Abstract
Determination of human papillomavirus (HPV) status has become clinically relevant for head and neck squamous cell carcinoma (HNSCC) patients. p16 immunohistochemistry is one of the recommended methods for classifying HPV status. However, long noncoding RNAs (lncRNAs) and related competing endogenous RNA (ceRNA) networks linked to different p16-status HNSCC are still absent.
In the present study, The Cancer Genome Atlas database provided RNA profiles as well as clinical information from 26 p16-positive HNSCC samples, 71 p16-negative HNSCC samples, and 44 adjacent normal control samples. Differentially expressed RNAs (DERNAs) between HNSCC samples and normal samples were identified by limma package in R. Functional enrichment analysis of differentially expressed mRNAs was performed using Clusterprofiler package in R. Survival analysis of DERNAs was carried out by survival package in R. The ceRNA network was constructed using GDCRNATools package in R.
A total of 102 lncRNAs, 196 microRNAs (miRNAs), and 2282 mRNAs were identified as p16-positive-specific DERNAs. There were 90 lncRNAs, 153 miRNAs, and 2038 mRNAs were identified as p16-negative-specific DERNAs. Functional enrichment analysis revealed that the differentially expressed mRNAs in the p16-positive and the p16-negative group were mainly enriched in the “DNA replication” and “extracellular matrix -receptor interaction” pathway, respectively. Among the top 25 DERNAs, there were 1 key lncRNA, 1 key miRNA, and 1 key messenger RNA in the p16-positive group and 2 key lncRNAs, 1 key miRNA, and 2 key mRNAs in the p16-negative group were significantly related to the overall survival. Then the ceRNA network in the p16-positive and p16-negative group was constructed. There were 5 lncRNAs, 16 miRNAs, and 66 mRNAs included in the p16-positive group ceRNA network and 1 lncRNA, 4 miRNAs, and 28 mRNAs included in the p16-negative group ceRNA network. Among the RNAs in the ceRNA network, 5 mRNAs were significantly related to the overall survival.
Taken together, we revealed the differential RNA expression profiling and the differential ceRNA network in the p16-positive and p16-negative group of HNSCC. Our findings provided a novel insight into this HPV-related cancer and potential biomarkers and therapeutic targets for HNSCC based on p16 status.
Keywords: competing endogenous RNA, head and neck squamous cell carcinoma, long noncoding RNA, p16, prognosis
1. Introduction
Head and neck squamous cell carcinoma (HNSCC) ranks as the sixth most common cancer worldwide. It has a yearly incidence of 660,000 cases with 40%–50% mortality.[1] The carcinogenesis relates to different etiologies and a large variety of molecular changes. Tobacco use and excessive alcohol consumption are the major classical risk factors for HNSCC.[2] Currently, human papillomaviruses (HPVs) have emerged as a novel risk factor for HNSCCs, especially for oropharyngeal squamous cell carcinoma (OPSCC).[3] In general, patients with HPV-positive OPSCC tend to be a relatively younger age than their HPV-negative counterparts and less likely to be smokers or excessive alcohol consumers.[4] Moreover, they tend to be more frequently male and have a better overall prognosis.[5] Although the effect of HPV in HNSCCs located outside the oropharynx is less well understood, HPV positivity is also associated with more favorable outcomes.[6] p16 is a tumor suppressor protein that is overexpressed in HPV-driven carcinomas.[7] Many research showed that p16 immunohistochemistry (IHC) is a cost-effective method with good sensitivity and negative predictive value. This method is recommended as a surrogate marker of HPV status by the Union for International Cancer Control and the College of American Pathologists.[8,9] However, the precise mechanisms of how p16-positive HNSCC and p-16 negative HNSCC occurs and progresses are still not elucidated.
Tumorigenesis and cancer development has been closely associated with the aberrant expression of RNAs. Less than 2% of the total genome encodes protein-coding genes, suggesting that noncoding RNAs (ncRNAs) represent most of the human transcriptome.[10] Long noncoding RNAs (lncRNAs) are defined as transcripts of more than 200 nucleotides and have been implicated in a diverse range of biological processes (BPs) in cancer.[11] MicroRNAs (miRNAs) are short ncRNAs, with a length of approximately 18–25 nucleotides. They are evolutionarily conserved and are pivotal post-transcriptional mediators of gene regulation.[12] In 2011, Salmena et al[13] hypothesized a novel regulatory mechanism between lncRNAs and messenger RNAs (mRNA), namely competing endogenous RNA (ceRNA) network. In this theory, ceRNAs regulate each other by competing specifically for shared miRNAs. miRNA competition thus extends beyond the noncoding transcriptome to protein-coding mRNAs. Increasing evidence has confirmed that the lncRNA-miRNA-mRNA ceRNA network plays key roles in human cancers, such as esophageal cancer,[14] lung cancer,[15] as well as HNSCC.[16] However, current knowledge for lncRNA-miRNA-mRNA in human cancers is not enough, including p16-positive and p16-negative HNSCC.
In the current study, to detect the aberrant expression profile of ncRNAs and mRNAs in p16-positive and negative HNSCC patients, the p16-positive and negative HNSCC tissue and normal tissue expression profiles from The Cancer Genome Atlas (TCGA) database were obtained. Subsequently, a functional enrichment analysis for these aberrantly expressed mRNAs was conducted. Furthermore, p16-positive group and p16-negative group ceRNA networks were constructed including differentially expressed RNAs (DERNAs). Kaplan-Meier analysis was performed to identify key DERNAs and ceRNA-related RNAs that are associated with overall survival (OS). They may also serve as promising diagnostic biomarkers and therapeutic targets for different p16 status HNSCC in the future.
2. Materials and Methods
2.1. Data collection and processing
Gene expression data (RNA sequencing profiles and miRNA profiling) and corresponding clinical data of HNSCC were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). The inclusion criteria were set as follows: the p16 IHC status of HNSCC samples were recorded and patients with complete data including age, gender, OS time, and vital status. The exclusion criteria were set as follows: the p16 IHC status of HNSCC samples were unrecorded and patients with incomplete clinicopathological data. According to the criteria, 97 primary HNSCC samples, including 26 patients with immunohistochemically p16-positive status (p16-positive group) HNSCC and 71 patients with immunohistochemically p16-negative status (p16-negative group), and 44 normal controls were collected in our analysis. The clinicopathological characters of the p16-positive group and p16-negative group HNSCC patients are shown in Table 1.
Table 1.
Clinicopathological characters of the p16 positive and p16 negative HNSCC patients.
| p16 positive HNSCC patients | p16 negative HNSCC patients | ||||
|---|---|---|---|---|---|
| Characteristics | Subtype | No. of cases (%) | Characteristics | Subtype | No. of cases (%) |
| Age | <60 | 18 (69.2) | Age | <60 | 35 (49.3) |
| ≥60 | 8 (30.8) | ≥60 | 36 (50.7) | ||
| Gender | Male | 23 (88.5) | Gender | Male | 54 (76.1) |
| Female | 3 (11.5) | Female | 17 (23.9) | ||
| Pathologic stage | Stage I | 1 (3.85) | Pathologic stage | Stage I | 5 (7.0) |
| Stage II | 3 (11.5) | Stage II | 5 (7.0) | ||
| Stage III | 1 (3.85) | Stage III | 9 (12.7) | ||
| Stage IV | 10 (38.5) | Stage IV | 44 (62.0) | ||
| NA | 11 (42.3) | NA | 8 (11.3) | ||
| Pathologic T | T1–T2 | 10 (38.5) | Pathologic T | T1–T2 | 20 (28.2) |
| T3–T4 | 7 (26.9) | T3–T4 | 43 (60.5) | ||
| Tx | 9 (34.6) | Tx | 8 (11.3) | ||
| Pathologic N | N0 | 3 (11.5) | Pathologic N | N0 | 16 (22.5) |
| N1–N3 | 12 (46.2) | N1–N3 | 48 (67.6) | ||
| Nx | 11 (42.3) | Nx | 7 (9.9) | ||
| Vital status | Alive | 22 (84.6) | Vital status | Alive | 46 (64.8) |
| Dead | 4 (15.4) | Dead | 25 (35.2) | ||
HNSCC = head and neck squamous cell carcinoma, NA, Nx, Tx = pathologic stage is unknown.
Among these data, mRNA and lncRNA data were collected from Illumina HiSeqRNASeq platforms, and miRNA data were collected from Illumina HiSeqmiRNASeq platforms. mRNAs and lncRNAs were encoded according to the ENSEMBL database version 89, European Bioinformatics Institute, Cambridge, UK (https://www.ensembl.org), miRNAs were annotated according to miRbase version 22, The University of Manchester, Manchester, UK (http://www.mirbase.org). The ethical approval was not necessary. This study conformed with the publication guidelines provided by TCGA and all the data are publicly available.
2.2. Analysis of differentially expressed lncRNAs, mRNAs, and miRNAs
The differentially expressed lncRNA (DElncRNA), mRNA (DEmRNA), and miRNA (DEmiRNA) in p16-positive HNSCC samples and normal controlled samples or p16-negative HNSCC samples and normal controlled samples were identified using “limma” package of R software. P value was adjusted to the false discovery rate (FDR). |log2 Fold Change|>1.0 and FDR <0.01 were used as statistically significant. In order to visualize the results of differential genetic analysis, the heatmaps were plotted using “pheatmap” packages of R software.
2.3. Functional enrichment analysis
To understand the potential mechanisms involved in the tumorigenesis of p16-positive and p16-negative HNSCC, Gene Ontology (GO) functional enrichment, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed on DEmRNAs using “ClusterProfiler” package of R software. A P < .01 was considered as enriched gene sets.
2.4. Survival analysis
To investigate the prognostic characteristics of DERNAs in different p16 status groups, the p16-positive group and p16-negative group HNSCC samples were divided into the high expression group or the low expression group (according to the median) in terms of the expression of lncRNAs, mRNAs, and miRNAs, separately. To correlate the DERNAs with the OS, the survival curves were plotted using “survival” package of R software based on the Kaplan-Meier method. A P < .05 was considered statistically significant.
2.5. Construction of lncRNA-miRNA-mRNA ceRNA network
Based on the theory that lncRNAs can invoke miRNA sponges to affect the expression of mRNA, we predict the interactions between DElncRNA and DEmiRNA and DEmiRNA. A ceRNA network was constructed by using “GDCRNATools” package of R software.[17] The ceRNA network was visualized using Cytoscape 3.6.1 software, Institute for Systems Biology, Seattle, USA.
3. Results
3.1. Screening of differentially expressed RNAs in the p16-positive group and p16-negative group HNSCC samples
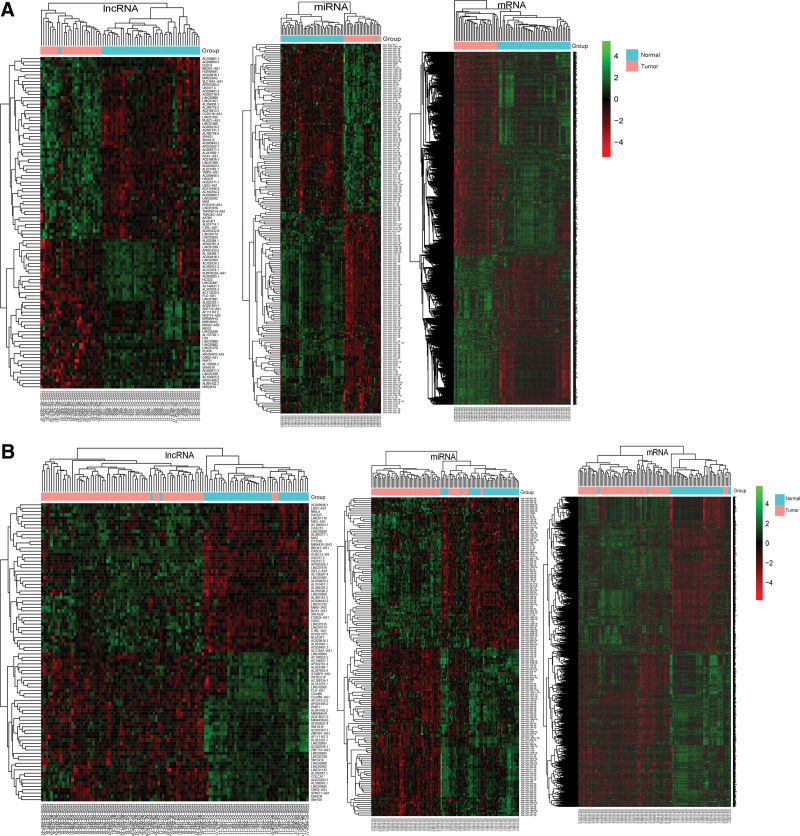
By comparing the expression of RNAs between the p16-positive (or negative) group HNSCC tissue samples with normal tissue samples, we identified some DERNAs. According to the cutoff criteria of |log2 Fold Change|>1.0 and an FDR <0.01, 102 DElncRNAs (56 upregulated and 46 downregulated), 196 DEmiRNAs (89 upregulated and 107 downregulated), and 2282 DEmRNAs (970 upregulated and 1312 downregulated) were identified in the p16-positive group. In the p16-negative group, 90 DElncRNAs (45 upregulated and 45 downregulated), 153 DEmiRNAs (72 upregulated and 81 downregulated), and 2038 DEmRNAs (1012 upregulated and 1026 downregulated) were identified. The variable expression of differentially expressed DElncRNAs, DEmiRNAs, and DEmRNAs between the p16-positive (Fig. 1A) group (or the p16-negative group [Fig. 1B]). HNSCC samples with the normal controls were visualized in the heatmap. The top 25 differentially expressed DElncRNAs/DEmiRNAs/DEmRNAs in the p16-positive group and the p16-negative group HNSCC samples compared with normal controls were listed. See Table S1, Supplemental Digital Content, http://links.lww.com/MD/H39, which illustrates the top 25 differentially expressed DElncRNAs in the p16-positive group. See Table S2, Supplemental Digital Content, http://links.lww.com/MD/H39, which illustrates the top 25 differentially expressed DElncRNAs in the p16- negative group. See Table S3, Supplemental Digital Content, http://links.lww.com/MD/H39, which illustrates the top 25 differentially expressed DEmiRNAs in the p16-positive group. See Table S4, Supplemental Digital Content, http://links.lww.com/MD/H39, which illustrates the top 25 differentially expressed DEmiRNAs in the p16-negative group. See Table S5, Supplemental Digital Content, http://links.lww.com/MD/H39, which illustrates the top 25 differentially expressed DEmRNAs in the p16-positive group. See Table S6, Supplemental Digital Content, http://links.lww.com/MD/H39, which illustrates the top 25 differentially expressed DEmRNAs in the p16-negative group.
Figure 1.
Heatmap of DElncRNAs, DEmiRNAs, and DEmRNAs in p16-positive HNSCC samples and normal control samples. (A); in p16-negative HNSCC samples and normal control samples (B). The pink and light blue colors represent the tumor group and the normal controls group, respectively. The red and green colors indicate higher expression levels and lower expression levels, respectively. The rows represent DERNAs and the columns represent the samples. DElncRNAs = differentially expressed lncRNAs, DEmiRNAs = differentially expressed miRNAs, DEmRNAs = differentially expressed mRNAs, DERNAs = differentially expressed RNAs, HNSCC = head and neck squamous cell carcinoma, lncRNAs = long noncoding RNAs, miRNA = microRNA, mRNA = messenger RNA.
3.2. Functional enrichment analysis for DEmRNAs in the p16-positive group and p16-negative group
To further analyze the biological function and corresponding pathways of DEmRNAs in the p16-positive group and the p16-negative group, GO and KEGG analysis was performed. GO annotation consists of cellular component, BP, and molecular function. The top 5 enrichment scores in GO analysis of the p16-positive group and the p16-negative group, DEmRNAs were shown in Figure 2A and B.
Figure 2.
The top 5 enrichment scores in GO enrichment analysis of DEmRNAs. (A) CC, BP, MF of DEmRNAs in the p16-positive group; (B) CC, BP, MF of DEmRNAs in the p16-negative group. ATPase = adenosine-triphosphate synthase, BP = biological process, CC = cellular component, DEmRNAs = differentially expressed mRNAs, GO = Gene Ontology, MF = molecular function.
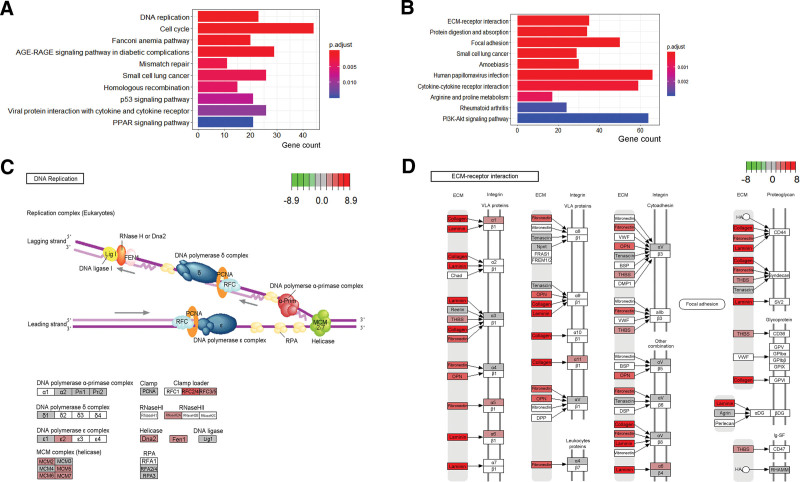
Additionally, the top 10 KEGG pathways of the p16-positive and the p16-negative group were shown in Figure 3A and B. The DNA replication pathway was found to have the lowest P value in the p16-positive group and the pathview was shown in Figure 3C. The extracellular matrix(ECM)-receptor interaction pathway was found to have the lowest P value in the p16-negative group and the pathview was shown in Figure 3D.
Figure 3.
The top 10 enrichment scores in KEGG pathway analysis of the DEmRNAs in the p16-positive group. (A) and in the p16-negative group (B). (C) DNA replication map from KEGG analysis; (D) ECM-receptor interaction map from KEGG analysis, the red colors represent the upregulated DEmRNAs green colors represent the downregulated DEmRNAs. AGE-RAGE = advanced glycosylation end-product specific receptor, BSP = binding sialoprotein, regulatoryDEmRNAs = differentially expressed mRNAs, DMP1 = dentin matrix acidic phosphoprotein 1, Dna2 = DNA replication helicase/nuclease 2, DPP = dipeptidyl peptidase, DSP = desmoplakin, ECM = extracellular matrix, FEN1 = flap structure-specific endonuclease 1, FRAS1 = fraser extracellular matrix complex subunit 1, FREM1/2 = FRAS1 related extracellular matrix ½, GPIbα = glycoprotein Ib platelet subunit alpha, GPIbβ = glycoprotein Ib platelet subunit beta, GPIX = glycoprotein IX platelet, GPV = glycoprotein V platelet, HA = glucose-6-phosphate isomerise, Ig-SF = junctophilin 4, Lig I = tumor protein P53, KEGG = Kyoto Encyclopedia of Genes and Genomes, MCM = minichromosome maintenance, Npnt = nephronectin, OPN = secreted phosphoprotein 1, PFNA = peroxisome proliferator activated receptor alpha, PI3K-Akt = phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha-AKT serine/threonine kinase, PPAR = peroxisome proliferator activated receptor, RFC = solute carrier family 19 member 1, RHAMM = hyaluronan mediated motility receptor, RNase = ribonuclease, RPA, RPA interacting protein, SV2 = SV2 related protein, THBS = thrombospondin 1, VLA = integrin subunit alpha 4, VWF = von willebrand factor, αDG = dystroglycan 1, α-Prim = DNA polymerase alpha 1, catalytic subunit, βDG = dolichyl-phosphate mannosyltransferase subunit 3, regulatory.
3.3. Prognostic OS assessment of key DElncRNAs, DEmiRNAs, and DEmRNAs in the p16-positive and p16-negative group
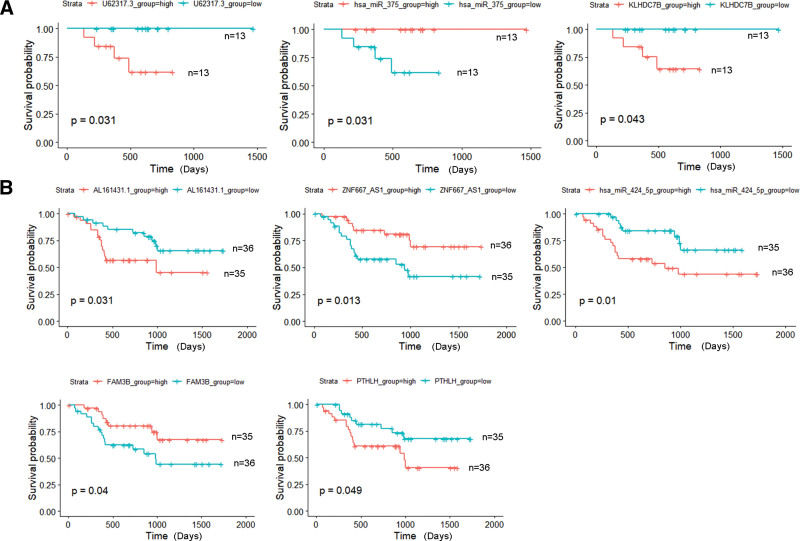
To evaluate the prognostic signatures of DElncRNAs, DEmiRNAs, and DEmRNAs, the survival analysis was performed and a P < .05 was considered as a cutoff. In the p16-positive group, a total of 9 DElncRNAs, 31 DEmiRNAs, and 167 DEmRNAs were significantly related to the OS. In the p16-negative group, a total of 10 DElncRNAs, 9 DEmiRNAs, and 106 DEmRNAs were significantly related to the OS. Combined with the results of survival analysis and expression analysis, we redefined the key DElncRNAs/DEmiRNAs/DEmRNAs as the top 25 upregulated or downregulated DElncRNAs/DEmiRNAs/DEmRNAs, which could also indicate the OS of HNSCC patients significantly. In the p16-positive group, key DElncRNA (U62317.3), key DEmiRNA (hsa-miR-375), and key DEmRNA (KLHDC7B) were found (Fig. 4A). Similarly, there were 2 key DElncRNAs (AL161431.1, ZNF667-AS1), 1 key DEmiRNA (has-miR-424-5p), and 2 key DEmRNAs (FAM3B, PTHLH) (Fig. 4B) in the p16-negative group.
Figure 4.
Kaplan-Meier analysis of key DElncRNAs, DEmiRNAs, DEmRNAs, and OS rate in the p16-positive group. (A) and p16-negative group (B) HNSCC patients. The median value of expression was set as cutoff point. The P values are shown on the diagrams (P < .05). DElncRNAs = differentially expressed lncRNA, DEmiRNAs = differentially expressed miRNAs, DEmRNAs = differentially expressed mRNAs, HNSCC = head and neck squamous cell carcinoma, OS = overall survival.
3.4. Construction and analysis of lncRNA-miRNA-mRNA ceRNA network in the p16-positive group and p16-negative group
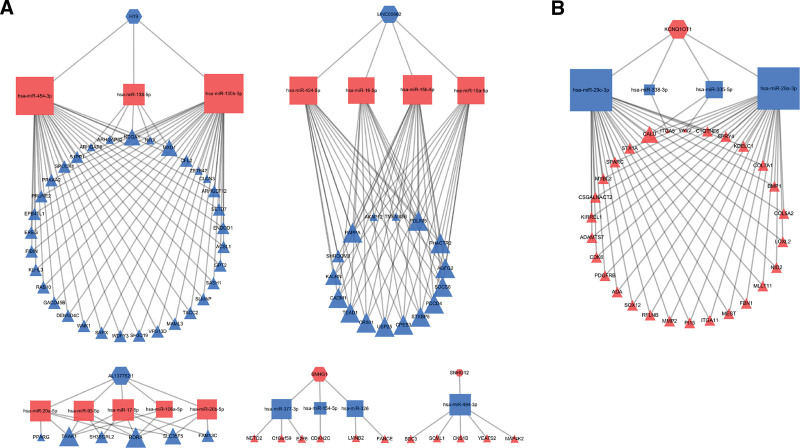
To further understand the interaction between DElncRNAs, DEmiRNAs, and DEmRNAs in the p16-positive group and the p16-negative group, the ceRNA networks were constructed. In the p16-positive group, 5 lncRNA nodes, 16 miRNA nodes, 66 mRNA nodes, and 162 edges were identified. In the p16-negative group, 1 lncRNA node, 4 miRNA nodes, 28 mRNA nodes, and 59 edges were identified. Cytoscape was used to visualize the p16-positive group (Fig. 5A) and the p16-negative group (Fig. 5B) ceRNA network.
Figure 5.
DElncRNAs/DEmiRNAs/DEmRNAs mediated ceRNA network in the p16-positive group. (A) and p16-negative group (B). The red hexagons represent upregulated lncRNAs, red rectangles represent upregulated miRNAs, red triangles represent upregulated mRNAs. The blue hexagons represent downregulated lncRNAs, blue rectangles represent downregulated miRNAs, blue triangles represent downregulated mRNAs. ceRNA = competing endogenous RNA, DElncRNAs = differentially expressed lncRNAs, DEmiRNAs = differentially expressed miRNA, DEmRNAs = differentially expressed mRNAs, lncRNAs = long noncoding RNAs, miRNA = microRNA, mRNA = messenger RNA.
3.5. Identify the RNAs in the p16-positive and p16-negative group ceRNA network associated with HNSCC patient OS
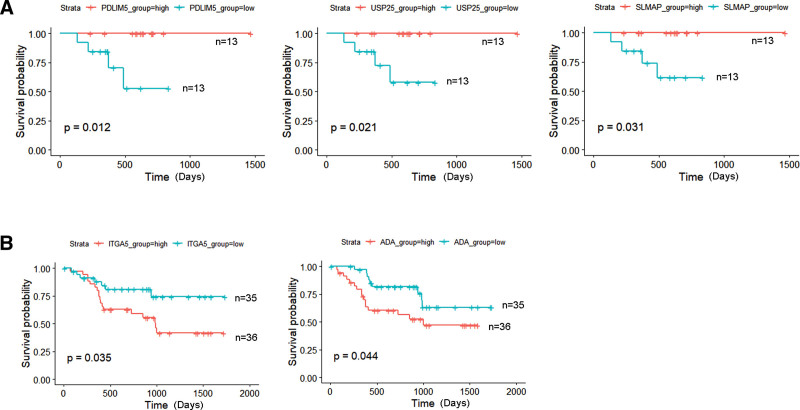
To identify critical RNAs that play important roles in both the BP and prognosis of the ceRNA network, Kaplan-Meier estimate and log-rank test were used to determine the relationship between OS and RNAs in the ceRNA network. In the p16-positive group, 3 mRNAs (PDLIM5, USP25, SLMAP) were observed to be significantly related to OS (P < .05) (Fig. 6A). In the p16-negative group, 2 mRNAs (ITGA5, ADA) were observed to be significantly related to OS (P < .05) (Fig. 6B).
Figure 6.
Kaplan-Meier analysis of ceRNA-related DElncRNAs, DEmiRNAs, DEmRNAs and OS rate in the p16-positive group. (A) and p16-negative group (B). The median value of expression was set as cutoff point. The P values are shown on the diagrams (P < .05). ceRNA = competing endogenous RNA, DElncRNAs = differentially expressed lncRNAs, DEmiRNAs = differentially expressed miRNA, DEmRNAs = differentially expressed mRNAs, OS = overall survival.
4. Discussion
HNSCCs affect more than 660,000 patients per year worldwide. Despite surgery, radiation, and chemotherapy, approximately half of patients will die of the disease.[18] Smoking is implicated in the rise of HNSCC in developing countries, and the infection of HPV is emerging as an important factor in the rise of HNSCCs affecting nonsmokers in developed countries.[19] HPV-positive HNSCC has unique epidemiological and clinicopathological features.[20] In HPV-driven tumorigenesis, the HPV E6 and E7 oncoproteins decrease the level of p53 and pRb by post-translational regulation, leading to aberrant overexpression of the cell-cycle protein p16 (CDKN2A).[21–24] HPV infection can be detected by various mechanisms. p16 IHC is a commonly used and well-established surrogate marker for detection HPV in HNSCC and has become the recommended standalone prognostic test for OPSCC patients. As underlined in the 8th edition American Joint Commission on Cancer staging guidelines, p16 IHC is required to stage OPSCC patients.[25] Some studies demonstrated that p16 IHC positivity is also an independent and positive prognostic factor in non-OPSCC.[26,27] Although some other studies are contradictory to the results,[28] the HPV status classifies 2 distinct entities of HNSCC and p16-positive and p16-negative HNSCC have significantly different disease profiles. We hope that the study of the molecular difference between p16-positive and p16-negative HNSCC will help with the exploration of HNSCC heterogeneity. However, integrated and comprehensive analysis of lncRNA, miRNA, mRNA, and associated ceRNA network in p16-positive and p16-positive HNSCC is still not enough. To the best of our knowledge, this is the first study to investigate the specific ceRNA network in HNSCC by p16-status. Inspiringly, novel lncRNA-miRNA-mRNA ceRNA networks were constructed and RNAs in those networks possessed significant prognostic value were highlighted.
In this present study, using the TCGA database, we identified a total of 102 DElncRNAs, 196 DEmiRNAs, and 2282 mRNAs as p16-positive-specific DERNAs. We also identified 90 DElncRNAs, 153 DEmiRNAs, and 2038 DEmRNAs as p16-negative-specific DERNAs based on the differential expression between tumor tissues and adjacent normal tissues. GO analysis is widely used as a functional enrichment analysis for a large number of genes.[29] The results of GO analysis demonstrated that the p16-positive-specific and p16-negative-specific DEmRNAs were significantly enriched in some different GO terms that were associated with cancer biological behaviors. In the p16-positive group, the DEmRNAs were significantly enriched in the condensed chromosome, DNA replication, and single-strand DNA-dependent ATPase activity. In the p16-negative group, the DEmRNAs were significantly enriched in the extracellular matrix, extracellular matrix organization, and extracellular matrix structural constituent. KEGG pathway enrichment analysis revealed that multiple different enriched pathways were obtained according to the p16-status. In the p16-positive group, the pathways involving in cancer mainly include DNA replication, cell cycle, and p53 signaling pathway. In the p16-negative group, the upregulated pathways related to cancer mainly enriched in ECM-receptor interaction, focal adhesion, and PI3k-Akt signaling pathway. Thus, these significant DEmRNAs may be involved in different modulation of HNSCC carcinogenesis and progression based on disparate p16-status.
For further identifying key DERNAs in p16-positive and p16-negative HNSCC, the top 25 upregulated and downregulated DERNAs were selected for further expression and survival analyses. The analytic results demonstrated that 2 upregulated DERNAs (lncRNA U62317.3, KLHDC7B) and 1 downregulated DERNA (has-miR-375) may act as the key p16-positive-specific RNAs. There were 3 upregulated DERNAs (lncRNA AL161431.1, hsa-miR-424-5p, PTHLH) and 2 downregulated DERNAs (lncRNA ZNF667-AS1, FAM3B) may act as the key p16-negative-specific RNAs. Intriguingly, most of these key RNAs have been well investigated in cancer. For example, downregulated lncRNA ZNF667-AS1 promoted the malignant progression and poor prognosis of cervical cancer, esophageal cancer, and laryngeal squamous cell carcinoma (LSCC)[30–32]; miR-375 was downregulated in LSCC and decreased miR-375 indicated a poor outcome[33]; miR-424-5p enhanced the aggressive progression of LSCC[34]; KLHDC7B participated the breast cancer cell proliferation[35]; PTHLH was a poor prognostic marker of HNSCC.[36] These publications partially support the accuracy of our informatics analyses.
LncRNAs and miRNAs are involved in the regulation of gene expression via the ceRNA mechanism as previously described. By constructing the ceRNA network, our research focused on the potentially different regulatory mechanisms of p16-positive and p16-negative HNSCCs. Several previous studies have already reported the RNAs and the interactions between RNAs in the 2 ceRNA networks. For example, SNHG1, an upregulated lncRNA in the p16-positive ceRNA network, was reported to regulate colorectal cancer cell growth through interactions with miR-154-5p and EZH2.[37] SNHG1 was also reported to regulate NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma.[38] These previous studies strongly demonstrated that the regulatory relationship in our ceRNA network was reliable. SNHG12 was another upregulated lncRNA in the p16-positive ceRNA network and was reported to promote the progression of clear cell renal cell carcinoma, hepatocellular carcinoma, and osteosarcoma by sponging miRNAs.[39–41] KCNQ1OT1 was an upregulated lncRNA in the p16-negative ceRNA network and was reported to act as the ceRNA to promote the progression of colorectal cancer and bladder cancer.[42,43] In tongue cancer, it was reported that KCNQ1OT1 not only promoted proliferation but also induced cisplatin resistance.[44] One of the reasons that the prognosis of p16-negative HNSCC patients is significantly worse than p16-positive patients were regarded as chemotherapy resistance.[45] Based on the analyses of our study, KCNQ1OT1 and the ceRNA network in the p16-negative HNSCC were the potential biomarkers and treatment targets of chemoresistance. Several reports suggested that H19, a downregulated lncRNA in the p16-positive ceRNA network, is linked to the poor prognosis of cancer patients. H19 was reported to interact with miR-138-5p and therefore promote tumor proliferation in cervical cancer. H19 could also regulate keratinocyte differentiation by targeting miR-130-3p.[46] Furthermore, H19 expression was reported strongly correlated with miR-675-5p in colorectal cancer. The binding between miR-675-5p and the 3’UTR in the TP53 mRNA resulted in the loss of the p53 protein and metastasis.[47] Another downregulated lncRNA in the p16-positive ceRNA network, LINC00662, was reported to promote hepatocellular carcinoma progression via competitively binding miR-15a and miR-16.[48] According to our study, the downregulation of H19 and LINC00662 in the p16-positive group and the corresponding ceRNA networks might be the potential reason why the p16-positive group HNSCC patients usually have a better prognosis than the patients in the p16-negative group. In the p16-positive ceRNA network, 3 key mRNAs (PDLIM5, USP25, SLMAP) were confirmed to be correlated with OS. PDLIM5 has been reported to be a potential biomarker of papillary thyroid carcinoma[49]; USP25 cancer-associated mutations led to activation in vitro and in vivo[50]; SLMAP was a target gene of miR-224 and regulated giant tumor-derived neoplastic stromal cells.[51] In the p16-negative group ceRNA network, 2 key mRNAs (ITGA5, ADA) were identified to be associated with OS. ITGA5 was reported to correlate with poor OS in colorectal adenocarcinoma,[52] pancreatic cancer, and inhibited the efficacy of chemotherapy in pancreatic cancer.[53] ADA exosomes in HNSCC plasma was regarded as a potential marker of tumor progression and immune competence.[54] These reports further imply the accuracy of our current analytic results. However, more researches and large-scale clinical trials are needed to explore the molecular mechanisms and the prognostic value of these key RNAs in p16-positive and p16-negative HNSCC.
5. Conclusions
In conclusion, HNSCC patients with p16-positive and p16-negative status have differential RNA expression profiling and differential ceRNA network. The findings of our analyses introduce new insights into HPV-related HNSCC and suggest that the key RNAs in the p16-positive and p16-negative group may be potential biomarkers and therapeutic targets for different p16-status patients. Further studies with multicenter data, larger clinical samples, and more lab experiments are needed to verify these findings.
Acknowledgments
The authors are thankful to the TCGA database (https://cancergenome.nih.gov/).
Author contributions
Conceptualization: Yifan Yang. Data curation: Ling Feng, Ru Wang. Methodology, Software: Yifan Yang, Ling Feng. Project administration and Supervision: Jugao Fang. Validation: Hongzhi Ma, Shizhi He. Writing—original draft: Yifan Yang. Writing—review and editing: Yifan Yang, Ling Feng, Ru Wang, Jugao Fang.
Supplementary Material
Abbreviations:
- AJCC =
- American Joint Commission on Cancer
- BP =
- biological process
- CC =
- cellular component
- ceRNA =
- competing endogenous RNA
- DElncRNA =
- differentially expressed lncRNA
- DEmiRNA =
- differentially expressed miRNA
- DEmRNA =
- differentially expressed mRNA
- DERNAs =
- differentially expressed RNAs
- FC =
- fold change
- FDR =
- false discovery rate
- GO =
- Gene Ontology
- HNSCC =
- head and neck squamous cell carcinoma
- HPV =
- human papillomavirus
- IHC =
- immunohistochemistry
- KEGG =
- Kyoto Encyclopedia of Genes and Genomes
- lncRNAs =
- long noncoding RNAs
- MF =
- molecular function
- miRNA =
- microRNA
- mRNA =
- messenger RNA
- ncRNAs =
- noncoding RNAs
- OPSCC =
- oropharyngeal squamous cell carcinoma
- OS =
- overall survival
- TCGA =
- The Cancer Genome Atlas
- UICC =
- Union for International Cancer Control
This study was funded by grants from Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (Grant No. KZ201910025034), Beijing Municipal Administration of Hospitals’ Ascent Plan (Grant No. DFL20180202), The Capital Health Research and Development of Special (Grant No. 2018-2-2054), and National Key R&D Program of China (Grant No. 2020YFB1312805).
The authors have no conflicts of interest to disclose.
All data generated or analyzed during this study are included in this published article [and its supplementary information files].
Supplemental Digital Content is available for this article.
How to cite this article: Yang Y, Feng L, Wang R, Ma H, He S, Fang J. Integrated analysis of lncRNA-associated ceRNA network in p16-positive and p16-negative head and neck squamous cell carcinoma. Medicine 2022;101:33(e26120).
Contributor Information
Yifan Yang, Email: yangyifan517@163.com.
Ling Feng, Email: floscar@126.com.
Ru Wang, Email: ruwang1989@yeah.com.
Hongzhi Ma, Email: mahzent@163.com.
Shizhi He, Email: shizhi_he@163.com.
References
- [1].Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- [2].Hashim D, Genden E, Posner M, et al. Head and neck cancer prevention: from primary prevention to impact of clinicians on reducing burden. Ann Oncol. 2019;30:744–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Castellsagué X, Alemany L, Quer M, et al. HPV Involvement in head and neck cancers: comprehensive assessment of biomarkers in 3680 patients. J Natl Cancer Inst. 2016;108:djv403. [DOI] [PubMed] [Google Scholar]
- [4].Chaturvedi AK, Engels EA, Anderson WF, et al. Incidence trends for human papillomavirus-related and -unrelated oral squamous cell carcinomas in the United States. J Clin Oncol. 2008;26:612–9. [DOI] [PubMed] [Google Scholar]
- [5].Rietbergen MM, Brakenhoff RH, Bloemena E, et al. Human papillomavirus detection and comorbidity: critical issues in selection of patients with oropharyngeal cancer for treatment de-escalation trials. Ann Oncol. 2013;24:2740–5. [DOI] [PubMed] [Google Scholar]
- [6].Li H, Torabi SJ, Yarbrough WG, et al. Association of human papillomavirus status at head and neck carcinoma subsites with overall survival. JAMA Otolaryngol Head Neck Surg. 2018;144:519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Wasylyk B, Abecassis J, Jung AC. Identification of clinically relevant HPV-related HNSCC: in p16 should we trust? Oral Oncol. 2013;49:e33–7. [DOI] [PubMed] [Google Scholar]
- [8].Huang SH, O’Sullivan B. Overview of the 8th edition TNM classification for head and neck cancer. Curr Treat Options Oncol. 2017;18:40. [DOI] [PubMed] [Google Scholar]
- [9].Fakhry C, Lacchetti C, Rooper LM, et al. Human papillomavirus testing in head and neck carcinomas: ASCO clinical practice guideline endorsement of the college of american pathologists guideline. J Clin Oncol. 2018;36:3152–61. [DOI] [PubMed] [Google Scholar]
- [10].ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172:393–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ferro E, Enrico Bena C, Grigolon S, et al. From endogenous to synthetic microrna-mediated regulatory circuits: an overview. Cells. 2019;8:1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146:353–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dong Z, Zhang A, Liu S, et al. Aberrant methylation-mediated silencing of lncRNA MEG3 functions as a ceRNA in esophageal cancer. Mol Cancer Res. 2017;15:800–10. [DOI] [PubMed] [Google Scholar]
- [15].Cao Q, Dong Z, Liu S, et al. Construction of a metastasis-associated ceRNA network reveals a prognostic signature in lung cancer. Cancer Cell Int. 2020;20:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen C, Jiang L, Zhang Y, et al. FOXA1-induced LINC01207 facilitates head and neck squamous cell carcinoma via up-regulation of TNRC6B. Biomed Pharmacother. 2020;128:110220. [DOI] [PubMed] [Google Scholar]
- [17].Li R, Qu H, Wang S, et al. GDCRNATools: an R/Bioconductor package for integrative analysis of lncRNA, miRNA and mRNA data in GDC. Bioinformatics. 2018;34:2515–7. [DOI] [PubMed] [Google Scholar]
- [18].Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- [19].Ang KK, Harris J, Wheeler R, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Maxwell JH, Grandis JR, Ferris RL. HPV-associated head and neck cancer: unique features of epidemiology and clinical management. Annu Rev Med. 2016;67:91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24:2810–26. [DOI] [PubMed] [Google Scholar]
- [22].Serrano M. The tumor suppressor protein p16INK4a. Exp Cell Res. 1997;237:7–13. [DOI] [PubMed] [Google Scholar]
- [23].Scheffner M, Werness B, Huibregtse J, et al. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36. [DOI] [PubMed] [Google Scholar]
- [24].Martinez-Zapien D, Ruiz F, Poirson J, et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature. 2016;529:541–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lydiatt WM, Patel SG, O’Sullivan B, et al. Head and neck cancers-major changes in the American Joint Committee on cancer eighth edition cancer staging manual. CA Cancer J Clin. 2017;67:122–37. [DOI] [PubMed] [Google Scholar]
- [26].Zhu Y, Xia X, Gross N, et al. Prognostic implications of human papillomavirus status and p16 expression in laryngeal squamous cell carcinoma. Head Neck. 2019;41:4151–63. [DOI] [PubMed] [Google Scholar]
- [27].Ko HC, Harari PM, Sacotte RM, et al. Prognostic implications of human papillomavirus status for patients with non-oropharyngeal head and neck squamous cell carcinomas. J Cancer Res Clin Oncol. 2017;143:2341–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].D’Souza G, Anantharaman D, Gheit T, et al. Effect of HPV on head and neck cancer patient survival, by region and tumor site: a comparison of 1362 cases across three continents. Oral Oncol. 2016;62:20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Zhao LP, Li RH, Han DM, et al. Independent prognostic factor of low-expressed LncRNA ZNF667-AS1 for cervical cancer and inhibitory function on the proliferation of cervical cancer. Eur Rev Med Pharmacol Sci. 2017;21:5353–60. [DOI] [PubMed] [Google Scholar]
- [31].Dong Z, Li S, Wu X, et al. Aberrant hypermethylation-mediated downregulation of antisense lncRNA ZNF667-AS1 and its sense gene ZNF667 correlate with progression and prognosis of esophageal squamous cell carcinoma. Cell Death Dis. 2019;10:930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Meng W, Cui W, Zhao L, et al. Aberrant methylation and downregulation of ZNF667-AS1 and ZNF667 promote the malignant progression of laryngeal squamous cell carcinoma. J Biomed Sci. 2019;26:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hu A, Huang JJ, Xu WH, et al. MiR-21/miR-375 ratio is an independent prognostic factor in patients with laryngeal squamous cell carcinoma. Am J Cancer Res. 2015;5:1775–85. [PMC free article] [PubMed] [Google Scholar]
- [34].Li Y, Liu J, Hu W, et al. miR-424-5p promotes proliferation, migration and invasion of laryngeal squamous cell carcinoma. Onco Targets Ther. 2019;12:10441–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jeong G, Bae H, Jeong D, et al. A Kelch domain-containing KLHDC7B and a long non-coding RNA ST8SIA6-AS1 act oppositely on breast cancer cell proliferation via the interferon signaling pathway. Sci Rep. 2018;8:12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chang WM, Lin YF, Su CY, et al. Parathyroid hormone-like hormone is a poor prognosis marker of head and neck cancer and promotes cell growth via RUNX2 regulation. Sci Rep. 2017;7:41131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Xu M, Chen X, Lin K, et al. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol Cancer. 2018;17:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Wang J, Cao L, Wu J, et al. Long non-coding RNA SNHG1 regulates NOB1 expression by sponging miR-326 and promotes tumorigenesis in osteosarcoma. Int J Oncol. 2018;52:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wu Z, Chen D, Wang K, et al. Long Non-coding RNA SNHG12 functions as a competing endogenous RNA to regulate MDM4 expression by sponging miR-129-5p in clear cell renal cell carcinoma. Front Oncol. 2019;9:1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lan T, Ma W, Hong Z, et al. Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12) promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in hepatocellular carcinoma. J Exp Clin Cancer Res. 2017;36:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Zhou S, Yu L, Xiong M, et al. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. 2018;495:1822–32. [DOI] [PubMed] [Google Scholar]
- [42].Bian Y, Gao G, Zhang Q, et al. KCNQ1OT1/miR-217/ZEB1 feedback loop facilitates cell migration and epithelial-mesenchymal transition in colorectal cancer. Cancer Biol Ther. 2019;20:886–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang J, Zhang H, Situ J, et al. KCNQ1OT1 aggravates cell proliferation and migration in bladder cancer through modulating miR-145-5p/PCBP2 axis. Cancer Cell Int. 2019;19:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhang S, Ma H, Zhang D, et al. LncRNA KCNQ1OT1 regulates proliferation and cisplatin resistance in tongue cancer via miR-211-5p mediated Ezrin/Fak/Src signaling. Cell Death Dis. 2018;9:742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Reid P, Marcu LG, Olver I, et al. Diversity of cancer stem cells in head and neck carcinomas: the role of HPV in cancer stem cell heterogeneity, plasticity and treatment response. Radiother Oncol. 2019;135:1–12. [DOI] [PubMed] [Google Scholar]
- [46].Li CX, Li HG, Huang LT, et al. H19 lncRNA regulates keratinocyte differentiation by targeting miR-130b-3p. Cell Death Dis. 2017;8:e3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cen B, Lang JD, Du Y, et al. Prostaglandin E(2) induces miR675-5p to promote colorectal tumor metastasis via modulation of p53 expression. Gastroenterology. 2020;158:971–84.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Tian X, Wu Y, Yang Y, et al. Long noncoding RNA LINC00662 promotes M2 macrophage polarization and hepatocellular carcinoma progression via activating Wnt/β-catenin signaling. Mol Oncol. 2020;14:462–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wei X, Zhang Y, Yu S, et al. PDLIM5 identified by label-free quantitative proteomics as a potential novel biomarker of papillary thyroid carcinoma. Biochem Biophys Res Commun. 2018;499:338–44. [DOI] [PubMed] [Google Scholar]
- [50].Sauer F, Klemm T, Kollampally RB, et al. Differential oligomerization of the deubiquitinases USP25 and USP28 regulates their activities. Mol Cell. 2019;74:421–35.e10. [DOI] [PubMed] [Google Scholar]
- [51].Fellenberg J, Saehr H, Lehner B, et al. A microRNA signature differentiates between giant cell tumor derived neoplastic stromal cells and mesenchymal stem cells. Cancer Lett. 2012;321:162–8. [DOI] [PubMed] [Google Scholar]
- [52].Lu L, Xie R, Wei R, et al. Integrin α5 subunit is required for the tumor supportive role of fibroblasts in colorectal adenocarcinoma and serves as a potential stroma prognostic marker. Mol Oncol. 2019;13:2697–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kuninty PR, Bansal R, De Geus SWL, et al. ITGA5 inhibition in pancreatic stellate cells attenuates desmoplasia and potentiates efficacy of chemotherapy in pancreatic cancer. Sci Adv. 2019;5:eaax2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Theodoraki MN, Hoffmann TK, Jackson EK, et al. Exosomes in HNSCC plasma as surrogate markers of tumour progression and immune competence. Clin Exp Immunol. 2018;194:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.