Abstract
The existence of a global gene regulatory system in the hyperthermophilic archaeon Sulfolobus solfataricus is described. The system is responsive to carbon source quality and acts at the level of transcription to coordinate synthesis of three physically unlinked glycosyl hydrolases implicated in carbohydrate utilization. The specific activities of three enzymes, an α-glucosidase (malA), a β-glycosidase (lacS), and an α-amylase, were reduced 4-, 20-, and 10-fold, respectively, in response to the addition of supplementary carbon sources to a minimal sucrose medium. Western blot analysis using anti-α-glucosidase and anti-β-glycosidase antibodies indicated that reduced enzyme activities resulted exclusively from decreased enzyme levels. Northern blot analysis of malA and lacS mRNAs revealed that changes in enzyme abundance arose primarily from reductions in transcript concentrations. Culture conditions precipitating rapid changes in lacS gene expression were established to determine the response time of the regulatory system in vivo. Full induction occurred within a single generation whereas full repression occurred more slowly, requiring nearly 38 generations. Since lacS mRNA abundance changed much more rapidly in response to a nutrient down shift than to a nutrient up shift, transcript synthesis rather than degradation likely plays a role in the regulatory response.
Microbial survival and proliferation in boiling acid environments has been accompanied by the evolution of a diversity of mechanisms for energy generation and carbon assimilation. Members of the domain Archaea, particularly the crenarchaeal subdivision, dominate these extreme environments. Many of the organisms assigned to this subdivision are members of the order Sulfolobales, which includes the genus Sulfolobus (3, 11). This genus is comprised of obligate aerobes which conduct both lithoautotrophic (3, 23) and chemoheterotrophic (11, 14) metabolism. One member of this genus, Sulfolobus solfataricus, uses a wide range of reduced organic compounds, including starch and its derivative maltose, as sole carbon and energy sources (20, 43).
Bacteria and eukaryotes typically employ transcriptional regulatory mechanisms to coordinate expression of genes involved in carbohydrate utilization. These types of genes generally are subject to a process termed the glucose effect, which includes three components: inducer exclusion, transient repression, and catabolite repression (27). Among the gram-negative bacteria, coordination of expression of such genes entails the action of cyclic AMP (cAMP) and cAMP receptor protein (CRP) acting to balance utilization of low- and high-quality carbon resources (46). In certain gram-positive bacteria, cAMP and CRP are absent and CcpA, a negative-acting transcription factor is used to affect promoter activity of target genes (12, 21). Coordinated expression in eukaryotes of genes involved in carbon catabolism also is accomplished by trans-acting transcription factors including the proteins CREA and CREB (9, 42). In all of these organisms, catabolite repression is mediated at the level of transcription initiation. However, a key feature distinguishing the gene regulatory mechanisms employed in bacterial prokaryotes from that in eukaryotes arises from their fundamentally distinct basal transcription systems. Interestingly, transcription in archaeal prokaryotes closely resembles that employed by eukaryotes and not bacteria. This includes conserved promoter sequences (17, 37), TATA binding protein homologs (29, 34, 45), TFIIB homologs (13, 35, 36), and an RNA polymerase II homolog (24). Transcriptional regulatory systems in archaea must therefore accommodate their eukaryotic-like basal transcription components.
Archaea do conduct metabolism and therefore must control the expression of genes encoding metabolic enzymes. However little is yet known about how this might occur. For example, it is unknown if the key features which distinguish bacterial and eukaryotic catabolite repression systems are present in the archaea such as global transcriptional gene regulation, signal molecules, or trans-acting factors. Gene regulatory systems have been found in the euryarchaea, which comprise the other major archaeal subdivision. In the halophilic archaea, examples include the regulation of synthesis of bacteriorhodopsin (bob [49]), halocins (6), gas vacuoles (vac [41, 52]), and the heat shock response (cct [31, 51]). In the methanogenic archaea, examples include the regulation of methane biosynthesis (22, 32), histones (47), carbon monoxide dehydrogenase (cdh [50]), and nitrogen fixation (nif [7]). However, for the hyperthermophilic archaea which lie in both archaeal subdivisions, gene regulatory studies are less common perhaps because many of these organisms are obligate anaerobes with fastidious growth requirements (10).
In the aerobic hyperthermophile S. solfataricus, starch utilization necessitates the inducible synthesis and secretion of a highly stable α-amylase (20). The resulting hydrolytic products including dextrins and maltodextrins are further hydrolyzed by the action of a cell-associated α-glucosidase encoded by malA (43). Expression of malA is modestly affected by carbon source type, while levels of the α-amylase are strongly influenced (20, 44). The variation in levels of these glycosyl hydrolases in response to carbon source type represents one of the hallmarks of catabolite repression, that is, carbon source preference (27). It remains unclear, however, if these changes occur through action at the level of enzyme activity, translation, or transcription. To better understand the catabolite repression-like response of S. solfataricus, a set of glycosyl hydrolase genes were characterized and used to probe the consequences of nutrient supplementation of a minimal medium. The observed pattern of gene expression indicates this organism employs a global transcriptional regulatory system to coordinate its response to carbon sources.
MATERIALS AND METHODS
Archaeal strains and cultivation.
S. solfataricus was grown as described previously (43) at 80°C at a pH of 3 in screw-cap flasks and aerated by vigorous shaking. The medium used contained 20 mM ammonium sulfate, 4 mM dibasic potassium phosphate, 4 mM magnesium sulfate, 1 mM calcium chloride, 0.2 mM iron chloride, 18 mM manganese chloride, 0.02 mM sodium borohydride, 1.5 μM zinc sulfate, 0.74 μM copper chloride, 0.25 μM sodium molybdenate, 0.37 μM vanadium sulfate, and 0.13 μM cobalt sulfate. Cyanocobalamin was used at a final concentration of 0.2 μg/liter, while all other vitamins were used at final concentrations of 50 μg/liter. Nucleosides and amino acids were used at final concentrations of 10 and 50 mg/liter, respectively. Sucrose was added at a final concentration of 0.2% (wt/vol), and yeast extract and tryptone were added at a final concentration of 0.1% (wt/vol) and 0.2% (wt/vol), respectively. The growth medium was adjusted with sufficient sulfuric acid to yield a pH of 3.0. Growth was monitored spectrophotometrically at a wavelength of 540 nm.
Molecular biology methods.
Restriction digestion and ligation of DNA were performed as described previously (2). Plasmid transformation was performed with E. coli DH5α as described previously (18). Plasmid DNA was isolated by the alkali lysis procedure (1). DNA sequence analysis was done as described elsewhere (39), and DNA alignment and analysis were performed with the fragment assembly programs of the Wisconsin Package (version 9.0; Genetics Computer Group, Inc.). All other manipulations of Escherichia coli strains were done as described previously (38).
Enzyme assays.
Assays for the α-glucosidase and the β-glycosidase used S. solfataricus cell extracts prepared by sonicating cells resuspended in 100 mM sodium acetate (pH 4.5) and 10 mM Tris hydrochloride (pH 7.0), respectively. The hydrolysis of p-nitrophenyl-α-glucopyranoside (α-PNPG) was used to measure the α-glucosidase as described previously (43). Substrate was used at a concentration of 10 mM in a reaction buffer consisting of 100 mM sodium acetate (pH 4.5). Reactions were initiated by the addition of crude cell sonicates or enzyme to prewarmed solutions and terminated by addition of 1 M sodium carbonate resulting in a sample pH of 10.0. Hydrolysis of p-nitrophenyl-β-d-glucopyranoside (β-PNPG) was used to measure the β-glycosidase, using the same procedure as employed for the α-glucosidase. The extent of substrate hydrolysis was determined by the absorbance of the sample at a wavelength of 420 nm with correction for spontaneous substrate hydrolysis. A unit of either α-glucosidase or β-glycosidase activity is defined as the amount of enzyme required to liberate 1 μmol of p-nitrophenol per min per mg of protein. Measurement of secreted α-amylase enzyme activity used cell-free culture supernatants concentrated by ultrafiltration as necessary as described previously (20). α-Amylase activity was determined as described elsewhere (20) by monitoring the loss of iodine binding to added starch (25, 28). The iodine binding assay was performed with a reaction mixture containing clarified culture supernatant, 2% (wt/vol) starch, 100 mM sodium acetate, and 2 mM calcium chloride (pH 3.0) at 80°C for 30 min. The reaction was terminated by cooling at 4°C. Color was developed by addition of 0.015 ml of an iodine solution (4% [wt/vol] potassium iodide, 1.25% [wt/vol] iodine). The sample absorbance was determined at a wavelength of 600 nm and was corrected for a sample lacking added substrate. One unit of α-amylase activity was equivalent to the amount of protein which hydrolyzed 1 μg of starch in 1 min. All samples were assayed in duplicate, and the averages of the sample results are reported.
Protein purification and antibody production.
Recombinant enzyme purification used transformants of E. coli DH5α (Gibco-BRL) harboring either the malA expression plasmid pBN56, a pLITMUS 29 (New England Biolabs) derivative (44), or the lacS expression plasmid pBN55 (this work). These strains were grown at 37°C with vigorous shaking in 4 liters of LB medium containing ampicillin (100 μg/ml) until they reached stationary phase. Cells were harvested by centrifugation, resuspended in 30 mM morpholinepropanesulfonic acid, pH 8.0 (MOPS buffer), and lysed by sonication at 4°C. The resulting lysates were clarified by centrifugation (3,000 × g for 30 min) and then heated at 85°C for 30 min and reclarified by centrifugation. The heating and centrifugation procedure was then repeated a second time. The heat-treated supernatants were concentrated by ultrafiltration using a YM3 (Amicon) membrane.
The concentrated supernatants were applied to a Mono Q FPLC (fast protein liquid chromatography) column (Pharmacia) previously equilibrated with MOPS buffer. The recombinant β-glycosidase and the recombinant α-glucosidase were eluted with linear gradients of sodium chloride in MOPS buffer. Active fractions for each enzyme were identified by enzyme assay, pooled, concentrated by ultrafiltration using a PM10 (Amicon) membrane, and dialyzed into 100 mM sodium phosphate buffer (pH 6.0). The dialyzed samples were applied to a Superdex 200 HR 10/30 FPLC column (Pharmacia) previously equilibrated with 100 mM sodium phosphate (pH 6.0). Active fractions were again pooled and concentrated by ultrafiltration.
The purified enzymes were hydrolyzed by using cyanogen bromide in 70% formic acid as described (16, 30). The resulting peptides for the recombinant α-glucosidase were dialyzed into 5 mM Tris-Cl (pH 7.0), lyophilized to dryness, and resuspended in water. The recombinant β-glycosidase was diluted to 0.07% formic acid with 5 mM Tris-Cl (pH 7.0) and lyophilized to near dryness, and the pH was adjusted to 7.0 with 10 M sodium hydroxide. Anti-α-glucosidase antibodies were raised in mice as described previously (2). Anti-β-glycosidase polyclonal antibodies were produced by injection of 0.1 mg of purified protein into New Zealand White rabbits as previously described (40). Polyclonal sera were further purified by precipitation with acetone powder as previously described (2).
Protein electrophoresis and Western blot analysis.
Proteins were resolved by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis under reducing conditions using unstained low- and high-molecular-weight markers (Bio-Rad). Prior to electrophoresis, samples were adjusted to 2% (wt/vol) SDS and 3 mM β-mercaptoethanol and boiled for 10 min. SDS-polyacrylamide gels were stained with Coomassie blue R250 to visualize protein. Chemiluminescent Western blot analysis using the Tropix system was performed as described elsewhere (40). The α-glucosidase and β-glycosidase protein standards were prepared as described above for use in the preparation of antibodies, with some modification. The recombinant E. coli extracts were subjected to only one heat treatment at 85°C for 1 h and then clarified at 14,000 × g at ambient temperature. The relative abundance of the two proteins in these extracts was determined by comparison to purified samples.
Isolation and DNA sequence analysis of lacS.
The S. solfataricus library was constructed by using genomic DNA prepared as described previously (44). Genomic DNA was partially digested with Sau3AI and then fractionated by electrophoresis; DNA between 3 and 5 kb in size was ligated into the BamHI site of pUC19 (New England Biolabs) and transformed into E. coli DH5α with selection for ampicillin resistance. Two thousand individual colonies were picked and propagated in 96-well microtiter plates in rich medium containing ampicillin. The S. solfataricus lacS gene was identified by screening these isolates, which had been preheated at 80°C for 1 h, for the ability to hydrolyze β-PNPG at 80°C. One such isolate was identified by this method, and its recombinant plasmid was called pBN55. The insert of pBN55 was subcloned for sequencing by restriction digestion with EcoRI. This digestion yielded three fragments, the first of which consisted of the 725-bp 5′ end of the insert and the pUC19 vector. It was religated to itself to generate the 5′-end subclone. The remaining two fragments produced by the EcoRI digestion were 491 and 552 bp and represented the central and 3′ portions of the insert, respectively. They were each ligated into the EcoRI site of pUC19 to generate the middle and 3′-end subclones. The inserts of all three subclones were then sequenced.
Northern blot analysis.
S. solfataricus total RNA was extracted as described previously (5) from wet cell paste obtained by filtration of cells at the mid-exponential phase of growth on the indicated carbon sources. Electrophoresis of RNA samples was as described elsewhere (4), and the RNA was electrophoretically transferred to Hybond N+ (Amersham) membranes and cross-linked by shortwave UV irradiation. RNA riboprobes were generated by using a riboprobe buffer kit (Promega) and the manufacturer’s protocol. Riboprobe templates were a 2,081-bp fragment encoding the malA region comprising base pair positions 141 to 2,265 relative to the malA start codon (44) and a 493-bp EcoRI lacS fragment including positions 544 to 1,037 relative to the lacS start codon. The lacS sense-strand RNA standard was synthesized by using the pBN55 lacS insert; the malA sense-strand RNA standard was synthesized by using the same fragment as employed for malA riboprobe synthesis. The DNA fragments used to generate the riboprobes and sense-strand RNA standards were cloned into plasmid pT7T3 18U (Pharmacia). Northern hybridizations were performed at 55°C with 50% formamide. Washed membranes were used to prepare autoradiograms with Kodak X-Omat film. Molecular weight standards were RNATranscripts (United States Biochemical).
Nucleotide sequence accession number.
The lacS sequence determined in this study has been deposited in GenBank under accession no. AF133096.
RESULTS
Glycosyl hydrolase levels vary in response to use of supplemental carbon sources.
Three enzymes were selected to help define the catabolite regulation-like system in S. solfataricus: the α-amylase (20), the α-glucosidase (43, 44), and the β-glycosidase (8, 15). Levels of each of the three enzymes were determined from cells in the mid-exponential phase of growth in a minimal sucrose medium with or without nutrient supplementation (Table 1). Cell-associated activities (α-glucosidase and β-glycosidase) were determined as specific activities, while activity levels of the secreted α-amylase were normalized to total cell protein present at the time of assay. Yeast extract is a common medium additive widely used for the growth of this organism (11, 14, 34, 37). Yeast extract supplementation of a minimal sucrose medium decreased cell generation times 13%, from 7.5 to 6.5 h, and increased cell yields 28%, from 4.5 to 5.8 OD540 (optical density at 540 nm) units/ml. This supplement simultaneously resulted in significant reductions in enzyme levels, approximately 4-fold for the α-glucosidase, 20-fold for the β-glycosidase, and at least 10-fold for the α-amylase (Table 1). No alterations in glycosyl hydrolase activities were noted when other methods were used to perturb cell growth rates and culture yields. Decreasing the temperature of incubation by 10°C (from 80 to 70°C) resulted in over a 50% increase in generation time with no concomitant alteration in the measured glycosyl hydrolase activities.
TABLE 1.
Steady-state levels of glycosyl hydrolases
| Carbon source(s) | Glycosyl hydrolase activity (mean ± SD)
|
||
|---|---|---|---|
| α-Glucosidase (μmol of PNP/min/mg) | β-Glycosidase (μmol of PNP/min/mg) | α-Amylase (U/ml) | |
| Sucrose | 46 ± 2 | 607 ± 55 | 121 ± 20 |
| Sucrose + yeast extract | 13 ± 1 | 32 ± 2 | <10 |
| Sucrose + tryptone | 35 ± 5 | 73 ± 5 | 37 ± 20 |
| Sucrose + amino acidsa | 10 ± 3 | 126 ± 7 | 55 ± 30 |
| Tryptone | 33 ± 2 | 151 ± 6 | 41 ± 4 |
The amino acids included alanine, arginine, asparagine, aspartate, glutamate, glutamine, glycine, leucine, and histidine.
Previous studies had indicated a role for certain amino acids acting as sole carbon and energy sources in glycosyl hydrolase expression (19, 20). When tryptone was used to supplement a sucrose minimal medium, levels of all three glycosyl hydrolase activities were reduced relative to levels in the unsupplemented sucrose medium (Table 1). However, comparison of glycosyl hydrolase activities produced during growth in tryptone without added sucrose indicates that sucrose does not act as an inducer of enzyme activities. The reduction in glycosyl hydrolase activities observed upon tryptone addition could be further ascribed to the action of a subset of nine amino acids. Addition of a pool comprising all other amino acids and excluding these nine amino acids had no effect on the levels of the measured activities. Medium supplementation with a pool of vitamins including biotin, folate, pyridoxine, cyanocobalamin, thiamine, riboflavin, nicotinate, pantothenate, p-aminobenzoate, and thioctate also had no effect on the measured activities. Similarly, medium supplementation with a pool of nucleosides including guanosine, adenosine, cytosine, and thymidine had no effect on glycosyl hydrolase activities. The effective amino acids could be further divided into two groups. A mixture of alanine, arginine, asparagine, and aspartate added to a sucrose minimal medium was inhibitory and resulted in a β-glycosidase specific activity 45% of maximum levels, while supplementation with a mixture of glutamate, glutamine, glycine, leucine, and histidine had no inhibitory effect. These results demonstrated that medium supplementation with certain amino acids was in part responsible for the observed change in glycosyl hydrolase levels caused by yeast extract addition. The magnitude of the effect of these amino acids as a group or as individual supplements, however, was significantly smaller than that observed with more complex additives. Since yeast extract addition produced the largest change in glycosyl hydrolase activities, it was used in subsequent studies as a medium supplement to facilitate analysis of the S. solfataricus regulatory response.
Variation in enzyme activities result from alterations in enzyme levels.
Western blot analysis was performed to test the possibility that the observed changes in enzyme levels resulted from changes in enzyme abundance rather than enzyme activity. Mouse polyclonal antibodies to the α-glucosidase were prepared by using hydrolyzed recombinant protein as an immunogen which was purified from an S. solfataricus malA E. coli expression system (44). Antibodies specific for the β-glycosidase were prepared in a similar manner using hydrolyzed protein purified from an S. solfataricus lacS E. coli expression system. The lacS gene initially was recovered from an S. solfataricus 98/2 plasmid-based E. coli expression library. The library was screened for isolates which produced thermostable β-glycosidase activity. One such isolate identified carried a plasmid expressing the S. solfataricus lacS gene under control of the lac promoter. The plasmid contained an insert of 1,768 bp and contained the entire lacS coding sequence of 1,467 bp as well as 176 bp 5′ to the start codon and 117 bp 3′ to the termination codon. The resulting deduced amino acid sequence exhibited one nonconservative change relative to the lacS sequence derived from S. solfataricus MT4 (8). This change resulted from three contiguous substitution mutations (CAT→GCA) beginning at bp 703 relative to the lacS start codon. The S. solfataricus 98/2 sequence encodes alanine at this position, while strain MT4 encodes a histidine. This DNA sequence difference also results in the elimination of an NdeI site and the creation of a BsrDI site, as indicated by restriction endonuclease analysis of the recombinant plasmid.
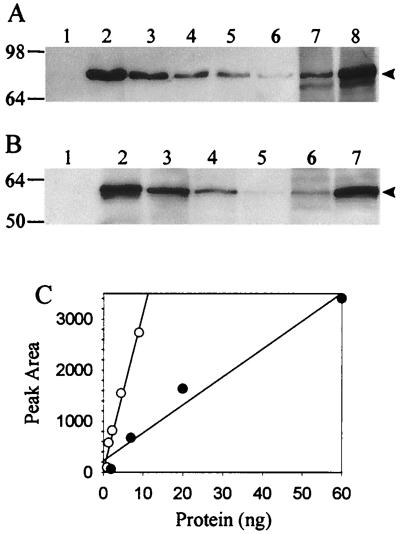
Chemiluminescent Western blot analysis of S. solfataricus cell extracts derived from cells growing in a sucrose minimal medium with or without added yeast extract revealed a large difference in levels of both the α-glucosidase and the β-glycosidase (Fig. 1). α-Glucosidase levels differed 4-fold (Fig. 1A, lanes 7 and 8), and β-glycosidase levels differed 17-fold (Fig. 1B, lanes 6 and 7). Protein abundance was determined by transmittance densitometry and comparison with recombinant α-glucosidase and β-glycosidase protein standards added as controls on the immunoblots. The intensity of the chemiluminescent signal obtained from the protein standards was linear in the range of the measured proteins (Fig. 1C). The observed variation in abundance of both enzymes as determined by Western blot analysis was directly proportional to the variation in enzyme activity detected in cell extracts (Table 1). These results indicate that allosteric control over enzyme activity or other forms of regulation operating at the posttranslational level are not significant factors in the observed regulatory response.
FIG. 1.
Western blot analysis of steady-state levels of α-glucosidase and β-glycosidase. (A) Levels of α-glucosidase. Lanes: 1, nonrecombinant E. coli extract; 2 to 6, recombinant α-glucosidase at (9, 4.5, 2.25, 1.4, and 1 ng, respectively); 7, S. solfataricus extract (30 μg) from cells grown in sucrose medium with yeast extract; 8, S. solfataricus extract (30 μg) from cells grown in sucrose medium. (B) Levels of β-glycosidase. Lanes: 1, nonrecombinant E. coli extract; 2 to 5, recombinant β-glycosidase (40, 13.4, 4.7, and 1.3 ng, respectively); 6, S. solfataricus extract (31 μg) from cells grown in sucrose medium with yeast extract; 7, S. solfataricus extract (18 μg) from cells grown in sucrose medium. Arrowheads on the right indicate positions of α-glucosidase and β-glycosidase; molecular mass markers in kilodaltons are shown on the left. (C) Densitometry of recombinant α-glucosidase (open circles) and recombinant β-glycosidase (closed circles) standards.
Variation in enzyme levels result from changes in mRNA abundance.
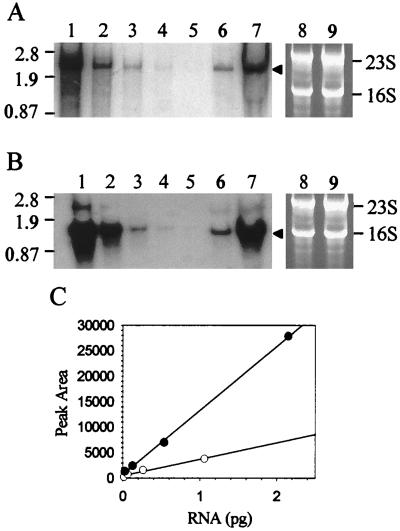
Northern blot analysis was conducted to determine if the observed variation in abundance of the α-glucosidase and β-glycosidase detected by Western blot analysis resulted from corresponding changes in levels of the malA and lacS transcripts. Riboprobes complementary to lacS and malA mRNA were used to probe blots of total S. solfataricus RNA derived from cells in the exponential phase of growth in a sucrose minimal medium with or without added yeast extract. In vivo levels of both mRNAs were significantly reduced as a consequence of yeast extract medium supplementation (Fig. 2). The levels of malA mRNA were reduced 4-fold (Fig. 2A, lanes 6 and 7), while levels of lacS mRNA were reduced 14-fold (Fig. 2B, lanes 6 and 7). Levels of total cellular RNA (5 μg/lane) were comparable for these samples, as indicated by ethidium bromide staining (Fig. 2A and B, lanes 8 and 9). The magnitudes of these changes in malA and lacS mRNA levels were proportional to those observed for differences in enzyme abundance and enzyme activity. To minimize the possibility of differential RNA transfer or hybridization in the blotting procedures, lacS and malA sense-strand RNA standards were prepared in vitro and were included as internal controls on each blot (Fig. 2A and B, lanes 1 to 5). The malA template for sense-strand RNA standard synthesis was 2,081 bp; however, transcription terminates approximately 500 bp into the vector backbone and results in an RNA close to the apparent size of 2.5 kb for the native malA mRNA (Fig. 2A and reference 44). The lacS sense-strand RNA standard has a predicted size of 1.7 kb but an apparent size of 1.8 kb and is therefore of a size greater than the apparent size of 1.5 kb for the natural lacS mRNA. This resulted from the use of a lacS riboprobe template which included 100 bp of lacS flanking sequence derived from regions located both 5′ and 3′ to the lacS open reading frame. The intensity of the autoradiographic signal obtained from the sense RNA standards was linear in the range of the measured natural lacS and malA mRNAs (Fig. 2C). The apparent variation in lacS and malA mRNA levels suggest that either mRNA synthesis or mRNA degradation, rather than protein stability or turnover, is the primary target for the S. solfataricus regulatory response.
FIG. 2.
Northern blot analysis of steady-state levels of malA and lacS mRNAs. Total cellular S. solfataricus RNA was loaded in 5-μg amounts per lane. (A) Levels of malA mRNA. Lanes: 1 to 5, dilution series of malA antisense standards (1.06, 0.26, 0.066, 0.017, and 0.004 pg of RNA); 6, RNA from cells grown on sucrose medium with yeast extract; 7, RNA from cells grown on sucrose medium; 8 and 9, identical to lanes 6 and 7 but stained with ethidium bromide and visualized by UV irradiation. (B) Levels of lacS mRNA. Lanes: 1 to 5, dilution series of lacS antisense standards (2.15, 0.54, 0.13, 0.033, and 0.008 pg of RNA); 6, RNA from cells grown on sucrose medium with added yeast extract; 7, RNA from cells grown on sucrose medium; 8 and 9, identical to lanes 6 and 7 but stained with ethidium bromide and visualized by UV irradiation. Arrowheads indicate positions of malA and lacS mRNAs; molecular weight markers in kilobases are shown on the left; 16S and 23S rRNAs are shown on the right. (C) Densitometry of malA (open circles) and lacS (closed circles) antisense standards.
A global gene regulatory mechanism.
Since mRNA levels of both malA and lacS were affected by presence of yeast extract in the growth medium, coordinated expression resulting from physical linkage such as the occurrence of these genes in a polycistronic operon might explain the observations. However, DNA sequence analysis of regions flanking malA exclude presence of lacS within 4.01 kb 5′ to malA and 0.96 kb 3′ to malA (44). In addition, sequence analysis of regions flanking lacS exclude presence of malA within 1.32 kb 5′ to lacS and 2.07 kb 3′ to lacS (8, 33). These results indicate that the minimum aggregate distance between lacS and malA is 2.28 kb. Further, Northern blot analysis presented here excludes presence of a large polycistronic mRNA which might encode both these genes. Instead the apparent sizes of the transcripts encoding both enzymes were close to the sizes of the corresponding open reading frames as expected for monocistronic mRNAs. In addition, levels of the α-amylase were also responsive to medium composition. Though the gene encoding the α-amylase in this organism has not been characterized, there are no open reading frames of the expected size based on the apparent subunit mass of the native enzyme (20) adjacent to either the lacS or malA gene. Therefore, there are at least three enzymes encoded by physically unlinked genes whose concentrations are coordinately regulated.
Induction of lacS expression.
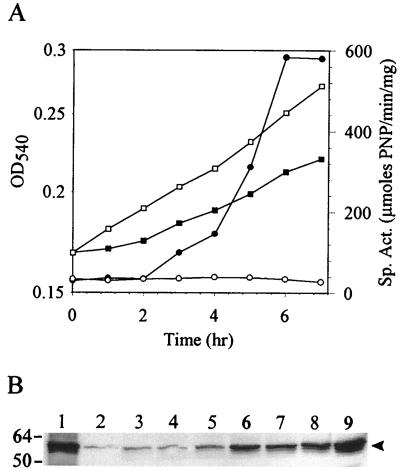
These results indicate there are steady-state differences in glycosyl hydrolase gene expression during balanced growth. Variations in levels of the lacS transcript under the two growth conditions, however, were noticeably more dramatic than for malA. The expression of lacS was therefore selected as a more sensitive measure of the S. solfataricus regulatory response. To better understand how this organism accomplishes changes in gene expression, the response time of the regulatory system was determined. Cells were subjected to a nutrient down shift and monitored for transient alterations in lacS expression. S. solfataricus was grown to mid-exponential phase in sucrose minimal medium supplemented with yeast extract. Cells were recovered by centrifugation and resuspended in sucrose medium with or without added yeast extract. The two cultures were then incubated, and the levels of β-glycosidase activity were monitored. β-Glycosidase activity remained unchanged in cells maintained in sucrose minimal medium supplemented with yeast extract but was quickly activated in cells which had been down shifted to the sucrose minimal medium without yeast extract addition (Fig. 3A). An increase was evident after 2 h of incubation and reached maximum levels 6 h after the shift. The induced levels of enzyme activity were equivalent to those observed during balanced growth in a sucrose minimal medium. The entire change in enzyme activity was accomplished within one generation. Simultaneous analysis of β-glycosidase levels was conducted by Western blot analysis using purified β-glycosidase protein standards (Fig. 3B). β-Glycosidase abundance increased in the downshifted cells in parallel to the observed changes in β-glycosidase activity, while enzyme levels remained unchanged in the unshifted culture.
FIG. 3.
Induction of β-glycosidase levels by nutrient down shift. (A) Growth of S. solfataricus in sucrose medium with yeast extract (open symbols) and in sucrose medium (closed symbols), OD540 (squares), and β-glycosidase specific activity (circles). (B) Western blot analysis of β-glycosidase levels. Lane 1 and 2 contain 40 and 4 ng recombinant β-glycosidase, respectively; lanes 3 to 9 contain S. solfataricus extracts following a nutrient down shift from growth in sucrose medium with yeast extract to a sucrose medium. Lanes and sample times after onset of the shift: 3, 0 h; 4, 1 h; 5, 2 h; 6, 3 h; 7, 4 h; 8, 5 h; 9, 7 h. Cell extracts were loaded in 24-μg amounts per lane. The arrowhead on the right indicates the position of β-glycosidase; molecular mass markers in kilodaltons are shown on the left.
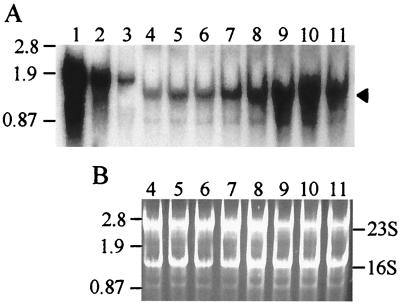
Variation in lacS mRNA abundance in response to removal of yeast extract was examined by Northern blot analysis to determine how rapidly lacS mRNA levels readjusted and how closely such readjustments might be to changes observed in β-glycosidase levels. Sense-strand lacS RNAs made in vitro were included on the blots to control for variation in transfer efficiency and for use in quantitative analysis of lacS mRNA. Levels of lacS mRNA began to increase within 2 h of the nutrient down shift and reached maximum levels in 6 h, slightly ahead of the maximum levels seen in β-glycosidase activity and protein levels (Fig. 4A). Transmittance densitometry of ethidium bromide-stained gels prior to Northern blot transfer indicated that the levels of both 16S rRNA and 23S rRNA were equivalent for all cell extract samples (Fig. 4B).
FIG. 4.
Northern blot analysis of lacS expression in response to nutrient down shift. Total cellular S. solfataricus RNA was loaded in 5-μg amounts per lane. (A) Northern blot. Lanes 1 to 3, lacS antisense RNA standards (2.2, 0.54, and 0.13 pg, respectively); lanes 4 to 11, total RNA from cells following a nutrient down shift from sucrose medium with yeast extract to sucrose medium. Lanes and sample times after onset of the shift: 4, 0 h; 5, 1 h; 6, 2 h; 7, 3 h; 8, 4 h; 9, 5 h; 10, 6 h; 11, 7 h. The arrowhead on the right indicates the position of lacS mRNA; molecular weight markers in kilobases are shown on the left. (B) Ethidium bromide-stained total RNA. Lanes 4 to 11 are identical to those shown in panel A but were stained with ethidium bromide and visualized by UV irradiation. The positions of the 16S and 23S rRNAs are indicated.
Repression of lacS expression.
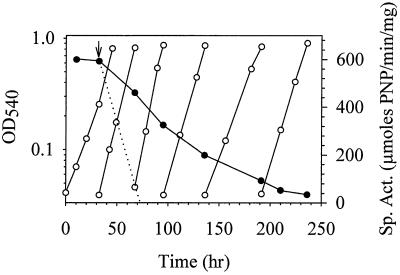
The change in lacS mRNA levels precipitated by nutrient down shift could result from increased lacS mRNA synthesis or decreased lacS mRNA degradation. To discern between these two mechanisms, nutrient up shift conditions were used to measure how rapidly lacS expression could readjust to the lower levels seen during steady-state growth on sucrose minimal medium containing yeast extract. Yeast extract was added to cells growing exponentially in a sucrose minimal medium, and levels of β-glycosidase activity were monitored until minimum levels were observed; cells were repeatedly subcultured to maintain conditions of balanced growth (Fig. 5). β-Glycosidase levels decreased in response to the nutrient up shift and after 245 h reached minimum levels. The elapsed time necessary for complete repression of β-glycosidase activity was 35 times that observed for the increase in enzyme activity produced in response to the nutrient down shift (Fig. 3A). For comparative purposes, the anticipated rate is presented for the change in β-glycosidase activity resulting from dilution due to cell division combined with ongoing repressed levels of enzyme production (Fig. 5).
FIG. 5.
Repression of β-glycosidase levels by nutrient up shift, determined by measurement of OD540 (open circles) and β-glycosidase specific activity (closed circles). Cells were maintained in exponential phase by repeated subculturing into prewarmed medium. At the time indicated by the arrow, the sucrose medium was supplemented with yeast extract resulting in a nutrient up shift. The theoretical rate of decrease in β-glycosidase activity resulting from an immediate reduction in synthesis to repressed levels combined with dilution of the activity by ongoing cell division is indicated by the dotted line.
The slow readjustment in β-glycosidase activity caused by nutrient up shift could occur independently of changes in lacS mRNA abundance. For example, β-glycosidase might be proteolytically insensitive, and thus changes in its levels would depend on dilution resulting from cell division. lacS mRNA could also be intrinsically stable and require dilution to achieve new cellular concentrations. Alternatively, lacS mRNA levels could readjust rapidly through an RNA degradation mechanism. To distinguish between these possibilities, levels of lacS mRNA were determined by Northern blot analysis in cell samples subjected to nutrient up shift (Fig. 6). Readjustment in lacS mRNA levels exhibited a pattern similar to that of β-glycosidase activity. Minimum levels of lacS mRNA were achieved only after 245 h of cell growth. The observed rate of readjustment in lacS mRNA abundance greatly lags behind that resulting from cell dilution due to ongoing cell division (Fig. 5). Since transcription inhibitors are not yet available for in vivo studies in hyperthermophilic archaea, measurements of the rate of change in gene expression were used to discern between possible gene regulatory mechanisms (Table 2). It is apparent from these values that the difference in the rate of change in lacS transcript abundance in response to the two nutrient shift conditions (down shift and up shift) was 85-fold, while that for differences in the rate of change of β-glycosidase enzyme activity was 43-fold. These results suggest that an active mechanism is employed for induction of lacS expression (down shift) whereas a passive mechanism involving cell dilution is used to repress lacS expression (up shift).
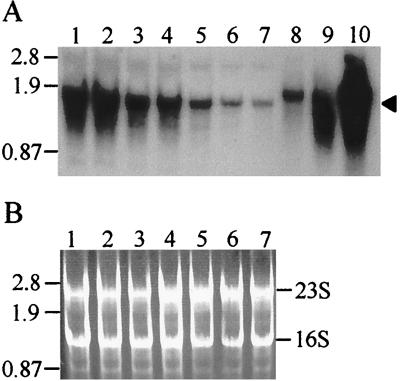
FIG. 6.
Northern blot analysis of lacS expression in response to nutrient up shift. Total cellular S. solfataricus RNA samples derived from cultures treated as shown in Fig. 5 were loaded in 5-μg amounts per lane. (A) Northern blot. Lanes 1 to 7 contain total RNA from cells shifted from sucrose medium with added yeast extract to sucrose medium. Lanes and sample times: 1, 0 h; 2, 35 h; 3, 63 h; 4, 103 h; 5, 159 h; 6, 178 h; 7, 204 h. Lanes 8 to 10 contain lacS antisense RNA standards (0.13, 0.54, and 2.2 pg, respectively). The arrowhead on the right indicates the position of lacS mRNA; molecular weight markers in kilobases are shown on the left. (B) Ethidium bromide-stained total RNA. Lanes 1 to 7 are identical to those in panel A but were stained with ethidium bromide and visualized by UV irradiation. Positions of the 16S and 23S rRNAs are indicated.
TABLE 2.
Rates of change in lacS expression during induction and repression
| Condition | Level (r2)
|
||
|---|---|---|---|
| β-Glycosidase activity (μmol of PNP/mg/h) | β-Glycosidase protein (μg/mg of total protein/h) | lacS mRNA (pg/mg/h) | |
| Induction (down shift) | 130 (0.89) | 0.23 (0.89) | 0.29 (0.92) |
| Repression (up shift) | 3 (0.96) | NDa | 0.0034 (0.87) |
ND, not determined.
DISCUSSION
The results presented here demonstrate the existence of a global gene regulatory system in the hyperthermophilic crenarchaeote S. solfataricus. The system exhibits features found previously in the catabolite repression systems of eukaryotes and bacterial prokaryotes such as the coordinate regulation of expression of genes involved in the metabolism of secondary carbon and energy sources. However, certain aspects such as inducer exclusion and transient repression (27) are not apparent. Though glycosyl hydrolase expression appears maximal during growth in a sucrose minimal medium, sucrose is not an inducer. Growth on sole carbon and energy sources other than sucrose resulted in near-maximal levels of glycosyl hydrolase activity (20, 44), and sucrose supplementation of a complex medium (tryptone) did not result in an elevation of glycosyl hydrolase activities relative to levels found during growth on tryptone alone. Similarly, yeast extract supplementation or removal led to a permanent and not a transient change in glycosyl hydrolase levels.
The S. solfataricus catabolite repression-like system acts at the level of mRNA abundance to coordinate levels of glycosyl hydrolases in response to changing medium nutrient status. Increased nutrient availability in the form of amino acids supplied primarily as yeast extract to a minimal sucrose growth medium increased the growth rate and cell yield of S. solfataricus, suggesting that these supplements support critical anabolic requirements. Amino acid supplementation therefore can be viewed as an increase in the quality of available nutrients. Previous work on this organism identified an important metabolic role for certain amino acids acting as sole carbon and energy sources on the production of the S. solfataricus α-amylase (2). These amino acids influenced production of the α-amylase in either a positive or a negative fashion. These effects could result from differences in their metabolic entry points into the citric acid cycle, again a reflection of some distinction in carbon source quality (20, 23). In the work presented here, however, amino acids and more complex additives were not used as sole carbon and energy sources but instead as supplements to an otherwise complete sucrose minimal medium. The effect of these supplements on glycosyl hydrolase expression is most likely a result of their consumption as alternative carbon and energy sources rather than as nitrogen sources. There is no apparent correlation between the identity of amino acids which exert a regulatory effect in the work presented here and the ability of these amino acids to act as nitrogen sources (14). In addition, nitrogen is supplied in the medium in the form of ammonium at a level (20 mM) which is in excess for maximal growth rates and cell yields. It should be noted that ammonium chloride is readily available in the hot springs environment and represents the only natural occurrence of this mineral (sal ammoniac). Ultimately, the ability of S. solfataricus to modulate gene expression and avoid unnecessary protein production in response to improved nutrient resources would help it conserve energy and promote survival.
Since growth rate and cell yields were correlated with altered gene expression, it remains possible that any condition altering growth leads to altered glycosyl hydrolase gene expression. This is apparently not true in at least some instances. Growth rate could be altered by varying culture incubation temperature without a noticeable effect on glycosyl hydrolase gene expression. Thus, the observed results using medium supplements are relatively specific in their effect on gene expression and appear to act through their role as carbon sources rather than as a result of their ability to change cell growth per se.
The regulatory mechanism that controls glycosyl hydrolase mRNA levels in response to medium composition could operate either at the level of mRNA synthesis or mRNA turnover. If transcript turnover were the primary mechanism, then a reduction in mRNA degradation resulting possibly by terminated synthesis of a putative RNase would lead to transcript accumulation. The rate of transcript accumulation should be proportional to the level of remaining RNase. If the RNase were stable, its levels would in turn be controlled by dilution resulting from ongoing cell division. Since the rate of increase in lacS mRNA precipitated by nutrient down shift is nearly 35 times faster than the rate of reduction in this transcript precipitated by nutrient up shift, synthesis rather than degradation is the more likely route for regulation of lacS expression. Additional approaches, however, will be necessary to more rigorously distinguish between these two possibilities. Interestingly these results also suggest that the lacS mRNA is unusually stable and possibly more so than most bacterial prokaryotic mRNAs. The stability of mRNA in archaeal prokaryotes in general is still largely unexamined, though some studies suggest that archaeal mRNAs may be quite stable (22).
The responsiveness of glycosyl hydrolase gene expression to medium composition could be readily distinguished at the transcriptional level. The difference in lacS mRNA was 14-fold, while the difference for malA mRNA was 4-fold. This indicates that if there is a common regulatory mechanism, it displays some preference in the degree of its action on target gene expression. Since the affected genes are physically unlinked, such a regulatory mechanism must be trans acting and therefore involve a diffusible factor such as a small molecule or a protein. In eukaryotes and gram-negative prokaryotes, cAMP is used as a trans-acting signal molecule to effect changes in gene transcription in response to changing nutrient status (27, 46). In both groups of organisms, cAMP-interacting proteins play critical roles. It is of interest, therefore, that this molecule has been found in archaea (26). However, the other well-characterized intracellular prokaryotic signal molecule, guanosine tetraphosphate, appears to be absent in these organisms (48).
The evolution of transcription systems now points to a common origin between archaea and eukaryotes. Basal transcription components including promoter sequence, promoter recognition factors, and RNA polymerase are closely related and clearly distinct in lineage from those of bacterial prokaryotes. How gene regulation is accomplished, and particularly how regulatory factors interact with the basal archaeal transcription system, is unknown. Ongoing studies extending the work presented here are focused on developing a better understanding of this process.
ACKNOWLEDGMENTS
This work was supported by grant MCB-9604000 to P.B. from the National Science Foundation.
We thank Mike Rolfsmeier, Jimmy Soto, Robyn Kaiser, and the other members of the Blum laboratory for help and encouragement.
REFERENCES
- 1.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blum P, Bauernfiend J, Ory J, Krska J. Physiological consequences of DnaK and DnaJ overproduction in Escherichia coli. J Bacteriol. 1992;174:7436–7444. doi: 10.1128/jb.174.22.7436-7444.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brock T D, Brock K M, Belly R T, Weiss R L. Sulfolobus: a genus of sulfur oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 4.Brown T, Mackey K. Analysis of RNA by Northern and slot blot hybridization. In: Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. pp. 4.9.1–4.9.16. [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Cheung J, Danna J K, O’Connor E M, Price L B, Shand R F. Isolation, sequence, and expression of the gene encoding halocin H4, a bacteriocin from the halophilic archaeon Haloferax mediterranei R4. J Bacteriol. 1997;179:548–551. doi: 10.1128/jb.179.2.548-551.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen-Kupiec R, Blank C, Leigh J A. Transcriptional regulation in Archaea: in vivo demonstration of a repressor binding site in a methanogen. Proc Natl Acad Sci USA. 1997;94:1316–1320. doi: 10.1073/pnas.94.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cubellis M V, Rozzo C, Montecucchi P, Rossi M. Isolation and sequencing of a new β-galactosidase-encoding archaebacterial gene. Gene. 1990;94:89–94. doi: 10.1016/0378-1119(90)90472-4. [DOI] [PubMed] [Google Scholar]
- 9.Cubero B, Scazzocchio C. Two different, adjacent and divergent zinc finger binding sites are necessary for CREA-mediated carbon catabolite repression in the proline gene cluster of Aspergillus nidulans. EMBO J. 1994;13:407–415. doi: 10.1002/j.1460-2075.1994.tb06275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danson M J. Central metabolism of the archaea. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of archaea. Amsterdam, The Netherlands: Elsevier; 1993. pp. 1–24. [Google Scholar]
- 11.De Rosa M, Gambacorta A, Bu’lock J D. Extremely thermophilic acidophilic bacteria convergent with Sulfolobus acidocaldarius. J Gen Microbiol. 1975;86:156–164. doi: 10.1099/00221287-86-1-156. [DOI] [PubMed] [Google Scholar]
- 12.Deutscher J, Kuster E, Bergstedt U, Charrier V, Hillen W. Protein kinase-dependent Hpr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol Microbiol. 1995;15:1049–1053. doi: 10.1111/j.1365-2958.1995.tb02280.x. [DOI] [PubMed] [Google Scholar]
- 13.Gohl H P, Grondahl B, Thomm M. Promoter recognition in archaea is mediated by transcription factors: identification of transcription factor aTFB from Methanococcus thermolithotrophicus as archaeal TATA-binding protein. Nucleic Acids Res. 1995;23:3837–3841. doi: 10.1093/nar/23.19.3837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grogan D W. Phenotypic characterization of the archaebacterial genus Sulfolobus: comparison of five wild-type strains. J Bacteriol. 1989;171:6710–6719. doi: 10.1128/jb.171.12.6710-6719.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grogan D W. Evidence that β-galactosidase of Sulfolobus solfataricus is only one of several activities of a thermostable β-d-glycosidase. J Bacteriol. 1991;57:1644–1649. doi: 10.1128/aem.57.6.1644-1649.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gross E. The cyanogen bromide reaction. Methods Enzymol. 1967;11:238–255. [Google Scholar]
- 17.Hain J, Reiter W-D, Hudepohl U, Zillig W. Elements of an archaeal promoter-defined by mutational analysis. Nucleic Acids Res. 1992;20:5423–5428. doi: 10.1093/nar/20.20.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 19.Haseltine C, Rolfsmeier M, Bini E, Carl A, Rodriguez-Montalvo R, Clark A, Blum P. Abstracts of the 98th General meeting of the American Society for Microbiology 1998. Washington, D.C: American Society for Microbiology; 1998. Transcriptional regulation of gene expression in the hyperthermophilic crenarchaeote, Sulfolobus solfataricus, abstr. I66; p. 319. [Google Scholar]
- 20.Haseltine C, Rolfsmeier M, Blum P. The glucose effect and regulation of α-amylase synthesis in the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 1996;178:945–950. doi: 10.1128/jb.178.4.945-950.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henkin T M, Grundy F J, Nicholson W L, Chambliss G H. Catabolite repression of an amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to Escherichia coli lacI and galR repressors. Mol Microbiol. 1991;5:575–584. doi: 10.1111/j.1365-2958.1991.tb00728.x. [DOI] [PubMed] [Google Scholar]
- 22.Hennigan A N, Reeve J. mRNAs in the methanogenic archaeon Methanococcus vannielii: numbers, half-lives and processing. Mol Microbiol. 1994;11:655–670. doi: 10.1111/j.1365-2958.1994.tb00344.x. [DOI] [PubMed] [Google Scholar]
- 23.Kandler O, Stetter K O. Evidence for autotrophic CO2 assimilation in Sulfolobus brierleyi via a reductive carboxylic acid pathway. Zentrbl Bakteriol Hyg I Abt Orig Reihe C. 1981;2:111–121. [Google Scholar]
- 24.Klenk H-P, Palm P, Lottspeich F, Zillig W. Component H of the DNA-dependent RNA polymerase of archaea is homologous to a subunit shared by the three eucaryal nuclear RNA polymerases. Proc Natl Acad Sci USA. 1992;89:407–410. doi: 10.1073/pnas.89.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laderman K A, Davis B R, Krutzsch H C, Lewis M S, Griko Y V, Privalov P L, Anfinsen C B. The purification and characterization of an extremely thermostable α-amylase from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Biol Chem. 1993;268:24394–24401. [PubMed] [Google Scholar]
- 26.Leichtling B H, Rickenberg H V, Seely R J, Fahrney D E, Pace N R. The occurrence of cyclic AMP in archaebacteria. Biochem Biophys Res Commun. 1986;136:1078–1082. doi: 10.1016/0006-291x(86)90443-2. [DOI] [PubMed] [Google Scholar]
- 27.Magasanik B, Neidhardt F C. Regulation of carbon and nitrogen utilization. In: Neidhardt F C, Ingrahm J L, Low K B, Magasanick B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 1318–1325. [Google Scholar]
- 28.Manning G B, Campbell L L. Thermostable α-amylase of Bacillus stearothermophilus. J Biol Chem. 1961;236:2952–2957. [PubMed] [Google Scholar]
- 29.Marsh T L, Reich C I, Whitelock R B, Olsen G J. Transcription factor IID in the Archaea: sequences in the thermococus celer genome would encode a product closely related to the TATA-binding protein of eukaryotes. Proc Natl Acad Sci USA. 1994;91:4180–4184. doi: 10.1073/pnas.91.10.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsudaira P. Limited N-terminal sequence analysis. Methods Enzymol. 1990;182:602–613. doi: 10.1016/0076-6879(90)82047-6. [DOI] [PubMed] [Google Scholar]
- 31.Palmer J R, Daniels C J. In vivo definition of an archaeal promoter. J Bacteriol. 1995;177:1844–1849. doi: 10.1128/jb.177.7.1844-1849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Palmer J R, Reeve J N. Structure and function of methanogen genes. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of archaea. Amsterdam, The Netherlands: Elsevier; 1993. pp. 497–534. [Google Scholar]
- 33.Prisco A, Moracci M, Rossi M, Ciaramella M. A gene encoding a putative membrane protein homologus to the major facilitator superfamily of transporters maps upstream of the β-glycosidase gene in the achaeaon Sulfolobus solfataricus. J Bacteriol. 1995;177:1614–1619. doi: 10.1128/jb.177.6.1614-1619.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qureshi S A, Baumann P, Rowlands T, Khoo B, Jackson S P. Cloning and functional analysis of the TATA binding protein from Sulfolobus shibatae. Nucleic Acids Res. 1995;23:1775–1781. doi: 10.1093/nar/23.10.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qureshi S A, Jackson S P. Sequence-specific DNA binding by the S. shibatae TFIIB homolog, TFB, and its effect on promoter strength. Mol Cell. 1998;1:389–400. doi: 10.1016/s1097-2765(00)80039-8. [DOI] [PubMed] [Google Scholar]
- 36.Qureshi S A, Khoo B, Baumann P, Jackson S P. Molecular cloning of the transcription factor TFIIB homolog from Sulfolobus shibatae. Proc Natl Acad Sci USA. 1995;92:6077–6081. doi: 10.1073/pnas.92.13.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reiter W-D, Hudepohl U, Zillig W. Mutational analysis of an archaebacterial promoter: essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proc Natl Acad Sci USA. 1990;87:9509–9513. doi: 10.1073/pnas.87.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rockabrand D, Arthur T, Korinek G, Livers K, Blum P. An essential role for the Escherichia coli DnaK protein in starvation-induced thermotolerance, H2O2 resistance, and reductive division. J Bacteriol. 1995;177:3695–3703. doi: 10.1128/jb.177.13.3695-3703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rockabrand D, Blum P. Multicopy plasmid suppression of stationary phase chaperone toxicity in Escherichia coli by phosphogluconate dehydratase and the N-terminus of DnaK. Mol Gen Genet. 1995;249:498–506. doi: 10.1007/BF00290575. [DOI] [PubMed] [Google Scholar]
- 40.Rockabrand D, Livers K, Austin T, Kaiser R, Jensen D, Burgess R, Blum P. Roles of DnaK and RpoS in starvation-induced thermotolerance of Escherichia coli. J Bacteriol. 1998;180:846–854. doi: 10.1128/jb.180.4.846-854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roder R, Pfeifer F. Influence of salt on the transcription of the gas vesicle genes of Haloferax mediterranei and identification of the endogenous activator gene. Microbiology. 1996;142:1715–1723. doi: 10.1099/13500872-142-7-1715. [DOI] [PubMed] [Google Scholar]
- 42.Roesler W J, Grahm J G, Kolen R, Klemm D J, McFie P J. The cAMP response element binding protein synergizes with other transcription factors to mediate cAMP responsiveness. J Biol Chem. 1995;270:8225–8232. doi: 10.1074/jbc.270.14.8225. [DOI] [PubMed] [Google Scholar]
- 43.Rolfsmeier M, Blum P. Purification and characterization of a maltase from the extremely thermophilic crenarchaeote Solfataricus solfataricus. J Bacteriol. 1995;177:482–485. doi: 10.1128/jb.177.2.482-485.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rolfsmeier M, Haseltine C, Bini E, Clark A, Blum P. Molecular characterization of the α-glucosidase gene (malA) from the hyperthermophilic archaeon Sulfolobus solfataricus. J Bacteriol. 1998;180:1287–1295. doi: 10.1128/jb.180.5.1287-1295.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowlands T, Baumann P, Jackson S P. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994;264:1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- 46.Saier M H, Jr, Ramseier T M, Reizer J. Regulation of carbon utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1318–1325. [Google Scholar]
- 47.Sandman K, Gralying R A, Dobrinski B, Lurz R, Reeve J N. Growth-phase-dependent synthesis of histones in the archaeon Methanothermus fervidus. Proc Natl Acad Sci USA. 1994;91:12624–12628. doi: 10.1073/pnas.91.26.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scoarughi G L, Cimmino C, Donini P. Lack of production of (p)ppGpp in Halobacterium volcanii under conditions that are effective in the eubacteria. J Bacteriol. 1995;177:82–85. doi: 10.1128/jb.177.1.82-85.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shand R F, Betlach M. Expression of the bop gene of Halobacterium halobium is induced by low oxygen tension and by light. J Bacteriol. 1991;173:4692–4699. doi: 10.1128/jb.173.15.4692-4699.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sowers K, Thai T, Gunsalus R. Transcriptional regulation of the carbon monoxide dehydrogenase gene (cdhA) in Methanosarcinia thermophila. J Biol Chem. 1993;268:23172–23178. [PubMed] [Google Scholar]
- 51.Thompson D, Daniels C. Heat shock inducibility of an archaeal TATA-like promoter is controlled by adjacent sequence elements. Mol Microbiol. 1998;27:541–551. doi: 10.1046/j.1365-2958.1998.00698.x. [DOI] [PubMed] [Google Scholar]
- 52.Yang C F, DasSarma S. Transcriptional induction of purple membrane and gas vesicle synthesis in the archaebacterium Halobacterium halobium is blocked by a DNA gyrase inhibitor. J Bacteriol. 1990;172:4118–4121. doi: 10.1128/jb.172.7.4118-4121.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]