Abstract
Erwinia chrysanthemi 3937 secretes into the external medium several pectinolytic enzymes, among which are eight isoenzymes of the endo-cleaving pectate lyases: PelA, PelB, PelC, PelD, and PelE (family 1); PelI (family 4); PelL (family 3); and PelZ (family 5). In addition, one exo-cleaving pectate lyase, PelX (family 3), has been found in the periplasm of E. chrysanthemi. The E. chrysanthemi 3937 gene kdgC has been shown to exhibit a high degree of similarity to the genes pelY of Yersinia pseudotuberculosis and pelB of Erwinia carotovora, which encode family 2 pectate lyases. However, no pectinolytic activity has been assigned to the KdgC protein. After verification of the corresponding nucleotide sequence, we cloned a longer DNA fragment and showed that this gene encodes a 553-amino-acid protein exhibiting an exo-cleaving pectate lyase activity. Thus, the kdgC gene was renamed pelW. PelW catalyzes the formation of unsaturated digalacturonates from polygalacturonate or short oligogalacturonates. PelW is located in the bacterial cytoplasm. In this compartment, PelW action could complete the degradation of pectic oligomers that was initiated by the extracellular or periplasmic pectinases and precede the action of the cytoplasmic oligogalacturonate lyase, Ogl. Both cytoplasmic pectinases, PelW and Ogl, seem to act in sequence during oligogalacturonate depolymerization, since oligomers longer than dimers are very poor substrates for Ogl but are good substrates for PelW. The estimated number of binding subsites for PelW is three, extending from subsite −2 to +1, while it is probably two for Ogl, extending from subsite −1 to +1. The activities of the two cytoplasmic lyases, PelW and Ogl, are dependent on the presence of divalent cations, since both enzymes are inhibited by EDTA. In contrast to the extracellular pectate lyases, Ca2+ is unable to restore the activity of PelW or Ogl, while several other cations, including Co2+, Mn2+, and Ni2+, can activate both cytoplasmic lyases.
The enterobacterium Erwinia chrysanthemi causes soft rot disease of various plants. Its pathogenicity is largely due to its ability to degrade pectin, a major constituent of plant cell walls and middle lamellae. The breakdown of pectin, a polysaccharide with a backbone consisting of partially esterified galacturonic acid, involves a battery of pectinases. The combination of different enzymatic activities allows E. chrysanthemi to efficiently degrade pectin and to subsequently use the liberated pectic oligomers as a carbon source for growth. The pectin esterases (pectin methylesterase and pectin acetylesterase) remove the ester groups of pectin and facilitate the action of depolymerases (pectate lyase and polygalacturonase) (12, 33). These depolymerases differ mainly in their reaction mechanisms (β-elimination or hydrolysis) and in the random or terminal mode of attack of the polymer (endo or exo).
The majority of pectinases are secreted by E. chrysanthemi into the extracellular medium through the type II secretion machinery, the Out system (35). In E. chrysanthemi 3937, eight endo-pectate lyases (PelA, PelB, PelC, PelD, PelE, PelI, PelL, and PelZ), the pectin methylesterase PemA, and the pectin acetylesterase PaeY are secreted by this system (12, 33). The endo-pectate lyases are the major pectinolytic enzymes produced by E. chrysanthemi, and they play an important role in the maceration of plant tissues. These enzymes randomly cleave, by β-elimination, internal glycosidic linkages in pectic polymers, preferentially polygalacturonate or pectins with small numbers of methoxyl groups, and generate a series of oligogalacturonates with a 4,5-unsaturated residue at the nonreducing end.
Besides these extracellular enzymes, several cell-bound pectinases have been identified in E. chrysanthemi: the outer-membrane pectin methylesterase PemB (31), the periplasmic exopolygalacturonate lyase PelX and exo-α-d-polygalacturonosidase PehX (2, 8, 34), and the cytoplasmic oligogalacturonate lyase Ogl (3, 29). The two periplasmic exopectinases act on different extremities of the polymeric chain: PelX catalyzes the formation of unsaturated digalacturonates by attack from the reducing end, while PehX releases digalacturonates by attack from the nonreducing end (4, 34). The cytoplasmic oligogalacturonate lyase Ogl, like the pectate lyases, cleaves glycosidic linkages by β-elimination, but in contrast to these enzymes, it is only active on short oligogalacturonates (22).
Pectate lyases are classified into five different families according to their primary amino acid sequences (15, 36). The E. chrysanthemi isoenzymes are distributed among these five families. Family 1 contains several bacterial pectate lyases, fungal pectin lyases, and plant proteins (9, 10, 36). It includes the E. chrysanthemi isoenzymes PelA to PelE, which are further separated into two subfamilies, PelADE and PelBC (39). Family 3 includes PelB/Pel-3 of Erwinia carotovora, PelI of E. chrysanthemi (36), and the four Pel proteins of the phytopathogenic fungus Nectria haematococca (Fusarium solani) (7). The endo-pectate lyase PelL (1, 18) and the exopolygalacturonate lyase PelX of E. chrysanthemi are the only members of family 4 (2, 34). PelZ of E. chrysanthemi is the sole characterized component of the fifth family (26). Family 2 contains the periplasmic pectate lyases PelB of Erwinia carotovora (11) and PelY of Yersinia pseudotuberculosis (20). Despite its homology with family 2 pectate lyases, no pectinolytic activity was observed for the product of the E. chrysanthemi kdgC gene (5).
In this study, we showed that the kdgC gene of E. chrysanthemi 3937 is longer than previously supposed (5). The corresponding protein was overproduced, and we demonstrated that it encodes an exopolygalacturonate lyase. Hence, the kdgC gene was renamed pelW. The biochemical properties of PelW and its cellular localization were analyzed. To clarify the role of this cytoplasmic enzyme in pectin degradation, we compared its enzymatic properties with those of the other E. chrysanthemi cytoplasmic pectinase, Ogl.
MATERIALS AND METHODS
Strains, media, and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. Cells were grown in either Luria-Bertani (LB) or M63 medium (21). When required, the media were solidified with agar (15 g · liter−1). E. chrysanthemi cells were usually incubated at 30°C, and Escherichia coli cells were generally incubated at 37°C. Pectate agar medium was used to detect pectinolytic activity (14). Carbon sources were added at 2 g · liter−1 except for polygalacturonate (PGA) (grade II; Sigma), which was added at 4 g · liter−1. When required, antibiotics were added at the following concentrations: kanamycin, 20 μg · ml−1; ampicillin, 50 μg · ml−1; and chloramphenicol, 20 μg · ml−1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| Bacterial strains | ||
| E. chrysanthemi 3937 derivatives | ||
| A350 | lacZ2 | 5 |
| A1077 | lacZ2 kdgR::Cm | Laboratory collection |
| A1671 | lacZ2 pelW::MudI1734 Km | 5 |
| A3439 | lacZ2 kdgR::Cm pelW::MudI1734 Km | This work |
| Escherichia coli | ||
| NM522 | Δ(lac-proAB) Δ(mcrB-hsdSM)5 supE thi (F′ proAB lacIqlacZΔM15) | Laboratory collection |
| BL21(DE3) | E. coli B; F−dcm ompT hsdS gal λ(DE3); T7 polymerase gene under the control of the lacUV5 promoter | Stratagene |
| K-38 | HfrC(λ) phoA4 pit-10 tonA22 ompF627 relA1 | Laboratory collection |
| Plasmids | ||
| pBS Ap | pBluescript SK(+); Apr | Stratagene |
| pET-20b(+) | Carries the N-terminal pelB signal sequence; Apr | Novagen |
| pGP1.2 | The T7 RNA polymerase gene under the control of the λPL promoter and the cI857 repressor; Kmr | 38 |
| pT7-6 | T7Φ10; Apr | 38 |
| pNV9 | pBR325 derivative carrying the 3.2-kb EcoRV-EcoRI fragment with pelW | 5 |
| pBS-KC | pBS derivative with the 2.9-kb EcoRV-ClaI fragment from pNV9; pelW+ | This work |
| pT7-KC | pT7-6 derivative with the 3-kb EcoRV-EcoRI fragment from pNV9; pelW+ | This work |
| pT7-KCsp | pET-20b(+) derivative with the 2.5-kb HpaI-HindIII ′pelW fragment under the control of the pelB signal sequence | This work |
| pOGL-10 | pBS derivative carrying ogl | 28 |
| pT7-OGL | pT7-5 derivative with the 1.6-kb PinAI-PstI fragment from pOGL-10; ogl+ | This work |
| pTaX | pT7-5 derivative with pelX; Apr | 34 |
| pT7-pelD | pT7-5 derivative with pelD; Apr | 39 |
Protein production and cell fractionation.
The pelW gene was overexpressed by using the T7 promoter-T7 RNA polymerase system (38). The 2.9-kb EcoRV-ClaI fragment of pelW was inserted into the pT7-6 expression vector. The resulting plasmid, pT7-KC, was introduced into E. coli BL21(DE3). The BL21(DE3)/pT7-KC cells were grown at 30°C in LB medium supplemented with ampicillin (150 μg · ml−1). At an optical density at 600 nm (OD600) of 0.8 to 1, 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added, and the cells were grown for an additional 2 to 3 h. The cells were harvested by centrifugation for 10 min at 5,000 × g and 4°C and were subjected to osmotic shock for extraction of the overproduced protein (6).
For Ogl overproduction, the E. coli BL21(DE3)/pT7-OGL cells were cultivated as described above, except that the culture was grown for 12 to 14 h after IPTG addition. The cells were harvested by centrifugation and disrupted in a French press in 50 mM Tris-HCl (pH 8.0) with Complete protease inhibitor (Boehringer). The crude cell envelope fraction was eliminated by centrifugation at 100,000 × g for 1 h, and the supernatant was used directly for enzyme analysis.
For the E. chrysanthemi subcellular fractionation, the cells were grown in LB medium supplemented with galacturonic acid at 2 g · liter−1. At an OD600 of 1.2 to 1.5, spheroplasts were prepared as described by Witholt et al. (40).
Protein labelling.
Plasmid-encoded proteins were exclusively labelled with [35S]cysteine-[35S]methionine by using the T7 promoter-T7 polymerase system (38). For the cell fractionation experiments, cells were labelled for 10 min. Inhibition of the signal sequence processing was performed by adding 0.1 mM carbonylcyanide-m-chlorophenylhydrazone (CCCP) into an induced cell culture 2 min after addition of [35S]cysteine-[35S]methionine. The labelled amino acids were chased 3 min later, and cells were lysed in Laemmli sample buffer.
Analytical procedures.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis was performed according to the procedure of Laemmli (16a). Isoelectric focusing (IEF) was performed in a pH 3.5 to 10 gradient by using Pharmalytes. For differential detection of various pectate lyase activities after IEF, the gel was incubated in 50 mM Tris-HCl (pH 8.5) plus 5 g of PGA · liter−1 with either 1 mM CaCl2 (for extracellular pectate lyase detection) or 1 mM CoCl2 (for PelW detection). After 10 to 60 min of incubation at 25°C, the gel was rinsed with water and stained with 0.05% ruthenium red. PelW activity is sensitive to the position of the sample application papers on the IEF gel; it must be placed in the area of neutral or slightly alkaline pH.
Thin-layer chromatography was used to identify the enzymatic degradation products of PGA and oligogalacturonates (18).
Enzyme assays.
Pectate lyase activity was determined by monitoring spectrophotometrically the formation of unsaturated products from substrate at 230 nm. Unless otherwise specified, the standard assay mixture consisted of 50 mM Tris-HCl (pH 8.5), 0.1 mM CaCl2, and 0.5 g of PGA · liter−1. The appearance of products was monitored at 37°C over a period of 1 min (at 6-s intervals). The molar extinction coefficient of unsaturated oligogalacturonates used was 5,200 (23). One unit of activity was defined as the amount of enzyme required to produce 1 μmol of unsaturated product per minute. To assay the PelW activity, 0.1 mM MnCl2 was substituted for the CaCl2 and 0.1 mM trigalacturonic acid (Sigma) was substituted for the PGA.
The alkaline phosphatase assay was performed with p-nitrophenyl phosphate as the substrate (19), and β-galactosidase activity was determined with o-nitrophenyl-β-d-galactoside as the substrate (21).
Recombinant DNA techniques.
Preparation of plasmid DNA, restriction digestions, ligations, DNA electrophoresis, and transformations were carried out as described by Sambrook et al. (30). Sequencing were performed by Genome Express SA (Grenoble, France).
Nucleotide sequence accession number.
The nucleotide sequence of the pelW (kdgC) gene has been submitted to the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. X62073.
RESULTS
Determination of the nucleotide sequence of the kdgC gene.
The 2.3-kb EcoRV-PstI fragment containing the kdgC gene had been previously sequenced (5). However, no pectinolytic activity was found to be associated with the protein encoded by this fragment even after overproduction in different expression systems (data not shown). We decided to verify the sequence of this DNA region, mainly in the 5′ and 3′ ends of the potential open reading frame. This analysis revealed an error at the 3′ end of the open reading frame which extends beyond the PstI site. The corrected gene is 1,659 nucleotides long and encodes a 553-amino-acid protein with a deduced molecular mass of 64,082 Da and a calculated pI of 7.7. Subcloning a 2.9-kb EcoRV-ClaI fragment downstream of the plac promoter of pBluescript (pBS-KC) allowed us to detect a moderate pectate agar pitting activity around the E. coli NM522 colonies carrying this plasmid. Assay of pectinolytic activities revealed a weak pectate lyase activity in the corresponding cell lysates. Thus, this pectate lyase gene was renamed pelW.
The deduced amino acid sequence of PelW shows 35% identity with PelY of Y. pseudotuberculosis and 34% identity with PelB of E. carotovora (Fig. 1). The level of similarity decreases in the C-terminal parts of these proteins. However, the most important difference between PelW and these two proteins concerns their N-terminal ends. While PelY and PelB possess a typical signal peptide sequence, no signal sequence was identified within the PelW protein sequence.
FIG. 1.
Comparison of the amino acid sequences of the family 2 pectate lyases. The PelY sequence of Y. pseudotuberculosis was determined by Manulis et al. (20), and PelB of E. carotovora was described by Hinton et al. (11). Vertical lines indicate identical residues. The putative signal sequence cleavage sites of PelY and PelB are indicated by triangles.
PelW identification and determination of its cellular localization.
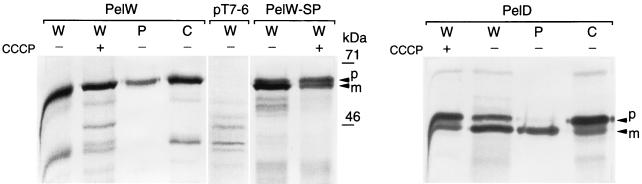
The PelW protein was overproduced in E. coli BL21(DE3)/pT7-KC cells (data not shown). A 65-kDa protein was detected by exclusive labelling of the plasmid-encoded proteins with [35S]cysteine-[35S]methionine (Fig. 2). Cell fractionation studies showed that this protein accumulated in the cytoplasm and that it was partially liberated by osmotic shock (Fig. 2).
FIG. 2.
Cellular localization of PelW in E. coli. The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the gels were autoradiographed. E. coli BL21(DE3)/pT7-KC (PelW), E. coli BL21(DE3)/pT7-6 (pT7-6), E. coli BL21(DE3)/pT7-KCsp (PelW-SP), and E. coli K38/pGP1.2/pT-pelD (PelD) cells were fractionated and then labelled with [35S]cysteine-[35S]methionine in the absence (−) or in the presence (+) of CCCP. W, whole-cell lysate; P, periplasmic fraction; C, osmotically shocked cell fraction. Precursor (p) and mature (m) forms of PelW-SP and PelD are indicated by arrowheads.
Since the two homologues of PelW, PelY and PelB, possess a signal peptide sequence and are periplasmic when expressed in E. coli (11, 20), we verified PelW localization in this host. No PelW was associated with the crude membrane fraction. Up to 30% of the total PelW enzymatic activity was released from E. coli cells by osmotic shock. However, the relative amount of osmotically released PelW depended on the strain used and on the protein production level and was significantly smaller than that of alkaline phosphatase (data not shown). This indicated that the partial extraction of PelW from the cells by osmotic shock was not due to a periplasmic localization of the protein but resulted from the partial leakage of overproduced PelW. Johnson and Hecht (13) showed that a nonspecific treatment of E. coli cells, e.g., repeated cycles of freezing and thawing, specifically released highly expressed recombinant protein from the bacterial cytoplasm without liberation of the bulk of the endogenous cytoplasmic E. coli proteins.
To exclude the possibility of the existence of a signal peptide for PelW, the protein was labelled and the cells were treated with CCCP, which stops signal peptide processing by dissipating the proton motive force (24). In contrast to PelD, which possesses a signal sequence, only one form of PelW was detected in the presence or in the absence of CCCP (Fig. 2). Moreover, PelW released by osmotic shock and PelW associated with the cells have the same molecular mass. Thus, as indicated by the sequence data, PelW does not possess a signal peptide and is located in the cytoplasm when produced in E. coli.
The differences observed among the N termini of PelW, PelB, and PelY led us to replace the 22 N-terminal amino acids of PelW with a classic signal peptide to construct a periplasmic derivative of PelW, PelW-SP. This chimeric protein was correctly maturated in E. coli cells and exported into the periplasm (Fig. 2 and data not shown). However, the resulting periplasmic PelW derivative was unstable, and no pectate lyase activity was found in association with this protein.
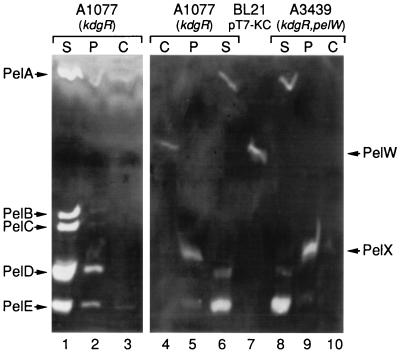
To determine the localization of PelW in E. chrysanthemi 3937, we used differential detection of PelW activity after IEF. This enzyme is activated in the presence of Co2+ cations, while the major E. chrysanthemi pectate lyases are inhibited under these conditions (39). Thus, to detect the PelW activity without interference from the major pectate lyases, we replaced Ca2+ with Co2+ for the detection of pectate lyase activities. A single band with an apparent pI of about 5.0 was detected in the extract of E. coli BL21(DE3)/pT7-KC cells overproducing PelW (Fig. 3). To increase PelW production in E. chrysanthemi, we used the kdgR mutant A1077, in which the pelW gene was shown to be derepressed (5). The band corresponding to PelW was detected in the cytoplasmic fraction of the A1077 strain but not in the periplasm or culture supernatant (Fig. 3). This band was absent from the pelW mutant A3439. Therefore, PelW is located in the cytoplasm of E. chrysanthemi 3937.
FIG. 3.
Cellular localization of PelW in E. chrysanthemi. IEF was followed by specific detection of pectate lyase activity in the presence of either CaCl2 for 10 min (lanes 1 to 3) or CoCl2 for 40 min (lanes 4 to 10). The osmotic-shock-released fraction of BL21(DE3)/pT7-KC (PelW+) (lane 7) was used as a control. Subcellular fractionation of E. chrysanthemi A1077 (kdgR) (lanes 1 to 6) and E. chrysanthemi A3439 (kdgR pelW) (lanes 8 to 10) was performed. The positions of the five major E. chrysanthemi pectate lyases (PelA to PelE), PelX, and PelW are indicated. S, culture supernatant; P, periplasm; C, spheroplast lysate.
Ogl production and assay.
Since PelW is a cytoplasmic pectate lyase, we decided to compare its properties with those of the other known cytoplasmic pectinase, the oligogalacturonate lyase Ogl. Overexpression of ogl in the E. coli recombinant strain BL21(DE3)/pT7-OGL yielded a high Ogl production level (about 0.4 mg · ml−1 of the culture), and the enzyme constituted about 90% of the cell protein content (data not shown). Therefore, the soluble cell fraction from BL21(DE3)/pT7-OGL was directly used for the enzyme analysis.
Ogl activity is usually estimated by the periodate-thiobarbiturate assay (22). We proposed a direct UV detection at 230 nm for measurement of Ogl activity. We observed that the action of Ogl on digalacturonic acid led to an increase in absorbance at 230 nm. Such an increase in absorbance is observed during the formation of 4,5-unsaturated oligogalacturonates by pectate lyases. However, the UV-detected product appeared unstable, and several minutes after the start of the enzymatic reaction, the increase in OD230 was followed by a successive decrease in absorbance. d-Galacturonic acid and 4-deoxy-l-threo-5-hexosulose uronic acid (DKI) have been identified as the Ogl products formed from digalacturonic acid (3, 22). Since these two products do not absorb at 230 nm, we proposed the following explanation for this phenomenon. The Ogl cleavage could give a 4,5-unsaturated pyranose monomer that, like 4,5-unsaturated digalacturonate, absorbs at 230 nm. The 4,5-unsaturated pyranose cycle could spontaneously produce a straight-chain 4,5-enolic form, which could then isomerize into the more stable 5-keto form (DKI). In spite of the relative instability of the first product formed by Ogl, it is possible to measure a linear increase in OD230 during short assay periods (20 to 90 s). Thus, a direct UV detection at 230 nm was routinely used to measure Ogl activity.
Substrate specificity of PelW and Ogl.
To analyze the enzymatic properties of PelW, we used the fraction released by osmotic shock from E. coli BL 21(DE3)/pT7-KC. Despite the smaller amount of PelW in this fraction than in the total-cell lysate, the enzyme constituted about 20% of the total protein content of this fraction. The PelW enzymatic activity was first detected by using PGA as a substrate in the standard pectate lyase assay medium. Like the other E. chrysanthemi pectate lyases, PelW is active within a range of alkaline pH values, with an optimum at pH 8.5 in Tris-HCl buffer. In accordance with the cellular localization of this enzyme, it seemed unlikely that the polymer could be its natural substrate. Therefore, we analyzed the PelW activity on short oligogalacturonates, di-, tri-, and tetragalacturonate (G2, G3, and G4, respectively), which are more likely to be present in the E. chrysanthemi cytoplasm (Table 2). PelW did not cleave G2, while G3 and G4 were good substrates. The maximal activity was observed with G3. Determination of the Km and Vmax indicated that PelW exhibits the highest activity and affinity for G3 (Table 2). PGA was cleaved by this enzyme less effectively then G3 or G4. In comparison with the other E. chrysanthemi pectate lyases, PelW exhibits a low specific activity on PGA but a good affinity for this substrate. The higher activity observed with G3, compared with G4 or the polymer, suggested that the number of binding subsites for PelW was three, since the array of subsites is completely covered by G3. PelW shows almost the same initial reaction rate on PGA and on pectins with degrees of methylation up to 60% (data not shown). The enzyme retains about 50% of its maximal activity on 75% methylated pectin.
TABLE 2.
Effect of substrate length on PelW and Ogl activities
| Substrate | Enzyme activitya ± SD
|
|||||
|---|---|---|---|---|---|---|
| PelW
|
Ogl
|
|||||
| Vob (U · mg−1) | Vmaxc (U · mg−1) | Kmc | Vod (U · mg−1) | Vmaxc (U · mg−1) | Kmc (μM) | |
| G2 | 0 | 57 ± 3 | 80 ± 5 | 150 ± 30 | ||
| G3 | 25 ± 2 | 30 ± 4 | 7 ± 2 μM | 10 ± 2 | 20 ± 7 | 120 ± 50 |
| G4 | 17 ± 2 | 23 ± 3 | 8 ± 2 μM | 4 ± 1.5 | NDe | ND |
| PGA | 14 ± 0.4 | 6 ± 2 | 0.01 ± 0.004 g liter−1 | 0 | ||
No enzymatic activity was detected on any of these substrates when E. coli extracts from vector-only-carrying strains were used.
The initial enzymatic rate of PelW was measured in 0.1 M Tris-HCl (pH 8.5) containing 0.1 mM MnCl2 and either 0.5 g of PGA · liter−1 or 50 μM oligogalacturonate (Gn, where n = 2 to 4).
For Vmax and Km determinations, PelW was incubated with PGA at concentrations of 0.005 to 0.025 g · liter−1 and with oligogalacturonates at concentrations ranging from 5 to 40 μM, while Ogl was incubated with oligogalacturonates at concentrations ranging from 50 to 400 μM.
The initial enzymatic rate of Ogl was measured in 0.1 M Tris-HCl (pH 7) containing 0.1 mM MnCl2 and either 0.5 g of PGA · liter−1 or 400 μM oligogalacturonate.
ND, not determined because the activity of Ogl with these substrates was too low, which led to a high level of variability in the experimental data.
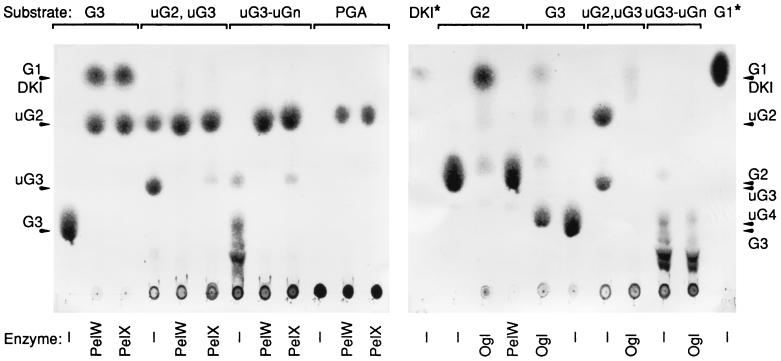
To determine precisely the mode of action of PelW, the reaction products obtained from the action of this enzyme on various substrates were characterized by thin-layer chromatography (Fig. 4). The activity of PelW was compared with those of the products obtained after action of the E. chrysanthemi periplasmic exopolygalacturonate lyase PelX. With PGA as a substrate, PelW catalyzes the formation of only one product, unsaturated digalacturonate. The same product was liberated by the action of PelX. Thus, PelW is a pectate lyase with an exo-cleaving mode. The smallest substrates cleaved by PelW or PelX are the saturated and unsaturated trimers.
FIG. 4.
Identification of PelW and Ogl reaction products by thin-layer chromatography. The reaction mixtures contained 0.1 M Tris-HCl (pH 8), 0.1 mM MnCl2, 5 U of enzyme ml−1, and 1.5 mg of the following substrates · ml−1: digalacturonic acid (G2), trigalacturonic acid (G3), a mixture of unsaturated di- and trigalacturonic acids (uG2, uG3), a mixture of unsaturated oligogalacturonic acids (uG3 to uGn), and polygalacturonic acid (PGA). Incubations were performed at 30°C for 3 h after addition of PelW, Ogl, or PelX or without enzyme (−). A 5-μl sample from each reaction was applied to a chromatogram sheet. The positions of the individual compounds are indicated by arrowheads. G1 and DKI are at the same position, but the staining method was not sensitive enough to detect the amounts of DKI applied to the chromatogram; 10 μg of DKI or G1 was applied to the lanes indicated by asterisks.
The enzymatic properties of Ogl were analyzed by using the soluble cell fraction of E. coli BL21(DE3)/pT7-OGL. The optimum pH of Ogl activity determined in 50 mM Tris-HCl buffer was between 7.3 and 7.7. Analysis of the action of Ogl on oligogalacturonates of different lengths (G2, G3, and G4) showed that the dimer was the best substrate for the enzyme (Table 2). Ogl cleaves both saturated and unsaturated dimers (Fig. 4). A large decrease in Ogl activity was observed for G3, while G4 was a very poor substrate (Table 2). Similarly, unsaturated oligogalacturonates larger than unsaturated G3 were not effectively cleaved by Ogl (Fig. 4). No enzymatic activity on polymeric substrates, such as PGA or pectins, could be detected.
Cation requirements of PelW and Ogl.
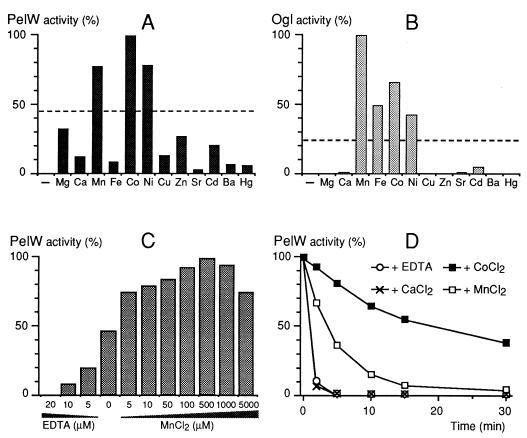
The characterized E. chrysanthemi pectate lyases have an absolute requirement for Ca2+. Addition of EDTA totally inhibits PelW, demonstrating its absolute cation requirement (Fig. 5A). PelW is weakly affected by the addition of Ca2+ but is strongly activated by Co2+, Mn2+, and Ni2+, with a maximum at concentrations of 0.1 to 1 mM (Fig. 5C and data not shown). Co2+ is the best cofactor for the enzyme. The effect of these cations is not additive, indicating that they have the same target on PelW and/or its substrate. It should be noted that the cation content of the commercially available G3 and PGA is high enough to give about 50% of the maximal PelW activity without addition of any cation (Fig. 5A and C).
FIG. 5.
Cation requirements of PelW and Ogl. The cation requirements of PelW (A) and Ogl (B) were tested by using 0.1 mM concentrations of their corresponding chloride forms, 25 μM EDTA, and 0.1 mM G3 in 0.1 M Tris-HCl (pH 8.5) as a substrate for PelW and 0.67 mM G2 in 0.1 M Tris-HCl (pH 7.0) as a substrate for Ogl. The dotted lines show the levels of enzyme activities in the absence of added EDTA or cations. −, no cation added. (C) The influence of Mn2+ concentration on PelW activity was tested in 0.1 M Tris-HCl (pH 8.5)–0.1 mM G3, using various concentrations of EDTA or MnCl2. (D) The thermostability of PelW was monitored at 45°C after various incubation times in 0.1 M Tris-HCl (pH 8.0) containing 0.4 mM EDTA, either alone or with a 1 mM concentration of CaCl2, CoCl2, or MnCl2. The residual activity is given as a percentage of the initial activity.
Binding of Ca2+ to the extracellular pectate lyases increases their stability (32, 39). We analyzed whether the stability of PelW is affected by the presence of cations. The stability of the protein was studied by measuring its thermal inactivation at 45°C. In the presence of EDTA, PelW lost 90% of its initial activity in 2 min (Fig. 5D). While addition of Ca2+ to PelW did not modify its stability, the presence of Mn2+ or Co2+ led to an essential increase in the enzyme’s stability, since the half-life of PelW increased to 4 or 15 min, respectively (Fig. 5D).
We tested Ogl’s cation requirement by using G2 as a substrate. Addition of 25 μM EDTA completely inhibited the Ogl activity (Fig. 5B). Among the divalent cations tested, Mn2+, Co2+, Fe2+, and Ni2+ strongly activated the enzyme. Mn2+ was the best cofactor for Ogl (Fig. 5B), with an optimal concentration of 0.1 mM (data not shown). Ca2+ had no effect on the enzyme activity.
DISCUSSION
This paper reports the characterization of PelW, the cytoplasmic exopolygalacturonate lyase of E. chrysanthemi 3937. It has been previously shown that the product of the kdgC gene is homologous to family 2 pectate lyases (5). However, an error in the gene sequence resulted in the failure of the authors to purify an active protein. We renamed this gene pelW after demonstrating that it encodes an exo-cleaving pectate lyase. It is not known whether the two other members of family 2, PelY of Y. pseudotuberculosis (20) and PelB of E. carotovora (11), are endo- or exo-cleaving enzymes. Their homology with PelW is not sufficient to infer a similar mode of substrate recognition. Indeed, pectate lyase family 3 includes both an exo- and an endo-cleaving enzyme, PelX and PelL, respectively (18, 34). Moreover, these enzymes are not situated in the same cellular compartment; PelL is extracellular, while PelX is located in the periplasm. Similarly, the cellular localization of the family 2 pectate lyases can differ. Both PelY and PelB possess a typical signal sequence and are periplasmic when expressed in E. coli (11, 20). We demonstrated that PelW has no signal sequence and is located in the cytoplasm. Thus, in addition to a set of at least eight extracellular endo-pectate lyases (36), E. chrysanthemi produces two intracellular exopolygalacturonate lyases, the periplasmic PelX and the cytoplasmic PelW. It is remarkable that these 10 pectate lyases are distributed among the five defined pectate lyase families, implying independent genetic acquisitions.
A cell-bound exo-pectate lyase activity was first reported in E. chrysanthemi CUCPB1237 (3). The pelX gene was then cloned from E. chrysanthemi EC16 (2) and E. chrysanthemi 3937 (34). Evidence that PelW, like PelX, is an exo-cleaving lyase arises from the fact that its action on a polymeric substrate generates a single product, unsaturated digalacturonate (Fig. 4). In contrast, an endo-cleaving enzyme would catalyze the formation of multiple products of various sizes (27). Despite the fact that the two E. chrysanthemi exopolygalacturonate lyases form the same product from the polymeric substrate PGA and show relatively good activity on this substrate, they exhibit some differences in activity on various oligomers. PelX shows a high level of activity on oligomers from G4 to G7, with a maximal activity on G4. G3 is a poor substrate for PelX (34). On the contrary, G3 is the best substrate of PelW, while this enzyme shows a lower level of activity on G4. The cellular localization of these two exopolygalacturonate lyases seems to be complementary. PelX could cleave oligomers when they pass through the periplasm, while PelW could cleave oligomers entering the cytoplasm. Thus, the two exopolygalacturonate lyases could ensure successive depolymerization of pectic substances, and their substrate preferences are linked to their cellular compartments. The data obtained with oligomers suggest that the number of subsites of PelW is only three (−1 to +2); PelW seems to recognize only three residues at the reducing end of the polysaccharide. In contrast to the other pectate lyases, PelW is highly tolerant of the methylation of the substrate. It shows almost the same activity on PGA as it does on pectins with a degree of methylation up to 60%. Two E. chrysanthemi pectin methylesterases, the extracellular PemA and the outer-membrane-located PemB, are responsible for the demethylation of pectic substances in the extracellular medium and in the periplasm, respectively (17, 31). Nevertheless, the cytoplasmic lyase PelW might be able to cleave methylated oligomers escaping from the demethylation by these pectin methylesterases.
To better understand the role of the cytoplasmic pectinases in the final steps of pectin depolymerization in the E. chrysanthemi cytoplasm, we compared the properties of PelW with those of the oligogalacturonate lyase Ogl, which was the only known Erwinia cytoplasmic pectinase (3, 22). Ogl from E. carotovora was previously purified, and its catalytic properties were analyzed (22). Although the E. chrysanthemi ogl gene was cloned and sequenced (28, 29), enzymatic properties of the corresponding protein have never been studied. Nevertheless, this enzyme seems to play a key role in the pectin catabolic pathway, since E. chrysanthemi ogl mutants are unable to grow on PGA or digalacturonates as a carbon source (3, 29). This enzyme links the extracellular and intracellular steps of the pectin catabolic pathway because it cleaves the dimers produced by the combined action of all the other pectinases. However, the role of PelW in pectin catabolism is not negligible, since the E. chrysanthemi pelW (kdgC) mutant shows reduced growth on PGA (5). The role of the two enzymes PelW and Ogl is clearer when their substrate specificities are compared. Oligomers longer than dimers are very poor substrates for Ogl, while the trimer is the best substrate for PelW, which retains a good activity on the tetramer. Thus, these two enzymes seem to act in sequence during oligogalacturonate depolymerization in the cytoplasm. The substrate specificity of PelW implies that trimers, and probably tetramers, enter the E. chrysanthemi cytoplasm. The transport of pectic oligomers into the bacterial cells is the least-well-known step of pectin catabolism. Numerous mutants of E. chrysanthemi were isolated for their inability to grow on medium with PGA as the carbon source. None of them appeared to be affected with regard to transport of pectic oligomers. This result led us to theorize that several systems could be used for oligogalacturonate uptake.
The extracellular E. chrysanthemi endo-pectate lyases demonstrate an absolute Ca2+ requirement for their enzymatic activity (18, 25, 36, 39). These enzymes either are not affected or are inhibited in the presence of other divalent cations, with the exception of PelZ, which uses Mn2+ as a cofactor more efficiently than it uses Ca2+ (25, 39). The periplasmic exopolygalacturonate lyase PelX also requires cations for its activity and is weakly activated by Ca2+, Mn2+, Co2+, Ni2+, or Cu2+ (34). It has been previously reported that the E. carotovora Ogl does not require calcium ions and is not inactivated by EDTA (22). Our results show that the activities of the two E. chrysanthemi cytoplasmic lyases, PelW and Ogl, are dependent on the presence of divalent cations, since both enzymes are inhibited by EDTA. However, Ca2+ is unable to restore their activity, while several other metal cations of similar size, such as Co2+, Mn2+, and Ni2+, can activate both PelW and Ogl with different efficiencies (Fig. 5A and B). The PelW thermoinactivation data indicate a direct binding of cation to the protein, resulting in the stabilization of the protein structure (Fig. 5D). It is remarkable that both cytoplasmic lyases are activated by almost the same spectrum of cations, which is very different from the cation requirement of the extracellular pectate lyases. Ca2+ binds to the extracellular pectate lyases and also interacts with the carboxyl groups of their substrate, PGA. These cations have been shown to be directly involved in catalysis (16). It is possible that the cation dependence of PelW and Ogl is related to specific features of the catalytic reactions carried out by these enzymes. However, the substrate specificities and the reaction products of PelW and Ogl are quite different. Moreover, except for the cation requirement, the enzymatic properties of PelW do not essentially differ from those of PelX or of the endo-pectate lyases. Another explanation for the similar cation requirement spectra of PelW and Ogl could be related to their cytoplasmic localization. The bacterial cells maintain an intracellular calcium level below that of the growth medium. For example, in E. coli, the intracellular calcium concentration is 0.1 μM, regardless of its extracellular concentration (37). On the other hand, several specific transport systems ensure the intracellular uptake of numerous other divalent metal cations (37). Thus, the cation content of the bacterial cytoplasm could probably constrain the bacteria to use other divalent cations in the place of Ca2+ for similar catalytic processes.
ACKNOWLEDGMENTS
Appreciation is expressed to Valerie James for reading the manuscript. We thank our colleagues Sylvie Reverchon and William Nasser for valuable discussions. We thank Copenhagen Pectin for the gift of well-characterized pectins and the Section of Molecular Genetics of Industrial Microorganisms, Wageningen, The Netherlands, for tetragalacturonate.
This work was supported by grants from the Centre National de la Recherche Scientifique (UMR 5577), from the Ministère de l’Education Nationale de la Recherche et de la Technologie, and from the Commission of the European Communities (AIR 2-CT-941345).
REFERENCES
- 1.Alfano J R, Ham J H, Collmer A. Use of Tn5tac1 to clone a pel gene encoding a highly alkaline, asparagine-rich pectate lyase isozyme from an Erwinia chrysanthemi EC16 mutant with deletions affecting the major pectate lyase isozymes. J Bacteriol. 1995;177:4553–4556. doi: 10.1128/jb.177.15.4553-4556.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooks A D, He S Y, Gold S, Keen N T, Collmer A, Hutcheson S W. Molecular cloning of the structural gene for exopolygalacturonate lyase from Erwinia chrysanthemi EC16 and characterization of the enzyme product. J Bacteriol. 1990;172:6950–6958. doi: 10.1128/jb.172.12.6950-6958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collmer A, Bateman D F. Impaired induction and self-catabolite repression of extracellular pectate lyase in Erwinia chrysanthemi mutants deficient in oligogalacturonide lyase. Proc Natl Acad Sci USA. 1981;78:3920–3924. doi: 10.1073/pnas.78.6.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collmer A, Whalen C H, Beer S V, Bateman D F. An exo-poly-α-d-galacturonosidase implicated in the regulation of extracellular pectate lyase production in Erwinia chrysanthemi. J Bacteriol. 1982;149:626–634. doi: 10.1128/jb.149.2.626-634.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Condemine G, Robert-Baudouy J. Analysis of an Erwinia chrysanthemi gene cluster involved in pectin degradation. Mol Microbiol. 1991;5:2191–2202. doi: 10.1111/j.1365-2958.1991.tb02149.x. [DOI] [PubMed] [Google Scholar]
- 6.Copeland B R, Richter R J, Furlong C E. Renaturation and identification of periplasmic proteins in two-dimensional gels of Escherichia coli. J Biol Chem. 1982;257:15065–15071. [PubMed] [Google Scholar]
- 7.González-Candelas L, Kolattukudy P E. Isolation and analysis of a novel inducible pectate lyase gene from the phytopathogenic fungus Fusarium solani f. sp. pisi (Nectria haematococca, mating population VI) J Bacteriol. 1992;174:6343–6349. doi: 10.1128/jb.174.20.6343-6349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He S Y, Collmer A. Molecular cloning, nucleotide sequence, and marker exchange mutagenesis of the exo-poly-α-d-galacturonosidase-encoding pehX gene of Erwinia chrysanthemi EC16. J Bacteriol. 1990;172:4988–4995. doi: 10.1128/jb.172.9.4988-4995.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heffron S, Henrissat B, Yoder M D, Lietzke S, Jurnak F. Structure-based multiple alignment of extracellular pectate lyase sequences. Mol Plant-Microbe Interact. 1995;8:331–334. doi: 10.1094/mpmi-8-0331. [DOI] [PubMed] [Google Scholar]
- 10.Henrissat B, Heffron S E, Yoder M D, Lietzke S E, Jurnak F. Functional implications of structure-based sequence alignment of proteins in the extracellular pectate lyase superfamily. Plant Physiol. 1995;107:963–976. doi: 10.1104/pp.107.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinton J C D, Sidebotham J M, Gill D R, Salmond G P C. Extracellular and periplasmic isoenzymes of pectate lyase from Erwinia carotovora subspecies carotovora belong to different gene families. Mol Microbiol. 1989;3:1785–1795. doi: 10.1111/j.1365-2958.1989.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 12.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 13.Johnson B H, Hecht M H. Recombinant proteins can be isolated from E. coli cells by repeated cycles of freezing and thawing. Bio/Technology. 1994;12:1357–1360. doi: 10.1038/nbt1294-1357. [DOI] [PubMed] [Google Scholar]
- 14.Keen N T, Dahlbeck D, Staskawicz B, Belser W. Molecular cloning of pectate lyase genes from Erwinia chrysanthemi and their expression in Escherichia coli. J Bacteriol. 1984;159:825–831. doi: 10.1128/jb.159.3.825-831.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim J F, Beer S V. HrpW of Erwinia amylovora, a new harpin that contains a domain homologous to pectate lyases of a distinct class. J Bacteriol. 1998;180:5203–5210. doi: 10.1128/jb.180.19.5203-5210.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kita N, Boyd C M, Garrett M R, Jurnak F, Keen N T. Differential effect of site-directed mutation in pelC on pectate lyase activity, plant tissue maceration, and elicitor activity. J Biol Chem. 1996;271:26529–26535. doi: 10.1074/jbc.271.43.26529. [DOI] [PubMed] [Google Scholar]
- 16a.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 17.Laurent F, Kotoujansky A, Labesse G, Bertheau Y. Characterization and overexpression of the pem gene encoding pectin methylesterase of Erwinia chrysanthemi strain 3937. Gene. 1993;131:17–25. doi: 10.1016/0378-1119(93)90664-o. [DOI] [PubMed] [Google Scholar]
- 18.Lojkowska E, Masclaux C, Boccara M, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Characterization of the pelL gene encoding a novel pectate lyase of Erwinia chrysanthemi 3937. Mol Microbiol. 1995;16:1183–1195. doi: 10.1111/j.1365-2958.1995.tb02341.x. [DOI] [PubMed] [Google Scholar]
- 19.Manoil C, Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci USA. 1985;82:8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manulis S, Kobayashi D Y, Keen N T. Molecular cloning and sequencing of a pectate lyase gene from Yersinia pseudotuberculosis. J Bacteriol. 1988;170:1825–1830. doi: 10.1128/jb.170.4.1825-1830.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller J H. Experiment in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 22.Moran F, Nasuno S, Starr M P. Extracellular and intracellular polygalacturonic acid trans-eliminase of Erwinia carotovora. Arch Biochem Biophys. 1968;123:298–306. doi: 10.1016/0003-9861(68)90138-0. [DOI] [PubMed] [Google Scholar]
- 23.Moran F, Nasuno S, Starr M P. Oligogalacturonide trans-eliminase of Erwinia carotovora. Arch Biochem Biophys. 1968;125:734–741. doi: 10.1016/0003-9861(68)90508-0. [DOI] [PubMed] [Google Scholar]
- 24.Palva E T, Hirst T R, Hardy S J S, Holmgren J, Randall L. Synthesis of a precursor to the B subunit of heat-labile enterotoxin in Escherichia coli. J Bacteriol. 1981;146:325–330. doi: 10.1128/jb.146.1.325-330.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pissavin C, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Biochemical characterization of the pectate lyase PelZ of Erwinia chrysanthemi 3937. Biochim Biophys Acta. 1998;1383:188–196. doi: 10.1016/s0167-4838(97)00191-x. [DOI] [PubMed] [Google Scholar]
- 26.Pissavin C, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Regulation of pelZ, a gene of the pelB-pelC cluster encoding a new pectate lyase in Erwinia chrysanthemi 3937. J Bacteriol. 1996;178:7187–7196. doi: 10.1128/jb.178.24.7187-7196.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preston J F, III, Rice J D, Ingram L O, Keen N T. Differential depolymerization mechanisms of pectate lyases secreted by Erwinia chrysanthemi EC16. J Bacteriol. 1992;174:2039–2042. doi: 10.1128/jb.174.6.2039-2042.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reverchon S, Huang Y, Bourson C, Robert-Baudouy J. Nucleotide sequence of the Erwinia chrysanthemi ogl and pelE genes, negatively regulated by the kdgR product. Gene. 1989;85:125–134. doi: 10.1016/0378-1119(89)90472-1. [DOI] [PubMed] [Google Scholar]
- 29.Reverchon S, Robert-Baudouy J. Molecular cloning of Erwinia chrysanthemi oligogalacturonate lyase gene involved in pectin degradation. Gene. 1987;55:125–133. doi: 10.1016/0378-1119(87)90255-1. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Shevchik V E, Condemine G, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Characterization of pectin methylesterase B, an outer membrane lipoprotein of Erwinia chrysanthemi 3937. Mol Microbiol. 1996;19:455–466. doi: 10.1046/j.1365-2958.1996.389922.x. [DOI] [PubMed] [Google Scholar]
- 32.Shevchik V E, Evtushenkov A N, Fomichev Y K. Isoenzymes of extracellular pectate lyases of bacteria of the genus Erwinia. Biokhimiya. 1988;53:1628–1638. . (In Russian.) [Google Scholar]
- 33.Shevchik V E, Hugouvieux-Cotte-Pattat N. Identification of a bacterial pectin acetyl esterase in Erwinia chrysanthemi. Mol Microbiol. 1997;24:1285–1301. doi: 10.1046/j.1365-2958.1997.4331800.x. [DOI] [PubMed] [Google Scholar]
- 34.Shevchik V E, Kester H C M, Benen J A E, Visser J, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Characterization of the exopolygalacturonate lyase PelX of Erwinia chrysanthemi 3937. J Bacteriol. 1999;181:1652–1663. doi: 10.1128/jb.181.5.1652-1663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shevchik V E, Robert-Baudouy J, Condemine G. Specific interaction between OutD, an Erwinia chrysanthemi outer membrane protein of the general secretory pathway, and secreted proteins. EMBO J. 1997;16:3007–3016. doi: 10.1093/emboj/16.11.3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shevchik V E, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. The pectate lyase PelI of Erwinia chrysanthemi 3937 belongs to a new family. J Bacteriol. 1997;179:7321–7330. doi: 10.1128/jb.179.23.7321-7330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Silver S. Transport of inorganic cations. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1091–1102. [Google Scholar]
- 38.Tabor S, Richardson C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tardy F, Nasser W, Robert-Baudouy J, Hugouvieux-Cotte-Pattat N. Comparative analysis of the five major Erwinia chrysanthemi pectate lyases: enzyme characteristics and potential inhibitors. J Bacteriol. 1997;179:2503–2511. doi: 10.1128/jb.179.8.2503-2511.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witholt B, Boekhout M, Brock M, Kingma J, van Heerikhuizen H, de Leij L. An efficient and reproducible procedure for the formation of spheroplasts from variously grown Escherichia coli. Anal Biochem. 1976;74:160–170. doi: 10.1016/0003-2697(76)90320-1. [DOI] [PubMed] [Google Scholar]