Abstract
Matrix metalloproteinase-1 (MMP-1) is a Zn(II) dependent endopeptidase involved in the degradation of collagen, the most abundant structural protein in the extracellular matrix of connective tissues and the human body. Herein we performed a multilevel computational analysis including molecular dynamics (MD), combined quantum mechanics/molecular mechanics (QM/MM), and quantum mechanics (QM) calculations to characterize the structure and geometry of the catalytic Zn(II) within the MMP-1 protein environment in comparison to crystallographic and spectroscopic data. The substrate’s removal fine-tuned impact on the conformational dynamics and geometry of the catalytic Zn(II) center was also explored. Finally, the study examined the effect of substituting catalytic Zn(II) by Co(II) on the overall structure and dynamics of the MMP-1 THP complex and specifically on the geometry of the catalytic metal center. Overall our QM/MM and QM studies were in good agreement with the MM description of the Zn(II) centers in the MD simulations.
Keywords: Zn(II) containing enzymes, Matrix metalloproteinases, Molecular dynamics, QM/MM calculations
Graphical Abstract

Multilevel MD, QM/MM and QM Studies Reveal the Effects of the Nature of the Metal Ion, Protein and Substrate on the Catalytic Center in Matrix Metalloproteinase-1.
INTRODUCTION
Zinc cofactor is essential for the activity of more than 300 metalloenzymes.[1–3] In enzymes, Zn(II) participates in a variety of biological processes by playing a catalytic, structural, or co-catalytic role.[4–6] Matrix metalloproteinases (MMPs) are a family of Zn(II) dependent hydrolases that contain two Zn(II) ions: a catalytic Zn(II), essential for catalysis, and a structural Zn(II) contributing to the overall structural stability of these enzymes.[7–9] MMPs are involved in the degradation of extracellular matrix (ECM) components essential for tissue homeostasis and participate in a wide range of processes including cell differentiation, migration, and proliferation.[10–12] However, MMPs dysregulation contributes to various pathological conditions, including arthritis and cancer.[13,14] The subject of this study was human MMP-1 also known as collagenase-1.[15]
The general structure of MMP-1 is made up of an N-terminal catalytic (CAT) domain, a C-terminal hemopexin (HPX) like domain, and a 16 amino acids proline-rich linker region connecting them (Figure 1).[8,16,17] The HPX domain consists of a Ca(II)-linked four-blade β-propeller fold.[16,18]
Figure 1.
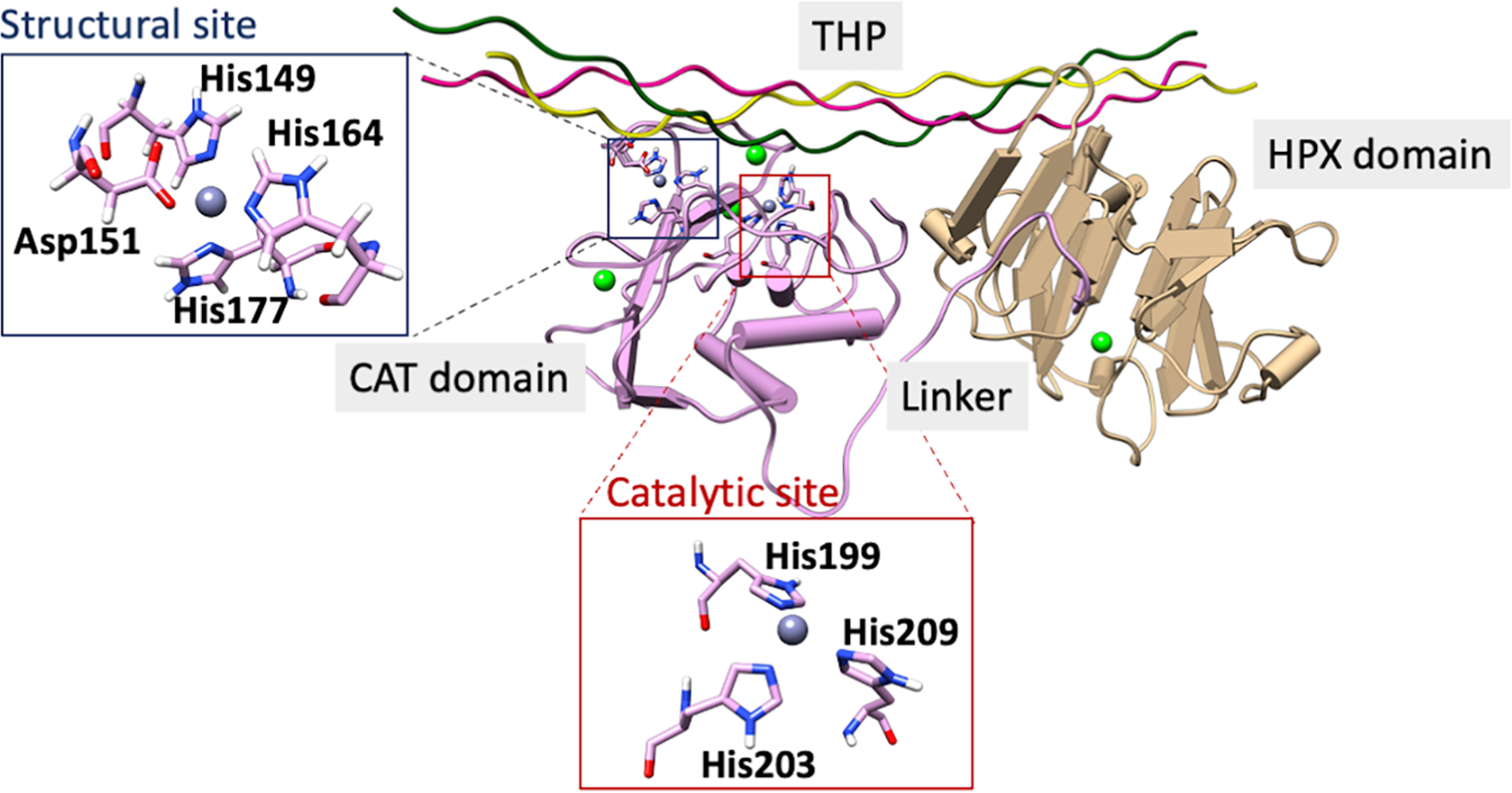
Substrate bound MMP-1 structure (PDB ID: 4AUO). The catalytic site is highlighted in a red box and the structural site in a blue box. Ca(II) ions in the CAT and HPX domains are colored in green.
The overall structure of the catalytic domain consists of five β-sheets (βI–V) and three α-helices (αI-III) connected by various loops that play important functional roles.[16,17] The CAT domain contains two Zn(II) ions, a structural one and a catalytic one, and various substrate-binding sites (known as specificity pockets or subsites designated as S and S’, respectively, with primed subsites positioned to the right of the catalytic Zn(II) and unprimed ones to the left)[16,19,20] (Figure S1). MMPs subsites differ in depth and hydrophobicity. Among the various subsites, the S1’ has been extensively utilized for the development of selective inhibitors owing to its sequence variation amongst MMPs.[21,22] In MMP-1 specifically, Arg195 (Figure 2a) serves as the S1’specificity pocket gatekeeper. This pocket is lined by a loop referred to as the S1’ specificity loop, the least conserved part in MMPs.[21,23] Other key connecting loops in the catalytic domain of MMP-1 include the V-B loop and the S-loop. The V-B loop, located between the fifth β-strand and the second α-helix of the catalytic domain, has a conserved sequence (183RWTNNFREY191) and has been considered essential for catalysis/collagenolytic activity.[24] A long S-shaped loop (called the S-loop) positioned above the catalytic Zn(II) site maintained by the structural Zn(II) ion lines the catalytic site.[25,26] The structural site in the CAT domain is formed by a Zn(II) ion that is tetra-coordinated to three histidine residues (His149, His164, His177) and an aspartate (Asp151).[17,27] Approximately 12 Å from the structural site is located the catalytic site.[28] In the catalytic site, a conserved Zn(II) binding motif ‘HEXXHXXGXXH’ contains the histidine residues (His199, His203, His209) coordinating the catalytic Zn(II) ion.[16] The same motif holds the conserved glutamate residue (Glu200), critical in the catalytic mechanism of MMP-1.[29,30] Below the Zn(II) catalytic site is the conserved 1,4 β-Met turn (Ala215, Leu216, Met217, Tyr218) (Figure 2b), forming a sheath that shields the bottom of the active site.[31,32] Apart from the Zn(II) ions, there are three Ca(II) ions stabilizing the different loops in the CAT domain.[33]
Figure 2.
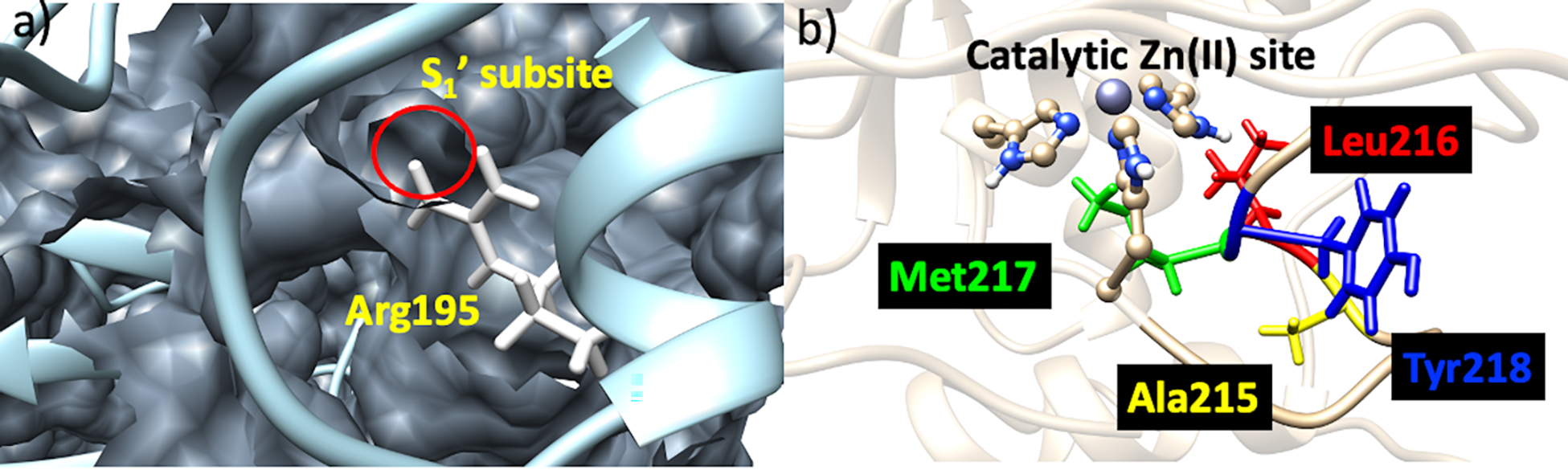
a) S1’ subsite (circled in red) and the gate-keeper residue Arg195 (white). b) 1,4 β-Met turn: Ala215 (yellow), Leu216 (red), Met217 (green), and Tyr218 (blue).
Triple-helical peptides (THPs) have been utilized as collagen model substrates.[34,35] Modeling studies on MMPs suggest that the enzyme prefers a looser triple-helical conformation to ease the cleavage process with assistance from the S10’ exosite in blade 1 and residues of blade 2 of the HPX domain.[36–38] Hydrolysis begins when the catalytic Zn(II) attacks the carbonyl group of the scissile bond resulting in a penta-coordinated Zn(II) with the fourth position occupied by a water molecule. The conserved Glu200 residue, which acts as a proton shuttle, strongly polarizes the Zn(II)-bound water molecule followed by hydrogen bond rearrangements, proton transfer, and finally, the C-N bond dissociation.[39–41]
A well-established experimental approach for probing Zn(II)-containing enzymes is a substitution of the Zn(II) ion with paramagnetically active Co(II).[42–44] Previous studies utilizing such metal replacement have implicated the necessity of Zn(II) for MMPs catalysis.[45,46] Since Co(II) and Zn(II) share similar covalent radii and have similar coordination geometry preferences, they are likely to interact in a similar fashion with the neighboring residues in the active site.[47–49] Co(II) substituted active site results in an enzyme that maintains a considerable fraction of the wild-type (WT) catalytic activity.[46,50] Yang and coworkers have probed the secondary structure of MMP-1 by a variety of methods including circular dichroism (CD), fluorescence, and X-ray absorption spectroscopies.[46] The spectroscopic studies suggested that the catalytic Zn(II) is in the five coordination (5C) state, and the structural Zn(II) is in the four coordination (4C) state in the free MMP-1. Co(II) substituted MMP-1 retained most of the secondary structural characteristics of WT MMP-1. Importantly, in the absence of structural Zn(II) in MMP-1, the coordination state of the catalytic Co(II) switches from 5C to 6C. The detailed atomic mechanism of this transition, however, remains unknown.
Computational methods have been extensively used to reveal structure-function correlation of MMP-1.[51–54] Modeling studies revealed aspects of the mechanism, dynamics, and binding in MMP-2, MMP-3, and MT1-MMP.[40,41,55] A recent study from our group explored the synergy between the catalytic and the structural Zn(II) ions in influencing the conformational dynamics of the MMP-1•THP complex.[56]
Despite the recent progress in understanding the structure and mechanisms of MMP-1, there is a substantial lack of essential knowledge in the following areas:
1) The role of the protein environment and the THP substrate in influencing the geometric and electronic structure of the catalytic Zn(II) site, and its further network of interactions with residues from the second coordination sphere. To explore this, we performed combined quantum mechanics and molecular mechanics (QM/MM) study using structures from recent MD simulations.[56] Furthermore, in order to evaluate the effect of the protein and substrate environment on the Zn(II) center, we need to compare the QM/MM structures with QM-optimized structures in the continuum solvent model.
2) Although prior investigations have revealed important conformational transitions of the already formed MMP-1•THP complex, there is a considerable lack of information about the long-range dynamics and correlated motions of the free MMP-1 after removal of the THP and how they relate to the dynamical properties of the MMP-1•THP complex. In order to reveal the effect of the THP substrate on the dynamics of the MMP-1•THP complex, we need to explore the local structural changes during the dynamics of the free MMP-1 upon removal of the THP from the structure.
3) Although earlier studies have indicated that Zn(II) replacement by Co(II) in the catalytic site results in the Co(II)-containing enzyme behaving in a similar fashion, we lack knowledge on how such a replacement affects the overall dynamics and long range-interactions in the protein and precisely how the geometric and electronic structure of the catalytic Co(II) center differs from the one of the catalytic Zn(II) center. To obtain this information, we need to perform MD simulation with the Co(II)-substituted catalytic site and subsequent QM/MM calculations of the geometric and electronic structure of the Co(II) center.
4) Importantly, we miss vital information about the molecular mechanism of the key experimental finding that the removal of the structural Zn(II) from MMP-1 changes the Co(II)-substituted catalytic site from 5C to 6C.[46] To obtain this knowledge, we need to perform QM/MM calculations.
To address all the above missing insights, we implemented a multilevel combination of QM/MM, QM, and MD methods on the following systems: (a) WT MMP-1 Zn1 Zn2•THP complex (notated as MMP-1•THP); (b) MMP-1 Co1 Zn2•THP complex (notated as MMP-1 Co1•THP); (c) substrate-free WT MMP-1 Zn1 Zn2 (notated as MMP-1•no THP); (d) substrate-free MMP-1 Co1 Zn2 (notated as MMP-1 Co1•no THP); and (e) substrate free MMP-1 Co1 without structural Zn(II) (notated as MMP-1 Co1•no Zn2 no THP).
METHODOLOGY
System Preparation
The X-ray crystallographic structure of MMP-1 (PDB ID: 4AUO)[16] bound to a triple-helical peptide (THP) substrate was used for modeling. This crystallographic structure contains an E200A mutation that renders the enzyme inactive, therefore, Ala200 was mutated in silico to Glu200. Missing hydrogen atoms were added using the leap module of AMBER 18,[57] and missing nonhydrogen atoms were added using the Swiss PDB viewer.[58] pKa values of the catalytic Zn(II) coordinated histidine residues were determined by the PROPKA 3.0 program[59] and their parameters were generated using python-based Metal Centre Parameter Builder MCPB.py v3.0.[60] Bond and angle parameters were derived using the Seminario method,[61] and the partial charges for the electrostatic potentials were obtained using the RESP charge fitting procedure[62] at the B3LYP/6-31G* level of theory. The same method was used for generating the parameters for the catalytic Co(II) system. The four-coordinated structural Zn(II) ion, bound to His149, Asp151, His164, and His177, was treated with the Zinc Amber Force Field (ZAFF).[63] The Ca(II) ions present in the CAT and HPX domains were included in all systems. The rest of the protein molecule, and the THP substrate, were modeled using the ff14SB force field.[64] The THP substrate was numbered with reference to our previous studies.[51] For parameter preparation of the enzyme without the substrate, we removed the substrate from the PDB, and the same procedures were applied.
Molecular Dynamics
The conformational changes in the various systems were studied using MD simulations.[56] Periodic boundary conditions were utilized to simulate a continuous system. The structures were enclosed in a periodic box with TIP3P water molecules that extended 10 Å away from the protein.[65] Cl− counter ions were added to neutralize the system. The MMP-1•noTHP simulation system contained 48915 atoms and the MMP-1 Co1•no THP contained 73272 atoms. Long range electrostatic interactions involving van der Waals interactions were incorporated via Particle Mesh Ewald Algorithm (PME) with a non-bonded cut-off of 10 Å.[66] Energy minimizations using the SANDER module of AMBER18 were carried out in two steps, with the steepest descent path for the initial 5000 steps and the conjugate gradient method applied for the final 5000 steps. The first minimization involved minimizing the solvent molecules while restraining the protein, whereas the complete system was minimized during the second minimization. This was followed by gradual annealing of the system from 0K to 300K for 100 ps in an NVT canonical ensemble. The temperature was maintained by Langevin thermostat[67] with a 1.0 ps−1 collision frequency. The SHAKE algorithm[68] was implemented to freeze the hydrogen containing bonds. During the process, solute molecules were restrained with a harmonic potential of 4.0 kcal/mol/Å. A uniform density was achieved through a weakly restrained MD simulation for 1 ns using an NPT ensemble. The system was then equilibrated without any restraints at 300K for 3 ns in NPT canonical ensemble. The pressure was controlled by Brendson barostat.[69] Finally, the 1 μs production MD was performed on the equilibrated structure with a time step of 2 fs in canonical ensemble NPT. The production simulations were done using the GPU enabled PMEMD version of AMBER18.[57]
The system stability was analyzed by Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), and Radius of Gyration (RoG), Solvent Accessible Surface Area (SASA) using the CPPTRAJ module[70] of AMBER. The collective correlated motions were studied using Dynamic Cross-Correlation Analysis (DCCA)[71] and Principal Component Analysis (PCA).[72] Both DCCA and PCA were done on the protein C-α atoms in the final 500 ns MD trajectory with Bio3D.[73] Three independent MD simulations, performed for each system, showed close average RMSD values (Figure S2), indicating for convergence of the simulations. In addition, as a criterion for convergence of a single trajectory, the cosine of the principal component (PC)[74,75] was evaluated using MODE-TASK[76] and showed satisfactory sampling (Table S1).
QM/MM and QM Calculations
To address the limited accuracy of the MM force field, QM/MM calculations were done to refine the catalytic Zn(II) site geometry. Structures taken from the equilibrated part of the MD trajectory with a stable RMSD value (> 400 ns) were used for the QM/MM optimizations. Due to enzyme internal flexibility, the choice of initial structure from the MD trajectory can influence the geometry and properties of the QM/MM optimized structure.[77–79] We therefore performed QM/MM optimizations of five different MD snapshots. All structures were randomly selected from the equilibrated part of the MD trajectory as follows: snapshot 1: 630.1 ns, snapshot 2: 568.0 ns, snapshot 3: 599.1 ns, snapshot 4: 606.5 ns, and snapshot 5: 654.3 ns. Water solvation was kept till 12 Å surrounding the protein. The rest of the solvation shell was truncated using the CPPTRAJ module[70] of AMBER. The QM/MM optimizations were performed in Chem-Shell software[80] combining TURBOMOLE program[81] for the QM region and DL-POLY[82] for the MM region. Chemshell default geometry convergence criteria of 0.00045 was used. Restricted B3LYP hybrid functional and def2-SVP basis set[83] was used for Zn(II) centered MMP-1 whereas unrestricted B3LYP functional was used for the catalytic Co(II) substituted MMP-1 with the same basis set.[84] Calculations on the Co(II) substituted MMP-1 was done on the high spin state (S=3/2).[40] Five-coordinated catalytic metal site was used for the calculations as previous studies suggested the presence of five-coordinated catalytic metal ion in the resting state of MMPs, including MMP-1.[46] The crystallographic structure of MMP-1 (PDB ID:4AUO) is unproductive one as the scissile peptide bond is not properly positioned for hydrolysis.[16] Considering the improper orientation of the THP in the crystallographic structure, two water molecules (Wat1 and Wat2) were included in the QM region coordinated to the catalytic Zn(II)/Co(II) in addition to the three histidine residues (His199, His203, His209). Next, we modeled the substrate free-MMP-1 with the structural Zn(II) removed. In the absence of structural Zn(II), a third water molecule (Wat3) was further added to the catalytic site of Co(II) substituted derivative of MMP-1, with regard to earlier studies.[46] The conserved Glu200 residue important for collagenolysis was also included in the QM region for QM/MM calculations. Thus, the QM region contained 49 atoms for the catalytic Zn(II)/Co(II) MMP-1 and 52 atoms for the Co(II) substituted MMP-1 without the structural Zn(II) having an overall charge of +1. QM subsystems were capped by the link atom (hydrogen atom), which approximates the QM/MM boundary. The rest of the system was defined as MM region and was described by the ff14SB AMBER force fields. We used the extended model for the QM/MM minimizations. Protein residues and water molecules within 8 Å from the QM region were allowed to move during the QM/MM minimizations, and the rest of the atoms were kept frozen. The polarization effect of the MM region of the protein on the QM part was incorporated by the electronic embedding scheme. To assure that variations in the size of the QM region do not influence the quality of the QM/MM calculation, we performed QM/MM optimizations for Zn(II)/Co(II) MMP-1 with the third water molecule included in the QM part of the system (i.e., 52 QM atoms). The results were consistent with the ones obtained with 49 QM atoms where the water molecule was in the MM part of the system (Figures S3a and S3b). The QM-continuum model calculations in water were performed using the Polarizable Continuum Model (PCM)[85] which is the default Self-Consistent Reaction Field (SCRF) method in Gaussian 16. The dielectric constant of the solvent (water) was 78.35. The B3LYP level of theory and the def2-SVP basis set was used for the calculations.
RESULTS AND DISCUSSION
This study first investigated the geometric and electronic structure of the catalytic Zn(II) center under the influence of the protein environment, additionally accounting for the effect of the large and conformationally flexible substrate with QM/MM and QM methods. The study then focused on assessing the effect of the substrate on local perturbations of the enzyme structure and the overall dynamics of substrate-free MMP-1. The next step was an exploration of the influence of a Co(II)-substituted catalytic site on the dynamics of the MMP-1•THP complex and further analysis of the geometric and electronic structure of the Co(II) catalytic site with QM/MM and QM methods. Finally, the study explored the atomic mechanism driving the experimentally observed transition from five-coordinated (5C) Co(II) to six-coordinated (6C) Co(II) center upon removal of the structural Zn(II) ion.
Geometric and Electronic Structure of the Catalytic Zn(II) Center in MMP-1•THP
The catalytic site of the MMP-1•THP complex was probed by means of MD, QM/MM, and QM methodologies (Figure 3, Figures S4, S5, S6, Table S2).
Figure 3.
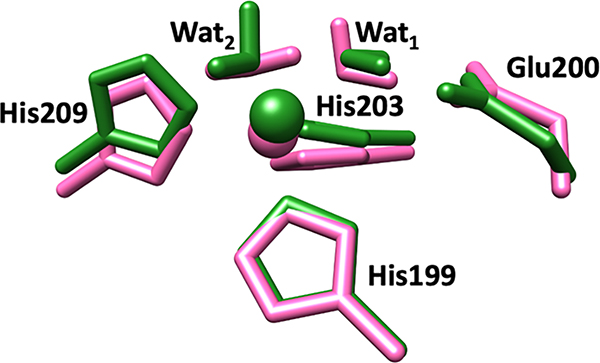
Superimposed catalytic Zn(II) sites of MMP-1•THP including coordinated histidine residues (His199, His203, His209), catalytic water (Wat1), auxiliary water (Wat2), and conserved glutamate residue (Glu200) in the X-ray crystallographic structure (PDB ID: 4AUO) (red), MD snapshot (blue), QM/MM optimized (green), and QM-continuum solvent (yellow) models.
An extended X-ray absorption fine structure (EXAFS) study by Yang et al. demonstrated five-coordinated (5C) catalytic Zn(II) for MMP-1.[46] Therefore, the QM region, for our QM/MM optimizations, was a five-coordinated catalytic Zn(II) with coordination to three His ligands and two water molecules (His199, His203, His209, and Wat1, Wat2), and the catalytically important non-coordinated glutamate (Glu200) residue.[46] QM/MM optimizations and MD simulations showed that the orientation of Glu200 was stabilized by interactions with the catalytic Zn(II) first-coordinating sphere residue His203, and a second sphere, Val196 residue. Catalytic water (Wat1) was strongly polarized by Glu200 carboxylate oxygens. In the QM-continuum solvent model calculations, the imidazole rings of Zn(II)-coordinated His199 and His209 residues rotated in respect to their orientation in the QM/MM optimized structure.
We characterized five QM/MM optimized structures from the MD trajectory. The average distances between the catalytic Zn(II) and the nitrogen atoms (NE2) of its first-sphere coordinating histidines His199, His203, and His209 were 2.06, 2.17, and 2.05 Å, respectively (Table 1). The QM-continuum solvent optimized structure showed similar bond lengths. In general, the bond lengths between catalytic Zn(II) and the two coordinated histidines (His199, His203) were larger for the MD, averaged QM/MM, and QM-continuum solvent model calculations compared to the X-ray crystallographic structure of MMP-1•THP.[16] A different trend was observed in the case of Zn(II) and the third coordinating His209. The bond length in the MD, averaged QM/MM, and QM- continuum solvent model was slightly shorter than the X-ray crystallographic structure, which may be due to the crystal packing effects. In the QM/MM optimized structures, the average bond angles between His199-Zn(II)-His203, His203-Zn(II)-His209, and His199-Zn(II)-His209 were 100.98°, 94.74°, and 108.90°, respectively (Table 2). Eliminating the protein effects, the bond angles between His203-Zn(II)-His209 and His199-Zn(II)-His209 increased to 99.45° and 121.75°, respectively, indicating perturbing effects on the enzyme’s environment. Greater deviation from the crystallographic structure was observed for the bond angle between His199-Zn(II)-His209 in the QM-continuum solvent model. Mulliken charges calculated for the QM/MM and the QM geometries (Table S3) were similar.
Table 1.
Distances between the catalytic Zn(II) and coordinated histidines in the X-ray crystallographic structure, MD trajectory, QM/MM snapshots, and QM-continuum solvent model.
| Atoms | Distance (Å) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| X-ray structure | MD | QM/MM Snapshots |
QM/continuum | |||||||
| 1 | 2 | 3 | 4 | 5 | Avg | |||||
|
| ||||||||||
| Zn(II) | His199-NE2 | 1.82 | 1.96 | 2.06 | 2.06 | 2.08 | 2.05 | 2.07 | 2.06 | 2.06 |
| Zn(II) | His203-NE2 | 1.93 | 1.98 | 2.19 | 2.18 | 2.14 | 2.14 | 2.18 | 2.17 | 2.12 |
| Zn(II) | His209-NE2 | 2.21 | 1.95 | 2.05 | 2.04 | 2.07 | 2.03 | 2.08 | 2.05 | 2.07 |
Table 2.
Angles between the catalytic Zn(II) ion and the coordinated histidines in the X-ray crystallographic structure, MD trajectory, QM/MM snapshots, and QM-continuum solvent model.
| Atoms | Angle (°) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| X-ray Structure | MD | QM/MM Snapshots |
QM/continuum | ||||||||
| 1 | 2 | 3 | 4 | 5 | Avg | ||||||
|
| |||||||||||
| His199-NE2 | Zn(II) | His203-NE2 | 104.43 | 108.70 | 103.33 | 98.46 | 102.97 | 101.29 | 98.85 | 100.98 | 100.75 |
| His203-NE2 | Zn(II) | His209-NE2 | 94.21 | 107.78 | 93.73 | 94.86 | 95.53 | 95.47 | 94.10 | 94.74 | 99.45 |
| His199-NE2 | Zn(II) | His209-NE2 | 108.47 | 111.90 | 108.67 | 111.08 | 106.08 | 111.52 | 107.10 | 108.90 | 121.75 |
To further explore the effect of the protein environment on the Zn(II) sites, hydrogen bonding interactions involving the catalytic Zn(II) and the structural Zn(II)-bound residues were analyzed (Table 3). The results show that both QM and QM/MM optimizations provided a good description of the geometric and electronic structure of the catalytic Zn(II) center with comparable accuracy.
Table 3.
Comparison of hydrogen bonding interactions in the catalytic and structural site of the X-ray crystallographic structure, averaged MD trajectory, and the QM/MM optimized geometry of MMP-1•THP.
| Atoms | Hydrogen bond length (Å) | ||||
|---|---|---|---|---|---|
|
| |||||
| Donor | DonorH | Acceptor | Crystal Structure | MD | QM/MM |
|
| |||||
| CATALYTIC SITE | |||||
|
| |||||
| His199-ND1 | His199-HD1 | Leu216-O | 3.17 | 2.81 | 2.82 |
| His209-N | His209-H | Ser208-OG | Not present | 2.82 | 2.85 |
| His203-N | His203-H | Glu200-O | Not present | 2.77 | 2.74 |
|
| |||||
| STRUCTURAL SITE | |||||
|
| |||||
| His177-N | His177-H | His164-O | 2.94 | 2.85 | 2.81 |
| His164-N | His164-H | His177-O | 3.10 | 2.90 | 2.92 |
| Ser153-N | Ser153-H | Asp151-O | 3.19 | 2.78 | 2.78 |
| His164-ND1 | His164-HD1 | Gln774-OE1 | 3.12 | 2.83 | 2.81 |
The hydrogen bond between His199 and Leu216 (a residue from the 1,4 β-Met turn (Figure 2b)) was relatively stronger in the QM/MM optimized structure and the MD trajectory (Table 3) compared to the X-ray structure. In the QM/MM optimized structure, His209 and His203 were stabilized by hydrogen bonding interactions with the nearby Ser208 and Glu200 residues (Ala200 in the X-ray crystallographic structure); however, these interactions were not observed in the X-ray crystallographic structure. Similarly, in the QM/MM optimized geometry of the structural site of MMP-1, stronger hydrogen bonds were observed between His164 and His177, Asp151 and Ser153, and His164 and Gln774 (T strand of the THP) in comparison to the X-ray crystallographic structure (Table 3).
The QM/MM optimization of the five randomly selected snapshots from the MD trajectory showed high structural similarity to one another and only minor deviations in the distances and angles (Figure 4, Tables 1, 2, S4 and S5).
Figure 4.
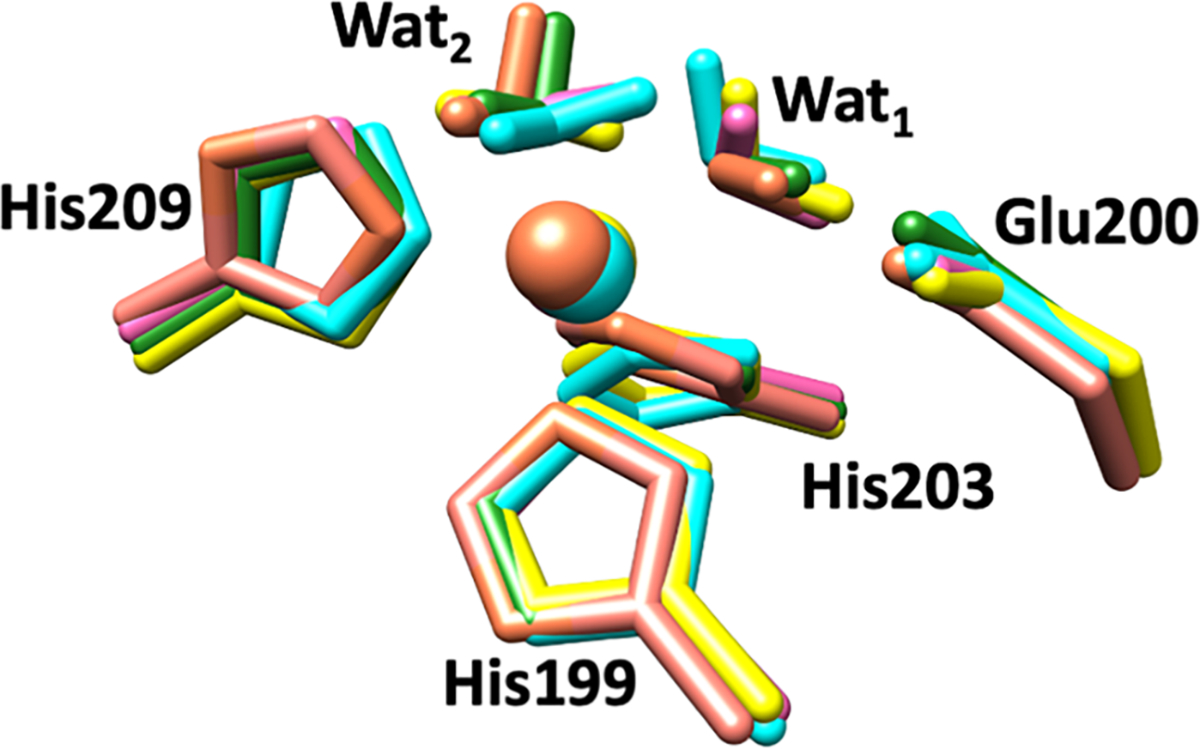
Superimposed catalytic sites of the QM/MM optimized structures from multiple snapshots of MMP-1•THP. Snapshot 1 (used for further calculations) is represented in green, while Snapshot 2 = cyan, Snapshot 3 = yellow, Snapshot 4 = pink, and Snapshot 5 = brown.
Effect of the THP Substrate on the Conformational Dynamics of MMP-1
MD studies of the MMP-1•THP complex revealed conformational features and long-range correlated motions between the CAT and the HPX domains and between the enzyme and THP.[56] However, it is important to elucidate the effect of THP removal on the overall dynamics and structural characteristics of MMP-1. Thus, we removed THP and performed MD simulations of substrate-free MMP-1 (MMP-1•no THP).
Overall Dynamics and Domain-Domain Interactions
Stable RMSDs were observed for both MMP-1•THP and the substrate-removed MMP-1[56] (Figure S7). The distance between the catalytic and structural sites in MMP-1•no THP (14.78 Å) did not change significantly from the one observed in MMP-1•THP (14.39 Å) (Figure 5a). THP removal affected the average distance between the CAT and HPX domains in MMP-1 (Figure 5b). The average distance between the CAT and HPX domains in MMP-1•THP was 38.94 Å (standard deviation of 0.36) which slightly increased to 40.50 Å for MMP-1•no THP with more significant fluctuations (standard deviation of 1.21). The difference of 1.46 Å between the mean interdomain distances with THP and upon its removal indicates that THP might potentially influence interactions between the CAT and the HPX domain compared to the ES complex. However, the interdomain interactions and reciprocal conformational dynamics are a general property of MMP1 as suggested by NMR and X-ray scattering[86] and may not be just a result of the THP unbinding. MMP-1•THP enzyme-substrate complex PCA showed a stable complex overall.[56] However, substrate removal induced some increased flexibility, primarily in the CAT domain of MMP-1 (Figure S8). DCCA of MMP-1•no THP shows a greater number of residues involved in anti-correlated motions compared to MMP-1•THP (Figure S9). Increased anti-correlated motions were observed between residues of the S1’ specificity loop, the S10’ exosite, and the blade 1 region of the HPX domain and the CAT domain. HPX blade 4 region residues were involved in a larger number of correlated motions coupled to the CAT domain residues when the THP was absent. Therefore, the PCA and DCCA further support the idea that the THP plays a key role in modulating the communication between the CAT and HPX domains.
Figure 5.
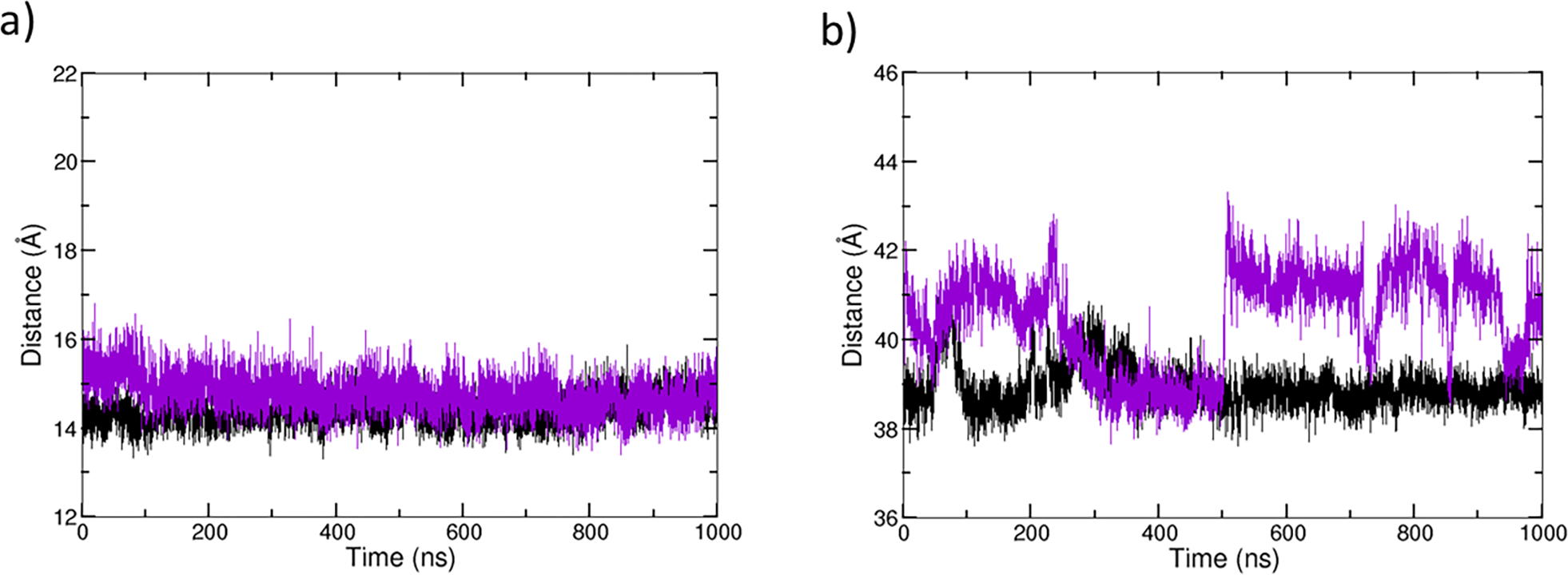
Distance plots of MMP-1•THP (black) and MMP-1•no THP (violet). a) Distance between center of mass of catalytic and structural site of MMP-1. b) Distance between center of mass of CAT and HPX domains of MMP-1.
How Does the THP Binding Change the Hydrogen Bonding Interactions Around Key Residues R195 and M217?
Prior studies from our group highlighted that the S1’ subsite gatekeeper residue, Arg195, was involved in a stable hydrogen bonding interaction with Tyr221 and Ser220, located at the entrance of the S1’ subsite in MMP-1•THP.[56] These interactions are essential for correct positioning of Arg195. As previously shown, the catalytic Zn(II) is a factor in maintaining these interactions.[56]
Analysis of Arg195 interactions with bound and removed THP indicate that the hydrogen bonds with Tyr221 and Ser220 are not present in MMP-1•no THP simulations (Figures 6a and 6b). Indeed, the hydrogen bonding interaction Arg195-NE···O-Tyr221 does not exist in the crystallographic structure of MMP-1•THP (4AUO).[16] This H-bond appeared in the MD simulation after 50 ns and remained relatively stable during the entire simulation. It is expected that in MD simulations, we can observe new interactions due to of the dynamic nature of proteins. In the MD simulation of the MMP-1•noTHP, however, this interaction does not exist which agrees with the 2CLT[17] crystallographic structure of the apo-MMP-1. The other characteristic interaction Arg195-NH2···O-Ser220, in contrast, exists in the crystallographic structure of MMP-1•THP (4AUO) and is also conserved during the MD simulation of the MMP-1•THP complex as well. Indeed, this interaction still exists in the crystallographic structure of apo-MMP-1, however, it disappears during the MD simulation without THP. This difference might be due to the time scale of the performed MD (1 μs), which is much shorter than the duration of the entire process of substrate unbinding. Indeed, after THP removal, we observe structural changes in the MD trajectory, but they are likely to be very transient and might not represent the final (equilibrated) state of the apo-MMP-1 after THP removal. Therefore, the simulation may capture only the initial events following substrate unbinding. The loss of these hydrogen bonds might be related to the disappearance of the interaction between Tyr221 and Gln779 (THP) upon THP removal. The loss of the two hydrogen bonds also agrees with the experimental study that Arg195, Ser220, and Tyr221 undergo induced fit upon THP binding.[87]
Figure 6.
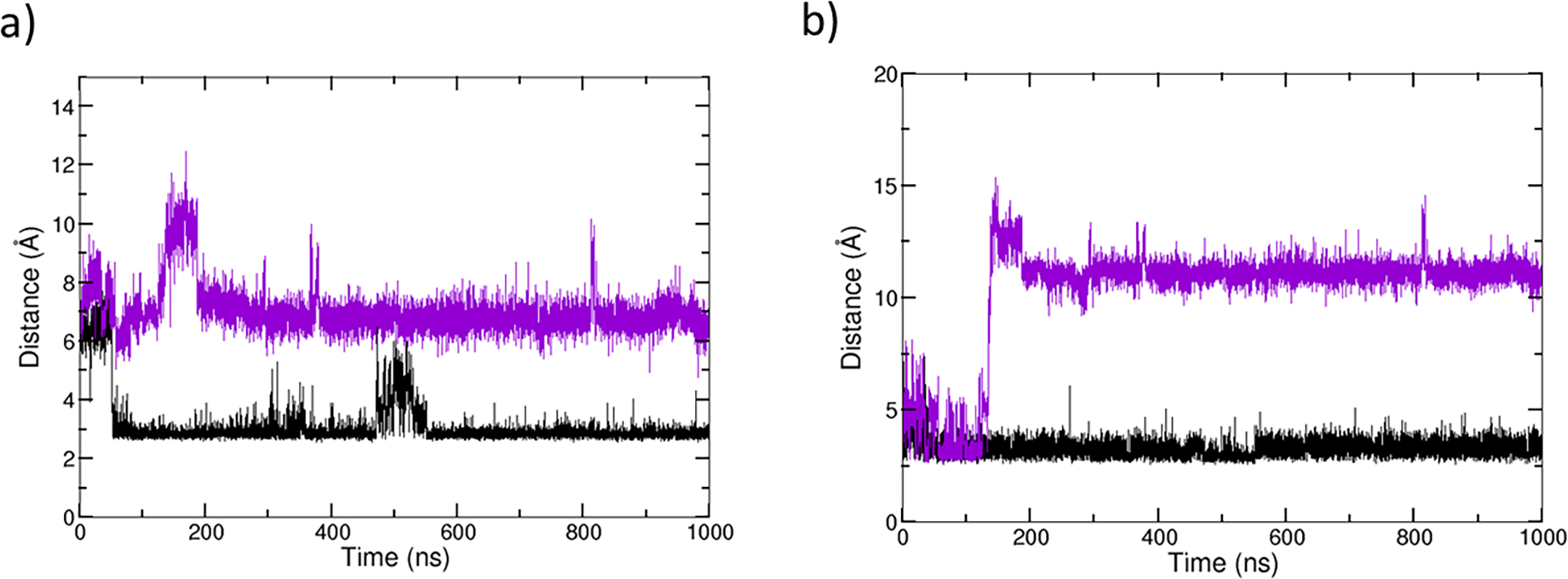
Hydrogen bonding interactions involving Arg195, Tyr221, and Ser220 for MMP-1•THP (black) and MMP-1•no THP (violet). a) Hydrogen bonding distances between the donor nitrogen (NE) of Arg195 and the acceptor oxygen (O) of Tyr221. b) Hydrogen bonding distance between donor nitrogen (NH2) of Arg195 with acceptor oxygen (O) of Ser220.
THP unbinding leads to two new hydrogen bonds for Arg195, which are not very stable: Arg195 - Ser224 (31%) and Arg195 - Thr222 (22%). Both the Ser224 and Thr222 residues belong to the S1’ specificity loop of MMP-1. In addition, the THP removal leads to a more stable hydrogen bond between Met217 and Ser210 (28% in the MMP-1•THP complex to 94% upon removal of THP). These results further suggest that along with the catalytic Zn(II), the THP might influence stabilizing interactions at the S1’ subsite; however, further studies are needed to define the extent of this effect.
Effect of the substrate THP on the QM/MM structure of the MMP-1 Catalytic Zn(II) Center
The substrate’s influence on the structure and interactions of the catalytic Zn(II) site in MMP-1 was examined by comparing the QM/MM optimized MMP-1•THP and MMP-1•no THP structures (Figure 7). The main difference was for the bond between the catalytic Zn(II) and Wat2. This bond was 2.42 Å in MMP-1•THP complex but in MMP-1•no THP decreased to 2.19 Å. In addition, Glu200 formed a single hydrogen bond with Wat1 in contrast to two hydrogen bonds in MMP-1•THP.
Figure 7.
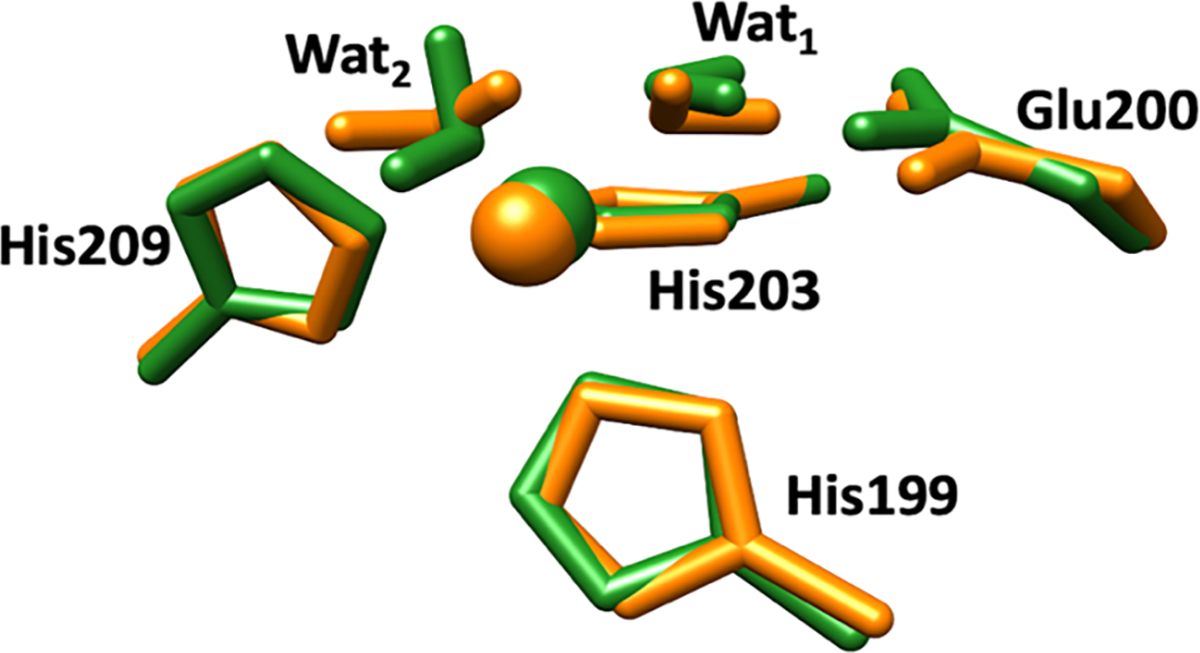
Superimposed catalytic Zn(II) sites of MMP-1•THP (green) and MMP-1•no THP (orange) in QM/MM optimized structure. Zn(II) is coordinated to three histidine residues, His199, His203, and His209, a catalytic water molecule (Wat1), and an auxiliary water molecule (Wat2).
There were no significant variations in the distances between Zn(II) and the coordinating residues for the catalytic site in MMP-1•no THP and MMP-1•THP complex (Table S6); however, the angle between His199-Zn(II)-His209 increased from 108.67° to 113.70° (Table S7).
Hydrogen bonding interactions in the catalytic site of MMP-1•no THP were studied to evaluate the effects of substrate removal (Table S8). Hydrogen bond between His209 and Ser208 was not observed for MMP-1•no THP structure; however, an additional hydrogen bonding interaction between His199 and Arg195 (that occludes the S1’ specificity pocket) appeared (with a donor-acceptor distance of 2.91 Å) upon substrate removal. This again demonstrates that substrate binding is essential for the proper orientation of Arg195 in the catalytic site.
How Does the Replacement of the Catalytic Zn(II) by Co(II) Influence the Conformational Dynamics of MMP-1•THP Complex?
Co(II) has often been used as a zinc substitute in metalloproteinase studies.[42,88] To probe the enzyme selectivity dependence on the nature of the catalytic metal ion, the catalytic Zn(II) was replaced by Co(II). MMP-1 Co1•THP showed a stable trajectory with an average RMSD of 2.43 Å, which is comparable to the 2.00 Å average RMSD of MMP-1•THP[56] (Figure S10a). Similar values were observed for the average distance between the center of mass of the catalytic Co(II) and structural Zn(II) coordinated residues and the center of mass of the catalytic site to the center of mass of the scissile bond (Figures S10b–d). The average inter-domain distances were nearly the same for Co(II) substituted MMP-1 and the WT MMP-1. As a relationship exists between MMP-1 collagenolytic activity and inter-domain dynamics,[54] our results are in agreement with the finding that Co(II) engineered enzyme retains catalytic activity.[48,50,89] Co(II) maintained the hydrogen bonding interactions of the WT MMP-1 in the catalytic site. The first coordination-sphere histidine residues for both the catalytic and structural Zn(II) sites of MMP-1 have a similar orientation (Figures 8a and 8b). A minor change was observed for the conformation of the S-loop and the V-B loop of MMP-1 Co1•THP compared to the MMP-1•THP complex (Figures 9a and 9b).
Figure 8.
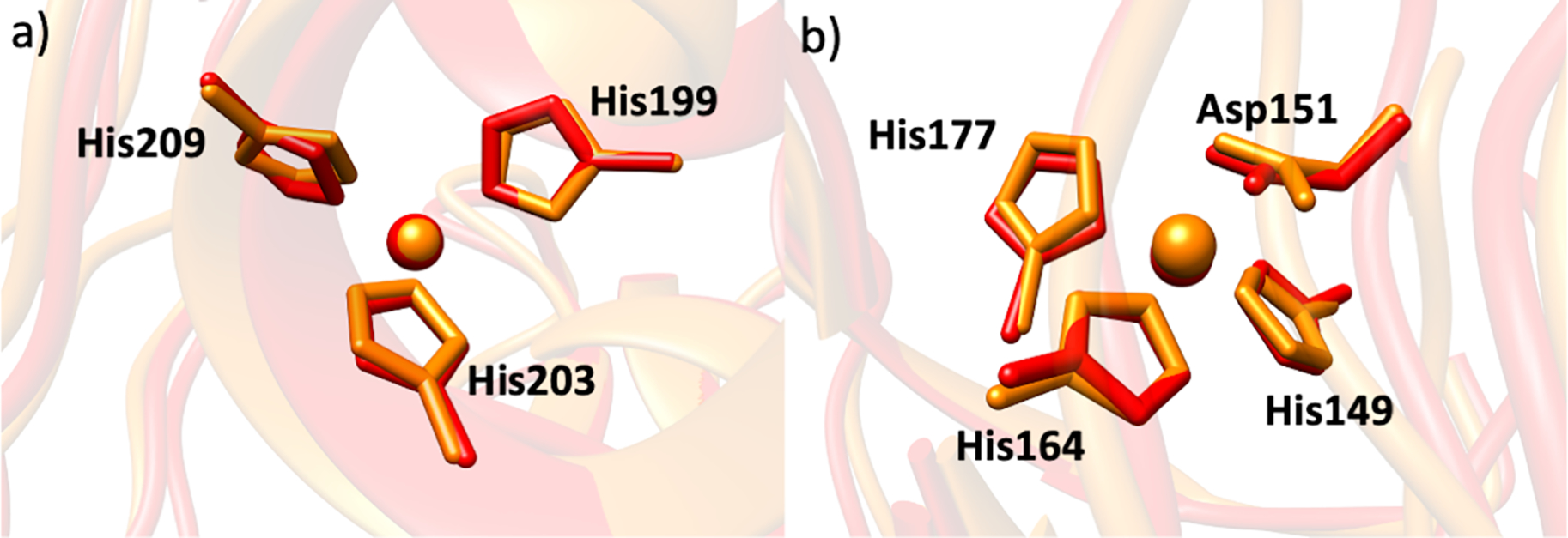
The superimposed a) catalytic Zn(II) site and b) structural Zn(II) site of MMP-1•THP (red) and MMP-1 Co1•THP (orange) from the averaged MD trajectory.
Figure 9.
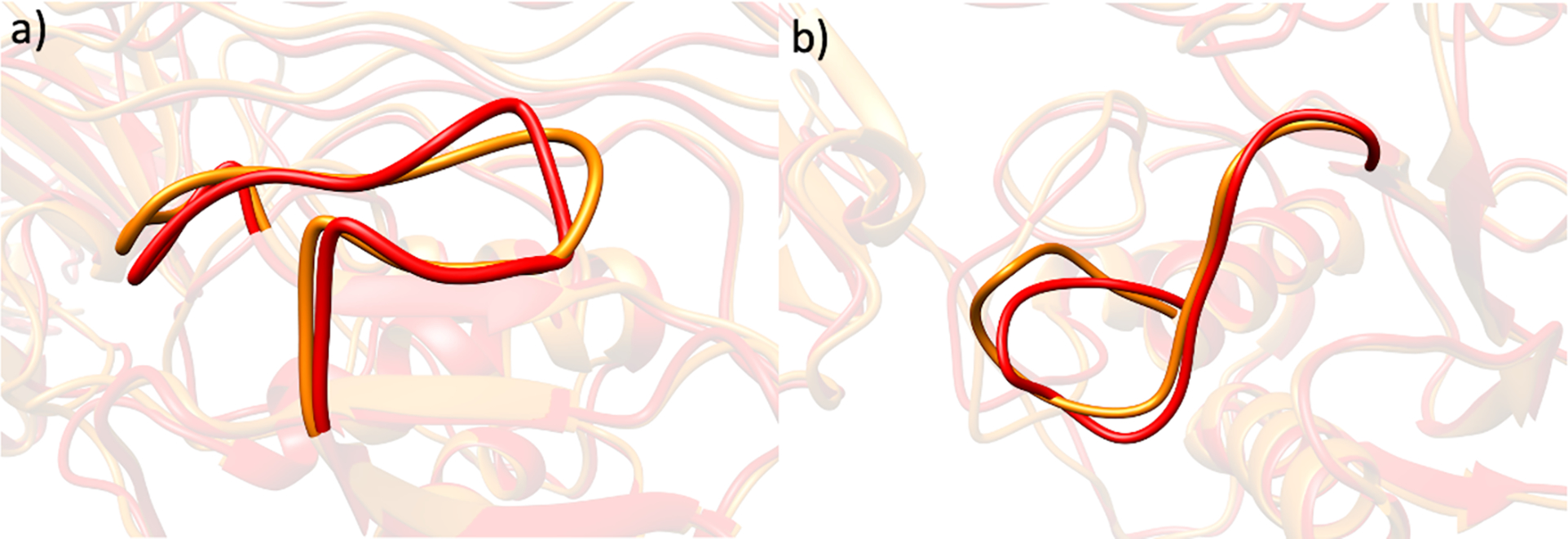
The superimposed a) S-loop and b) V-B loop of MMP-1•THP (red) and MMP-1 Co1•THP (orange) from the averaged MD trajectory.
RMSD and RMSF studies of the CAT and HPX domains and the key loops of both Zn(II) and Co(II) substituted MMP-1 further confirm the similarities of their overall dynamics (Figures S11, S12). Furthermore, DCCA of MMP-1 Co1•THP complex showed a positive correlation between the S10’ exosite and the S1’ specificity loop (Figure S13). However, the correlations included fewer exosite residues (282–283 and 292–300) compared to MMP-1•THP. A loss of anti-correlation was observed between the THP scissile bond residues- Gly775 and Leu776 and the structural Zn(II). Upon Zn(II) substitution with Co(II), the THP leading (L) strand residues, 767–774 participated in a positive correlation with residues 324–330 of HPX blade 2. PCA of MMP-1 Co1•THP complex indicated that the HPX domain shows more flexibility compared to the CAT one (Figure S14).
Geometric Structure of the Catalytic Metal Center in MMP-1 Co1•THP Compared to MMP-1•THP
We further probed the effect of Co(II) substitution of the catalytic Zn(II) ion on the geometric and electronic structure of the catalytic site of MMP-1 Co1•THP in comparison to MMP-1•THP using QM/MM.
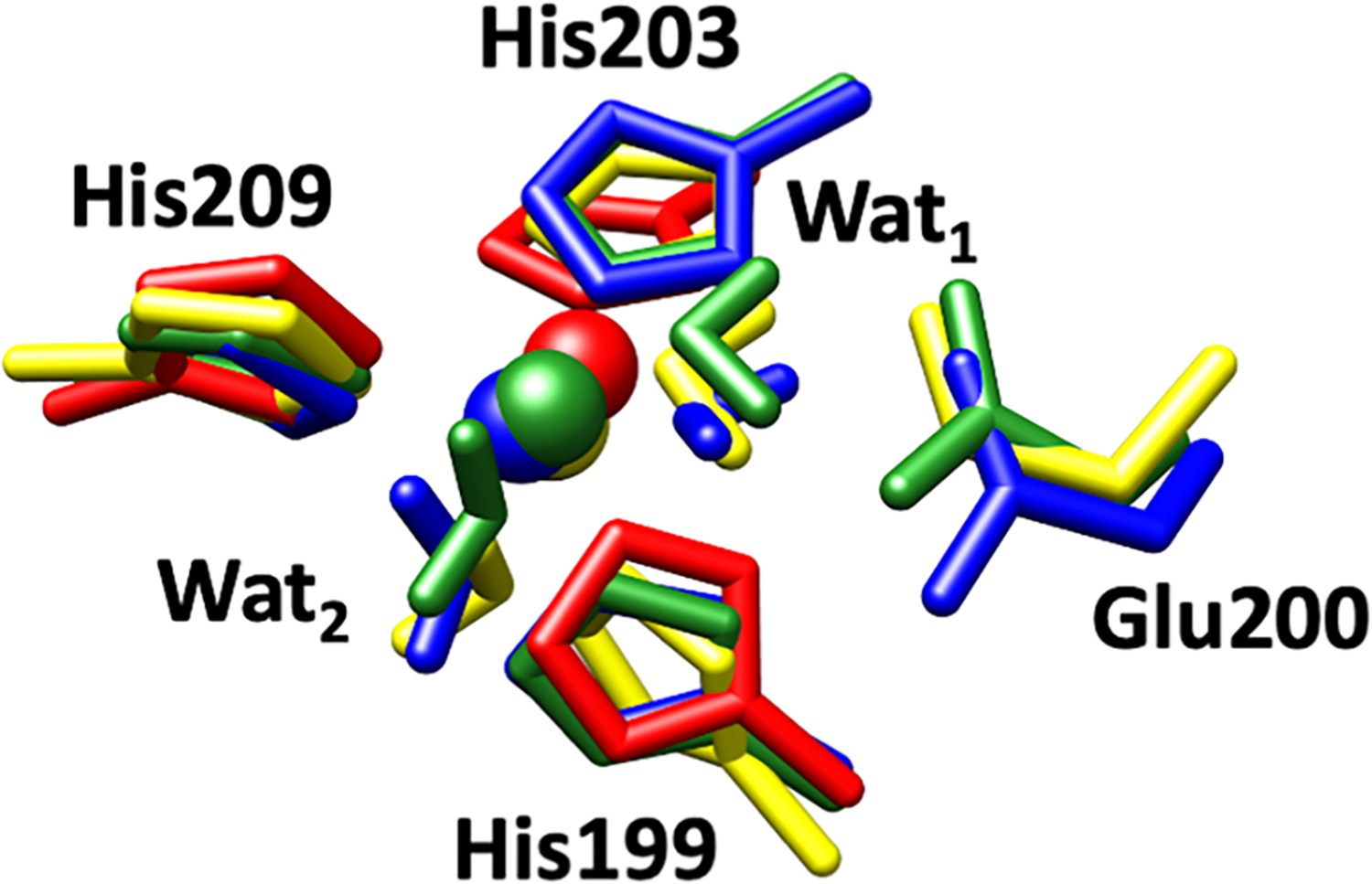
Spectroscopic studies of mixed-metal MMP-1 by Yang et al. reported similar coordination number for Zn(II) and Co(II) in the catalytic site.[46] In agreement with these studies, the QM/MM optimized structure of MMP-1 Co1•THP was similar to that of MMP-1•THP (Figure 10). Glu200 interacted with Wat1 via a single hydrogen bond in MMP-1 Co1•THP compared to two hydrogen bonding interactions in MMP-1•THP. A very minor change was observed for the metal-His209 and metal-His203 bond lengths (Table 4). The unpaired electron spin densities are localized exclusively on the Co(II) ion (Table S9). Co(II) replacement of Zn(II) in the catalytic site of MMP-1 did not show a substantial effect on angles between the metal center and its coordinated residues (Table 5).
Figure 10.
Superimposed QM/MM optimized structures of MMP-1•THP (green) and MMP-1 Co1•THP (pink). Zn(II)/Co(II) is coordinated to three histidine residues, His199, His203, and His209, a catalytic water molecule (Wat1), and an auxiliary water molecule (Wat2).
Table 4.
Distances between Zn(II) and the coordinated histidine residues in the QM/MM optimized geometry of MMP-1•THP and MMP-1 Co1•THP.
| Atoms | Distance (Å) (QM/MM) |
||
|---|---|---|---|
| MMP-1•THP | MMP-1 Co1•THP | ||
|
| |||
| Zn(II)/Co(II) | His199-NE2 | 2.06 | 2.07 |
| Zn(II)/Co(II) | His203-NE2 | 2.19 | 2.11 |
| Zn(II)/Co(II) | His209-NE2 | 2.05 | 2.09 |
Table 5.
Angles between Zn(II)/Co(II) and the coordinated histidine residues in the QM/MM optimized geometry of MMP-1•THP and MMP-1 Co1•THP.
| Atoms | Angle (Å) (QM/MM) |
|||
|---|---|---|---|---|
| MMP-1•THP | MMP-1 Co1•THP | |||
|
| ||||
| His199-NE2 | Zn(II)/Co(II) | His203-NE2 | 103.33 | 105.39 |
| His203-NE2 | Zn(II)/Co(II) | His209-NE2 | 93.73 | 96.69 |
| His199-NE2 | Zn(II)/Co(II) | His209-NE2 | 108.67 | 107.70 |
How the Structural Zn(II) Center Influences the Change of the Coordination State of the Catalytic Co(II) Center from 5C to 6C in Substrate-Free MMP-1
Removal of THP substrate does not dramatically influence the geometry of the catalytic Co(II) site (Tables S10, S11). Nonetheless, studies by Yang et al. showed that upon removal of structural Zn(II) in MMP-1, the catalytic Co(II) shows a change in coordination number from 5C to 6C with sixth coordination position occupied by a water molecule (Wat3)[46] (Figure 11). To evaluate the effect of the structural Zn(II) ion on the catalytic Co(II) coordinating geometry, we performed QM/MM studies after 1 μs MD of two computationally engineered enzyme forms – with structural Zn(II) ion (MMP-1 Co1•no THP) and with removed structural Zn(II) ion (MMP-1 Co1•no Zn2 no THP) (Figure 11). An increased number of water molecules were observed near the catalytic Co(II) site for MMP-1 Co1•no Zn2 no THP complex (Figure 12a). The availability of more water molecules in the enzyme active site may be the possible reason for a Co(II) six coordination position.
Figure 11.

Superimposed QM/MM optimized structures of MMP-1 Co1•no THP (magenta) and MMP-1 Co1•no Zn2 no THP (yellow).
Figure 12.
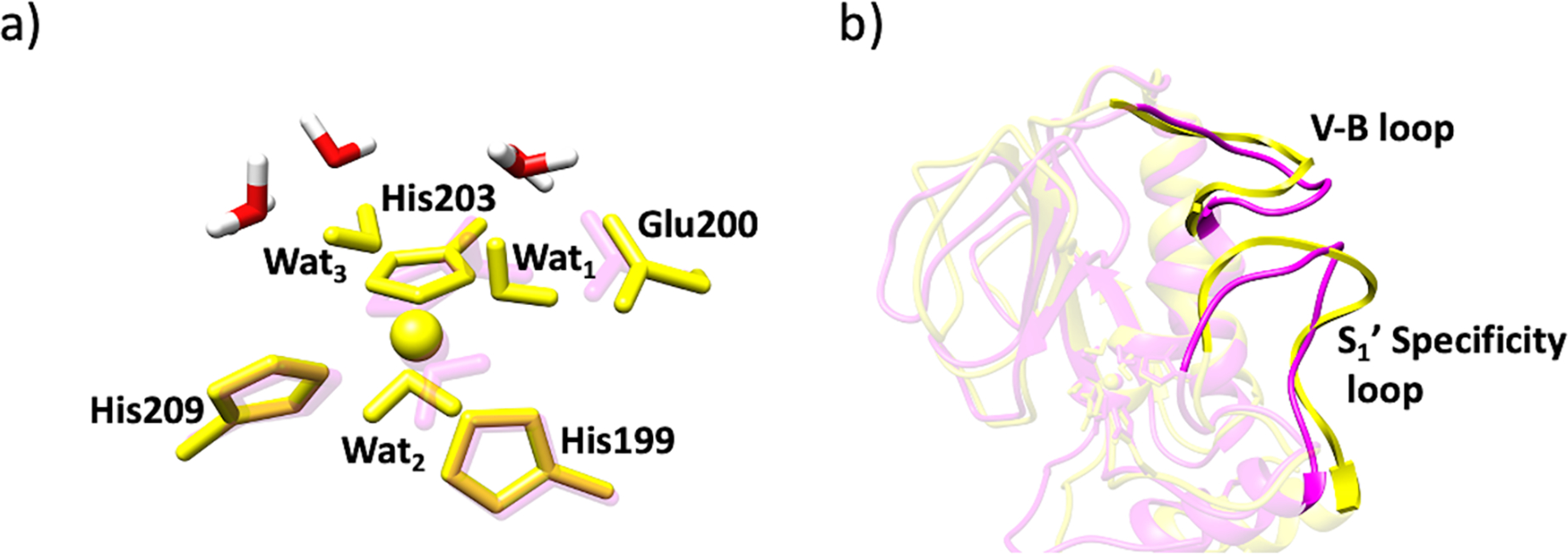
a) New water molecules (in red and white) in the catalytic site of MMP-1 Co1•no Zn2 no THP (yellow) and MMP-1 Co1•no THP (magenta). b) Open S1’ specificity loop of MMP-1 Co1•no Zn2 no THP (yellow) and MMP-1 Co1•no THP (magenta).
To evaluate the effect of including the third water molecule in the MM or QM part on the optimized QM/MM geometry, we performed additional QM/MM optimizations with the third water molecule included in the QM region for two systems: i) MMP-1 Co1•no THP and ii) MMP-1•no THP. We compared the QM/MM optimized structures containing the third water molecule either in the QM or in the MM part for MMP-1 Co1•no THP and MMP-1•no THP. High structural similarity was observed in the presence and absence of the third water, and thus the third water had an insignificant effect in the MM or in the QM part (Figures S3a and S3b).
Exploring the enzyme structure for the cause of increased water molecules in the catalytic site closely, we found a more open conformation of the S1’ specificity loop which may allow for water molecules to enter into the catalytic site of MMP-1 Co1•no Zn2 no THP (Figure 12b). SASA based on the MD simulations revealed that upon the structural Zn(II) removal, the V-B loop showed reduced solvent accessibility, in contrast, the S1’ specificity loop demonstrated increased solvent accessibility (Figures S15a and S15b).
Networks of stabilizing hydrogen bonding interactions were analyzed to gain insights into the effect exerted by the structural Zn(II) on the Co(II)-substituted enzyme. Overall, the absence of structural Zn(II) did not result in new hydrogen bonding interactions in the catalytic site; however, in MMP-1 Co1•no Zn2 no THP, His149 (bound to structural Zn(II)) formed a new hydrogen bond with nearby Asp151- a new interaction that was not observed in the presence of structural Zn(II) (Table S12). This finding suggests the cooperativity between the two metal centers and provides a mechanistic explanation of the structural Zn(II) influence on the catalytic Co(II) center.
CONCLUSIONS
This study provides complementary insights to the X-ray crystallographic studies on the catalytic Zn(II) center in the MMP-1•THP complex, emphasizing that both QM and QM/MM optimizations provided a good description of the geometric and electronic structure of the catalytic Zn(II) center. The research further revealed the impact of substrate THP removal on the dynamics, inter-domain interactions, and correlated motions of the MMP-1•THP complex. It demonstrated that catalytic Zn(II) substitution by Co(II) leads to subtle but measurable changes in the geometric structure of the catalytic metal center and the overall dynamics of the ES complex. Notably, the study provided atomic-level mechanistic insight into the experimentally observed transition from 5C to 6C of the catalytic Co(II) center that occurs upon structural Zn(II) removal. While MM characterization of a Zn(II) center in proteins might not be accurate due to parametrization procedures, QM/MM and QM methods provide an accurate geometric and electronic structure description of Zn(II) sites. However, the performed QM/MM and QM calculations in this study were in good agreement with the MM calculations of the Zn(II) centers in our MD simulations. Overall, the present investigation demonstrates the power of multilevel computational methods (MD, QM/MM, and QM) to provide advanced insights into MMP-1 structure-function relationships.
Supplementary Material
Acknowledgments
This research was supported by NIH grant GM132873 (to TKC and GBF).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
REFERENCES
- [1].McCall KA, Huang C, Fierke CA, J. Nutr 2000, 130, 1437S–1446S. [DOI] [PubMed] [Google Scholar]
- [2].Parisi AF, Vallee BL, Am. J. Clin. Nutr. 1969, 22, 1222–1239. [DOI] [PubMed] [Google Scholar]
- [3].Andreini C, Bertini I, Cavallaro G, PLoS ONE 2011, 6, e26325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Vallee BL, Auld DS, Biochemistry 1993, 32, 6493–6500. [DOI] [PubMed] [Google Scholar]
- [5].Cui N, Hu M, Khalil RA, in Prog. Mol. Biol. Transl Sci, Elsevier, 2017, pp. 1–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Askary VR, Jahan NA, Sabbagh A, Jahani FS, Dourandish N, Kamachali ARK, Clin. Biochem. 2011, 44, S323–S324. [Google Scholar]
- [7].Birkedal-Hansen H, Moore WGI, Bodden MK, Windsor LJ, Birkedal-Hansen B, DeCarlo A, Engler JA, Crit. Rev. Oral Biol. Med. 1993, 4, 197–250. [DOI] [PubMed] [Google Scholar]
- [8].Laronha H, Caldeira J, Cells 2020, 9, 1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Bode W, Fernandez-Catalan C, Grams F. HT, Maskos K. HN, Cell. Mol. Life Sci. 1999, 55, 639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weaver VM, Roskelley CD, Trends Cell Biol. 1997, 7, 40–42. [DOI] [PubMed] [Google Scholar]
- [11].Marneros AG, Olsen BR, Matrix Biol. 2001, 20, 337–345. [DOI] [PubMed] [Google Scholar]
- [12].Streuli C, Curr. Opin. Cell Biol. 1999, 11, 634–640. [DOI] [PubMed] [Google Scholar]
- [13].Rosenberg GA, Lancet Neurol. 2009, 8, 205–216. [DOI] [PubMed] [Google Scholar]
- [14].Burrage S P, Front Biosci. 2006, 11, 529. [DOI] [PubMed] [Google Scholar]
- [15].Pardo A, Selman M, Int. J. Biochem. Cell Biol. 2005, 37, 283–288. [DOI] [PubMed] [Google Scholar]
- [16].Manka SW, Carafoli F, Visse R, Bihan D, Raynal N, Farndale RW, Murphy G, Enghild JJ, Hohenester E, Nagase H, Proc. Natl. Acad. Sci. 2012, 109, 12461–12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Iyer S, Visse R, Nagase H, Acharya KR, J. Mol. Biol. 2006, 362, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Murphy G, Knäuper V, Matrix Biol. 1997, 15, 511–518. [DOI] [PubMed] [Google Scholar]
- [19].Vargova V, Pytliak M, Mechirova V, in Matrix Metalloproteinase Inhibitors: Specificity of Binding and Structure-Activity Relationships (Eds.: Gupta S) Springer; Basel, AG, 2012. [PubMed] [Google Scholar]
- [20].Rao B, Curr. Pharm. Des. 2005, 11, 295–322. [DOI] [PubMed] [Google Scholar]
- [21].Browner MF, Lovejoy B, Welch AR, Carr S, Luong C, Broka C, Hendricks RT, Campbell JA, Walker KAM, Martin R, Van Wart H, Nat. Struct Biol. 1999, 6, 217–221. [DOI] [PubMed] [Google Scholar]
- [22].Gimeno A, Beltrán-Debón R, Mulero M, Pujadas G, Garcia-Vallvé S, Drug Discov. Today 2020, 25, 38–57. [DOI] [PubMed] [Google Scholar]
- [23].Jacobsen JA, Major Jourden JL, Miller MT, Cohen SM, Biochim. Biophys. Acta. Mol. Cell Res 2010, 1803, 72–94. [DOI] [PubMed] [Google Scholar]
- [24].Chung L, Shimokawa K, Dinakarpandian D, Grams F, Fields GB, Nagase H, J. Biol. Chem 2000, 275, 29610–29617. [DOI] [PubMed] [Google Scholar]
- [25].Tallant C, Marrero A, Gomis-Rüth FX, Biochim. Biophys. Acta. Mol. Cell Res 2010, 1803, 20–28. [DOI] [PubMed] [Google Scholar]
- [26].Maskos K, Biochimie 2005, 87, 249–263. [DOI] [PubMed] [Google Scholar]
- [27].Massova I, Kotra LP, Mobashery S, Bioorg. Med. Chem. Lett. 1998, 8, 853–858. [DOI] [PubMed] [Google Scholar]
- [28].Massova I, Kotra LP, Fridman R, Mobashery S, FASEB J. 1998, 12, 1075–1095. [PubMed] [Google Scholar]
- [29].Cha J, Auld DS, Biochemistry 1997, 36, 16019–16024. [DOI] [PubMed] [Google Scholar]
- [30].Johnson LL, Pavlovsky AG, Johnson AR, Janowicz JA, Man CF, Ortwine DF, Purchase CF, White AD, Hupe DJ, J. Biol. Chem. 2000, 275, 11026–11033. [DOI] [PubMed] [Google Scholar]
- [31].Bode W, Gomis-Rüth FX, Stöckler W, FEBS Lett. 1993, 331, 134–140. [DOI] [PubMed] [Google Scholar]
- [32].Stöcker W, Grams F, Baumann U, Reinemer P, Gomis-Rüth FX, McKay DB, Bode W, Protein Sci. 1995, 4, 823–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Díaz N, Suarez D, Biochemistry 2007, 46, 8943–8952. [DOI] [PubMed] [Google Scholar]
- [34].Brodsky B, Thiagarajan G, Madhan B, Kar K, Biopolymers 2008, 89, 345–353. [DOI] [PubMed] [Google Scholar]
- [35].Koide T, Phil. Trans. R. Soc. B 2007, 362, 1281–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lauer-Fields JL, Juska D, Fields GB, Biopolymers 2002, 66, 19–32. [DOI] [PubMed] [Google Scholar]
- [37].Brandstetter H, Grams F, Glitz D, Lang A, Huber R, Bode W, Krell H-W, Engh RA, J. Biol. Chem. 2001, 276, 17405–17412. [DOI] [PubMed] [Google Scholar]
- [38].Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H, EMBO J 2004, 23, 3020–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Díaz N, Suárez D, J. Phys. Chem. B 2008, 112, 8412–8424. [DOI] [PubMed] [Google Scholar]
- [40].Vasilevskaya T, Khrenova MG, Nemukhin AV, Thiel W, J. Comput. Chem. 2015, 36, 1621–1630. [DOI] [PubMed] [Google Scholar]
- [41].Decaneto E, Vasilevskaya T, Kutin Y, Ogata H, Grossman M, Sagi I, Havenith M, Lubitz W, Thiel W, Cox N, Phys. Chem. Chem. Phys. 2017, 19, 30316–30331. [DOI] [PubMed] [Google Scholar]
- [42].Cha J, Sørensen MV, Ye Q-Z, Auld DS, J. Biol. Inorg Chem. 1998, 3, 353–359. [Google Scholar]
- [43].Holland DR, Hausrath AC, Juers D, Matthews BW, Protein Sci. 1995, 4, 1955–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Plugis NM, Rudd ND, Krzystek J, Swenson DC, Telser J, Larrabee JA, J. Inorg. Biochem. 2020, 203, 110876. [DOI] [PubMed] [Google Scholar]
- [45].Underwood CK, Min D, Lyons JG, Hambley TW, J. Inorg. Biochem. 2003, 95, 165–170. [DOI] [PubMed] [Google Scholar]
- [46].Yang H, Makaroff K, Paz N, Aitha M, Crowder MW, Tierney DL, Biochemistry 2015, 54, 3631–3639. [DOI] [PubMed] [Google Scholar]
- [47].Kremer-Aach A, Kläui W, Bell R, Strerath A, Wunderlich H, Mootz D, Inorg. Chem. 1997, 36, 1552–1563. [DOI] [PubMed] [Google Scholar]
- [48].Vallee WMBL, in Methods Enzymol, Elsevier, 1993, pp. 52–71. [DOI] [PubMed] [Google Scholar]
- [49].Jacobsen FE, Breece RM, Myers WK, Tierney DL, Cohen SM, Inorg. Chem. 2006, 45, 7306–7315. [DOI] [PubMed] [Google Scholar]
- [50].Salowe SP, Marcy AI, Cuca GC, Smith CK, Kopka IE, Hagmann WK, Hermes JD, Biochemistry 1992, 31, 4535–4540. [DOI] [PubMed] [Google Scholar]
- [51].Singh W, Fields GB, Christov CZ, Karabencheva-Christova TG, RSC Adv. 2016, 6, 23223–23232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Karabencheva-Christova TG, Christov CZ, Fields GB, J. Phys. Chem. B 2018, 122, 5316–5326. [DOI] [PubMed] [Google Scholar]
- [53].Singh W, Fields G, Christov C, Karabencheva-Christova T, Int. J. Mol. Sci. 2016, 17, 1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kumar L, Nash A, Harms C, Planas-Iglesias J, Wright D, Klein-Seetharaman J, Sarkar SK, Biophys. J. 2020, 119, 360–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Pelmenschikov V, Siegbahn PEM, Inorg. Chem. 2002, 41, 5659–5666. [DOI] [PubMed] [Google Scholar]
- [56].Varghese A, Chaturvedi SS, Fields GB, Karabencheva-Christova TG, J. Biol. Inorg Chem. 2021, 26, 583–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Case DA, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham TE III, Cruzeiro VWD, Darden TA, Duke RE, Ghoreishi D, Gilson MK, Gohlke H, Goetz AW, Greene D, R Harris N Homeyer, Huang Y, Izadi S, Kovalenko A, Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Mermelstein DJ, Merz KM, Miao Y, Monard G, Nguyen C, Nguyen H, Omelyan I, Onufriev A, Pan F, Qi R, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Smith J, SalomonFerrer R, Swails J, Walker RC, Wang J, Wei H, Wolf RM, Wu X, Xiao L, York DM and Kollman PA, AMBER 2018, University of California, San Francisco, 2018. [Google Scholar]
- [58].Guex N, Peitsch MC, Electrophoresis 1997, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- [59].Olsson MHM, Søndergaard CR, Rostkowski M, Jensen JH, J. Chem. Theory Comput. 2011, 7, 525–537. [DOI] [PubMed] [Google Scholar]
- [60].Li P, Merz KM, J. Chem. Inf. Model. 2016, 56, 599–604. [DOI] [PubMed] [Google Scholar]
- [61].Seminario JM, Int. J. Quantum Chem. 1996, 60, 1271–1277. [Google Scholar]
- [62].Cieplak P, Cornell WD, Bayly C, Kollman PA, J. Comput. Chem. 1995, 16, 1357–1377. [Google Scholar]
- [63].Peters MB, Yang Y, Wang B, Füsti-Molnár L, Weaver MN, Merz KM, J. Chem. Theory Comput. 2010, 6, 2935–2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C, J. Chem. Theory Comput. 2015, 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML, J. Chem. Phys. 1983, 79, 926–935. [Google Scholar]
- [66].Darden T, York D, Pedersen L, J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar]
- [67].Davidchack RL, Handel R, Tretyakov MV, J. Chem. Phys. 2009, 130, 234101. [DOI] [PubMed] [Google Scholar]
- [68].Ryckaert J-P, Ciccotti G, Berendsen HJC, J. Comput. Phys. 1977, 23, 327–341. [Google Scholar]
- [69].Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR, J. Chem. Phys. 1984, 81, 3684–3690. [Google Scholar]
- [70].Roe DR, Cheatham TE, J. Chem. Theory Comput. 2013, 9, 3084–3095. [DOI] [PubMed] [Google Scholar]
- [71].Arnold GE, Ornstein RL, Biophys. J. 1997, 73, 1147–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Genheden S, Reymer A, Saenz-Méndez P, Eriksson LA, in Computation tools for Chemical Biology (Eds.: Martin-Santamaria S), Royal Society of Chemistry, Cambridge, 2017, pp. 1–38. [Google Scholar]
- [73].Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD, Bioinformatics 2006, 22, 2695–2696. [DOI] [PubMed] [Google Scholar]
- [74].Maisuradze GG, Liwo A, Scheraga HA, J. Mol. Biol. 2009, 385, 312–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hess B, Phys. Rev. E 2002, 65, 031910. [DOI] [PubMed] [Google Scholar]
- [76].Ross C, Nizami B, Glenister M, Sheik Amamuddy O, Atilgan AR, Atilgan C, Tastan Bishop Ö, Bioinformatics 2018, 34, 3759–3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Chaturvedi SS, Ramanan R, Lehnert N, Schofield CJ, Karabencheva-Christova TG, Christov CZ, ACS Catal. 2020, 10, 1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Chaturvedi SS, Ramanan R, Waheed SO, Ainsley J, Evison M, Ames JM, Schofield CJ, Karabencheva-Christova TG, Christov CZ, Chem. Eur. J. 2019, 25, 5422–5426. [DOI] [PubMed] [Google Scholar]
- [79].Christov CZ, Lodola A, Karabencheva-Christova TG, Wan S, Coveney PV, Mulholland AJ, Biophys. J. 2013, 104, L5–L7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Metz S, Kästner J, Sokol AA, Keal TW, Sherwood P, WIREs Comput. Mol. Sci. 2014, 4, 101–110. [Google Scholar]
- [81].Ahlrichs R, Bär M, Häser M, Horn H, Kölmel C, Chem. Phys. Lett. 1989, 162, 165–169. [Google Scholar]
- [82].Smith W, Forester TR, J. Mol. Graph. 1996, 14, 136–141. [DOI] [PubMed] [Google Scholar]
- [83].Li H, Zheng X, Jia Z, Wang X, J. Mol. Model. 2020, 26, 39. [DOI] [PubMed] [Google Scholar]
- [84].Wick CR, Smith DM, J. Phys. Chem. A 2018, 122, 1747–1755. [DOI] [PubMed] [Google Scholar]
- [85].Cossi M, Barone V, Cammi R, Tomasi J, Chem. Phys. Lett. 1996, 255, 327–335. [Google Scholar]
- [86].Bertini I, Fragai M, Luchinat C, Melikian M, Mylonas E, Sarti N, Svergun DI, J. Biol. Chem. 2009, 284, 12821–12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].O’Farrell TJ, Guo R, Hasegawa H, Pourmotabbed T, Biochemistry 2006, 45, 15411–15418. [DOI] [PubMed] [Google Scholar]
- [88].Bertini I, Luchinat C, Adv Inorg Biochem. 1984, 6, 71–111. [PubMed] [Google Scholar]
- [89].Gomis-Rüth FX, Grams F, Yiallouros I, Nar H, Küsthardt U, Zwilling R, Bode W, Stöcker W, J. Biol. Chem. 1994, 269, 17111–17117. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.