participants with the GG vs AA genotype was observed at time 10 minutes (10.76 pmol/L; 95% CI, 5.58-15.93, P = 6.9 × 10-5), whereas for active GLP-1, the largest difference was observed at time 60 minutes (10.64 pmol/L; 95% CI, 3.96-17.33; P = 0.002; Fig. 2B). The largest difference in glucose levels was also observed at time 60 minutes Fig. 2A), although this association did not reach statistical significance (19.8 mg/dL; 95% CI, -1.18 to 41.4; P = 0.07). Genotypes were not associated with insulin levels at any of the time points (P > 0.1). None of the GLP1R polymorphisms tested were associated with the active/total GLP-1 ratio (Supplemental Table 7) (17).
Figure 2.
Time graph for glucose, insulin, and incretin levels during an oral glucose tolerance test by rs761387 genotypes. Vertical bars represent the standard error. Total sample size was of 868 for insulin and glucose and 143 for the incretin hormones.
We found statistical interaction between rs761387 and age as a continuous variable on total GLP-1 levels (P value for interaction = 0.01) (Supplemental Table 8) (17). To evaluate for effect modification, we performed a stratified analysis by age group using the median age (50 years) as the cut point and found that the genetic effect on total GLP-1 levels appeared slightly larger among older individuals. Older individuals had higher blood pressure, BMI, FG, and fasting insulin levels than younger individuals in the study (Supplemental Tables 9 and 10) (17).
Time graphs for laboratory of the other 4 variants during the OGTT can be found in the Supplemental Figure 2 and Supplemental Figure 3, and effect estimates of every timepoint for all 5 variants are in Supplemental Table 11 (17).
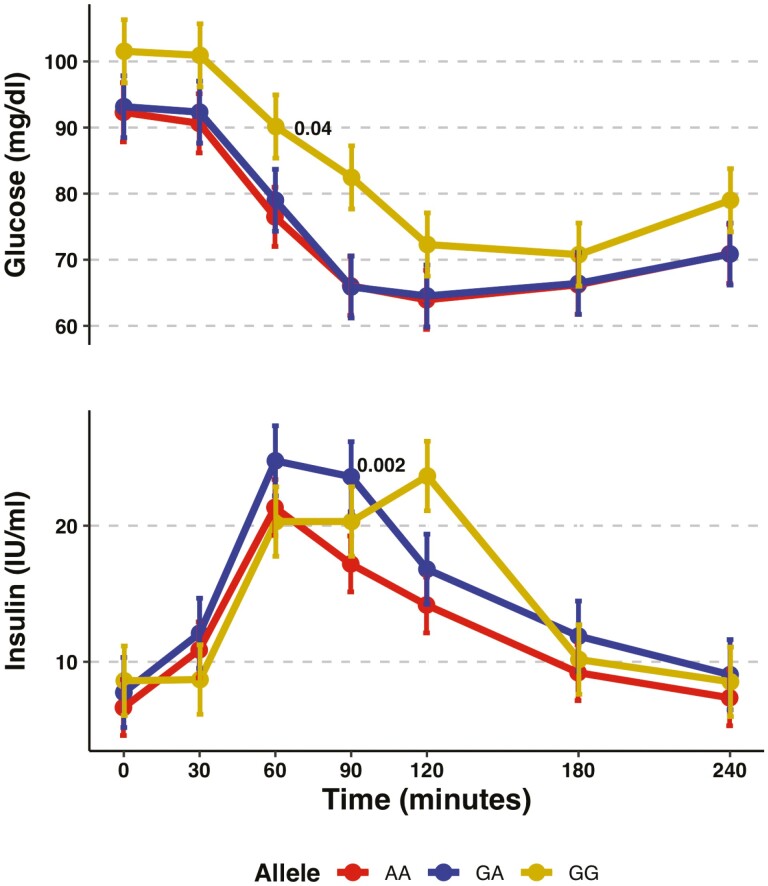
During the glipizide challenge, rs761387 was associated with a larger insulin response (2.01 IU per minor allele; 95% CI, 0.26-3.76; P = 0.02) in the additive model from the mixed effects model (Fig. 3). Although carriers of the GG genotype, compared with the AA genotype, had higher glucose levels throughout the glipizide challenge and a delayed peak insulin at time 120 minutes and 60 minutes when compared with the other genotypes (Fig. 4), changes in glucose or insulin levels from baseline (Supplemental Table 12) (17) or any of the 6 prespecified pharmacological endpoints for glipizide or metformin did not differ by genotype (Table 2).
Figure 3.
Association of polymorphisms in GLP1R variants with glucose and insulin during a glipizide challenge. (A) Glucose and (B) insulin. Beta coefficients were obtained from additive linear mixed models for measurements at all timepoints between 0 and 120 minutes. *Statistically significant.
Figure 4.
Time measurements for glucose and insulin for rs761387 during the glipizide challenge. Vertical bars represent the standard error. Total sample size was of 868.
Table 2.
Pharmacological endpoints for rs761387
| Endpoint | Beta | 95% CI | P value |
|---|---|---|---|
| Time to glucose trough, min | 5.71 | -3.88 to 15.3 | 0.24 |
| Time to glucose trough adjusted for baseline, min | -0.008 | -0.04 to 0.02 | 0.55 |
| Insulin peak adjusted for baseline, mU/L | -0.022 | -0.1 to 0.06 | 0.58 |
| Glucose recovery period | -0.01 | -0.04 to 0.02 | 0.5 |
| Glucose trough adjusted for baseline, mg/dL | 0.978 | -0.24 to 2.19 | 0.11 |
| Fasting glucose at OGTT adjusted for baseline, mg/dL | -0.397 | -1.59 to 0.79 | 0.51 |
Abbreviations: AUC, area under the curve; GIP, gastric inhibitory polypeptide; GLP-1, glucagon-like peptide-1; OGTT, oral glucose tolerance test.
Given the known relationship between incretins and appetite regulation, we looked up the association between GLP-1 polymorphisms and BMI in one of the largest published GWAS of BMI (33). Although rs761387 was not associated with BMI, rs742762 was nominally associated with BMI (P = 0.04, Supplemental Table 13) (17, 33), potentially suggesting pleiotropy at GLP1R on incretin secretion and body weight.
In the Diabetes Epigenome Atlas, rs761387 is intronic and overlaps a weak enhancer in islets of Langerhans. To determine whether the variant is associated with mRNA expression, we interrogated the Translational Human Pancreatic Islet Genotype Tissue-Expression Resource (34, 35) and the Genotype-Tissue Expression and found that the variant was a pancreatic tissue splicing quantitative trait locus of SAYSD1, a gene approximately 15 kb from the 3′ end of GLP1R (P = 5.8 × 10-8) and a pancreatic islet expression QTL of SAYSDI (P = 0.005) and GLP1R (P = 0.048) (36).
Conclusions
Polymorphisms at GLP1R have been associated with the risk of T2D and other glycemic traits in GWAS, but studies to demonstrate whether they modify the incretin effect or antidiabetic drug response in human subjects are limited. Using a prospective human pharmacogenetic resource—SUGAR-MGH—we performed a physiologic and hormonal evaluation of 6 variants in GLP1R that had been reported to be associated with T2D in GWAS. During the OGTT, we found that the G allele at rs761387 was associated with higher incretin levels and a trend toward higher glucose levels but was not associated with insulin levels. These findings provide evidence for altered incretin signaling as a potential mechanism by which polymorphisms at GLP1R increase the risk of T2D.
Although multiple genetic discovery efforts of glycemic traits have been performed over the past decade, large-scale GWAS of incretin levels are lacking. In our study, the G allele at rs761787 was associated with higher levels of GLP-1 and glucose but similar levels of insulin during the OGTT, consistent with incretin resistance. This may be explained by lower intracellular calcium mobilization invoked by GLP-1 receptor signaling, resulting in a reduction in the first phase of insulin secretion from preformed storage granules within the beta cell (37). Glucose absorption in the small intestine may trigger additional GLP-1 secretion that overcomes receptor resistance (38, 39), allowing for a more robust second phase of insulin secretion from increased insulin biosynthesis and mobilization of insulin granules (40).
Other genetic variants located in or near other relevant genes reported to be associated with T2D may also influence incretin pathways (15, 41-43). The association of the A allele at the low-frequency nonsynonymous coding variant in GLP1R (A316T, rs10305492) with lower fasting glucose but higher 2-hour glucose reported by Wessel et al (6) was not replicated in our study, possibly because of the small sample size. Saxena et al found that carriers of the A allele at rs10423928, an intron variant in GIPR, had higher levels of glucose and lower insulin secretion after an OGTT (44). Previous work in SUGAR-MGH has shown that carriers of the T-risk allele at TCF7L2 had higher levels of total and active GLP-1, albeit with no differences in glucose or insulin levels across all timepoints of the OGTT. Moreover, although rs742762, a common variant in the GLP1R locus, was highly associated with T2D (P = 3.99 × 10-10) in large-scale GWAS (45), the variant was not associated with the incretin response in our study. Notably, the variant was only associated with GLP1R expression in endothelial tissue (tibial artery; P = 1.0 × 10-7), and not the pancreatic tissue (P = 0.97) (46), suggesting that the mechanism through this variant modifies the risk of T2D may not involve beta-cell function (46).
Studies evaluating the association of polymorphisms at GLP1R with insulin levels following a hyperglycemic stimulus have found a delayed response (47-50). In our study, the G allele at rs761387 was associated with a delayed insulin response to an oral glucose load, as well as during the glipizide challenge. This may be explained by lower intracellular calcium mobilization invoked by GLP-1 receptor signaling, resulting in a reduction in the first phase of insulin secretion from preformed storage granules within the beta cell (51, 52). Rising glycemia may trigger additional GLP-1 secretion, leading to higher circulating GLP-1 that overcomes receptor resistance, and a more robust second phase of insulin secretion from increased insulin biosynthesis and mobilization of insulin granules (40).
A recent clinical study found that, in patients with and without diabetes, inter-individual variations in the GLP-1 response were influenced by glucose, proteins, and lipids that reach the small intestines possibly partly because of differences in gastric emptying (53). Additionally, we acknowledge that multiple factors contribute to inter-individual differences in incretin secretion, including the intrinsic rate of gastric emptying, small intestinal transit upon stimulation of enteroendocrine K and L cells, and the rate of nutrient absorption in the small intestine that are coupled with incretin secretion via autocrine regulation (37). It is possible that the observed association of GLP1R genetic variation with differences in GLP-1 levels following an oral glucose load may be modulated through these pathways. Furthermore, although we observed major differences in total GIP levels by genotype over several time points during the OGTT, differences in GIP levels did not follow an allele-dose response relationship. Thus, it remains unclear whether genetic variation in GLP1R influences GIP secretion. Measurements of both active and total GIP levels may clarify whether the association between GLP1R polymorphisms with the incretin response to oral glucose was specific to GLP-1.
Previous clinical studies to evaluate the effect of genetic variation at GLP1R on drug exposure in patients with T2D have focused on incretin mimetics or DPP-4 inhibitors. Sathananthan et al found that the A allele in rs6923761, a missense variant (Gly168Ser), was associated with a decrease in insulin secretion in patients without diabetes who received a 2-hour infusion of GLP-1 (50). Javrosky et al reported that the A allele in rs6923761 was associated with a smaller reduction in HbA1c levels after 6 months of treatment with DPP-4 inhibitors (54). Han et al found a greater reduction in HbA1c associated with the A allele at the missense variant rs3765467 (Arg131Gln) in patients receiving DPP-4 for 6 months (55). In our study, neither rs6923761 nor rs3765467 was associated with metformin response, glipizide response, or incretin levels during the OGTT.
Our study has limitations. Recent evidence has pointed out that metformin stimulates incretin hormone secretion (8, 9). Not conducting an OGTT in the absence of metformin, a logistical decision adopted when the study was originally designed, prevented our ability to isolate a genotype-driven effect of metformin over incretin hormones. We acknowledge that our small sample was not able to examine rare variants and had limited power to detect modest effects of low-frequency variants on the incretin response. We performed a power analysis to calculate the minimum effect size that can be detected in our sample using a significance threshold of P = 0.01 to account for 5 comparisons (Supplemental Table 14) (17). Our study was only able to detect differences in glucose levels that were larger than 15 mg/dL and differences in incretin levels that exceeded 1 SD for low-frequency variants (minor allele frequency = 1%). Although such large genetic effects are unlikely, they are still physiologically plausible and would be clinically significant. We recommend caution when interpreting results from our secondary analyses that included multiple models. Replication of these results in larger samples are needed to confirm or refute these findings. Although our study illuminates a potential role of GLP1R with immediate incretin response, the impact of polymorphisms at GLP1R on drug effects over the long term remains unclear. Longer assessments are needed to provide more impactful results that could be used in clinical practice (56). The effect of rs781387 on GLP-1 levels appeared to be slightly larger in older compared with younger individuals. It is possible that the impact of GLP1R genetic variation on incretin secretion may be larger in older individuals because of their higher underlying risk of diabetes compared with younger individuals. Because our study recruited mostly participants without diabetes, a future study could focus on determining whether these genetic effects on GLP-1 secretion and glucose homeostasis are more pronounced in older individuals with evidence of dysglycemia.
Our study, SUGAR-MGH, extends on previous knowledge implicating genetic variation at GLP1R as a contributor to an increased risk for T2D. It enabled us to provide a detailed characterization of the effect of genetic variation at GLP1R, ascertained through systematic mining of the Type 2 Diabetes Knowledge Portal, on the incretin effect, through multiple measurements of incretin, glucose, and insulin levels over time during an OGTT. In sum, we found an association between GLP1R polymorphisms and higher GLP-1 and glucose levels following an oral glucose load but found no association with insulin levels; this pattern of association is consistent with impaired incretin signaling that may, in turn, raise diabetes risk. A pharmacogenetic evaluation over multiple examinations in people with diabetes is needed to confirm our findings and assess the potential relevance of GLP1R genetic variation in patient care.
Acknowledgments
We acknowledge the contributions of the TIGER Data Portal and the Type 2 Diabetes Portal.
Glossary
Abbreviations
- AUC
area under the curve
- BMI
body mass index
- CV
coefficient of variation
- FG
fasting glucose
- GIP
glucose-dependent insulinotropic polypeptide
- GLP-1
glucagon-like peptide-1
- GWAS
genome-wide association studies
- OGTT
oral glucose tolerance test
- QC
quality control
- SUGAR-MGH
Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans
- T2D
type 2 diabetes
Contributor Information
Edgar G Dorsey-Trevino, Department of Medicine, Harvard Medical School, Boston, MA 02115, USA; Division of General Internal Medicine, Massachusetts General Hospital, Boston, MA 02114, USA.
Varinderpal Kaur, Diabetes Unit and Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Programs in Metabolism and Medical & Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Josep M Mercader, Department of Medicine, Harvard Medical School, Boston, MA 02115, USA; Diabetes Unit and Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Programs in Metabolism and Medical & Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Jose C Florez, Department of Medicine, Harvard Medical School, Boston, MA 02115, USA; Diabetes Unit and Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Programs in Metabolism and Medical & Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Aaron Leong, Department of Medicine, Harvard Medical School, Boston, MA 02115, USA; Division of General Internal Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Diabetes Unit and Center for Genomic Medicine, Massachusetts General Hospital, Boston, MA 02114, USA; Programs in Metabolism and Medical & Population Genetics, Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
Funding
This project is partly funded by the American Diabetes Association Innovative and Clinical Translational Award 1-19-ICTS-068. A.L. is supported by grant 2020096 from the Doris Duke Charitable Foundation. J.C.F. is supported by NIDDK K24 DK110550. SUGAR-MGH was conducted with support from National Institutes of Health/NIDDK awards R01 DK088214, R03 DK077675, and P30 DK036836; from the Joslin Clinical Research Center from its philanthropic donors; and the Harvard Catalyst: The Harvard Clinical and Translational Science Center (National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health Awards M01-RR-01066, 1 UL1 RR025758-04 and 8UL1TR000170-05 and financial contributions from Harvard University and its affiliated academic health care centers). The funders played no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Authors Contributions
A.L., J.C.F., and E.G.D.T. conceptualized the study design, hypothesis, and research project. V.K. and J.C.F. led the data collection. J.M.M. and V.K. led data management and performed the genetic analysis. E.G.D.T. and A.L. analyzed and interpreted the data. E.G.D.T. drafted the initial manuscript. A.L., J.C.F., V.K., and J.M.M. reviewed and edited the manuscript. All authors have reviewed and approved the final version of the manuscript. J.C.F. is the guarantor.
Disclosures
J.C.F. has received honoraria from Novo Nordisk and AstraZeneca. The rest of the authors report having no conflicts of interests.
Data Availability
The dataset analyzed during the current study are available from the corresponding author upon reasonable request.
References
- 1. Palmer SC, Mavridis D, Nicolucci A, et al. Comparison of clinical outcomes and adverse events associated with glucose-lowering drugs in patients with type 2 diabetes: a meta-analysis. JAMA. 2016;316(3):313-324. [DOI] [PubMed] [Google Scholar]
- 2. Florez JC. Pharmacogenetics in type 2 diabetes: precision medicine or discovery tool? Diabetologia. 2017;60(5):800-807. [DOI] [PubMed] [Google Scholar]
- 3. Fitipaldi H, McCarthy MI, Florez JC, Franks PW. A global overview of precision medicine in type 2 diabetes. Diabetes. 2018;67(10):1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vujkovic M, Keaton JM, Lynch JA, et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet. 2020;52(7):680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wessel J, Chu AY, Willems SM, et al. ; The E-IC. Low-frequency and rare exome chip variants associate with fasting glucose and type 2 diabetes susceptibility. Nature Communications. 2015;6(1):5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ezcurra M, Reimann F, Gribble FM, Emery E. Molecular mechanisms of incretin hormone secretion. Curr Opin Pharmacol. 2013;13(6):922-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bahne E, Sun EWL, Young RL, et al. Metformin-induced glucagon-like peptide-1 secretion contributes to the actions of metformin in type 2 diabetes. JCI Insight. 2018;3(23):e93936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. DeFronzo RA, Buse JB, Kim T, et al. Once-daily delayed-release metformin lowers plasma glucose and enhances fasting and postprandial GLP-1 and PYY: results from two randomised trials. Diabetologia. 2016;59(8):1645-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mulherin AJ, Oh AH, Kim H, Grieco A, Lauffer LM, Brubaker PL. Mechanisms underlying metformin-induced secretion of glucagon-like peptide-1 from the intestinal L cell. Endocrinology. 2011;152(12):4610-4619. [DOI] [PubMed] [Google Scholar]
- 11. Kappe C, Patrone C, Holst JJ, Zhang Q, Sjöholm A. Metformin protects against lipoapoptosis and enhances GLP-1 secretion from GLP-1-producing cells. J Gastroenterol. 2013;48(3):322-332. [DOI] [PubMed] [Google Scholar]
- 12. Maida A, Lamont BJ, Cao X, Drucker DJ. Metformin regulates the incretin receptor axis via a pathway dependent on peroxisome proliferator-activated receptor-α in mice. Diabetologia. 2011;54(2):339-349. [DOI] [PubMed] [Google Scholar]
- 13. Cordiner RLM, Mari A, Tura A, Pearson ER. The impact of low-dose gliclazide on the incretin effect and indices of beta-cell function. J Clin Endocrinol Metab. 2021;106(7):2036-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christensen AS, Hædersdal S, Storgaard H, et al. GIP and GLP-1 potentiate sulfonylurea-induced insulin secretion in hepatocyte nuclear factor 1α mutation carriers. Diabetes. 2020;69(9):1989-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Srinivasan S, Kaur V, Chamarthi B, et al. TCF7L2 genetic variation augments incretin resistance and influences response to a sulfonylurea and metformin: the Study to Understand the Genetics of the Acute Response to Metformin and Glipizide in Humans (SUGAR-MGH). Diabetes Care. 2018;41(3):554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walford GA, Colomo N, Todd JN, et al. The study to understand the genetics of the acute response to metformin and glipizide in humans (SUGAR-MGH): design of a pharmacogenetic resource for type 2 diabetes. PLoS One. 2015;10(3):e0121553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dorsey-Trevino EG, Kaur V, Mercader JM, Florez JC, Leong A. Supplemental Material. Deposited 23 April 2021. http://doi.org/18.361/generic.846s2
- 18. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Delaneau O, Zagury JF, Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10(1):5-6. [DOI] [PubMed] [Google Scholar]
- 21. Abecasis GR, Auton A, Brooks LD, et al. ; 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McCarthy S, Das S, Kretzschmar W, et al. ; Haplotype Reference Consortium. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kowalski MH, Qian H, Hou Z, et al. ; Consortium NT-OfPM. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 2019;15(12):e1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Das S, Forer L, Schonherr S, et al. Next-generation genotype imputation service and methods. Nat Genet. 2016;48(10):1284-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Type 2 Diabetes Knowledge Portal. 2022. https://t2d.hugeamp.org.
- 26. National Cancer Institute Division of Cancer Epidemiology & Genetics. 2022. https://ldlink.nci.nih.gov/?tab=home.
- 27. Ge T, Chen C-Y, Ni Y, Feng Y-CA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10(1):1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuznetsova A, Brockhoff PB, Christensen RHB. lmerTest package: tests in linear mixed effects models. 2017;82:26. 2017-11-29 ed2017. [Google Scholar]
- 30. Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. 2015;67:48. 2015-10-07 ed2015. [Google Scholar]
- 31. Meier JJ, Nauck MA. Is the diminished incretin effect in type 2 diabetes just an epi-phenomenon of impaired beta-cell function? Diabetes. 2010;59(5):1117-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen J, Spracklen CN, Marenne G, et al. The trans-ancestral genomic architecture of glycemic traits. Nat Genet. 2021;53(6):840-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pulit SL, Stoneman C, Morris AP, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28(1):166-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alonso L, Piron A, Morán I, et al. TIGER: The gene expression regulatory variation landscape of human pancreatic islets. Cell Rep. 2021;37(2):109807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Translational human pancreatic Islet Genotype tissue-Expression Resource (TIGER) Data Portal. 2016. http://tiger.bsc.es.
- 36. Broad Institute: GTEx Portal. 2021. https://gtexportal.org/home/.
- 37. Pais R, Gribble FM, Reimann F. Stimulation of incretin secreting cells. Ther Adv Endocrinol Metab. 2016;7(1):24-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang X, Young RL, Bound M, et al. Comparative effects of proximal and distal small intestinal glucose exposure on glycemia, incretin hormone secretion, and the incretin effect in health and type 2 diabetes. Diabetes Care. 2019;42(4):520-528. [DOI] [PubMed] [Google Scholar]
- 39. Zhang X, Cheng Z, Dong S, et al. Effects of ileal glucose infusion on enteropancreatic hormone secretion in humans: relationship to glucose absorption. Metabolism. 2022;131:155198. [DOI] [PubMed] [Google Scholar]
- 40. Fehmann HC, Habener JF. Insulinotropic hormone glucagon-like peptide-I(7-37) stimulation of proinsulin gene expression and proinsulin biosynthesis in insulinoma beta TC-1 cells. Endocrinology. 1992;130(1):159-166. [DOI] [PubMed] [Google Scholar]
- 41. Pilgaard K, Jensen CB, Schou JH, et al. The T allele of rs7903146 TCF7L2 is associated with impaired insulinotropic action of incretin hormones, reduced 24 h profiles of plasma insulin and glucagon, and increased hepatic glucose production in young healthy men. Diabetologia. 2009;52(7):1298-1307. [DOI] [PubMed] [Google Scholar]
- 42. Villareal DT, Robertson H, Bell GI, et al. TCF7L2 variant rs7903146 affects the risk of type 2 diabetes by modulating incretin action. Diabetes. 2010;59(2):479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schäfer SA, Tschritter O, Machicao F, et al. Impaired glucagon-like peptide-1-induced insulin secretion in carriers of transcription factor 7-like 2 (TCF7L2) gene polymorphisms. Diabetologia. 2007;50(12):2443-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saxena R, Hivert MF, Langenberg C, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42(2):142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spracklen CN, Horikoshi M, Kim YJ, et al. Identification of type 2 diabetes loci in 433,540 East Asian individuals. Nature. 2020;582(7811):240-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alonso L, Piron A, Morán I, et al. TIGER: The gene expression regulatory variation landscape of human pancreatic islets. Cell Rep. 2021;37(2):109807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koole C, Wootten D, Simms J, et al. Polymorphism and ligand dependent changes in human glucagon-like peptide-1 receptor (GLP-1R) function: allosteric rescue of loss of function mutation. Mol Pharmacol. 2011;80(3):486-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fortin JP, Schroeder JC, Zhu Y, Beinborn M, Kopin AS. Pharmacological characterization of human incretin receptor missense variants. J Pharmacol Exp Ther. 2010;332(1):274-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scrocchi LA, Brown TJ, MaClusky N, et al. Glucose intolerance but normal satiety in mice with a null mutation in the glucagon-like peptide 1 receptor gene. Nat Med. 1996;2(11):1254-1258. [DOI] [PubMed] [Google Scholar]
- 50. Sathananthan A, Man CD, Micheletto F, et al. Common genetic variation in GLP1R and insulin secretion in response to exogenous GLP-1 in nondiabetic subjects: a pilot study. Diabetes Care. 2010;33(9):2074-2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Nauck MA, Bartels E, Orskov C, Ebert R, Creutzfeldt W. Additive insulinotropic effects of exogenous synthetic human gastric inhibitory polypeptide and glucagon-like peptide-1-(7-36) amide infused at near-physiological insulinotropic hormone and glucose concentrations. J Clin Endocrinol Metab. 1993;76(4):912-917. [DOI] [PubMed] [Google Scholar]
- 52. Nauck M, Schmidt WE, Ebert R, et al. Insulinotropic properties of synthetic human gastric inhibitory polypeptide in man: interactions with glucose, phenylalanine, and cholecystokinin-8. J Clin Endocrinol Metab. 1989;69(3):654-662. [DOI] [PubMed] [Google Scholar]
- 53. Xie C, Huang W, Watson LE, et al. Plasma GLP-1 response to oral and intraduodenal nutrients in health and type 2 diabetes - impact on gastric emptying. J Clin Endocrinol Metab. 2021;107(4):e1643-e1652. [DOI] [PubMed] [Google Scholar]
- 54. Javorský M, Gotthardová I, Klimčáková L, et al. A missense variant in GLP1R gene is associated with the glycaemic response to treatment with gliptins. Diabetes Obes Metab. 2016;18(9):941-944. [DOI] [PubMed] [Google Scholar]
- 55. Han E, Park HS, Kwon O, et al. A genetic variant in GLP1R is associated with response to DPP-4 inhibitors in patients with type 2 diabetes. Medicine (Baltim). 2016;95(44):e5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Meigs JB. The genetic epidemiology of type 2 diabetes: opportunities for health translation. Curr Diab Rep. 2019;19(8):62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The dataset analyzed during the current study are available from the corresponding author upon reasonable request.