Significance
We report the de novo microbial biosynthesis of (S)-tetrahydropapaverine (THP) and the semisynthesis of papaverine using microbially biosynthesized THP. We used protein engineering to develop variants of two enzymes with improved nonnative activity on pathway intermediates. The biosynthesis of THP demonstrates the ability to use protein homologs and protein engineering to replace the activity of unknown enzymes in heterologous biosynthetic pathways. We improved pathway flux by knocking out two yeast multidrug resistance (MDR) transporters, which reduces the export of pathway intermediates. MDR knockouts may be applied to increase flux through other heterologous pathways. The strain engineering in this work provides a demonstration of fermentation-based production of the clinically significant molecules THP and papaverine, which have experienced recent supply chain shortages.
Keywords: tetrahydropapaverine, papaverine, metabolic engineering, plant natural products, protein engineering
Abstract
Tetrahydropapaverine (THP) and papaverine are plant natural products with clinically significant roles. THP is a precursor in the production of the drugs atracurium and cisatracurium, and papaverine is used as an antispasmodic during vascular surgery. In recent years, metabolic engineering advances have enabled the production of natural products through heterologous expression of pathway enzymes in yeast. Heterologous biosynthesis of THP and papaverine could play a role in ensuring a stable supply of these clinically significant products. Biosynthesis of THP and papaverine has not been achieved to date, in part because multiple pathway enzymes have not been elucidated. Here, we describe the development of an engineered yeast strain for de novo biosynthesis of THP. The production of THP is achieved through heterologous expression of two enzyme variants with activity on nonnative substrates. Through protein engineering, we developed a variant of N-methylcoclaurine hydroxylase with activity on coclaurine, enabling de novo norreticuline biosynthesis. Similarly, we developed a variant of scoulerine 9-O-methyltransferase capable of O-methylating 1-benzylisoquinoline alkaloids at the 3′ position, enabling de novo THP biosynthesis. Flux through the heterologous pathway was improved by knocking out yeast multidrug resistance transporters and optimization of media conditions. Overall, strain engineering increased the concentration of biosynthesized THP 600-fold to 121 µg/L. Finally, we demonstrate a strategy for papaverine semisynthesis using hydrogen peroxide as an oxidizing agent. Through optimizing pH, temperature, reaction time, and oxidizing agent concentration, we demonstrated the ability to produce semisynthesized papaverine through oxidation of biosynthesized THP.
Plant secondary metabolism is a rich source of structurally unique and functionally diverse bioactive compounds. These secondary metabolites, called plant natural products (PNPs), serve crucial functions in plant development, communication, and defense (1). In addition, many PNPs exhibit therapeutic activities and have been used as antibiotics, analgesics, antivirals, neurocognitive therapies, and chemotherapeutics (2). Isolation of PNPs from plant biomass requires laborious extraction and purification procedures, and the yields are subject to both seasonal and long-term instability due to weather and climate change. Furthermore, because of the significant lead time required for planting and maturation of crops, the supply of plant-derived PNPs is less able to respond to sudden shocks in demand.
Benzylisoquinoline alkaloids (BIAs) are a family of PNPs with diverse structures and significant medicinal value. Medicinally important BIAs include the analgesics morphine and codeine, the antimicrobials sanguinarine and berberine, and the cough suppressant noscapine (3). Due to often complex stereochemistry, many BIAs continue to be produced through extraction of the active ingredient, or its precursors, from commercially grown plant material followed by any necessary semisynthetic tailoring steps (4, 5). (S)-Tetrahydropapaverine (THP) and papaverine are BIAs with established clinical significance that are derived from the opium poppy (Papaver somniferum). THP is a direct precursor to papaverine and a precursor in the production of the neuromuscular blocking agents atracurium and cisatracurium (6). Atracurium and cisatracurium are often administered during anesthesia to facilitate intubation (7). Due to an increased incidence of patient intubation during the COVID-19 pandemic, atracurium and cisatracurium have experienced recent global supply shortages (8). Papaverine is used directly in the clinic as a vasodilator and antispasmodic (9–11). In addition, recent studies have demonstrated other potential clinical applications of papaverine due to its anticancer (12) and antiviral (13, 14) activities. Despite the availability of several chemical syntheses of papaverine dating back to the early 1900s (15–17), shortages of papaverine in the 2010s forced many vascular surgeons to seek replacements for the drug due to the disruption in supply (18).
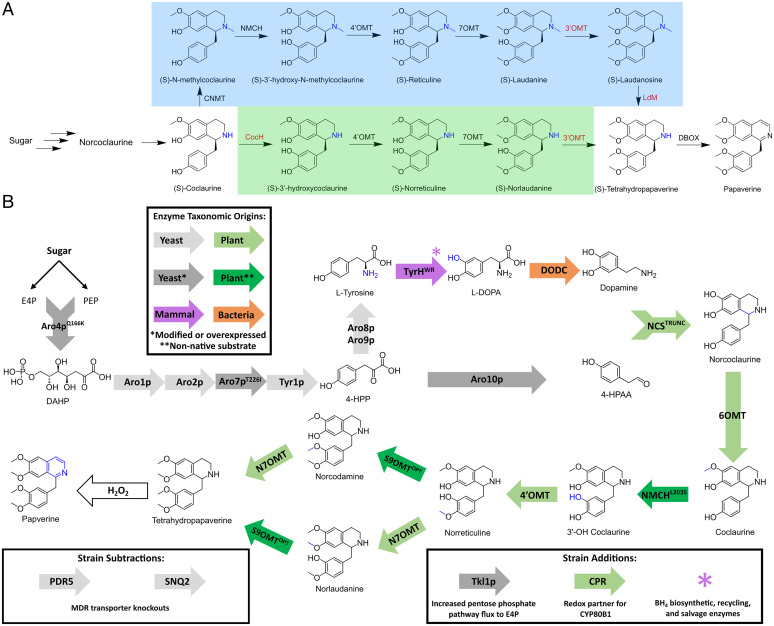
The natural biosynthetic pathway for papaverine in opium poppy has not been fully elucidated, and differing hypotheses have been suggested (19). Evidence has been presented for the existence of two biosynthetic pathways for papaverine production—the N-desmethylated route involving norreticuline (20, 21) and the N-methylated route involving reticuline (22) (Fig. 1A). Despite rigorous study on both potential pathways, key enzymes required for reactions in the pathway remain uncharacterized. In the N-desmethylated route, an enzyme to hydroxylate the 3′ position of the 1-BIA scaffold remains unknown. In the N-methylated route, an enzyme to N-demethylate the 1-BIA scaffold remains unknown(19). In addition, for both routes, no enzyme is known to efficiently O-methylate the 3′ position (23). While the enzyme dihydrobenzophenanthridine oxidase from P. somniferum (PsDBOX) has been shown to carry out the final oxidation step in papaverine synthesis (24), it has not been confirmed as the main contributor to papaverine production in opium poppy (25).
Fig. 1.
De novo production of THP and papaverine is achieved following the N-desmethylated routes. (A) Structures and names of each molecule in the proposed N-methylated (blue background) and N-desmethylated (green background) routes to papaverine production. The nitrogen molecule, whose methylation distinguishes the two paths from one another, is highlighted in blue. Truncated enzyme names accompany the arrows between molecules, and presently uncharacterized enzymes are highlighted in red. CocH, coclaurine hydroxylase; NMCH, N-methylcoclaurine hydroxylase; LdM, laudanosine demethylase. (B) De novo production of THP is achieved with enzymes from four kingdoms, overexpressed yeast genes, and multiple engineered variants of plant enzymes. Light-gray arrows, native yeast enzymes; dark-gray arrows, native yeast enzymes that have been modified to improve activity in the context of this pathway or that are overexpressed with the addition of multiple copies to the strain; purple arrows; mammalian enzymes; light-green arrows, wild-type plant enzymes; dark-green arrows; plant enzymes that have been modified to improve their activity on a nonnative substrate; orange arrows, bacterial enzymes. Engineered enzymes modified from their wild-type sequence are noted with a superscript identifying the mutation if a single mutation or with an abbreviation if multiple mutations. Moieties added by a single reaction in the pathway are highlighted in blue. E4P, erythrose 4-phosphate; PEP, phosphoenolpyruvate; DAHP, 3-deoxy-d-arabino-2-heptulosonic acid 7-phosphate; 4-HPP, 4-hydroxyphenylpyruvate; Tkl1p, transketolase; CPR, cytochrome P450 reductase; Aro4pQ166K, DAHP synthase; Aro1p, pentafunctional arom enzyme; Aro2p, bifunctional chorismate synthase and flavin reductase; Aro7pT226I, chorismate mutase; Tyr1p, prephenate dehydrogenase; Aro8p, aromatic aminotransferase I; Aro9p, aromatic aminotransferase II; Aro10p, phenylpyruvate decarboxylase; TyrHWR, feedback inhibition–resistant tyrosine hydroxylase; DODC, L-DOPA decarboxylase; NCSTRUNC, truncated norcoclaurine synthase; 6OMT, norcoclaurine 6-OMT; NMCHL203S, N-methylcoclaurine hydroxylase; N7OMT, norreticuline 7-OMT; S9OMT, scoulerine 9-OMT; PDR5, pleiotropic drug resistance transporter; SNQ2, sensitivity to 4-nitroQuinoline-N-oxide transporter.
Saccharomyces cerevisiae (baker’s yeast) has served as a platform for enzyme characterization, pathway elucidation (26–28), and de novo metabolite production for various PNPs (29). Compared to plant hosts, S. cerevisiae is readily genetically engineered (days versus months to make genetic changes), is compatible with fermentative production (conserving land, water, and time when strains are engineered to produce high titers of a target molecule), and lacks many specialized secondary metabolite pathways, providing for easier downstream isolation of target molecules (30–33). As a result, S. cerevisiae has been used as a platform for the construction of several complex PNP pathways, including opioids (34), noscapinoids (27, 35), artemisinin (36), strictosidine (37), and tropane alkaloids (28), with yields as high as 40 g/L for artemisinin precursor amorphadiene (38) and 4.6 g/L for the key BIA branchpoint compound reticuline (39). Significant work on BIAs has already been accomplished in yeast, including the de novo production of key intermediates of THP and papaverine production (40).
Here, we report the biosynthetic production of THP and the semisynthetic production of papaverine via a reconstructed N-desmethylated route in yeast (Fig. 1B). We built upon a previously reported platform yeast strain designed to produce reticuline (34, 40). The strain was modified by removing the enzyme responsible for N-methylation of the 1-BIA scaffold, P. somniferum coclaurine N-methyltransferase (PsCNMT). We then engineered a variant of Eschscholzia californica N-methyl-coclaurine hydroxylase (EcNMCH), capable of accepting the nonnative substrate (S)-coclaurine to produce (S)-3′-hydroxy-coclaurine at a 40-fold higher concentration compared to the wild-type enzyme. These modifications resulted in a platform yeast strain that produced (S)-norreticuline de novo instead of reticuline. Two additional O-methyltransferases (OMTs), namely, P. somniferum norreticuline 7-O-methyltransferase (PsN7OMT) and Thalictrum flavum scoulerine 9-O-methyltransferase (TfS9OMT), were added to the strain to O-methylate norreticuline at the 7 and 3′ positions resulting in de novo yeast biosynthesis of THP. We engineered a variant of TfS9OMT with seven amino acid mutations, which resulted in a 35-fold increase in THP titers compared to the original TfS9OMT isoform. Two multidrug resistance (MDR) transporters, namely, SNQ2 and PDR5, were knocked out in the strain to increase flux through the THP production pathway, resulting in a 15-fold increase in THP titers. The highest titers of THP were achieved by expressing the engineered TfS9OMT from a high-copy plasmid with galactose as a carbon source, resulting in 121 µg/L THP (± 3.00 µg/L). Finally, we demonstrated that hydrogen peroxide is capable of oxidizing THP to produce papaverine in a one-step, aqueous oxidation reaction that converts the biosynthesized THP to papaverine with a yield of ∼15%.
Results
A Reconstructed Biosynthetic Pathway for Reticuline Serves as the Starting Point for a De Novo–Reconstructed THP Pathway.
We used a previously reported de novo (S)-reticuline-producing yeast platform strain as the starting point for a de novo THP biosynthetic pathway (34, 35, 40). This reticuline-producing platform strain incorporates 5 modules with a total of 17 heterologously expressed enzymes to produce reticuline. Each of these modules was integrated into distinct chromosomal regions in the wild-type, haploid yeast strain Cen.PK2-1D (41).
Module I encodes the expression of four yeast enzymes to improve the production of the intermediate metabolites tyrosine and 4-hydroxyphenylacetaldehyde (4-HPAA). Copies of 3-deoxy-D-arabino-2-heptulosonic acid 7-phosphate synthase (Aro4pQ166K) and chorismate mutase (Aro7pT226I), which were modified to decrease feedback inhibition from tyrosine (40, 42, 43), and transketolase (TKL1p) and phenylpyruvate decarboxylase (Aro10p) are overexpressed. Module II encodes expression of four Rattus norvegicus enzymes to synthesize and recycle the mammalian redox cofactor tetrahydrobiopterin (34, 35), which enables the activity of the R. norvegicus tyrosine hydroxylase (RnTyrH); sepiapterin reductase (RnSepR) and 6-pyruvoyl tetrahydrobiopterin synthase (RnPTPS) increase tetrahydrobiopterin production, while quinonoid dihydropteridine reductase (RnQDHPR) and pterin carbinolamine dehydratase (RnPCD) improve cofactor recycling. Module III encodes expression of four enzymes that increase titers of the BIA precursor (S)-norcoclaurine (34, 35); R. norvegicus dihydrofolate reductase (RnDHFR) improves tetrahydrobiopterin (BH4) salvage, a mutant (R37E, R38E, and W166Y) of the R. norvegicus tyrosine hydroxylase (RnTyrHWR) produces L-3,4-dihydroxyphenylalanine (L-DOPA) with improved resistance to negative feedback (40), DOPA decarboxylase from Pseudomonas putida (PpDODC) produces dopamine (40), and norcoclaurine synthase from Coptis japonica (CjNCS) catalyzes the reaction between dopamine and 4-HPAA to produce norcoclaurine (44). Module IV encodes the expression of five plant proteins that convert norcoclaurine to reticuline (34, 35); P. somniferum norcoclaurine 6-O-methyltransferase (Ps6OMT), PsCNMT, cytochrome P450 reductase (PsCPR), and 4’-O-methyltransferase (Ps4′OMT) and EcNMCH. Lastly, module V encodes the overexpression of three proteins that were previously identified as bottlenecks in the pathway (RnTyrHWR, Ps4′OMT, CjNCS), resulting in strain CSY1060 (34, 35, 40).
Reticuline production in the platform strains was assayed as a benchmark for flux through the native benzylisoquinoline pathway in yeast. The strains were grown for three days at 30 °C in synthetic complete (SC) media. Reticuline concentrations in the growth media were assayed by liquid chromatography coupled with tandem mass spectroscopy (LC-MS/MS). The base platform strain CSY1060 produces 19.1 µg/L reticuline (±2.23 µg/L). We further incorporated two previously reported modifications to increase reticuline production (35). Specifically, a variant of NCS with the first 24 amino acids truncated (CjNCStrunc) and a yeast codon-optimized variant of RnTyrHWR were replaced in the appropriate modules, resulting in strain CSY1171 (35), which produces 1.14 mg/L reticuline (±47.3 µg/L). The platform strain CSY1171 was used as the starting point for constructing the THP-producing yeast strains.
An Engineered EcNMCH Variant with Higher Activity on the Nonnative Substrate Coclaurine Increases Norreticuline Titer.
In order to construct the N-desmethylated biosynthetic route in yeast to THP and papaverine, we first constructed a strain to produce norreticuline. The reticuline-producing platform strain CSY1171 produces the intermediate norcoclaurine, which is subsequently O-methylated and N-methylated by Ps6OMT and PsCNMT, respectively, to produce N-methylcoclaurine. In order to produce only N-demethylated products, the coding region for PsCNMT was deleted from strain CSY1171 yielding CSY1172. With the removal of PsCNMT from the biosynthetic pathway, norreticuline becomes the final pathway product (Fig. 1A).
We confirmed norreticuline production in the base strain CSY1172. Strains were grown for 3 d at 30 °C in SC media, and norreticuline concentrations in the growth media were assayed by LC-MS/MS. The results demonstrate that CSY1172 produces norreticuline at a concentration of 5.85 µg/L (±1.04 µg/L), and the reticuline concentration drops below the limit of detection of our assay. The concentration of norreticuline produced by CSY1172 is 200-fold lower than the concentration of reticuline produced by CSY1171 (the reticuline-producing platform strain) on a molar basis. We hypothesized that the lower titer of norreticuline results from a lower activity of EcNMCH on its nonnative substrate coclaurine in the reconstructed norreticuline pathway (20, 35, 40). Since Ps4′OMT has been shown to modify both N-methylated and N-demethylated substrates (20), we predicted that EcNMCH activity would be limiting norreticuline production in CSY1172. This hypothesis was supported by a relative accumulation of pathway intermediates in the norreticuline-producing strain CSY1172. The concentration of the substrate of EcNMCH, coclaurine, was estimated to be one to two orders of magnitude higher than the concentration of the product 3′OH-coclaurine. By comparison, the concentration of the substrate of Ps4′OMT, 3′OH-coclaurine, was estimated to be an order of magnitude lower than the concentration of its product, norreticuline (SI Appendix, Fig. S1).
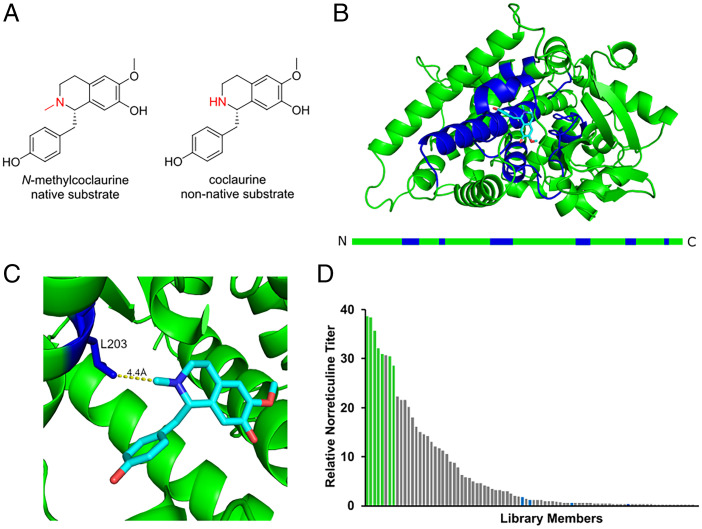
We focused on improving EcNMCH activity on coclaurine to increase norreticuline production in the coclaurine-producing yeast strain. Because of the structural similarity between coclaurine and N-methylcoclaurine (Fig. 2A), site-directed, random mutagenesis was used to improve the catalytic activity of the enzyme on the nonnative substrate coclaurine. A homology model of EcNMCH was developed using RaptorX (45), and the substrate binding pocket for the native substrate N-methylcoclaurine, was predicted using SwissDock (46). Six contiguous chains of amino acids (amino acid positions—C1: 94 to 109, C2: 202 to 205, C3: 271 to 294, C4: 344 to 356, C5: 416 to 434, C6: 465 to 468) were identified as being in proximity to the substrate binding pocket (Fig. 2B). We hypothesized that the amino acids in these six regions may play roles in substrate binding and orientation and selected them as targets for mutagenesis to improve the binding of the nonnative substrate coclaurine. Each of the chains was individually targeted for mutagenesis using a combination of error-prone PCR and CRISPR-targeted (30) integration. Colonies containing one to two mutations were picked and grown for 3 d at 30 °C in SC media supplemented with 2 mM L-DOPA and 10 mM ascorbic acid to increase pathway flux. The spent medium was subsequently assayed for norreticuline production by LC-MS/MS. After screening 1,000 colonies, a variant of EcNMCH that increased norreticuline production 10-fold was identified. Subsequent sequencing of this top variant identified a mutation at the L203 position.
Fig. 2.
Structure-guided semitargeted mutagenesis helps identify NMCH variant with improved activity on the N-desmethylated substrate coclaurine. (A) Comparison between native substrate for EcNMCH, N-methylcoclaurine, and the nonnative substrate in the reconstructed THP biosynthetic pathway, coclaurine. Key differences are highlighted in red. (B) Homology model of EcNMCH with regions adjacent to the binding pocket highlighted. EcNMCH is shown in green; binding pocket adjacent residues targeted for mutagenesis are highlighted in blue; native enzyme substrate, N-methylcoclaurine, is shown in cyan. The green bar below represents the linearized form of the enzyme with the binding pocket adjacent regions highlighted in blue and identified as regions for targeted mutagenesis. (C) Detailed view of N-methylcoclaurine (cyan) docked in the binding pocket of EcNMCH (green). The L203 position is highlighted in blue, demonstrating its proximity to the nitrogen that is differentially methylated between the desired substrate, coclaurine, and the native substrate N-methylcoclaurine. (D) Relative norreticuline production of different EcNMCH variants in an NNK library including all possible amino acids at the L203 position, tested in CSY1172 with L-DOPA feeding. Wild-type negative controls are shown in blue, and norreticuline titers are normalized to the average of the wild type being equal to one. Glycine and serine mutants are shown in green; other library members are shown in gray.
We used site-saturation mutagenesis to further improve the activity of the EcNMCH enzyme on the nonnative substrate coclaurine. A library using the degenerate codon NNK, was used to screen every possible amino acid substitution at the L203 position. In the resulting library, 42 of 93 colonies screened resulted in higher than wild-type norreticuline production, indicating that several amino acid substitutions at this position are able to improve the activity on the nonnative substrate. The L203S variant exhibited the largest improvement to norreticuline production at ∼40-fold over the wild-type variant with L-DOPA feeding (Fig. 2D). The previously generated homology model with docked substrate shows proximity between the wild-type leucine at this position and the N-methyl group of the native substrate (Fig. 2C). The improved potential for hydrogen bonding between a serine at this position and the unmethylated nitrogen in the nonnative substrate provides a possible mechanistic explanation for this improvement in activity. An expression cassette encoding the engineered variant EcNMCHL203S was integrated into strain CSY1172, replacing the wild-type EcNMCH in the strain resulting in CSY1173, which produces 72.0 µg/L norreticuline (±11.3 µg/L). Overall, we developed an engineered EcNMCH variant that increased norreticuline production 12-fold over the wild-type enzyme in engineered yeast grown in SC without L-DOPA feeding.
An Engineered TfS9OMT Variant with Higher Activity on Nonnative Substrates Norreticuline and Norlaudanine Increases THP Titer.
In order to produce THP via the N-desmethylated biosynthetic route, norreticuline must be O-methylated at the 7 and 3′ positions on the 1-BIA scaffold (Fig. 1B). An enzyme that can carry out O-methylation at the 7 position of the 1-BIA scaffold, PsN7OMT, has been previously reported (47). The enzyme responsible for O-methylating the 3′ position of the 1-BIA scaffold in planta had not been reported at the time of this study. A number of plant OMTs have been extensively characterized that perform similar reactions (48–50), and thus, we took an enzyme engineering approach leveraging known plant OMTs as a starting point to complete the reconstructed THP pathway.
We first examined whether incorporation of the PsN7OMT enzyme into our norreticuline biosynthetic pathway would yield a yeast strain capable of producing (S)-norlaudanine via O-methylation of the 7 position of norreticuline. Previous characterization of PsN7OMT showed that it efficiently methylates the 7 position and has narrow substrate specificity (47). An expression cassette encoding PsN7OMT was integrated into the URA3 locus of the norreticuline-producing strain CSY1173, resulting in strain CSY1174. Norlaudanine production was assayed by LC-MS/MS after 3 d of growth at 30 °C in SC media. The results demonstrate that strain CSY1174 produced norlaudanine at an estimated titer of 20 µg/L, indicating that PsN7OMT catalyzes the conversion of norreticuline to norlaudanine in the engineered yeast strain.
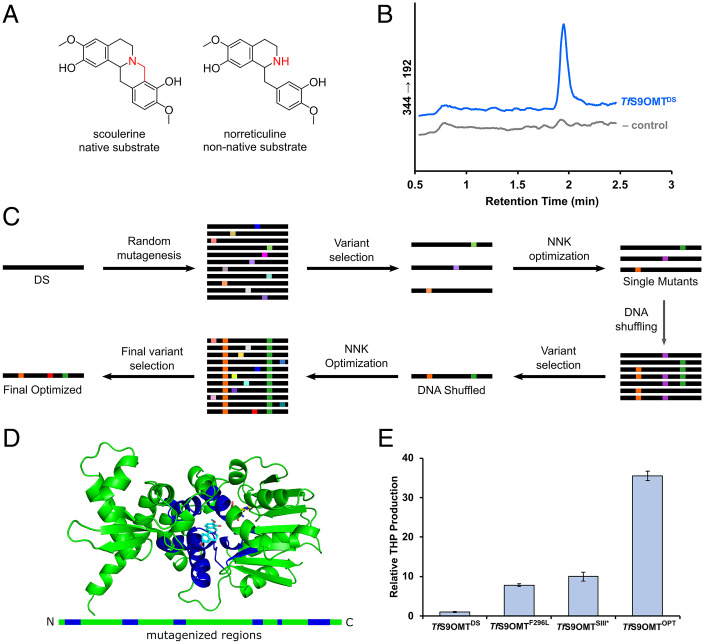
To complete the reconstructed THP biosynthetic pathway, we next focused on the O-methylation of the 1-BIA scaffold at the 3′ position. We screened a variety of plant OMTs, including some reported to perform 3′-O-methylation in vitro or in vivo in other organisms (48), and none were able to efficiently O-methylate the 3′ position in the context of a norlaudanine-producing yeast strain. The only enzyme we screened with detectable 3′-O-methylation activity was the DS isoform of TfS9OMT (50), which produced low titers of (S)-norcodamine and THP. TfS9OMT was included in our 3′-O-methylation screen because of the structural similarity between norreticuline and the enzyme’s native substrate scoulerine (Fig. 3A). Scoulerine and norreticuline are similar molecules except for the presence of a carbon bridge between the isoquinoline nitrogen and the 2’ benzyl moiety in scoulerine. Screens for 3′OMT activity were performed in the norlaudanine-producing strain CSY1174. The strain was grown for 3 d in synthetic defined (SD) media with the appropriate dropout for plasmid-based expression, and THP and norcodamine were assayed using LC-MS/MS. THP was produced at a concentration of 372 ng/L (± 52 ng/L) in CSY1174 with a high-copy plasmid encoding TfS9OMTDS, whereas it was not detected in negative-control samples with a similar plasmid encoding green fluorescent protein (GFP) (Fig. 3B). The results indicate that TfS9OMTDS can exhibit the desired 3′ OMT activity for THP production, but this activity is low on the nonnative substrates in the reconstructed THP pathway.
Fig. 3.
Multiple protein engineering strategies were applied to generate variants of TfS9OMT with an improved capacity to produce de novo THP. (A) Comparison between the native substrate of TfS9OMT, scoulerine, and the nonnative substrate in the context of this pathway, norreticuline. Key differences are highlighted in red. (B) LC-MS/MS traces for the MRM transition 344 → 192 used to detect the presence of THP. CSY1174 was grown with a high-copy plasmid encoding expression of either the DS variant of TfS9OMT (blue) or with GFP as a negative control (gray), and the medium was analyzed for the presence of THP. Traces show a single sample that was representative of tests in triplicate. (C) Schematic of the protein engineering strategy used on TfS9OMT. Targeted mutagenesis over individual chains of the enzyme, DNA shuffling, and targeted NNK libraries were used to construct and identify a variant of the enzyme with substantially improved 3′-O-methylation activity. (D) Crystal structure of TfS9OMT (green) with the substrate binding pocket adjacent region highlighted in blue and the native substrate scoulerine in cyan. The green bar below represents the linearized form of the enzyme with the binding pocket adjacent regions highlighted in blue. (E) Relative THP concentration produced by CSY1354 when expressing different TfS9OMT variants: TfS9OMTDS; the best single mutant identified, TfS9OMTF296L; the DNA shuffled variant, TfS9OMTSIII*; the final optimized mutant incorporating additional NNK library selections, TfS9OMTOPT. Error bars represent the SD of triplicate samples.
We next focused on improving TfS9OMT activity at the 3′ position on the nonnative substrates norreticuline and norlaudanine. We took a site-directed, random mutagenesis approach with TfS9OMT, similar to the approach taken with EcNMCH. The previously elucidated crystal structure (50) of the TfS9OMT enzyme was used to identify six contiguous chains of amino acids (amino acid positions—C1: 10 to 20, C2: 108 to 120, C3: 161 to 168, C4: 250 to 256, C5: 281 to 283, C6: 293 to 314) with proximity to the substrate binding pocket of the enzyme (Fig. 3D). Each of the chains was individually targeted for mutagenesis using an error-prone PCR approach. Because of the low signal of THP detection at the titers produced by genomically integrated TfS9OMT, variants generated by error-prone PCR were expressed from a high-copy plasmid in CSY1174, in contrast to the EcNMCH engineering experiments. Colonies were picked and grown for 3 d at 30 °C in SD media with the appropriate dropout and subsequently assayed for THP production. A total of 1,000 colonies were screened for THP titers, and 10% of colonies screened exhibited at least a 50% increase in THP titer over a control strain expressing the TfS9OMTDS. Enzyme variants from 12 colonies with improved THP titers were sequenced, and the sequencing data indicated that mutations at several off-target residues (S47, S77, T83, C98) and on-target residues (A119, F296, L304) are present in enzyme variants exhibiting up to threefold higher THP titers compared to TfS9OMTDS.
In order to identify the optimal amino acid at each identified residue location, NNK libraries were generated and screened at each residue where mutations were shown to improve 3′ OMT activity in the initial site-directed mutagenesis screen. The TfS9OMT NNK libraries were tested by expression from high-copy plasmids in CSY1174. For each NNK library, 92 colonies were screened in order to reach a 95% chance of full library coverage. NNK libraries allowed us to identify mutations superior to those found in the initial screen at five of the seven amino acid residues (SI Appendix, Table S5). For example, the initial F296S mutation, which yielded a threefold higher THP titer than TfS9OMTDS, was improved to F296L, which produced a fivefold higher THP concentration than TfS9OMTDS (SI Appendix, Fig. S2).
To further improve the 3′ OMT activity, we applied DNA shuffling to screen for synergistic combinations of the identified amino acid mutations. The site-directed mutagenesis and NNK libraries identified mutations at seven residues that individually improved THP production (S47, T83, S77, C98, A119, F296, L304). A library to test both optimized and unoptimized mutations in combination was generated via DNA shuffling (51) with three input templates, as follows: TfS9OMTDS, TfS9OMTV1 (S47F, T83A, C98R), and TfS9OMTV2 (S77W, C98P, A119L, F296L, L304Y). Fully reassembled TfS9OMT shuffled variants were similarly characterized by expressing the variants from high-copy plasmids transformed into strain CSY1174. A total of 720 colonies were screened for THP titers, and 36% of these colonies produced THP titers higher than TfS9OMTDS. The three top variants increased THP titers more than 10-fold compared to TfS9OMTDS. TfS9OMTSI incorporates A119L and the off-target mutation V281I and increases the THP titer 12-fold; TfS9OMTSII incorporates four on-target mutations, namely, S47P, T83A, C98R, and A119L, and increases the THP titer 17-fold; and TfS9OMTSIII incorporates T83A, C98P, A119L, and the off-target mutation P208Q and improves THP titer 16-fold (SI Appendix, Fig. S3).
We achieved additional improvements of 3′ OMT activity in the engineered TfS9OMT variant with additional NNK optimization. In previous experiments, libraries were generated using TfS9OMTDS as a template, but with the availability of improved variants, we used a pool of TfS9OMTSI, TfS9OMTSII, TfS9OMTSIII, and TfS9OMTSIII* (a version of TfS9OMT with the off-target P208Q reverted to its wild-type identity) as templates for further improvements. We generated and tested NNK libraries at V281, F296, N309, and S160, using the highest performing variant as the template for each subsequent library. These mutagenesis experiments screened colonies by expression from a low-copy plasmid in CSY1174 to better match the protein titers that can be achieved through genomic integration. This iterative optimization resulted in TfS9OMTOPT, which contains mutations T83A, C98P, A119L, S160V, V281I, F296L, and N309T. We compared the TfS9OMTDS, TfS9OMTF29L (the highest performing single mutant), TfS9OMTSIII*, and TfS9OMTOPT by expressing them from a low-copy plasmid in CSY1354 (strain described in the next section) grown for 3 d in SD media with appropriate dropout. Under these conditions, TfS9OMTOPT produced 35-fold more THP than TfS9OMTDS (Fig. 3E). To confirm that the improved THP titer was the result of improved enzyme function, TfS9OMTDS and TfS9OMTOPT were purified by nickel-nitrilotriacetic acid (Ni-NTA) affinity chromatography and their activities examined in vitro. TfS9OMTOPT produced 55-fold more norcodamine than TfS9OMTDS when incubated with norreticuline as a substrate, indicating that the observed improvements in THP titer are the result of improved catalytic efficiency. TfS9OMTOPT was incorporated into the LYP locus of CSY1174, resulting in CSY1359, which produces THP at 271 ng/L THP (±24.2 ng/L). Overall, engineering a TfS9OMT variant capable of 3′ O-methylating the 1-benzylisoquinoline scaffold for de novo THP production included site-directed mutagenesis, individual NNK optimization, and DNA shuffling (Fig. 3C), resulting in a variant harboring seven mutations that supported a 35-fold increase in THP production from yeast.
During the preparation of this manuscript, two enzymes were reported in separate studies to methylate the 3′-hydroxyl group of norreticuline. In one study, a putative 3′OMT from P. somniferum (Ps3′OMT) associated with papaverine production was identified (23) via virus-induced gene silencing. In vitro characterization experiments demonstrated methylation of a structural homolog, dihydroxyacetophenone, but 3′OMT activity on norreticuline and norlaudanine was not demonstrated. In a second study, an engineered variant of an OMT from Glaucium flavum [GfOMT1, referred to elsewhere as GFLOMT1 (49)] was demonstrated to exhibit 3′OMT activity on the 1-BIA scaffold (52). The authors of this study reported a variant of GfOMT1 with the ability to methylate four different positions on the 1-BIA scaffold, including the 3′ position, to produce THP from fed norlaudanosaline in Escherichia coli. We examined the 3′OMT activity of both enzymes in the context of our engineered pathway by expressing them from a low-copy plasmid in the norreticuline-producing strain CSY1173 and in the norlaudanine-producing strain CSY1174. Under these conditions, norcodamine and THP, the expected products of 3′-O-methylation, were not detected in the media at levels above background.
Knocking Out MDR Transporters Improves Pathway Flux.
Despite the improvements to TfS9OMT activity in the THP biosynthetic pathway, the incorporation of two engineered enzymes (EcNMCHL203S, TfS9OMTOPT) modifying nonnative substrates in the pathway results in a strain that exhibits relatively low THP titers compared to the titer of reticuline in CSY1171. We sought to further improve the titer of THP produced de novo by making complementary strain modifications.
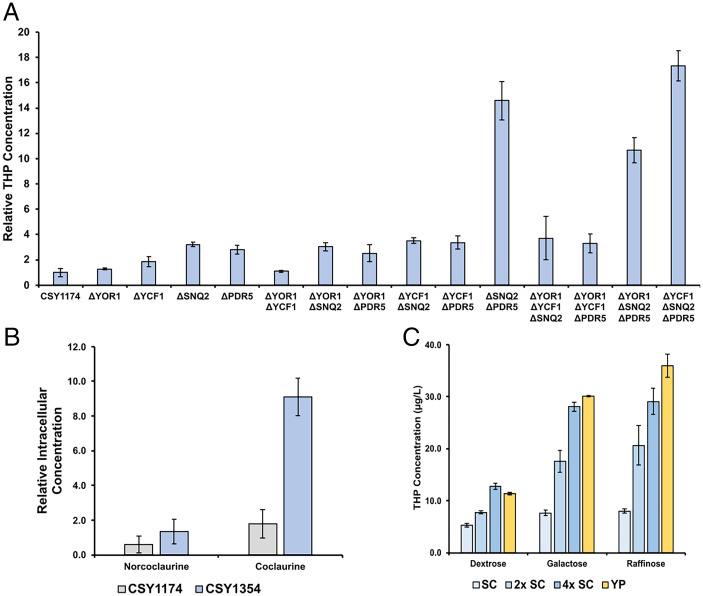
We first examined whether the deletion of MDR transporters can lead to increased pathway flux. Biosynthetic intermediates in our pathway must remain within cells in order to be accepted and functionalized by pathway enzymes, so premature export of pathway intermediates may be limiting our titer of THP. We hypothesized that removal of specific MDRs may result in higher intracellular concentrations of pathway intermediates, resulting in increased pathway flux. Four ATP-utilizing MDRs were selected as knockout targets, as follows: PDR5, SNQ2, YOR1, and YCF1 (53). PDR5, SNQ2, and YOR1 are the main drivers of pleiotropic drug resistance in S. cerevisiae (54–56). YCF1 was included to provide increased MDR diversity; YCF1 has been shown to provide MDR (55) and cadmium resistance, and uniquely localizes to the vacuole (57). MDR proteins were knocked out by introducing a premature stop codon within the first 50 amino acids of the protein using a CRISPR-based approach. The MDRs were knocked out individually and in combination in the norlaudanine-producing strain CSY1174. The engineered TfS9OMTOPT was expressed in each of the resulting knockout strains from a low-copy plasmid. The strains were grown for 3 d in SD media with the appropriate dropout, and THP levels were assayed using LC-MS/MS. PDR5 and SNQ2 knockouts improved the overall production of THP compared to equivalent strains with the transporters expressed. The strain harboring the double knockout of SNQ2 and PDR5, namely, CSY1354, exhibited a 15-fold increase in THP titer (Fig. 4A). The strain resulting from the triple knockout of SNQ2, PDR5, and YCF1 (CSY1358) produced the highest THP titer increase at 17-fold higher than CSY1174. We chose to proceed with the double knockout CSY1354 because additional strain modification can negatively impact strain health (58) and because the YCF1 knockout was not shown to be statistically significant in a head-to-head comparison between CSY1354 and CSY1358 using a t test (P = 0.119).
Fig. 4.
MDR transporter knockouts affect the transport of pathway intermediates into the media and contribute to higher THP-producing strains. (A) Relative THP titers produced in CSY1174 and related strains with one to three MDR transporters knocked out. All strains are expressing TfS9OMTOPT from a low-copy plasmid. Relative THP concentration is normalized to the THP concentration in CSY1174. (B) Relative intracellular concentrations of pathway metabolites in CSY1174, the base norlaudanine-producing strain, and CSY1354, which is identical to CSY1174 except the MDR transporters SNQ2 and PDR5 are knocked out. Both strains contain a low-copy plasmid expressing TfS9OMTOPT. Intracellular concentration is reported as relative to the extracellular concentration of that metabolite in the same sample. (C) THP concentration in the media when CSY1361 is grown at the indicated medium conditions. 2× SC and 4× SC indicate twice and four times the standard concentration of amino acid supplements when preparing SC media, respectively. Carbon sources indicated on the x-axis were present at 2% (wt/vol). Error bars represent the SD of triplicate samples in all panels.
We next examined the potential mechanism by which CSY1354 results in higher THP titers compared to CSY1174. Based on our initial hypothesis, we expected that export of one or more THP pathway intermediates would be diminished in CSY1354. To better understand the production and export of pathway intermediates in CSY1174 and CSY1354, we expressed TfS9OMTOPT from a low-copy plasmid, and we measured the optical density (SI Appendix, Fig. S4) and concentrations of norcoclaurine, coclaurine, 3′-hydroxycoclaurine, norreticuline, norlaudanine, norcodamine, and THP at 6-h intervals (SI Appendix, Figs. S5–S11). These metabolites were analyzed as we predicted they may be targets for yeast MDR transporters due to their size and unique scaffold when compared to native yeast metabolites. For each metabolite, the concentration measured in the media with CSY1354 was higher than with CSY1174 at every timepoint, except for the 12- and 18-h timepoints for coclaurine. The increase in metabolite titers resulting from the removal of SNQ2 and PDR5 varied significantly (SI Appendix, Table S6). For example, at the 54-h timepoint, coclaurine concentration in CSY1354 was 1.4-fold higher than CSY1174, whereas the concentration of THP in CSY1354 was 10.1-fold higher. Since the overall pathway flux is increased, we predicted that metabolites with smaller increases in extracellular concentration, like coclaurine, may be exported less efficiently with SNQ2 and PDR5 knocked out.
To further understand the impact of MDR transporter knockouts on pathway metabolite transport, we grew cultures of CSY1354 and CSY1174 with TfS9OMTOPT expressed from a low-copy plasmid for 66 h and compared intracellular and extracellular metabolite concentrations. The LC-MS/MS integration areas from extracted intracellular metabolites were multiplied by a corrective factor in attempt to compare metabolite concentrations intracellularly and extracellularly (Materials and Methods). For the furthest downstream metabolites (3′-hydroxycoclaurine, norreticuline, norcodamine, norlaudanine, THP), intracellular concentrations were too low to accurately quantify (SI Appendix, Fig. S12). For the upstream metabolites (norcoclaurine, coclaurine), the ratio of intracellular concentration to extracellular concentration was higher in CSY1354 than it was for CSY1174 (Fig. 4B). In CSY1354, the concentration of coclaurine intracellularly was 9.1-fold higher than in the media, whereas in CSY1174, the intracellular coclaurine concentration was only 1.8-fold higher than in the media. The intracellular accumulation of coclaurine resulting from SNQ2 and PDR5 knockouts suggests that these MDR transporters may play a role in exporting coclaurine in CSY1174. The decreased rate of coclaurine export in CSY1354 may play a role in increasing flux through the THP production pathway and improving THP titers.
Optimization of Carbon Source and TfS9OMTOPT Expression Results in Increased THP Production in Yeast.
We next sought to increase the titer of THP produced via optimization of growth media and TfS9OMTOPT expression in the engineered yeast strain. We began by integrating a copy of TfS9OMTOPT into the LYP locus of CSY1354, the norlaudanine producing strain with SNQ2 and PDR5 knocked out, to generate CSY1360, which produces 4.38 µg/L THP (±0.25 µg/L). Adding a second copy of TfS9OMTOPT at the TRP locus of CSY1360 yielded CSY1361, which produces more than double the amount of THP at a concentration of 10.1 µg/L (±0.21 µg/L). Based on the results of previous studies on the biosynthesis of 1-BIAs (35, 59), we grew CSY1361 with multiple medium bases and carbon sources to improve pathway flux. For medium bases, we used a yeast nitrogen base to prepare a standard SC medium and more amino acid–rich versions of SC with two and four times the standard amino acid concentrations (2xSC and 4xSC, respectively). We also prepared media using a yeast peptone (YP) base for a richer nutrient medium option. For carbon sources, we tested the standard carbon source, dextrose, along with galactose, sucrose, trehalose, glycerol, and raffinose. Carbon sources were tested individually and in combination with dextrose. Each medium was tested at a concentration of 2% (wt/vol) in the final preparation of the medium unless otherwise noted. Overall, the highest titer of THP was observed with the conditions of YP with raffinose as a carbon source, which produced 36.0 µg/L THP (±2.20 µg/L) (Fig. 4C and SI Appendix, Figs. S13–S15).
As the introduction of a second copy of TfS9OMTOPT more than doubled THP production, we examined whether increasing copies of this bottleneck enzyme via plasmid-based expression would lead to additional increases in THP titers. Expression of TfS9OMTOPT from a high-copy plasmid in CSY1354 resulted in higher THP titers than any fully genomically integrated strain, with a THP concentration of 68.9 µg/L (± 304 ng/L) when grown in SC with appropriate dropout. To maximize the production of THP with a high-copy plasmid, we tested galactose and raffinose as alternates to the carbon source dextrose. Galactose and raffinose were chosen based on the results of the carbon source optimization using CSY1361, where they consistently yielded the best results compared to other carbon sources regardless of medium base. SC with both galactose and raffinose resulted in more than 2-fold higher THP concentration compared to SC with dextrose (SI Appendix, Figs. S16 and S17). In this context, expression of TfS9OMTOPT from a high-copy plasmid in CSY1354 with raffinose as a carbon source yielded the highest THP titer of any conditions tested at 121 µg/L (± 3.00 μg/L). Taken together, the combination of optimizing MDR transporter knockouts, TfS9OMT engineering, TfS9OMT expression, and carbon source improved the THP titer 600-fold compared to our lowest THP-producing strain (i.e., low-copy plasmid expression of TfS9OMTDS in CSY1174).
One-Step Chemical Oxidation of Biosynthesized THP Yields Semisynthetic Papaverine.
The final step in papaverine production biosynthesis is the four-electron oxidation of THP to produce papaverine. To accomplish this final step, we first explored enzymatic oxidation of THP to produce papaverine. PsDBOX from P. somniferum has been proposed to facilitate this reaction in planta and has demonstrated THP oxidase activity when tested in vitro. We grew the THP-producing strain CSY1361 harboring a low-copy plasmid encoding a PsDBOX expression cassette in SD media with appropriate dropouts for 3 d. Despite the presence of the substrate, THP, and PsDBOX, papaverine was not detected in the growth media. We made several attempts to establish PsDBOX activity in this context including feeding high concentrations of THP, localizing PsDBOX to subcellular compartments, improving culture oxygenation by growth in baffled flasks, and tethering PsDBOX to GFP to confirm protein expression via fluorescence microscopy (SI Appendix, Fig. S18). Despite significant troubleshooting efforts, PsDBOX activity on THP to produce papaverine in yeast strain CSY1361 was not observed.
In the absence of viable enzymatic oxidation, we explored chemical oxidation of THP. Initial screens, which included numerous oxidizing agents, demonstrated that both hydrogen peroxide and potassium persulfate were able to oxidize THP to produce papaverine in an aqueous setting, and we selected these two oxidizing agents for further testing. To improve reaction yields, pH, oxidizing agent concentration, reaction time, and temperature were adjusted to improve reaction yield. First, we tested the effect of pH on the oxidation of THP by hydrogen peroxide and potassium persulfate by varying the pH of Tris HCl buffer. Oxidation reactions were performed at 42 °C for 3 h before being quenched with an equal volume of 1 M sodium thiosulfate. For both hydrogen peroxide and potassium persulfate, the highest yields were achieved at a pH of 9.5 (SI Appendix, Fig. S19). Tris⋅HCl buffer with a pH of 9.5 was used as the base for all subsequent reactions. We next examined reaction yield in the context of reaction time, reaction temperature, and oxidizing agent concentration. These variables were adjusted in combination because of their potential effect on one another. For hydrogen peroxide, we measured the reaction yield at nine conditions with variable temperatures (55 °C, 70 °C, 85 °C) and concentrations (0.1%, 0.3%, 0.5%, 0.75%, 1.0%). For potassium persulfate, we measured reaction yield in six conditions at three temperatures (30 °C, 40 °C, 55 °C) and two concentrations (15 mM, 30 mM). To determine optimal conditions, we also tracked reaction specificity, defined as the product produced divided by substrate consumed (SI Appendix, Table S7). The highest yield achieved with potassium persulfate as an oxidizing agent was 7.41% at a temperature of 40 °C, a concentration of 30 mM, and a reaction time of 6 h (SI Appendix, Fig. S20). The highest yield achieved overall was 16.3% at a temperature of 85 °C, a hydrogen peroxide concentration of 0.5%, and a time of 60 min (SI Appendix, Fig. S21).
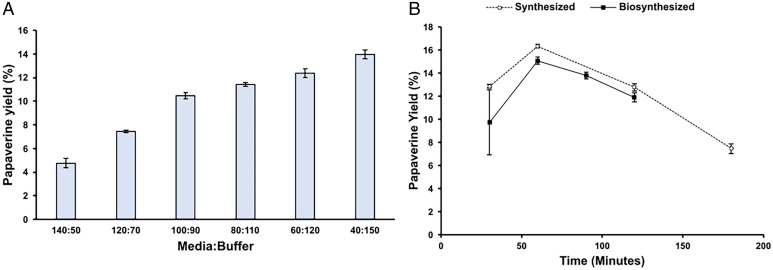
We next used these THP oxidation conditions with biosynthesized THP to produce de novo semisynthesized papaverine. Biosynthesized THP was produced in CSY1354 with TfS9OMTOPT expressed from a high-copy plasmid as previously described. The spent medium from this experiment containing 121 µg/L THP was used as the substrate for oxidation reactions. We first tested how the ratio of substrate-containing media to Tris⋅HCl buffer would affect the yield and specificity of the reactions. We tested six different ratios of media-to-buffer (40:150, 60:130, 80:110, 100:90, 120:70, 140:50) under the two sets of conditions that produced the highest yield in the previously described oxidation experiments using synthesized THP—60 min with 0.5% hydrogen peroxide at 85 °C and 3 h with 0.3% hydrogen peroxide at 70 °C (SI Appendix, Fig. S22). In both sets of reaction conditions, we found that the yield of papaverine was highest when the media-to-buffer ratio was lowest (Fig. 5A and SI Appendix, Fig. S23). At the lowest media-to-buffer ratio, the yield of reactions with biosynthesized THP in the presence of media was similar to the yield of reactions with pure, chemically synthesized THP (Fig. 5B). Using biosynthesized THP, the yield of papaverine was 15.1% (SI Appendix, Fig. S24)—versus 16.3% with synthesized THP. Overall, we were able to use synthesized substrate to find a set of aqueous reaction conditions capable of oxidizing THP to produce papaverine and validated that these same reaction conditions can be used on biosynthesized THP produced in the media of our yeast strains.
Fig. 5.
Chemical oxidation of biosynthesized THP enables semisynthetic papaverine production. (A) Yield of papaverine in chemical oxidation reactions with biosynthesized THP. All reactions were carried out with 0.5% hydrogen peroxide at 85 °C for 1 h. The media-to-buffer ratio of each reaction is indicated on the x-axis. Error bars represent the SD of triplicate samples. (B) Yield of papaverine in chemical oxidation reactions with 0.5% hydrogen peroxide at 85 °C using chemically synthesized (white circles) and biosynthesized (black squares) THP. The synthesized THP reaction used 500 nM THP and 75 mM Tris⋅HCl buffer. Biosynthesized THP reaction used spent media from CSY1354 grown with TfS9OMTOPT expressed from a high-copy plasmid. The reactions had a medium-to-buffer ratio of 40:150 and had a final Tris⋅HCl concentration of 75 mM. Error bars represent the SD of triplicate samples in all panels.
Discussion
In this work, we demonstrate the de novo microbial biosynthesis of THP and semisynthetic production of papaverine. We utilized a reticuline-producing platform yeast strain that produces 1.14 mg/L reticuline. We modified the strain to produce norreticuline by removing PsCNMT and engineering a variant of EcNMCH with improved activity on the nonnative substrate coclaurine. In the absence of a known 1-BIA 3′-OMT, we identified that TfS9OMT has 3′-OMT activity on this scaffold. We used a combination of site-directed, random mutagenesis, DNA shuffling, and site-saturation mutagenesis to identify TfS9OMTOPT, a variant with improved catalytic efficiency. We improved the overall flux through the pathway by knocking out two MDR transporters, with results indicating that these transporters reduce the transport of the intermediate coclaurine out of the cell. THP production was further improved by increasing TfS9OMTOPT copy number and altering the carbon source of the media. Overall, these improvements to pathway flux increased THP titers in the media 600-fold resulting in 121 µg/L THP production. We also identified reaction conditions that allow for the aqueous oxidation of THP to produce papaverine using hydrogen peroxide, demonstrating reported semisynthesis of papaverine from biosynthesized THP.
Despite extensive prior research, the enzymes for the in planta papaverine production pathway have remained elusive (19, 60). While evidence for the N-methylated route toward papaverine production has been presented (22), most recent studies have presented data and evidence favoring the N-desmethylated route (19–21). Along the N-desmethylated route, there are three major challenges for microbial production of papaverine, as follows: coclaurine hydroxylation, norreticuline or norlaudanine 3′-O-methylation, and THP oxidation. Earlier studies of EcNMCH in which coclaurine was tested as a potential substrate reported no activity (61). Studies of potential norreticuline 3′OMTs have either lacked in vitro tests to confirm activity (23) or 3′-O-methylation has been a low-conversion side reaction compared to the enzyme’s main activity (48). We tested both the proposed Ps3′OMT and a GfOMT1 mutant (52) that has been shown to O-methylate at several positions of the 1-BIA scaffold, including the 3′ position, and we observed no activity in the context of our constructed THP pathway in yeast. While the putative Ps3′OMT activity was not verified on the pathway substrates, the difference in the observed activity could result from a number of factors, including differences in protein folding conditions, intracellular pH [6.8 for yeast (62, 63) vs. 7.7 for E. coli (64)], or other pathway and cellular contexts. Finally, earlier in vitro characterization studies of PsDBOX demonstrated that the conversion of THP to papaverine was 7% of the conversion of dihydrosanguinarine to sanguinarine (24). The relatively low activity of PsDBOX on THP taken together with the localization of PsDBOX to plant roots and the accumulation of papaverine in the aerial organs implies either that another enzyme is involved in papaverine synthesis in planta or that papaverine is transported from the roots, where PsDBOX is present, to the aerial organs.
This work does not evaluate or address the in planta synthesis route for papaverine. Instead, we sought to construct a biosynthetic route to THP and papaverine despite existing gaps in knowledge. The three main tools we used to address these gaps were protein engineering to alter the substrate specificity of enzymes, yeast MDR transporter knockouts to increase the flux through the pathway, and the addition of a chemical oxidation reaction in aqueous conditions to replace the final enzymatic oxidation step. Many enzymes essential to the production of key PNPs have yet to be elucidated—as for 3′-hydroxylation and O-methylation in this pathway—or cannot be functionally expressed in a heterologous microbial production platform—as we found to be the case for PsDBOX. Similar gaps in knowledge currently face heterologous microbial production systems for important PNP drugs like paclitaxel (65) and vinca alkaloids (66). The discovery of new plant enzymes has accelerated as genome and transcriptome sequencing become less expensive and more robust (26). Additionally, protein engineering strategies such as truncations to improve activity in a heterologous host and modifications to prevent product inhibition have been shown to significantly boost production of PNPs in microbial hosts (26). The production of THP in yeast with two enzymes engineered for altered substrate specificity demonstrates the utility of applying protein engineering on known enzymes that act on related secondary metabolites to replace the activity of unknown pathway enzymes.
In our work, we also show significant increases to THP production as a result of MDR transporter knockouts. These host strain modifications may have broad applications in optimizing flux through other complex PNP pathways reconstructed in yeast. Yeast MDR transporters have broad substrate specificity and generally export compounds with drug-like structures that may be harmful to the cell viability (67). Export by MDR transporters of pathway intermediates from the cytosol to the media results in those metabolites no longer being colocalized with pathway enzymes, thereby decreasing pathway flux. We hypothesized that knocking out one or more MDR transporters could increase the intracellular concentration of intermediate pathway metabolites without decreasing product export. Metabolite analyses showing elevated intracellular coclaurine concentrations and a 15-fold increase in THP titer when SNQ2 and PDR5 are knocked out aligned with that prediction. SNQ2 and PDR5 disruption could improve flux through pathways that include coclaurine, and the concept of knocking out MDR transporters in combination could be applicable to intermediates in other heterologous PNP pathways in yeast.
Our work on this biosynthetic pathway began, in part, because of the supply shortages of clinically significant products. Papaverine, cisatracurium, and atracurium, which are pharmaceuticals produced from THP, have important medicinal uses, but the supply of these drugs has been interrupted over the past decade. Our work to enable the microbial biosynthesis of THP and semisynthesis of papaverine provides an initial proof of concept of a microbial fermentation route that can be further advanced to improve the supply chain for these vital drugs. The final titer of biosynthesized THP in this work is only 10.6% as high as the titer of reticuline in the starting strain, underscoring the challenges of PNP production in heterologous systems with multiple enzymes modified for substrate specificity and the importance of future research focused on continued enzyme engineering improvements. We expect that further functional genomic discovery efforts may elucidate additional enzymes capable of performing the final oxidation step in papaverine production. Protein engineering of PsDBOX, or another functionally similar oxidase, could enable fully biosynthetic papaverine production. In the absence of an enzyme capable of carrying out the final step, the chemical oxidation reaction that we describe can be improved upon. Further investigation to identify improved oxidizing agents or buffer conditions could improve yields and conditions for commercial production. Finally, incorporating the advances of this work with other efforts to improve 1-BIA titers (39), as well as fermentation process development, could enable the efficient production of THP via yeast fermentation and ultimately improve the supply chain stability of several important pharmaceutical compounds.
Materials and Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized. The investigators were not blinded to allocation during experiments and outcome assessment.
Media, Chemicals, and Materials.
Difco yeast nitrogen base (YNB) without amino acids and ammonium sulfate, Bacto peptone, Bacto yeast extract, Luria Broth (LB), LB agar, dextrose, and galactose were obtained from BD (Becton, Dickinson and Company). Adenine hemisulfate, kanamycin monosulfate, and ampicillin were obtained from Sigma Chemicals. G418 (sc-29065A) used for yeast integration selections was obtained from Santa Cruz Biotechnology. Amino acid dropout medium was obtained from Takara Bio (product #630400 and #630431). A frozen-EZ transformation kit for yeast was obtained from Zymo Research. E. coli was selected on LB agar plates with 50 mg/L kanamycin and 75 mg/L carbenicillin and grown in LB liquid media with the appropriate antibiotic. A yeast 10× drop out (DO) supplement was prepared from Takara as a SC supplement with desired DO component omitted. S. cerevisiae strains were selected on YNB-DO (0.17% yeast nitrogen base, 0.5% ammonium sulfate, 2% dextrose, and 1× DO) agar or on YPD-G418 (1% yeast extract, 2% peptone, and 2% dextrose, 250 µ/mL geneticin). Yeast was grown in selective YNB-DO.
(S)-Reticuline and THP were purchased from Toronto Research Chemicals. (S)-Norreticuline was purchased from Alfa Chemistry. Papaverine hydrochloride, S-adenosylmethionine (SAM), hydrogen peroxide (30% wt/wt), and potassium persulfate were purchased from Sigma-Aldrich. Tris base was purchased from Fisher Scientific International, Inc. The 5 N hydrochloric acid was purchased from VWR International. BL21(DE3) E. coli cells were purchased from Invitrogen. Ni-NTA resin was purchased from Fisher Scientific International, Inc. Amicon 30-kDa cutoff spin filters were purchased from EMD Millipore.
Plasmid Construction.
Strains and plasmids used and constructed in this work are described in SI Appendix, Tables S1–S3. E. coli strain TOP10 (Life Technologies) was used for cloning and amplification of plasmids. Plasmids were recovered using Econospin columns (Epoch Life Sciences) according to manufacturer’s instructions. Oligonucleotides were synthesized by the Stanford Protein and Nucleic Acid Facility. PCR was carried out using Q5 DNA Polymerase (New England Biolabs) unless otherwise specified, and all restriction enzymes, the T4 DNA ligase, and deoxynucleotides were purchased from New England Biolabs. Heterologous gene sequences were cloned from previously published plasmids, obtained from Addgene, or synthesized by Twist Bioscience. Cloning was exclusively performed using Gibson assembly (68) followed by transformation into E. coli or by gap-repair directly into yeast or direct genomic integration. Guide RNA (gRNA) integration plasmids were constructed from Addgene plasmid pCAS, which was a gift from Jamie Cate (Addgene plasmid # 60847) (69). pCAS was modified by Gibson assembly to create SpCas9 expression vector pCS3410, which was digested with PacI followed by Gibson assembly of each gRNA fragment. All gRNA sequences used in this work are listed in SI Appendix, Table S3.
Mutagenesis Experiments.
To generate genomically integrated libraries of the EcNMCH, error-prone PCR was performed using Taq polymerase and TriLink Biotechnologies mutagenic dNTPs (item #2748 and #2746) according to the manufacturer’s instructions. Error-prone PCR was used to amplify ∼100-base pair (bp) DNA fragments encoding the peptide chain of interest and overhangs for homologous recombination using conditions generating, on average, one to two mutations per fragment. The fragments were transformed into CSY1172 along with a plasmid encoding CRISPR-Cas9 and a gRNA targeting the corresponding chain. The CRISPR gRNA targets double-stranded breaks in the DNA of each chain, and when the breaks are repaired by the error-containing PCR fragments via homologous recombination, further cutting is inhibited by mutations in the region targeted by the CRISPR gRNA. The transformation therefore results in colonies with genomically integrated mutations in the desired chains. Libraries of TfS9OMT were generated by PCR amplification using Taq polymerase and TriLink Biotechnologies mutagenic dNTPs (item #2748 and #2746) according to the manufacturer’s instructions. Three separate PCR reactions were performed at 20 cycles with varying concentrations of equimolar nucleotide analogs according to manufacturer instructions. Amplicons from mutagenic PCR generally were at least 75 bp long and included up to 10 bp upstream and downstream of the indicated region when necessary to meet that length. Products from each mutagenic PCR were amplified further using Taq polymerase. For plasmid-based expression, the amplified DNA was incorporated into the full TfS9OMT gene using overlap-extension PCR and incorporated into plasmids using Gibson assembly. TfS9OMT NNK libraries were generated with primers containing NNK-randomized regions at the desired codon. These primers were used to amplify two fragments, as follows: the front of the gene up to the NNK codon with 15 additional bp of overhang and the back of the gene from 15 bp before the NNK codon to the end of the gene. These two amplicons were amplified, and Gibson overhangs were attached using overlap-extension PCR. The DNA purification columns, Zymoclean gel DNA recovery kit, and yeast genomic DNA prep kit (D2002) were all obtained from Zymo Research. All routine sequencing was performed using Quintara Biosciences.
DNA Shuffling.
Variants combined using DNA shuffling were amplified using PCR and pooled in an equimolar mixture with a final DNA concentration of 200 ng/µL. Separately, a DNase I (Zymo Research E1010) solution was prepared with 167 mM Tris⋅HCl buffer, 83.3 mM manganese chloride, and 1.67 U/mL DNase I. A total of 30 µL DNase I solution was added to 70 µL of the equimolar amplicon mixture, and the reactions were incubated for 60 s at 15 °C before being quenched with 6 µL of 500 mM EDTA. The resulting DNA fragments were separated using an agarose gel. Fragments between 50 bp and 150 bp in length were cut from the gel and purified using the Zymoclean Gel DNA Recovery Kit (Zymo Research). DNA fragments were reassembled using the following cycling procedure: 1×: 96 °C for 2 min; 35×: 95 °C for 15 s → 65 °C for 30 s → 62 °C for 30 s → 59 °C for 30 s → 56 °C for 30 s → 53 °C for 30 s → 50 °C for 30 s → 47 °C for 30 s → 44 °C for 30 s → 41 °C for 30 s → 72 °C for 1 min; 1×: 72 °C for 3 min. A second PCR with terminal primers was used to lift full-length sequences and attach overhangs for Gibson assembly.
Growth Conditions for Metabolite Production.
For this study, all strains were grown for 3 d at 30 °C unless otherwise noted, shaking at either 250 rpm (for 1-mL cultures or larger) or 460 rpm (for 300-µL cultures). Yeast was grown in YP base (10 g yeast extract, 20 g peptone, and 80mg adenine hemisulfate for 1 L media) or YNB (1.7 g yeast nitrogen base, 5 g ammonium sulfate for 1 L media, pH to 5.8 using NaOH). The concentration of the carbon source was 2% (wt/vol) in all media with a single carbon source present. Dextrose was the carbon source unless otherwise noted. For samples with multiple carbon sources, the media contained 2% (wt/vol) dextrose and the concentration of the secondary carbon source was also 2% (wt/vol). SC medium was prepared by combining seven parts YNB, one part 10× amino acid supplement mix, one part water, and one part dextrose. SD medium was prepared similarly using 10× amino acid supplement mixes lacking either uracil or tryptophan depending on the auxotrophic marker on the plasmid being used.
Intracellular Metabolite Extraction.
For intracellular metabolite analysis, 50-mL cultures were grown for 3 d at 30 °C before being centrifuged at 3,500 g for 10 min at 4 °C. A total of 1 mL was collected for extracellular metabolite analysis. The cells were then washed twice by resuspending in 5 mL water, centrifuging at 3,500 g for 10 min at 4 °C, and removing the supernatant. A sample of the wash after centrifugation was collected each time. The cells were resuspended in 1 mL 50% methanol, and 250 µL 0.5 mm glass beads (USA Scientific #7400-2405) were added. Samples were then vortexed at maximum speed for 2 h with an ambient temperature of 4 °C (Scientific Industries Vortex-Genie 2, SI-0236; TurboMix attachment, SI-0564) before being centrifuged at 20,000 g for 90 min. The supernatant was collected for analysis by LC-MS/MS. To compare intracellular and extracellular concentrations, we calculated, as follows, the total intracellular volume of cells in a 50-mL culture using the average volume of a yeast cell (70), number of yeast cells per optical density at 600 nm (OD600) unit (71), and the OD600 measurement of the culture:
Our estimate for an intracellular volume of 105 µL indicates that our procedure of extracting lysing 50 mL of cells in a volume of 1 mL results in a 1:9.5 dilution of the intracellular metabolites. We multiplied intracellular metabolite concentrations by 9.5 to more accurately compare them to the metabolite concentrations measured in the media.
Chemical Oxidation Reactions.
Chemically synthesized THP was used in initial experiments to determine favorable reaction conditions. Final concentrations were 500 nM THP and 75 mM Tris⋅HCl buffer (pH as indicated), and oxidizing agent concentrations were as indicated in a total reaction volume of 200 µL. Reactions were incubated at the indicated temperature with shaking at 600 rpm using an Eppendorf Thermomixer F1.5. Reactions were quenched by adding an equal volume of 1 M sodium thiosulfate and stored at 4 °C before analysis via LC-MS/MS. Reactions with biosynthetically produced THP contained three components, as follows: spent yeast media containing THP, Tris⋅HCl buffer, and hydrogen peroxide. The sum of the volume of the first two components was always 190 µL at the ratio indicated (e.g., 40:150 media-to-buffer ratio means 40 µL media and 150 µL buffer were used). A total of 10 µL of concentrated a hydrogen peroxide solution was then added for a total reaction volume of 200 µL. These reactions were similarly incubated, quenched, and stored at 4 °C prior to analysis via LC-MS/MS.
Supplementary Material
Acknowledgments
Funding for this work was provided by NIH Grant AT007886 to C.D.S. We thank Prashanth Srinivasan and Deze Kong for invaluable discussion and guidance during the preparation of this project.
Footnotes
Competing interest statement: C.D.S. is a founder and CEO of Antheia, Inc; there is a pending patent application on aspects of the work.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2205848119/-/DCSupplemental.
Data, Materials, and Software Availability
All data are included in the manuscript and/or SI Appendix.
References
- 1.Theis N., Lerdau M., The evolution of function in plant secondary metabolites. Int. J. Plant Sci. 164, S93–S102 (2003). [Google Scholar]
- 2.Atanasov A. G., et al. , Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 33, 1582–1614 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagel J. M., Facchini P. J., Benzylisoquinoline alkaloid metabolism: A century of discovery and a brave new world. Plant Cell Physiol. 54, 647–672 (2013). [DOI] [PubMed] [Google Scholar]
- 4.Pferschy-Wenzig E.-M., Bauer R., The relevance of pharmacognosy in pharmacological research on herbal medicinal products. Epilepsy Behav. 52, 344–362 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Sneader W., Drug Discovery: A History (John Wiley & Sons, 2005). [Google Scholar]
- 6.Hill D. A., Turner G. L., Neuromuscular blocking agents US Patent 5,453,510 (1995) Accessed 3 November, 2021.
- 7.Renew J. R., Clinical use of neuromuscular blocking agents in anesthesia. UpToDate (2021). https://www.uptodate.com/contents/clinical-use-of-neuromuscular-blocking-agents-in-anesthesia#:~:text=Neuromuscular%20blocking%20agents%20(NMBAs)%20are,or%20to%20improve%20surgical%20conditions. Accessed 12 February 2022.
- 8.Bohand X., Jordan D., Dubois F., Managing the risk of shortages and medication errors with curares during the COVID-19 pandemic: A hospital pharmacy experience. Eur. J. Hosp. Pharm. 31, ejhpharm-2020-002605 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bell S. G., Use of a papaverine solution in peripheral arterial lines. Neonatal Netw. 27, 65–66 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Baltsavias G., Yella S., Al Shameri R. A., Luft A., Valavanis A., Intra-arterial administration of papaverine during mechanical thrombectomy for acute ischemic stroke. J. Stroke Cerebrovasc. Dis. 24, 41–47 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Girard D. S., et al. , Papaverine delivery to the internal mammary artery pedicle effectively treats spasm. Ann. Thorac. Surg. 78, 1295–1298 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Sajadian S., et al. , Cell cycle arrest and apoptogenic properties of opium alkaloids noscapine and papaverine on breast cancer stem cells. Toxicol. Mech. Methods 25, 388–395 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Aggarwal M., Leser G. P., Lamb R. A., Repurposing papaverine as an antiviral agent against influenza viruses and paramyxoviruses. J. Virol. 94, e01888-e19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wyde P. R., Gilbert B. E., Ambrose M. W., Comparison of the anti-respiratory syncytial virus activity and toxicity of papaverine hydrochloride and pyrazofurin in vitro and in vivo. Antiviral Res. 11, 15–26 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Kang D., Qiang G.-F., Du L.-D., Du G.-H., “Papaverine” in Natural Small Molecule Drugs from Plants, Du G.-H., Ed. (Springer, 2018), pp. 109–114. [Google Scholar]
- 16.United Nations Office on Drugs and Crime. “Bulletin on narcotics” (U. N. Off. Drugs Crime, Issue 3, 1952) Accessed 2 August, 2021.
- 17.Dawson W., An examination of synthetic routes to papaverine, and, synthetic studies in 1:4 oxazine chemistry, ProQuest Dissertations and Theses. https://theses.gla.ac.uk/78871/1/13838418.pdf. Accessed 2 August 2021.
- 18.Ricci J. A., et al. , Comparing the outcomes of different agents to treat vasospasm at microsurgical anastomosis during the papaverine shortage. Plast. Reconstr. Surg. 138, 401e–408e (2016). [DOI] [PubMed] [Google Scholar]
- 19.Singh A., Menéndez-Perdomo I. M., Facchini P. J., Benzylisoquinoline alkaloid biosynthesis in opium poppy: An update. Phytochem. Rev. 18, 1457–1482 (2019). [Google Scholar]
- 20.Desgagné-Penix I., Facchini P. J., Systematic silencing of benzylisoquinoline alkaloid biosynthetic genes reveals the major route to papaverine in opium poppy. Plant J. 72, 331–344 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Pathak S., et al. , Comparative transcriptome analysis using high papaverine mutant of Papaver somniferum reveals pathway and uncharacterized steps of papaverine biosynthesis. PLoS One 8, e65622 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han X., et al. , The biosynthesis of papaverine proceeds via (S)-reticuline. Phytochemistry 71, 1305–1312 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agarwal P., et al. , 3‘O-Methyltransferase, Ps3’OMT, from opium poppy: Involvement in papaverine biosynthesis. Plant Cell Rep. 38, 1235–1248 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Hagel J. M., et al. , Characterization of a flavoprotein oxidase from opium poppy catalyzing the final steps in sanguinarine and papaverine biosynthesis. J. Biol. Chem. 287, 42972–42983 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agarwal P., et al. , Short-chain dehydrogenase/reductase, PsDeHase, from opium poppy: Putative involvement in papaverine biosynthesis. Plant Cell Tissue Organ Cult. Plant Cell Tissue Organ Cult. 143, 431–440 (2020). [Google Scholar]
- 26.Cravens A., Payne J., Smolke C. D., Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 10, 2142 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y., Smolke C. D., Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat. Commun. 7, 12137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Srinivasan P., Smolke C. D., Biosynthesis of medicinal tropane alkaloids in yeast. Nature 585, 614–619 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun W., Zhao Y., Li C., De Novo Synthesis of Plant Natural Products in Yeast (IntechOpen, 2019). [Google Scholar]
- 30.Ryan O. W., et al. , Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife 3, e03703 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo L., et al. , The opium poppy genome and morphinan production. Science 362, 343–347 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Goffeau A., et al. , Life with 6000 genes. Science 274, 546, 563–567 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Sedeek K. E. M., Mahas A., Mahfouz M., Plant genome engineering for targeted improvement of crop traits. Front Plant Sci 10, 114 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galanie S., Thodey K., Trenchard I. J., Filsinger Interrante M., Smolke C. D., Complete biosynthesis of opioids in yeast. Science 349, 1095–1100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y., Li S., Thodey K., Trenchard I., Cravens A., Smolke C. D., Complete biosynthesis of noscapine and halogenated alkaloids in yeast. Proc. Natl. Acad. Sci. USA 115, E3922–E3931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paddon C. J., et al. , High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Brown S., Clastre M., Courdavault V., O’Connor S. E., De novo production of the plant-derived alkaloid strictosidine in yeast. Proc. Natl. Acad. Sci. U.S.A. 112, 3205–3210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Westfall P. J., et al. , Production of amorphadiene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin. Proc. Natl. Acad. Sci. U.S.A. 109, E111–E118 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pyne M. E., et al. , A yeast platform for high-level synthesis of tetrahydroisoquinoline alkaloids. Nat. Commun. 11, 3337 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trenchard I. J., Siddiqui M. S., Thodey K., Smolke C. D., De novo production of the key branch point benzylisoquinoline alkaloid reticuline in yeast. Metab. Eng. 31, 74–83 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Entian K.-D., Kötter P., 25 yeast genetic strain and plasmid collections. Methods Microbiol 36, 629–666 (2007). [Google Scholar]
- 42.Schmidheini T., Sperisen P., Paravicini G., Hütter R., Braus G., A single point mutation results in a constitutively activated and feedback-resistant chorismate mutase of Saccharomyces cerevisiae. J. Bacteriol. 171, 1245–1253 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fukuda K., Watanabe M., Asano K., Ouchi K., Takasawa S., A mutated ARO4 gene for feedback-resistant DAHP synthase which causes both o-fluoro-DL-phenylalanine resistance and beta-phenethyl-alcohol overproduction in Saccharomyces cerevisiae. Curr. Genet. 20, 453–456 (1991). [DOI] [PubMed] [Google Scholar]
- 44.Nishihachijo M., et al. , Asymmetric synthesis of tetrahydroisoquinolines by enzymatic Pictet-Spengler reaction. Biosci. Biotechnol. Biochem. 78, 701–707 (2014). [DOI] [PubMed] [Google Scholar]
- 45.Xu J., Mcpartlon M., Li J., Improved protein structure prediction by deep learning irrespective of co-evolution information. Nat. Mach. Intell. 3, 601–609 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grosdidier A., Zoete V., Michielin O., SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acids Res. 39, W270–W277 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pienkny S., Brandt W., Schmidt J., Kramell R., Ziegler J., Functional characterization of a novel benzylisoquinoline O-methyltransferase suggests its involvement in papaverine biosynthesis in opium poppy (Papaver somniferum L). Plant J. 60, 56–67 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Dang T.-T. T., Facchini P. J., Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol. 159, 618–631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chang L., Hagel J. M., Facchini P. J., Isolation and characterization of O-methyltransferases involved in the biosynthesis of glaucine in Glaucium flavum. Plant Physiol. 169, 1127–1140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Valentic T. R., Payne J. T., Smolke C. D., Structure-guided engineering of a scoulerine 9-O-methyltransferase enables the biosynthesis of tetrahydropalmatrubine and tetrahydropalmatine in yeast. ACS Catal. 10, 4497–4509 (2020). [Google Scholar]
- 51.Joern J. M., “DNA shuffling” in Directed Evolution Library Creation: Methods and Protocols, Arnold F. H., Georgiou G., Eds. (Methods in Molecular Biology, Humana Press, 2003), pp. 85–89. [DOI] [PubMed] [Google Scholar]
- 52.d’Oelsnitz S., et al. , Using structurally fungible biosensors to evolve improved alkaloid biosyntheses bioXriv [Preprint] (2021). 10.1101/2021.06.07.447399. Accessed 15 January 2022. [DOI] [PMC free article] [PubMed]
- 53.Decottignies A., Goffeau A., Complete inventory of the yeast ABC proteins. Nat. Genet. 15, 137–145. (2021). [DOI] [PubMed] [Google Scholar]
- 54.Demir A. B., Koc A., High-copy overexpression screening reveals PDR5 as the main doxorubicin resistance gene in yeast. PLoS One 10, e0145108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rogers B., et al. , The pleitropic drug ABC transporters from Saccharomyces cerevisiae. J. Mol. Microbiol. Biotechnol. 3, 207–214 (2001). [PubMed] [Google Scholar]
- 56.Harris A., et al. , Structure and efflux mechanism of the yeast pleiotropic drug resistance transporter Pdr5. Nat. Commun. 12, 5254 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L. S., Szczypka M., Lu Y. P., Thiele D. J., Rea P. A., The yeast cadmium factor protein (YCF1) is a vacuolar glutathione S-conjugate pump. J. Biol. Chem. 27, 6509–6517 (1996). [DOI] [PubMed] [Google Scholar]
- 58.Lalwani M. A., Zhao E. M., Avalos J. L., Current and future modalities of dynamic control in metabolic engineering. Curr. Opin. Biotechnol. 52, 56–65 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Payne J. T., Valentic T. R., Smolke C. D., Complete biosynthesis of the bisbenzylisoquinoline alkaloids guattegaumerine and berbamunine in yeast. Proc. Natl. Acad. Sci. U.S.A. 118, e2112520118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han J., Wu Y., Zhou Y., Li S., Engineering Saccharomyces cerevisiae to produce plant benzylisoquinoline alkaloids. Abiotech 2, 264–275 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pauli H. H., Kutchan T. M., Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J. 13, 793–801 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Imai T., Ohno T., Measurement of yeast intracellular pH by image processing and the change it undergoes during growth phase. J. Biotechnol. 38, 165–172 (1995). [DOI] [PubMed] [Google Scholar]
- 63.Valli M., et al. , Intracellular pH distribution in Saccharomyces cerevisiae cell populations, analyzed by flow cytometry. Appl. Environ. Microbiol. 71, 1515–1521 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Booth I. R., “The regulation of intracellular pH in bacteria” in Novartis Foundation Symposium 221 - Bacterial Responses to PH (John Wiley & Sons, Ltd), Edited by: Derek J. Chadwick and Gail Cardew pp. 19–37 (2007). [DOI] [PubMed]
- 65.Li Y., Zhang G., Pfeifer B. A., Current and emerging options for taxol production. Adv. Biochem. Eng. Biotechnol. 148, 405–425 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Zhang D., Kanakkanthara A., Beyond the paclitaxel and vinca alkaloids: Next generation of plant-derived microtubule-targeting agents with potential anticancer activity. Cancers (Basel) 12, 1721 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kolaczkowski M., Kolaczowska A., Luczynski J., Witek S., Goffeau A., In vivo characterization of the drug resistance profile of the major ABC transporters and other components of the yeast pleiotropic drug resistance network. Microb. Drug Resist. 4, 143–158 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Gibson D. G., et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Ryan O. W., Cate J. H. D., Multiplex engineering of industrial yeast genomes using CRISPRm. Methods Enzymol. 546, 473–489 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Jorgensen P., Nishikawa J. L., Breitkreutz B.-J., Tyers M., Systematic identification of pathways that couple cell growth and division in yeast. Science 297, 395–400 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Sherman F., [1] Getting started with yeast. Methods Enzymol. 194, 3–21 (1991). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and/or SI Appendix.