Significance
The molecular mechanisms that regulate the “choice” for singular gene activation among multicopy gene families remains an unsolved mystery in eukaryotic gene expression. Immune evasion of Plasmodium falciparum, the parasite responsible for the deadliest form of human malaria, depends on its ability to switch between variable surface antigens, expressed in a mutually exclusive manner by the var gene family. We found that long-noncoding RNAs that mark the single active var gene are associated with redox sensor, PfTPx-1, at the site of active transcription, in close proximity to the enzyme responsible for providing methyl groups for epigenetic memory. We demonstrate that PfTPx-1 plays a pivotal role in var gene expression, possibly by providing optimal conditions for their epigenetic regulation.
Keywords: malaria, Plasmodium falciparum, var genes, lncRNA, thioredoxin peroxidase
Abstract
The virulence of Plasmodium falciparum, which causes the deadliest form of human malaria, is attributed to its ability to evade the human immune response. These parasites “choose” to express a single variant from a repertoire of surface antigens called PfEMP1, which are placed on the surface of the infected red cell. Immune evasion is achieved by switches in expression between var genes, each encoding a different PfEMP1 variant. While the mechanisms that regulate mutually exclusive expression of var genes are still elusive, antisense long-noncoding RNAs (lncRNAs) transcribed from the intron of the active var gene were implicated in the “choice” of the single active var gene. Here, we show that this lncRNA colocalizes with the site of var mRNA transcription and is anchored to the var locus via DNA:RNA interactions. We define the var lncRNA interactome and identify a redox sensor, P. falciparum thioredoxin peroxidase I (PfTPx-1), as one of the proteins associated with the var antisense lncRNA. We show that PfTPx-1 localizes to a nuclear subcompartment associated with active transcription on the nuclear periphery, in ring-stage parasite, when var transcription occurs. In addition, PfTPx-1 colocalizes with S-adenosylmethionine synthetase (PfSAMS) in the nucleus, and its overexpression leads to activation of var2csa, similar to overexpression of PfSAMS. Furthermore, we show that PfTPx-1 knockdown alters the var switch rate as well as activation of additional gene subsets. Taken together, our data indicate that nuclear PfTPx-1 plays a role in gene activation possibly by providing a redox-controlled nuclear microenvironment ideal for active transcription.
Malaria remains a serious global health issue with 241 million cases and an estimated 627,000 deaths per year (1), mostly caused by the protozoan parasite, Plasmodium falciparum. The virulence of P. falciparum is attributed to its ability to modify the infected red blood cells (iRBCs), causing them to adhere to endothelial receptors, thereby removing the iRBCs from the circulation and preventing their clearance by the spleen and at the same time causing local inflammation and blockage of blood vessels. The major ligand responsible for the cytoadhesive properties of the iRBCs is P. falciparum erythrocyte membrane protein 1 (PfEMP1), which is expressed by the parasite, placed on the iRBC surface, and binds to host endothelial receptors. PfEMP1 is encoded by a multicopy gene family named var. Each var gene encodes a different PfEMP1 variant, and the var gene repertoire in different parasite isolates may vary between 47 and 90 genes per parasite genome (2). To avoid unnecessary exposure of its antigenic repertoire, var gene expression has evolved so that each parasite expresses a single var gene at a time and maintains the rest of the gene family transcriptionally silent. Once the human immune system recognizes PfEMP1, antibodies are generated to target the invading pathogen and often significantly reduce the number of infected cells. However, a small subset of parasites switches expression to a different PfEMP1 variant, thereby avoiding antibody recognition and continuing the infection. Thus, the ability of the parasite to evade the antibody-mediated response against PfEMP1 relies on mutually exclusive var gene expression and antigenic switches in expression between different var genes.
The mechanism of mutually exclusive var gene expression in human infections is not completely understood; however, it is epigenetically regulated and involves histone modifications, promoter pairing, and long-noncoding RNAs (lncRNAs) (3). The promoter found within the var intron is bidirectional and is responsible for the production of long-noncoding transcripts, which are capped but not polyadenylated (4), suggesting that they are retained and function in the nucleus. The antisense lncRNA is transcribed by the single active var gene at the ring stage when the var mRNA is produced. Ectopic expression of var-specific antisense lncRNAs activates a silent gene in a dose-dependent manner, while interference with the endogenous lncRNA leads to transcriptional down-regulation of the active var gene, switching, and loss of epigenetic memory (5). However, the mechanism by which the lncRNA facilitates var gene activation is not understood. In other organisms, lncRNAs are implicated in recruitment of protein complexes important for chromatin modifications and transcriptional control (6). We hypothesized that the var antisense lncRNA could similarly function to recruit factors important for var expression.
We uncovered the antisense lncRNA interactome, which includes thioredoxin peroxidase I (PfTPx-1), a well-studied redox protein that is responsible for H2O2 homeostasis and redox relays to downstream proteins (7–14). During the intraerythrocytic developmental cycle, the parasite is under constant oxidative stress from both inside and outside the RBC. As the parasite matures, it produces reactive oxygen species (ROS) from mitochondrial electron transport and from digestion of hemoglobin as a food source (15, 16). The parasite also encounters high levels of ROS from antimalarial compounds, high fevers, and immune cells. However, little is known about redox sensing and regulation of gene expression in P. falciparum. We found that in addition to its cytoplasmic role, PfTPx-1 is also located in a subcellular nuclear compartment of active transcription that includes the var expression site. Furthermore, alterations in PfTPx-1 expression change var gene switching and influence transcriptional activation of additional gene subsets. Taken together, these results demonstrate a role for PfTPx-1 in gene regulation at a particular subnuclear compartment, potentially by creating an optimal niche for transcription.
Results
var Antisense lncRNAs Colocalize with the Site of var mRNA Transcription and Are Anchored via DNA:RNA Interactions.
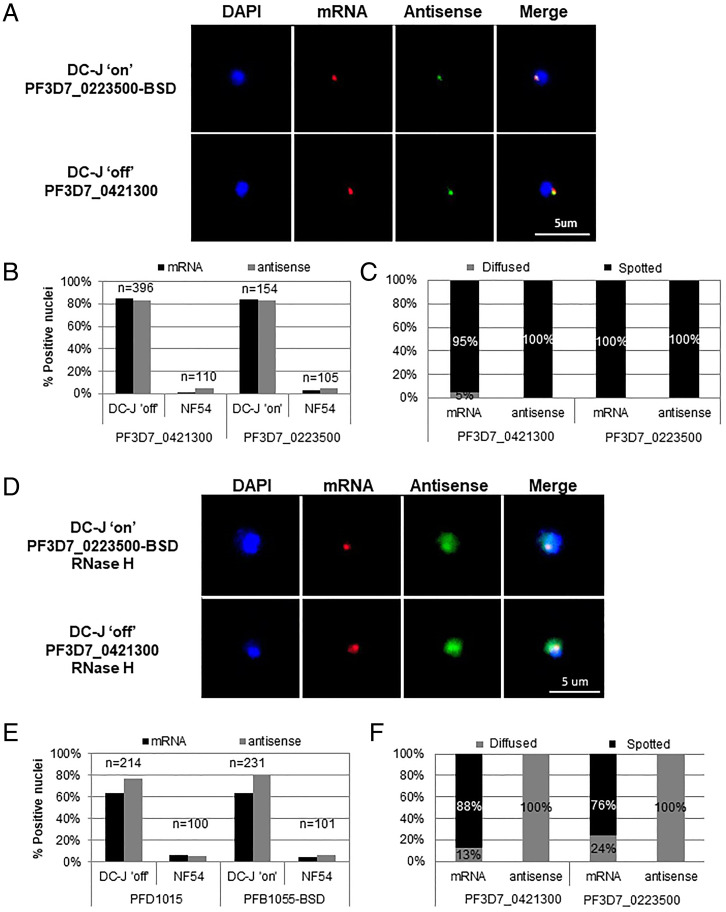
We have previously shown that the antisense lncRNA is expressed from the promoter found within the intron of the active var gene (5). This lncRNA is incorporated into the chromatin and plays an important role in the activation of the single var gene that is expressed (5). To localize the antisense lncRNA transcript within the parasite nucleus, we performed two-color RNA-fluorescence in situ hybridization (FISH) with strand-specific RNA probes in the DC-J transgenic parasite line (17). This line allows for activation of a specific var gene (PF3D7_0223500-BSD) by culturing the parasites in the presence of blasticidin-S-HCl (DC-J “ON”), whereas a clonal parasite population grown in the absence of blasticidin (DC-J “OFF”) expresses the PF3D7_0421300 var gene. Previous work showed that var transcripts localize to a specific nuclear site, which is distinct from the location of transcripts of unrelated genes (18). We observed colocalization of the var mRNA and antisense lncRNA at the nuclear periphery during the ring stage in both parasite populations (DC-J ON and OFF), indicating that the lncRNA is found in close proximity to the nascent var mRNA at the var expression site (Fig. 1 A and B). The vast majority of the signals observed were characteristically condensed spots with fewer than 5% showing a diffused pattern (Fig. 1C). However, when performing similar RNA-FISH using RNase H treatment to break up DNA:RNA hybrids, we found that 100% of the antisense lncRNA signals became diffused, while the majority of the var mRNA signals remained in condensed spots (Fig. 1 D–F). This result supports the idea that the antisense lncRNA is anchored to the var expression site via DNA:RNA interaction, possibly labeling the active locus.
Fig. 1.
var antisense lncRNAs colocalize with the site of var mRNA transcription and are anchored via DNA:RNA interactions. Dual color RNA-FISH of the var antisense lncRNA and the var mRNA of two different active var genes in tightly synchronized ring stage parasites at ∼18 h postinvasion. Nuclei are stained with DAPI (blue). (A) Dual color RNA-FISH showing the nuclear positioning of messenger RNA (red) and antisense ncRNA (green) actively transcribed from the PF3D7_0223500-BSD (Upper) or PF3D7_0421300 (Lower) var genes. (B) Quantification of positive nuclei shown in A. (C) Quantification of fluorescent signal appearance (diffused vs. spotted) of positive nuclei shown in A. (D) Nuclear positioning of mRNA (red) and antisense ncRNA (green) actively transcribed from the PF3D7_0223500-BSD (Upper) or PF3D7_0421300 (Lower) var genes after treatment with 120U of RNase-H. (E) Quantification of positive nuclei shown in D. (F) Quantification of fluorescent signal appearance (diffused vs. spotted) of positive nuclei shown in D. (Scale bars, 5 µm.)
The Conserved Region of the var Antisense lncRNA Is Predicted to Form Secondary RNA Structures.
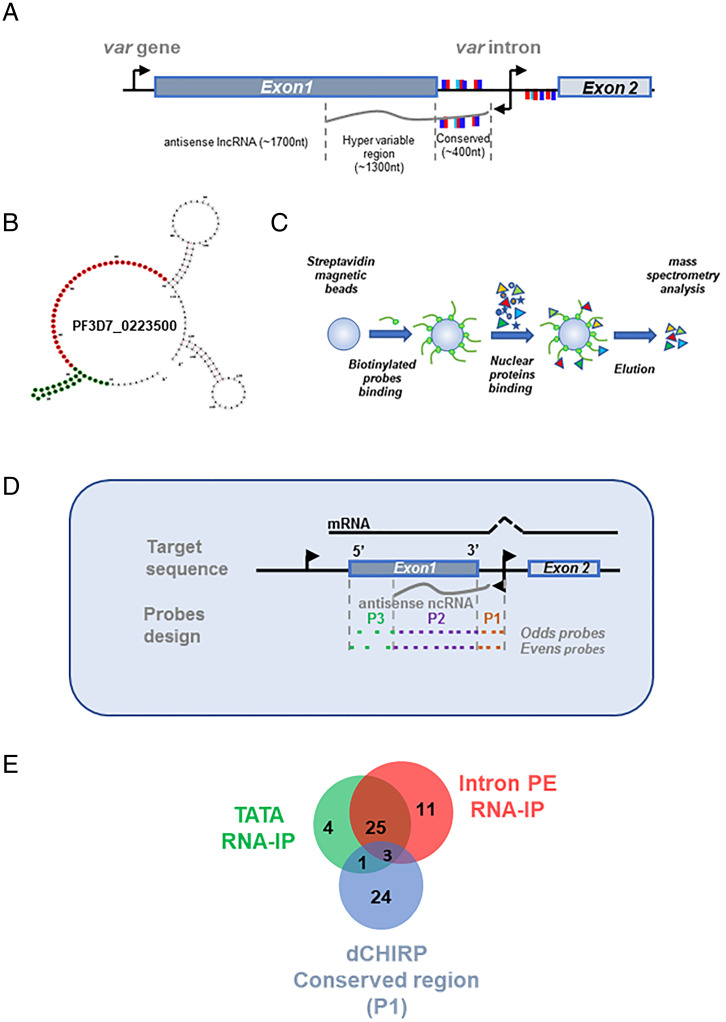
The antisense lncRNA transcribed by var introns is 1.7- to 2.5-kb long and contains sequences from the intron as well as the 3′ region of exon 1 (4) (Fig. 2A). The intronic part of the RNA transcript is conserved among the var gene family and contains the sequence of the pairing elements, which were shown to be essential for var gene silencing and mutually exclusive expression (19). On the other hand, the transcript encoded by the 3′ of exon 1 contains hypervariable sequences that may contribute to the “choice” for specific var gene activation. We hypothesized that the var antisense lncRNA may contribute to var gene activation by forming a DNA:RNA hybrid with its gene-specific sequence and by recruiting activation factors to the “chosen” locus through its conserved region. Using a bioinformatic tool for structural RNA prediction (RNAalifold Server of the ViennaRNA WEB), we analyzed the antisense lncRNA conserved region from the entire var gene family. As expected, given its sequence conservation, the analysis revealed a unique secondary structure composed of a loop and a highly preserved stem at the first 20 nt, which contains a repetitive TATA sequence (4) (Fig. 2B, green). This basic structure of the loop and the conserved stem is maintained among all var genes, even though the number of stems vary (SI Appendix, Fig. S1).
Fig. 2.
The conserved region of the var antisense lncRNA is predicted to form secondary RNA structure and binds specific nuclear proteins. (A) Schematic diagram of var gene structure. The antisense lncRNA (gray line, ∼1.7 kb in length) is transcribed from a bidirectional promoter located at the var intron (black arrows) and can be divided into two regions: conserved region complimentary to the var intron (∼400 nt in length) and a hypervariable region complimentary to exon 1 (∼1,300 nt in length). The pairing element (PE) repeats of the var intron are marked with light blue, dark blue, and red rectangles. The presentation of the PE motifs above or below the genes represents their orientation on the DNA strands and on the lncRNA. (B) Secondary structure of PF3D7_0223500-BSD var antisense conserved region. Positions marked in green served for the antisense UA-RNA stem-loop probe (TATA) and positions marked in red served for the antisense PE RNA probe (IntPE). Prediction of secondary structures was done using the RNAfold Server of the ViennaRNA WEB. (C) Schematic diagram of affinity chromatography assay using streptavidin coated magnetic beads (blue circle), biotinylated nucleic acid probes (green circle and orange line, respectively), and nuclear extract (colored shapes). Purified proteins were analyzed using MS. Affinity chromatography was performed with nuclear extract produced from DC-J ON parasite line in which the chromosomal bsd cassette (PF3D7_0223500-BSD) is active. (D) dChIRP was performed using the DC-J OFF parasite line in which the var gene, PF3D7_0421300, is active. Two sets of probes were designed to bind the antisense lncRNA of PF3D7_0421300 (odds and evens) and were divided to three pools; probes designed to bind the conserved region of the antisense lncRNA (P1), probes designed to bind the hypervariable region of the antisense lncRNA (P2), and probes that are complementary to the 5′ edge of exon 1 in which the var lncRNA antisense does not reach (P3). Tightly synchronized ring-stage parasites at ∼18 h postinvasion were cross-linked and sonicated. Biotinylated probes were incubated with parasite extract and were bound to streptavidin magnetic beads. Proteins bound in each pool were identified using LC-MS/MS. (E) Venn diagram depicting the overlap in proteins identified using the three experimental approaches: TATA-stem loop affinity chromatography, IntPE single-stranded RNA affinity chromatography, and dCHIRP.
The Conserved Region of var Antisense lncRNA Binds Specific Nuclear Proteins.
As a first step to identify the nuclear proteins that are recruited to the active var gene via the antisense lncRNA, we used biotinylated RNA probes specific to either the UA-RNA stem-loop (TATA) region or to the pairing element sequences (IntPE) of the conserved region of the antisense lncRNA of PF3D7_0223500 (Fig. 2B and SI Appendix, Table S1). A non-Plasmodium RNA sequence (HSL RNA) was used as a control. These probes were incubated with nuclear extract taken from ring-stage parasites (Fig. 2C). Identification of bound proteins by liquid chromatography tandem mass spectrometry (LC-MS/MS) revealed 33 and 39 proteins that were exclusively bound, or enriched more than 10-fold, to the TATA and PE probes, respectively (SI Appendix, Table S2 and S3). Interestingly, 28 proteins were shared between the two experiments, and among them are proteins involved in DNA:RNA binding, cellular metabolism, and translation (Table 1). To confirm that the proteins identified in the in vitro pull-down are present at the active var locus in cultured parasites, we performed domain-specific chromatin isolation by RNA purification, (dChIRP) (20, 21). RNA probes were designed for the conserved-intron region (P1), the hypervariable region (P2), and the 5′ region of exon 1 (P3), a region that does not have a complementary antisense transcript associated with it (Fig. 2D and SI Appendix, Table S4). It is worth noting that in this experiment, the variable P2 region is much larger than the conserved P1 region (206 bp vs. 1,663 bp). Nevertheless, P1 pulled down more proteins than P2 (55 vs. 19), and most of the P2-associated proteins were histones. These results support our hypothesis that P1 might be involved in recruiting proteins to the active var gene locus. We identified 28 proteins that were uniquely associated with the conserved P1 domain of the antisense lncRNA (Fig. 2E and SI Appendix, Tables S5 and S6). To narrow down our analysis, we looked at the overlap between the proteins identified by all three approaches and found three proteins: thioredoxin peroxidase I (PfTPx-1, PF3D7_1438900), ornithine aminotransferase (PfOAT, PF3D7_0608800), and elongation factor 2 (PF3D7_1451100) (Table 1, boldface). Given the reasons below, we chose to focus on PfTPx-1.
Table 1.
Nuclear proteins identified by affinity chromatography in both the TATA-stem loop and PE sequences of the antisense lncRNA
| Cellular process | PlasmoDB ID | Annotation | Enrichment | |
|---|---|---|---|---|
| IntPE | TATA | |||
| DNA/RNA binding | PF3D7_0919000 | Nucleosome assembly protein | * | * |
| PF3D7_1006200 | DNA/RNA-binding protein Alba 3 | * | * | |
| PF3D7_1011800 | PRE-binding protein | * | * | |
| PF3D7_1105100 | Histone H2B | 63-fold | 55-fold | |
| PF3D7_1202900 | High mobility group protein B1 | * | * | |
| PF3D7_1224300 | Polyadenylate-binding protein 1, putative | * | * | |
| PF3D7_1347500 | DNA/RNA-binding protein Alba 4 | * | * | |
| PF3D7_1426100 | Transcription factor BTF3, putative | * | * | |
| PF3D7_1438900 | Thioredoxin peroxidase 1 | 50-fold | 42-fold | |
| Metabolism | PF3D7_0513300 | Purine nucleoside phosphorylase | * | * |
| PF3D7_0608800 | Ornithine aminotransferase | * | * | |
| PF3D7_0922500 | Phosphoglycerate kinase | * | * | |
| PF3D7_1324900 | L-lactate dehydrogenase | * | * | |
| PF3D7_1444800 | Fructose-bisphosphate aldolase | * | * | |
| PF3D7_1462800 | Glyceraldehyde-3-phosphate dehydrogenase | * | * | |
| Translation | PF3D7_0309600 | 60S acidic ribosomal protein P2 | * | * |
| PF3D7_0517000 | 60S ribosomal protein L12, putative | * | * | |
| PF3D7_0913200 | Elongation factor 1-β | * | * | |
| PF3D7_1357100 | Elongation factor 1-α | 22-fold | 12-fold | |
| PF3D7_1451100 | Elongation factor 2 | * | * | |
| Conserved unknown function | PF3D7_0721100 | Conserved protein, unknown function | * | * |
| PF3D7_0813300 | Conserved protein, unknown function | * | * | |
| PF3D7_1115800 | Conserved Plasmodium protein, unknown function | * | * | |
| Other | PF3D7_0505800 | Small ubiquitin-related modifier | * | * |
| PF3D7_0826700 | Receptor for activated c kinase | * | * | |
| PF3D7_1115600 | Peptidyl-prolyl cis-trans isomerase | * | * | |
| PF3D7_1129100 | Parasitophorous vacuolar protein 1 | * | * | |
| PF3D7_1229400 | Macrophage migration inhibitory factor | * | * | |
Proteins were grouped by function. The three proteins shaded in boldface were also identified using the dCHIRP methodology. A full list of proteins identified in each method can be found in SI Appendix, Tables S2, S3, S5, and S6. Asterisks (*) denotes proteins exclusively found in antisense specific versus control probes.
PfTPx-1 Is Localized to the Active var Expression Site in Ring-Stage Parasites.
PfTPx-1 belongs to the 2-Cys peroxiredoxin family that is responsible for detoxifying ROS and reactive nitrogen species from the parasite’s oxygen-rich environment (14). In addition to their peroxidase activity, peroxiredoxins are now recognized as sensors and transducers of H2O2 status via the transfer of their oxidation state to effector proteins (22). Plasmodium parasites possess five peroxiredoxins localizing to various organelles, including the nucleus, cytoplasm, and mitochondria (23). PfTPx-1 is abundantly expressed in the trophozoite stage and is described as a cytoplasmic protein (7). We were intrigued that we repeatedly identified PfTPx-1 as a nuclear protein associated with the lncRNA transcribed by the active var gene, and decided to further investigate the nuclear role of PfTPx-1, specifically looking at var gene regulation. As a first step, we created a recombinant PfTPx-1 and confirmed its ability to directly bind the var antisense lncRNA by RNA EMSA (SI Appendix, Fig. S2).
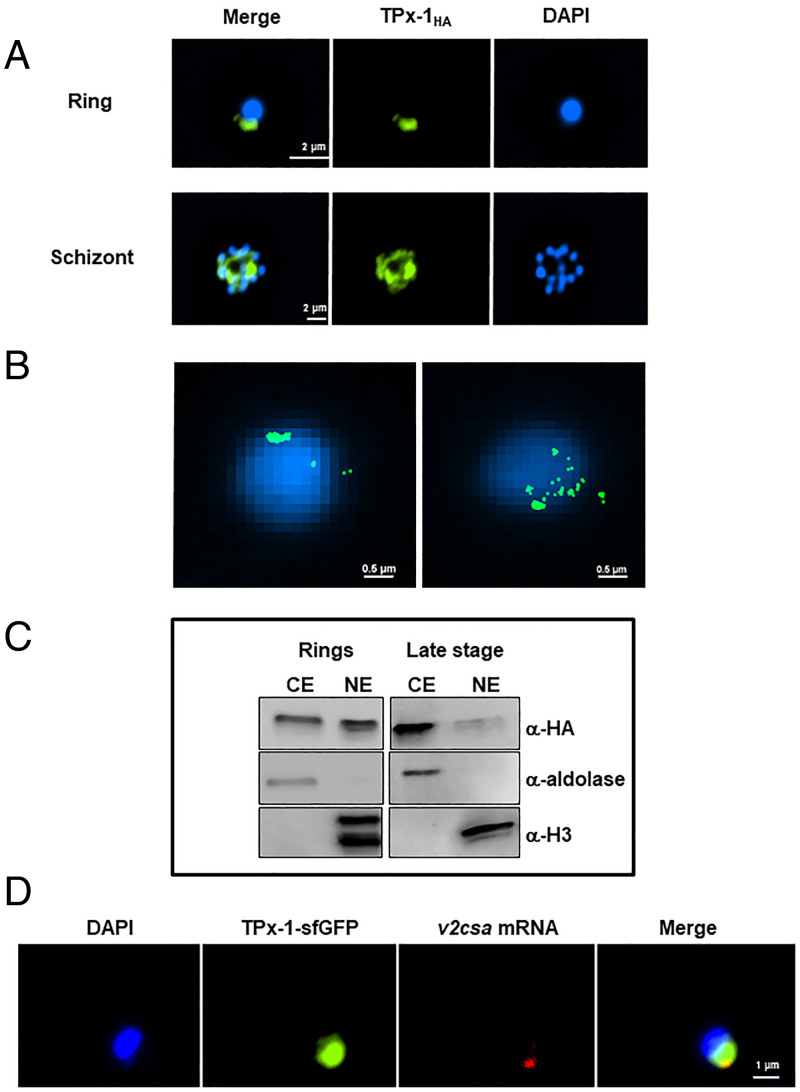
We then created a transgenic line using the selection-linked integration (pSLI) system (24), where PfTPx-1 was endogenously tagged with 3× HA-tags and the glmS ribozyme was fused with the mRNA transcript to allow for inducible knockdown of the gene upon the addition of glucosamine (25) (SI Appendix, Fig. S3). Immunofluorescence analysis (IFA) revealed that PfTPx-1 localizes to a distinct focus at the nuclear periphery in ring-stage parasites and is more diffused in late stages (Fig. 3A and SI Appendix, Fig. S4). Using superresolution stochastic optical reconstruction microscopy (STORM) imaging, we were able to visualize the nuclear signal at higher resolution and obtained two patterns of localization: one in which the PfTPx-1 signal is clustered to one concentrated spot on the nuclear periphery (Fig. 3 B, Left), and another where the signal is in patches along one side of the nuclear periphery (Fig. 3 B, Right). While this difference is likely due to differences in orientation of the parasite, it seems that PfTPx-1 occupies a larger nuclear region than just the var expression site. To confirm that PfTPx-1 is indeed nuclear in ring-stage parasites, we performed subcellular fractionation and detected PfTPx-1 in both nuclear and cytoplasmic compartments in ring-stage parasites, while in later stages it is mostly cytoplasmic (Fig. 3C). This is in agreement with a previous proteomic analysis that also found PfTPx-1 in the nucleus (26). To determine whether the nuclear PfTPx-1 observed in ring-stage parasites localizes to the var expression site, we performed RNA-FISH in parasites exclusively expressing var2csa (CSA line). We visualized the active var gene using var2csa-specific probes and observed colocalization of PfTPx-1 with the var mRNA signal in all the cells where both signals were present (n = 111), confirming the results of our pull-down experiments that PfTPx-1 is found at the var expression site (Fig. 3D and SI Appendix, Fig. S5). Remarkably, while the signal of the var2csa transcript overlaps with PfTPx-1 (Mander’s overlap coefficient 0.97), the PfTPx-1 signal occupies a larger nuclear area (Mander’s overlap coefficient 0.34), indicating that PfTPx-1 may have additional nuclear functions that are not limited to the var expression site.
Fig. 3.
PfTPx-1 is localized to the active var expression site in ring-stage parasites. (A) Immunofluorescence microscopy of PfTPx-1, endogenously tagged with HAx3 in ring (Upper) and schizont (Lower) parasites. Nuclei are stained with DAPI. (Scale bar, 2 µm.) (B) Superresolution STORM imaging of tightly synchronized ring-stage parasites (∼18 h postinfection) expressing PfTPx-1HAx3. Nuclei stained with YOYO1 were imaged by conventional epifluorescence for cellular orientation. (Scale bar, 0.5 µm.) (C) Cellular fractionation of synchronized ring and late-stage parasites was performed on the PfTPx-1HAglmS line and nuclear and cytoplasmic extracts (NE and CE, respectively). PfTPx-1 was detected with anti-HA, antialdolase was used as a cytoplasmic marker, and antihistone H3 as a nuclear marker. (D) RNA-FISH, of var2csa-expressing parasites that episomally express sfGFP-PfTPx-1 fusion (green), using custom-designed Stellaris probes that hybridize to var2csa (red). In all nuclei (n = 111) PfTPx-1 was found to overlap with the active var mRNA. (Scale bar, 1 µm.)
PfTPx-1 Localizes to a Transcriptionally Active Nuclear Region in Ring-Stage Parasites.
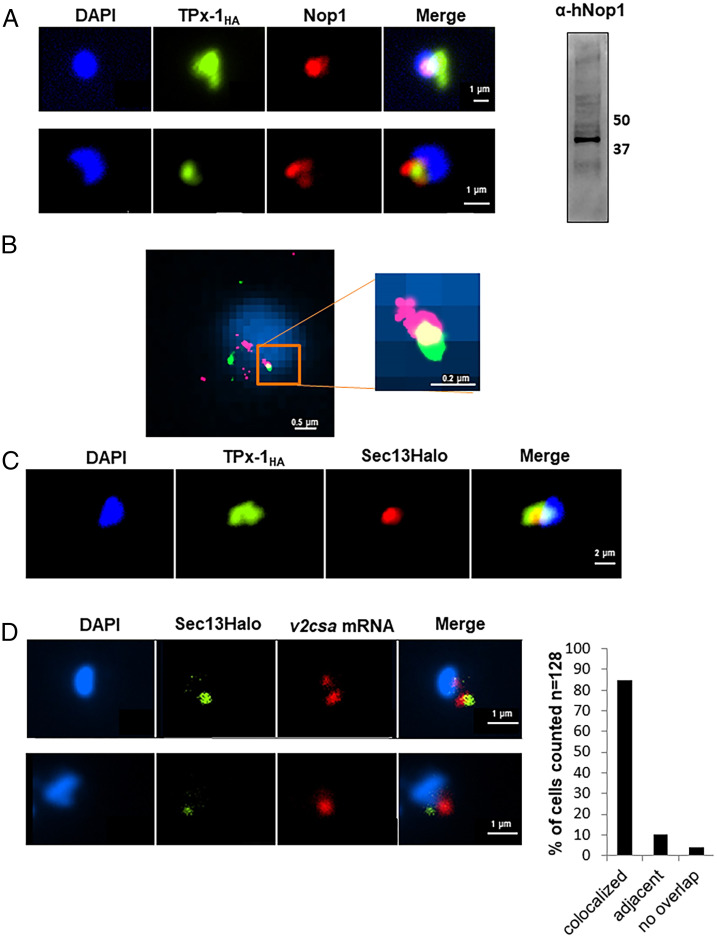
We were further interested in checking whether additional genes transcribed at a particular subnuclear compartment are associated with PfTPx-1. One of the most abundant RNA species transcribed in ring-stage parasites are rRNAs located in a peri-nuclear compartment overlapping with the nucleolar marker fibrillarin (PF3D7_1407100), also referred to as PfNop1 (27). Although a defined nucleolus was never observed in Plasmodium, PfTPx-1 has been shown to interact with PfNop1 in vitro (8, 10). Using IFA analysis, we found that in ring-stage parasites PfTPx-1 and PfNop1 were adjacent at the nuclear periphery, with some regions of overlap (Fig. 4A). Superresolution STORM imaging confirmed this observation and more clearly distinguished regions where the two proteins colocalized (Fig. 4B), indicating that PfTPx-1 is also present in the area referred to as the nucleolus where active transcription of rDNA takes place. An additional subnuclear compartment associated with active transcription is the region near the nuclear pore complexes (NPC). Gene positioning near the NPC is thought to enhance transcription and aid in immediate mRNA transport (28, 29). P. falciparum shows a unique clustering of the NPC at a distinct region of the nuclear envelope in ring-stage parasites, as well as in late schizonts, where these clusters are located adjacent to euchromatic regions (30, 31). Given that PfTPx-1 is present in a region of active transcription, we tested whether active transcription could take place near the NPC. We used PfSec13, which was shown to be an essential nucleoporin, as a marker for the NPC (30). We found that in ring-stage parasites, PfTPx-1 colocalizes with PfSec13, with PfTPx-1 once again occupying a larger region than PfSec13 (Fig. 4C). Furthermore, since we found that the active var gene colocalized with PfTPx-1 (Fig. 3D), we tested the association of the var expression site and the NPC. We performed RNA-FISH in the var2csa expressing line, ectopically expressing a PfSec13-Halo fusion protein, and found that PfSec13 colocalizes with the active var gene in over 80% of the parasites (n = 128) (Fig. 4D). Altogether, these observations indicate that PfTPx-1 is located in regions of active transcription near the NPC during the ring stage.
Fig. 4.
PfTPx-1 localizes to a transcriptionally active nuclear region in ring-stage parasites. (A, Left) Immunofluorescence microscopy of PfTPx-1HAglms parasites where PfTPx-1 (green) is associated with the nucleolar protein, PfNop1 (red). Nuclei are stained with DAPI. (Scale bar, 1 µm.) (Right) An anti hNop1/Fibrillarin antibody specifically detects PfNop1 in parasite extract. (B) Superresolution STORM imaging of PfTPx-1 (green) and PfNop1 (red) in synchronized ring-stage PfTPx-1HA parasites. Nuclei stained with YOYO1 were imaged by conventional epifluorescence for cellular orientation. An area of overlap is enlarged (square Inset). (Scale bar, 0.5 and 0.2 µm, respectively.) (C) Immunofluorescence of ring-stage PfTPx-1HAglms (PfTPx-1 in green) transfected with a Sec13-Halo plasmid (red). (Scale bar, 2 µm.) (D) RNA-FISH of CSA expressing parasites that carry a Sec13-Halo plasmid (green). RNA-FISH was performed using custom-designed Stellaris probes that hybridize to var2csa transcripts (red). Representative images of colocalized signals (Upper) and adjacent signals (Lower) are shown. (Scale bar, 1 µm.) Quantification of signals (n = 128) that were found to be overlapping or adjacent to each other are shown to the right.
PfTPx-1 Is Involved in var Gene Switching.
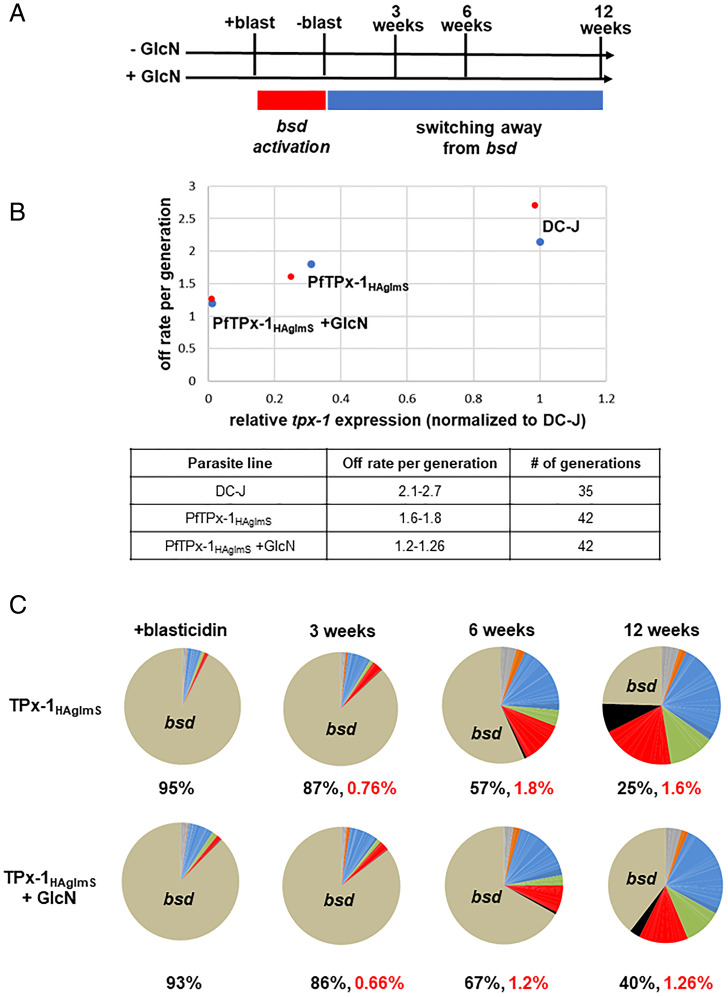
Given that PfTPx-1 is found at the var expression site, we reasoned that altering expression of PfTPx-1 may lead to changes in var gene expression. The PfTPx-1HAglmS transgenic line was created in the DC-J background to allow for selective activation of the var-bsd gene (PF3D7_0223500-BSD), which also enables controlled monitoring of var switching patterns over time upon PfTPx-1 knockdown. We chose a clone in which the var-bsd transgene was silent and another var gene was active (PF3D7_1240300/1240600) (SI Appendix, Fig. S6D), and knocked down PfTPx-1 expression using 5 mM GlcN (Fig. 5A and SI Appendix, Fig. S6A). We observed that upon the addition of blasticidin, RNA levels of the active var gene go down as the var-bsd transgene (PF3D7_0223500-BSD) becomes the dominant transcript in the parasite culture (Fig. 5C and SI Appendix, Fig. S8B), indicating that switching can occur despite PfTPx-1 knockdown. We then removed blasticidin from the cultures and followed var switching patterns over time (Fig. 5A). We found that the switch rate per generation in parasites in which PfTPx-1 was knocked down by GlcN was reduced by 40 to 55% (1.2 to 1.26 off rate per generation) compared to the DC-J parent line where PfTPx-1 is expressed and displayed a 2.1 to 2.7% off rate per generation (32) (Fig. 5B). PfTPx-1HAglmS parasites growing on regular media displayed a 15 to 41% reduction in switch rate (1.6 to 1.8 off rate per generation) compared to the parent line. Interestingly, we found that PfTPx-1 expression levels, measured by RT-qPCR, were reduced in the PfTPx-1HAglmS transgenic line even without addition of GlcN (SI Appendix, Fig. S6B), which is associated with the reduction in var switch rate (Fig. 5B). In addition, we observed unusually high levels of the active var transcript in our PfTPx-1HAglmS transgenic line, where PfTPx-1 transcription levels are low as compared to the DC-J control (SI Appendix, Figs. S6B, S7, and S8A). This increase in transcript abundance is even more pronounced upon PfTPx-1 knockdown, suggesting that switching away from the active var gene is altered when PfTPx-1 levels are low (SI Appendix, Fig. S7). Taken together, these data suggest that var gene switching is delayed without sufficient PfTPx-1 expression.
Fig. 5.
PfTPx-1 is involved in var gene switching. (A) Schematic of experimental design. Parasites were grown in the absence or presence of 5 mM glucosamine for the duration of the experiment (±GlcN). Blasticidin was added continuously for 3 wk to induce switching to the var-bsd (+blast, red rectangle). Blasticidin was then removed from the cultures to monitor switching away from var-bsd (−blast, blue rectangle). Parasites were collected 3, 6, or 12 wk following blasticidin removal. Synchronous ring-stage parasite (20 to 22 h postsynchronization) cultures were collected for RNA, and RT-qPCR for the var gene family was performed on cDNA at the times indicated. (B) Off switch rates per generation were calculated and plotted against the relative PfTPx-1 mRNA levels of the different parasite lines. The results of the two biological replicates are shown in blue or red dots. The ranges of the calculated switch rates per generation, for each of the parasite lines, are presented in the lower table. (C) Dynamics in var switching patterns over time measured by RT-qPCR. Each pie graph represents the total of var gene transcripts with each slice of the pie representing the abundance of an individual var gene within the pool of var transcripts. The percentage of bsd transcript present in the total var gene pool is written below each pie graph and the off rate of bsd is written in red.
Overexpression of PfTPx-1 Induces Switching to var2csa.
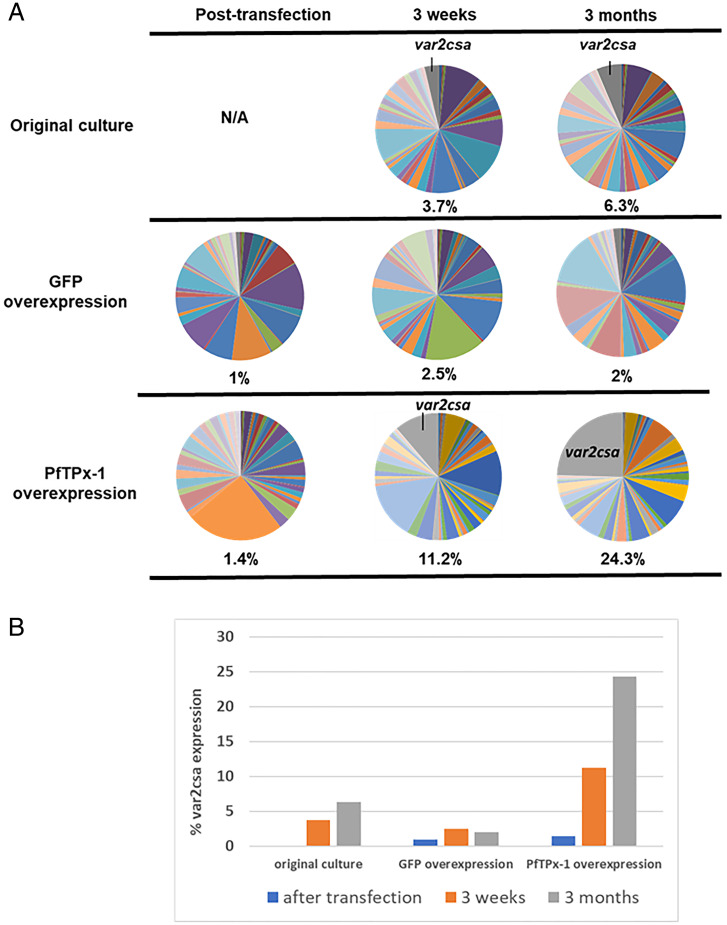
As a complementary approach to PfTPx-1 knockdown, we used a regulatable system (33) to overexpress PfTPx-1 in an NF54 clonal population (C3) by increasing blasticidin concentrations (SI Appendix, Fig. S9A). To control for any off-target effects of blasticidin, a similar plasmid expressing GFP was transfected into the same parasite line. We monitored the expression of the entire var gene family by RT-qPCR (34) over time to detect changes caused by PfTPx-1 overexpression. The initial C3 culture displayed heterogeneous var gene expression with no dominant var gene being transcribed in the parasite population (Fig. 6 A, Top, and SI Appendix, Fig. S9B). After the transfected parasite lines were established by selection with 2 µg/mL blasticidin, we performed RT-qPCR to obtain a baseline var transcription profile (Fig. 6), since transfection presents a bottleneck where var gene expression could change simply due to isolation of subpopulations from the original culture. The amount of blasticidin in the media was then increased to 10 µg/mL and RNA samples of ring-stage parasites were collected after 3 wk and 3 mo. We noticed that over time there was a gradual shift toward expression of var2csa in the line overexpressing PfTPx-1, while var2csa levels remained low in the GFP overexpression control (11.2% vs. 2.5% at 3 wk and 24.3% vs. 2% at 3 mo) (Fig. 6A and SI Appendix, Fig. S9 C and D). Both the original culture and the GFP control exhibited var gene switching, as is typical for a heterogenous culture, but the rate of var2csa activation remained low compared to the PfTPx-1 overexpression line (Fig. 6 and SI Appendix, Fig. S9 B and C). This result suggests that overexpression of PfTPx-1 leads to transcriptional changes that cause parasites within the population to switch to var2csa at an accelerated rate.
Fig. 6.
PfTPx-1 overexpression induces activation of var2csa. (A) var gene-expression profiles were determined by RT-qPCR for each culture posttransfection at low levels of PfTPx-1 or GFP (control) overexpression and then 3 wk and 3 mo after increasing levels of overexpression by increasing the dose of blasticidin from 2 to 10 µg/mL. var gene expression was also determined from the original parent culture, C3, at 3 wk and 3 mo in culture. Each pie graph represents the total of var gene transcripts with each slice of the pie representing the abundance of an individual var gene within the pool of var transcripts. The percentage of var2csa transcript present in the total var gene pool is written below each pie graph. (B) Quantification of the percentage of var2csa transcript within the total var transcript pool for each parasite population.
PfTPx-1 Colocalizes with PfSAMS in the Nucleus of Ring-Stage Parasites.
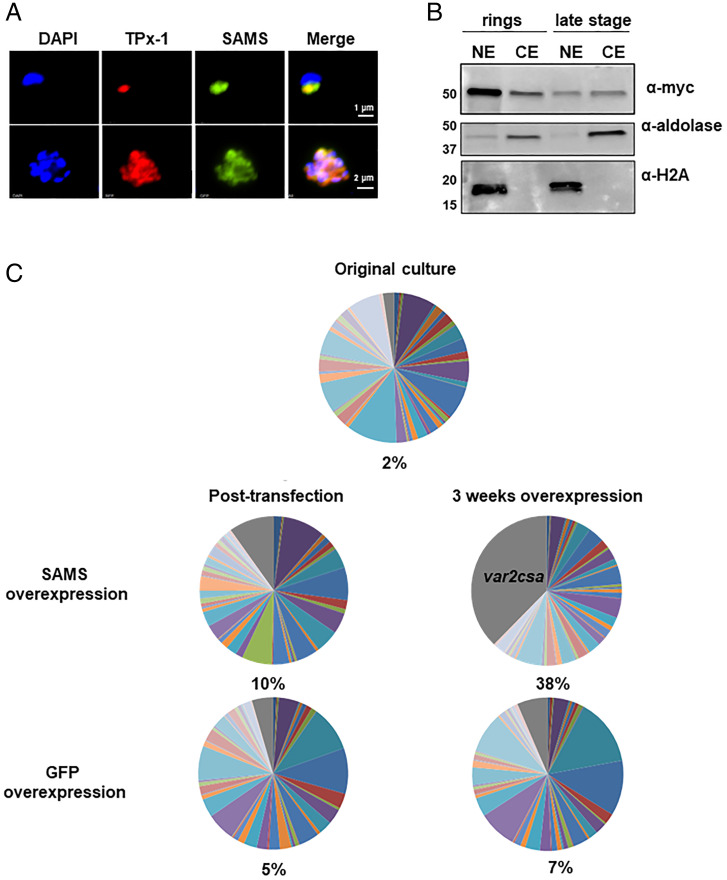
Accelerated activation of var2csa upon PfTPx-1 overexpression is similar to a phenotype previously observed when a dominant–negative version of the histone methyl-transferase PfSET2 was overexpressed (35). Histone methylation requires a constant supply of methyl groups, primarily supplied by the methyl donor, S-adenosylmethionine (SAM). PfTPx-1 has been shown to interact with enzymes involved in SAM metabolism in vitro, including SAM-synthetase (SAMS) (PF3D7_0922200) (8, 10), and has experimentally been shown to act as an oxidoreductase and enhance SAMS activity in vitro (36). Although SAM is needed for DNA and RNA methylation in the nucleus, SAM metabolism is generally thought to take place in the cytoplasm. To test whether the nuclear PfTPx-1 found at the var expression site could possibly interact with SAMS in the nucleus, we transfected a plasmid expressing PfSAMS-3 X MYC into the transgenic PfTPx-1HA-glmS line. By IFA, we found that PfSAMS localized to a distinct region on the nuclear periphery in ring-stage parasites and showed partial overlapping occupancy with PfTPx-1 (Fig. 7A). Nuclear localization of PfSAMS was confirmed by cellular fractionation (Fig. 7B). The proximity of PfTPx-1 to PfSAMS in the nucleus in ring-stage parasites suggests that they may act together at the var expression site to epigenetically regulate var gene expression. Interestingly, when we overexpressed PfSAMS in the PfTPx-1 line, var2csa was activated, similar to the phenotype observed when PfTPx-1 was overexpressed (Fig. 7C). This similarity further supports the idea that PfTPx-1 may be recruited to the active var gene to support optimal redox conditions for PfSAMS enzymatic activity that appears to be important for epigenetic regulation.
Fig. 7.
PfTPx-1 colocalizes with PfSAMS in the nucleus of ring stage parasites. (A) Immunofluorescence microscopy of PfTPx-1HAglmS parasites (PfTPx-1 in red) transfected with a plasmid expressing PfSAM-synthetase with a mycx3 tag (green) in ring (Upper) and schizont (Lower) parasites. Nuclei are stained with DAPI. (Scale bar, 1 µm.) (B) PfSAMS is primarily nuclear in ring-stage parasites. Cellular fractionation of synchronized ring- and late-stage parasites was performed and the nuclear and cytoplasmic extracts (NE and CE, respectively) were subjected to Western blot analysis. PfSAMS was detected using anti-myc antibody; antialdolase antibody was used to confirm cytoplasmic extraction, while antihistone H2A was used as a nuclear marker. (C) var gene-expression profiles were determined by RT-qPCR for PfTPx-1HAglmS ring-stage parasites overexpressing PfSAMS or GFP control after 1 mo of overexpression (5 µg/mL blasticidin). The percentage of var2csa transcript present in the total var gene pool was calculated and is written below each pie graph.
PfTPx-1 Knockdown Causes Transcriptional Changes of Several Gene Subsets.
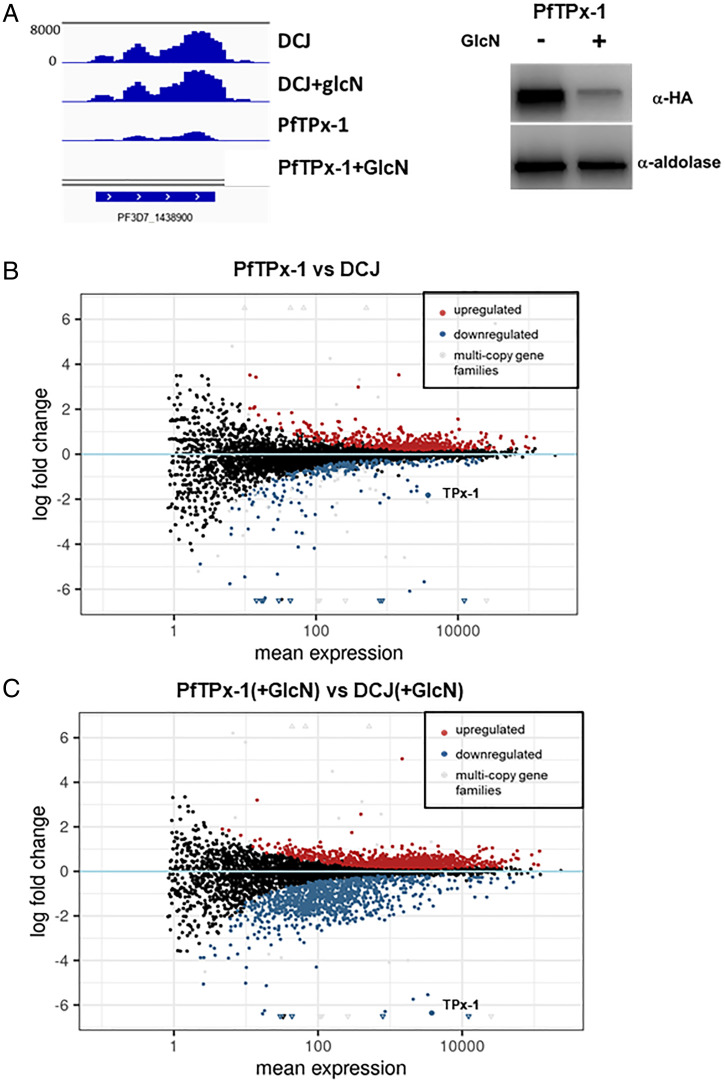
To look at the global transcriptional changes caused by PfTPx-1 down-regulation, RNA sequencing (RNA-seq) was performed on tightly synchronized late ring stages of the DC-J line and a clone of the PfTPx-1HAglmS line grown in the absence or presence of 5 mM GlcN for 96 h. Performing this comparison two cycles after knockdown, at late ring stages when PfTPx-1 is mostly nuclear, maximizes our ability to potentially detect both primary and secondary transcriptional changes. We found that PfTPx-1 mRNA levels were 3.5-fold down-regulated in the PfTPx-1HAglmS compared to the parent DC-J line, and 78-fold down-regulated upon GlcN treatment (Fig. 8). We ensured that the RNA-seq data from all samples corresponded to hour 16 of the cell cycle, indicating that any differences observed are due to the treatment condition (GlcN/PfTPx-1 knockdown) and not due to differences in cell-cycle timing. Our analysis indicated that there are few transcriptomic differences between DC-J and PfTPx-1HAglmS growing on regular media (Fig. 8B); however, upon PfTPx-1 knockdown (+GlcN), 692 transcripts were significantly changed (Padj < 0.05) by at least twofold as compared to the DC-J control grown on GlcN (Fig. 8C). Strikingly, 91% (632 of 692) of these transcripts were down-regulated and only 9% were up-regulated, indicating that PfTPx-1 may function as a nuclear factor that is involved in providing the optimal conditions for transcriptional activation. Our data showing that PfTPx-1 is present in the part of the nucleus where active transcription takes place is in line with this observation. The up-regulated genes consisted primarily of genes from multicopy gene families (PfEMP-1, stevor, rifins, PfMC-2TM), which are often distinct between two clonal populations. Despite the almost 80-fold reduction in PfTPx-1 transcript, the resulting down-regulation of other transcripts was more modest (average 3.24-fold down-regulation, range log2fold −10 to −1), yet significant. Gene ontology and metabolic pathway enrichment analysis of down-regulated genes showed significant enrichment in the categories of nucleotide biosynthesis, mitochondrial electron transport, DNA binding and replication, and oxidoreductase activity (SI Appendix, Fig. S10). Overall, these data suggest that PfTPx-1 is needed for transcriptional activation of particular gene subsets.
Fig. 8.
PfTPx-1 knockdown causes transcriptional changes of several gene subsets. (A) Expression profile for the PfTPx-1 transcript (PF3D7_1438900) (bedgraph coverage illustration of relative expression from IGV) showing the knockdown of the PfTPx-1 in the transgenic line compared to the parent DC-J line. Western blot analysis demonstrating the reduction of PfTPx-1 protein levels in the transgenic lines are shown on the right. (B and C) MAplots of PfTPx-1HAglmS vs. DC-J control without (B) and with GlcN (C), show genes that are differentially regulated upon down-regulation of PfTPx-1 with GlcN. Within the plots, black dots represent nonsignificant genes. Significantly up-regulated and down-regulated genes (Padj < 0.05) are shown in red and blue, respectively. Significantly changed genes of the multicopy gene families are shown in gray. Triangles are used to represent values that are beyond the axes limit.
Discussion
The var antisense lncRNA transcripts were previously shown to be involved in var gene activation (5); however, the mechanism is unknown. In this study, we provide evidence that these lncRNAs are located at the var expression site, in close proximity to the nascent var mRNA, and that they are held in place by DNA:RNA interactions. These transcripts contain a hypervariable sequence that can confer gene specificity by marking a single gene for activation, while the conserved intronic sequence could be involved in an activation mechanism common to the entire var gene family. Indeed, in our dChIRP analysis, the majority of protein–RNA interactions were obtained with the conserved part of the var lncRNA, while histones were the primary proteins pulled down with the gene-specific sequence of the lncRNA. These data support our observation that the lncRNA is anchored by DNA:RNA interactions and is incorporated into chromatin (5). Therefore, it is reasonable to suggest a model by which the gene-specific sequence of the antisense transcript marks the locus by forming a DNA:RNA hybrid, while the conserved region is involved in protein recruitment. lncRNAs can mediate transcriptional and epigenetic changes by recruiting transcriptional activators or chromatin-modifying enzymes (6). We further used the conserved region of the antisense lncRNA as bait and identified the proteins that interact with these transcripts. The var lncRNA interactome contained DNA/RNA binding proteins, such as Alba3/4 and polyadenylate binding protein, which have previously been mapped to var gene loci (37, 38), validating our methodologies. However, while it is well established that var genes are epigenetically regulated (39), we did not pull down any histone modifiers. Such interactions could be transient, or alternatively histone modifiers may act later in the cell cycle when the epigenetic imprint for the next cycle is needed (37). Though PfTPx-1 was previously believed to be cytoplasmic, we consistently observed its association with var lncRNAs within the nucleus. While it is difficult to discriminate between direct RNA–protein interaction or via protein complexes, using affinity chromatography and dChIRP, our RNA-EMSA on recombinant PfTPx-1 supports direct interaction. Similarly, TPx-1 from humans (40) and Plasmodium knowlesi (PkTPx-1), exhibits DNA and RNA binding in vitro, and PkTPx-1 was shown to be able to melt an RNA stem-loop structure upon binding and act as an RNA chaperone (41), indicating that PfTPx-1 might also be involved in molecular mechanisms in the nucleus that influence gene expression.
Interestingly, down-regulation of endogenous PfTPx-1 slowed the var switch rate, while its overexpression accelerated activation of var2csa. var2csa is unique among the var gene family; it is highly conserved among different isolates and has an upstream open reading frame, which leads to its translational repression (42). Due to its conserved sequence and translational control, var2csa could be the ideal switch intermediate; var2csa is present in all parasite isolates and its activation would not necessarily initiate an immune response. This hypothesis is supported by studies that observed selective activation of var2csa when var switching was induced through inhibition of histone modifiers (35, 43), as well as computational modeling that identified an important role for a common switch intermediate (or “sink node”), a function that could be fulfilled by var2csa (35, 44). In our experiments PfTPx-1 was constitutively overexpressed and caused var2csa to remain activated for long periods, rather than the transient activation that would be predicted for a switch intermediate during a natural infection.
Activation of var2csa was previously reported upon overexpression of a dominant–negative PfSET2 construct and treatment with the histone methyltransferase inhibitor chaetocin (35, 43), like the phenotype observed here when PfTPx-1 is overexpressed. In addition, overexpression of SAM synthase (PfSAMS) resulted in var2csa activation similar to overexpression of dominant–negative PfSET2 and PfTPx-1. However, var2csa activation occurred more rapidly when PfSAMs is overexpressed possibly because SAMS enzymatic activity is the limiting factor for providing the methyl groups essential for var2csa activation. Here we show that PfTPx-1 and PfSAMS occupy a subnuclear compartment where active transcription takes place. The nuclear localization of PfSAMS may provide the methyl groups necessary for nearby histone methyltransferases to epigenetically mark their target genes. Several histone methylation marks are implicated in the epigenetic regulation of var gene expression (39). H3K36me3 is present at both active and silent var loci (35, 43), while H3K9me3 is found only at silent members of variant gene families and serves to recruit PfHP1 (45–47). H3K4me3, on the other hand, is only present at the active var gene and is thought to be responsible for keeping the active var poised for the next cell cycle (37). Interestingly, in model organisms the histone methyltransferase that deposits H3K4me3 is sensitive to fluctuations in SAM levels (48). Studies have shown that cellular metabolism can influence gene expression via changes in histone methylation (49). In view of the interaction between PfSAMS and PfTPx-1 (10), the demonstrated role of PfTPx-1 on the enzymatic activity of PfSAMS (8), and their colocalization in the nucleus, it is possible that PfSAMS and PfTPx-1 play a role in providing methyl groups to methyltransferases at a specific nuclear subcompartment to influence gene expression.
The nuclear localization of PfTPx-1 supports its association with transcriptional activity. It appears to reside in a perinuclear region that contains the var expression site, a portion of the nucleolus defined by Nop1, and the NPC marked by the nucleoporin PfSec13. The association of the var expression site with the NPC, as demonstrated here, is contrary to a previous report that used an antibody raised against a Plasmodium protein (PF3D7_1473700) that shares sequence similarity to the yeast ScNup116 (34% identity, 50% similarity), that did not colocalize with the var expression site (50). It remains to be determined whether the identified P. falciparum Nup116 is a bona fide nucleoporin. Interestingly, Nup116 is absent from Plasmodium berghei and is the only known putative nucleoporin that is not conserved among the different Plasmodium species (51). The spatial positioning of the var active site near the nuclear pore is in agreement with what is known from many model organisms and may contribute to efficient nuclear export of the var mRNA (28, 29).
In vivo var expression switching is postulated to occur in response to the adaptive immune attack against the expressed PfEMP1 variant. However, the mechanistic process that governs such switching is not understood. Immune complexes consisting of antibody–antigen pairs are powerful stimulators of immune cells, which can release a burst of ROS (52, 53). Interestingly, we observed an increase in PfTPx-1 expression following exposure to different sources of oxidative stress (SI Appendix, Fig. S11) suggesting that PfTPx-1 can sense and respond to stress conditions present in a human infection. It has been demonstrated that peroxiredoxins act as sensors that influence activities of their target proteins based on the cell’s redox status (22, 54), and it is tempting to suggest that PfTPx-1 could mediate transcriptional changes in response to changing environmental conditions and immune response.
While cellular metabolism is generally thought to occur outside the nucleus, the presence of nuclear metabolic enzymes is now widely accepted. Many of these enzymes function both in their canonical metabolic role and also take on additional roles in transcriptional regulation, providing a way for organisms to adapt their transcriptional program in response to metabolic stress (55). Therefore, it is not surprising that metabolic enzymes were found in the var lncRNA interactome. The recently published var interactome, using CRIPSR-dCas9 to target both active and silent var genes, also uncovered metabolic enzymes associated with var genes (38). Intriguingly, four glycolytic enzymes (Table 1) were found to interact with the nuclear var lncRNA. One of the main products of glycolysis is NADH/NAD+, which is impermeable to the nuclear membrane. Since many transcription factors require NADH to function, nuclear production of NADH is linked to transcriptional control (55). Interestingly, the NAD+-dependent histone deacetylases, PfSir2a and PfSir2b, have been implicated in heterochromatin formation and var gene silencing (56, 57), although their functions may be strain-dependent (58). Aside from their canonical roles, in other organisms, glycolytic enzymes were shown to have noncanonical functions in the nucleus participating in transcription, DNA replication, and telomere maintenance (59). Interestingly, some of these glycolytic enzymes were also shown to interact with PfTPx-1, suggesting that their functions may be redox-controlled (10). In addition, OAT was present in all three pull-down methodologies and has previously been shown to interact with PfTPx-1 (8). Although there are reports of OAT being present in the nucleus in other organisms (60), to the best of our knowledge the nuclear roles of OAT are still elusive.
In conclusion, PfTPx-1’s association with var lncRNAs and its presence, adjacent to PfSAMS, in the transcriptionally active region of the nucleus, provides a mechanistic clue as to how the parasite may sense and react to the changing conditions within the human host and switch var gene expression in response to malaria progression and immune attack.
Materials and Methods
Detailed materials and methods are found in the SI Appendix. Parasite culture, transfection, and selection was previously described (61–63). Genomic DNA extraction, RNA extraction, and cDNA synthesis were done as described previously (17, 64, 65). Transcript copy numbers was measured by RT-qPCR and were determined using the equation 2−ΔΔCT as described in the Applied Biosystems User Bulletin and previously published (17). TSA and Stellaris RNA FISH, nuclear fractionation, and affinity chromatography were performed as described previously with some modifications (66–68). Imaging analysis was performed using JACoP ImageJ plugin to calculate the Mander’s overlap coefficient (69). dChIRP was performed as described previously (20, 21, 70). Probes were designed using Biosearch technologies Stellaris FISH Probe Designer. Probes were added to cross-linked and sonicated parasite samples, and after reverse cross-linking, protein samples were sent to LC-MS/MS. A detailed protocol can be found in SI Appendix, Supplementary Methods. STORM imaging and analysis was performed as previously described (71). Calculation of off rate for individual var loci were calculated as previously described (32), where off rate is defined as the decrease of transcript from a particular var locus per generation. Parasite collection and RNA preparation for RNA-seq, as well as data analysis, is described in details in SI Appendix. Cell-cycle progression of our samples was normalized by comparing our RNA-seq data to Otto et al. (72). Significance threshold was taken as Padj < 0.05.
Supplementary Material
Acknowledgments
This work was supported partially by the Israeli Academy for Science, Israel Science Foundation Grant 1523/18, and in part by European Research Council (erc.europa.eu) Consolidator Grant 615412 (to R.D.). R.D. is also supported by the Dr. Louis M. Leland and Ruth M. Leland Chair in Infectious Diseases. V.M. was supported by Dr Emanuel & Mrs. Olga Lourie PhD Scholarships in Tropical Medicine.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2201247119/-/DCSupplemental.
Data, Materials, and Software Availability
All study data are included in the article and supporting information.
References
- 1.(WHO) World Malaria Report 2020 (2020). https://www.who.int/publications/i/item/9789240015791. Accessed 31 July 2022.
- 2.Otto T. D., et al. , Long read assemblies of geographically dispersed Plasmodium falciparum isolates reveal highly structured subtelomeres. Wellcome Open Res. 3, 52 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dzikowski R., Deitsch K. W., Genetics of antigenic variation in Plasmodium falciparum. Curr. Genet. 55, 103–110 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epp C., Li F., Howitt C. A., Chookajorn T., Deitsch K. W., Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA 15, 116–127 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amit-Avraham I., et al. , Antisense long noncoding RNAs regulate var gene activation in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 112, E982–E991 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer T. R., Dinger M. E., Mattick J. S., Long non-coding RNAs: Insights into functions. Nat. Rev. Genet. 10, 155–159 (2009). [DOI] [PubMed] [Google Scholar]
- 7.Kawazu S., et al. , Molecular characterization of a 2-Cys peroxiredoxin from the human malaria parasite Plasmodium falciparum. Mol. Biochem. Parasitol. 116, 73–79 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Brandstaedter C., et al. , The interactome of 2-Cys peroxiredoxins in Plasmodium falciparum. Sci. Rep. 9, 13542 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahbari M., Rahlfs S., Jortzik E., Bogeski I., Becker K., H2O2 dynamics in the malaria parasite Plasmodium falciparum. PLoS One 12, e0174837 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sturm N., et al. , Identification of proteins targeted by the thioredoxin superfamily in Plasmodium falciparum. PLoS Pathog. 5, e1000383 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kimura R., Komaki-Yasuda K., Kawazu S., Kano S., 2-Cys peroxiredoxin of Plasmodium falciparum is involved in resistance to heat stress of the parasite. Parasitol. Int. 62, 137–143 (2013). [DOI] [PubMed] [Google Scholar]
- 12.Yano K., et al. , Disruption of the Plasmodium berghei 2-Cys peroxiredoxin TPx-1 gene hinders the sporozoite development in the vector mosquito. Mol. Biochem. Parasitol. 159, 142–145 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Yano K., et al. , 2-Cys peroxiredoxin TPx-1 is involved in gametocyte development in Plasmodium berghei. Mol. Biochem. Parasitol. 148, 44–51 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Komaki-Yasuda K., Kawazu S., Kano S., Disruption of the Plasmodium falciparum 2-Cys peroxiredoxin gene renders parasites hypersensitive to reactive oxygen and nitrogen species. FEBS Lett. 547, 140–144 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Deslauriers R., Butler K., Smith I. C., Oxidant stress in malaria as probed by stable nitroxide radicals in erythrocytes infected with Plasmodium berghei. The effects of primaquine and chloroquine. Biochim. Biophys. Acta 931, 267–275 (1987). [DOI] [PubMed] [Google Scholar]
- 16.Becker K., et al. , Oxidative stress in malaria parasite-infected erythrocytes: Host-parasite interactions. Int. J. Parasitol. 34, 163–189 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Dzikowski R., Frank M., Deitsch K., Mutually exclusive expression of virulence genes by malaria parasites is regulated independently of antigen production. PLoS Pathog. 2, e22 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang L., et al. , PfSETvs methylation of histone H3K36 represses virulence genes in Plasmodium falciparum. Nature 499, 223–227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avraham I., Schreier J., Dzikowski R., Insulator-like pairing elements regulate silencing and mutually exclusive expression in the malaria parasite Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 109, E3678–E3686 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinn J. J., et al. , Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat. Biotechnol. 32, 933–940 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn J. J., Chang H. Y., In situ dissection of RNA functional subunits by domain-specific chromatin isolation by RNA purification (dChIRP). Methods Mol. Biol. 1262, 199–213 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Netto L. E., Antunes F., The roles of peroxiredoxin and thioredoxin in hydrogen peroxide sensing and in signal transduction. Mol. Cells 39, 65–71 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Müller S., Redox and antioxidant systems of the malaria parasite Plasmodium falciparum. Mol. Microbiol. 53, 1291–1305 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Birnbaum J., et al. , A genetic system to study Plasmodium falciparum protein function. Nat. Methods 14, 450–456 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Prommana P., et al. , Inducible knockdown of Plasmodium gene expression using the glmS ribozyme. PLoS One 8, e73783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oehring S. C., et al. , Organellar proteomics reveals hundreds of novel nuclear proteins in the malaria parasite Plasmodium falciparum. Genome Biol. 13, R108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mancio-Silva L., Zhang Q., Scheidig-Benatar C., Scherf A., Clustering of dispersed ribosomal DNA and its role in gene regulation and chromosome-end associations in malaria parasites. Proc. Natl. Acad. Sci. U.S.A. 107, 15117–15122 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akhtar A., Gasser S. M., The nuclear envelope and transcriptional control. Nat. Rev. Genet. 8, 507–517 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Dieppois G., Stutz F., Connecting the transcription site to the nuclear pore: A multi-tether process that regulates gene expression. J. Cell Sci. 123, 1989–1999 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Dahan-Pasternak N., et al. , PfSec13 is an unusual chromatin-associated nucleoporin of Plasmodium falciparum that is essential for parasite proliferation in human erythrocytes. J. Cell Sci. 126, 3055–3069 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Kapishnikov S., et al. , Oriented nucleation of hemozoin at the digestive vacuole membrane in Plasmodium falciparum. Proc. Natl. Acad. Sci. U.S.A. 109, 11188–11193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank M., Dzikowski R., Amulic B., Deitsch K., Variable switching rates of malaria virulence genes are associated with chromosomal position. Mol. Microbiol. 64, 1486–1498 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Epp C., Raskolnikov D., Deitsch K. W., A regulatable transgene expression system for cultured Plasmodium falciparum parasites. Malar. J. 7, 86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Salanti A., et al. , Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol. Microbiol. 49, 179–191 (2003). [DOI] [PubMed] [Google Scholar]
- 35.Ukaegbu U. E., et al. , A unique virulence gene occupies a principal position in immune evasion by the malaria parasite Plasmodium falciparum. PLoS Genet. 11, e1005234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pretzel J., et al. , Characterization and redox regulation of Plasmodium falciparum methionine adenosyltransferase. J. Biochem. 160, 355–367 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Volz J. C., et al. , PfSET10, a Plasmodium falciparum methyltransferase, maintains the active var gene in a poised state during parasite division. Cell Host Microbe 11, 7–18 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Bryant J. M., et al. , Exploring the virulence gene interactome with CRISPR/dCas9 in the human malaria parasite. Mol. Syst. Biol. 16, e9569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deitsch K. W., Dzikowski R., Variant gene expression and antigenic variation by malaria parasites. Annu. Rev. Microbiol. 71, 625–641 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Kim J. H., et al. , RNA-binding properties and RNA chaperone activity of human peroxiredoxin 1. Biochem. Biophys. Res. Commun. 425, 730–734 (2012). [DOI] [PubMed] [Google Scholar]
- 41.Hakimi H., et al. , Plasmodium knowlesi thioredoxin peroxidase 1 binds to nucleic acids and has RNA chaperone activity. Parasitol. Res. 113, 3957–3962 (2014). [DOI] [PubMed] [Google Scholar]
- 42.Amulic B., Salanti A., Lavstsen T., Nielsen M. A., Deitsch K. W., An upstream open reading frame controls translation of var2csa, a gene implicated in placental malaria. PLoS Pathog. 5, e1000256 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ukaegbu U. E., et al. , Recruitment of PfSET2 by RNA polymerase II to variant antigen encoding loci contributes to antigenic variation in P. falciparum. PLoS Pathog. 10, e1003854 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Recker M., et al. , Antigenic variation in Plasmodium falciparum malaria involves a highly structured switching pattern. PLoS Pathog. 7, e1001306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flueck C., et al. , Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 5, e1000569 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lopez-Rubio J. J., et al. , 5′ Flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol. Microbiol. 66, 1296–1305 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chookajorn T., et al. , Epigenetic memory at malaria virulence genes. Proc. Natl. Acad. Sci. U.S.A. 104, 899–902 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ding W., et al. , Stress-responsive and metabolic gene regulation are altered in low S-adenosylmethionine. PLoS Genet. 14, e1007812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shiraki N., et al. , Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 19, 780–794 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Guizetti J., Martins R. M., Guadagnini S., Claes A., Scherf A., Nuclear pores and perinuclear expression sites of var and ribosomal DNA genes correspond to physically distinct regions in Plasmodium falciparum. Eukaryot. Cell 12, 697–702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kehrer J., et al. , Nuclear pore complex components in the malaria parasite Plasmodium berghei. Sci. Rep. 8, 11249 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vermeren S., Karmakar U., Rossi A. G., Immune complex-induced neutrophil functions: A focus on cell death. Eur. J. Clin. Invest. 48 (suppl. 2), e12948 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Mayadas T. N., Tsokos G. C., Tsuboi N., Mechanisms of immune complex-mediated neutrophil recruitment and tissue injury. Circulation 120, 2012–2024 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sobotta M. C., et al. , Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat. Chem. Biol. 11, 64–70 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Boukouris A. E., Zervopoulos S. D., Michelakis E. D., Metabolic enzymes moonlighting in the nucleus: Metabolic regulation of gene transcription. Trends Biochem. Sci. 41, 712–730 (2016). [DOI] [PubMed] [Google Scholar]
- 56.Freitas-Junior L. H., et al. , Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell 121, 25–36 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Merrick C. J., Duraisingh M. T., Plasmodium falciparum Sir2: An unusual sirtuin with dual histone deacetylase and ADP-ribosyltransferase activity. Eukaryot. Cell 6, 2081–2091 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Merrick C. J., et al. , Functional analysis of sirtuin genes in multiple Plasmodium falciparum strains. PLoS One 10, e0118865 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huangyang P., Simon M. C., Hidden features: Exploring the non-canonical functions of metabolic enzymes. Dis. Model. Mech. 11, dmm033365 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ginguay A., Cynober L., Curis E., Nicolis I., Ornithine aminotransferase, an important glutamate-metabolizing enzyme at the crossroads of multiple metabolic pathways. Biology (Basel) 6, E18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Aley S. B., Sherwood J. A., Howard R. J., Knob-positive and knob-negative Plasmodium falciparum differ in expression of a strain-specific malarial antigen on the surface of infected erythrocytes. J. Exp. Med. 160, 1585–1590 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Calderwood M. S., Gannoun-Zaki L., Wellems T. E., Deitsch K. W., Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J. Biol. Chem. 278, 34125–34132 (2003). [DOI] [PubMed] [Google Scholar]
- 63.Deitsch K., Driskill C., Wellems T., Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 29, 850–853 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dzikowski R., Deitsch K. W., Active transcription is required for maintenance of epigenetic memory in the malaria parasite Plasmodium falciparum. J. Mol. Biol. 382, 288–297 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kyes S., Pinches R., Newbold C., A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol. Biochem. Parasitol. 105, 311–315 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Ronander E., Bengtsson D. C., Joergensen L., Jensen A. T., Arnot D. E., Analysis of single-cell gene transcription by RNA fluorescent in situ hybridization (FISH). J. Vis. Exp. 68, 4073 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Voss T. S., Mini T., Jenoe P., Beck H. P., Plasmodium falciparum possesses a cell cycle-regulated short type replication protein A large subunit encoded by an unusual transcript. J. Biol. Chem. 277, 17493–17501 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Jutras B. L., Verma A., Stevenson B., Identification of novel DNA-binding proteins using DNA-affinity chromatography/pull down. Curr. Protoc. Microbiol., Chapter 1: Unit1F.1 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bolte S., Cordelières F. P., A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232 (2006). [DOI] [PubMed] [Google Scholar]
- 70.Chu C., Quinn J., Chang H. Y., Chromatin isolation by RNA purification (ChIRP). J. Vis. Exp. 61, 3912 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Fastman Y., et al. , An upstream open reading frame (uORF) signals for cellular localization of the virulence factor implicated in pregnancy associated malaria. Nucleic Acids Res. 46, 4919–4932 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Otto T. D.et al., . New insights into the blood-stage transcriptome of Plasmodium falciparum using RNA-Seq. Mol. Microbiol. 76, 12–24 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and supporting information.