Significance
Long pentraxins are soluble pattern-recognition molecules. There is a wealth of functional data linking them to processes as varied as innate immune defense, female fertility, and neurobiology. However, structural information is missing. Here, we present a complete high-resolution model of a long pentraxin, PTX3, using a hybrid structural biology approach combining cryoelectron microscopy, mass spectrometry, and AlphaFold-based artificial intelligence structure prediction, which was able to model flexible regions of the complex. This gives insights into the functions of PTX3, such as immune defense, as well as exemplifying a method to resolving flexible domains of protein complexes.
Keywords: Long pentraxin, complement, AlphaFold, cryoEM, COVID19
Abstract
Pattern recognition molecules (PRMs) form an important part of innate immunity, where they facilitate the response to infections and damage by triggering processes such as inflammation. The pentraxin family of soluble PRMs comprises long and short pentraxins, with the former containing unique N-terminal regions unrelated to other proteins or each other. No complete high-resolution structural information exists about long pentraxins, unlike the short pentraxins, where there is an abundance of both X-ray and cryoelectron microscopy (cryo-EM)-derived structures. This study presents a high-resolution structure of the prototypical long pentraxin, PTX3. Cryo-EM yielded a 2.5-Å map of the C-terminal pentraxin domains that revealed a radically different quaternary structure compared to other pentraxins, comprising a glycosylated D4 symmetrical octameric complex stabilized by an extensive disulfide network. The cryo-EM map indicated α-helices that extended N terminal of the pentraxin domains that were not fully resolved. AlphaFold was used to predict the remaining N-terminal structure of the octameric PTX3 complex, revealing two long tetrameric coiled coils with two hinge regions, which was validated using classification of cryo-EM two-dimensional averages. The resulting hybrid cryo-EM/AlphaFold structure allowed mapping of ligand binding sites, such as C1q and fibroblast growth factor-2, as well as rationalization of previous biochemical data. Given the relevance of PTX3 in conditions ranging from COVID-19 prognosis, cancer progression, and female infertility, this structure could be used to inform the understanding and rational design of therapies for these disorders and processes.
The pentraxin family of proteins are a group of humoral pattern recognition molecules (PRMs) and comprise both short and long pentraxins (1). The canonical long pentraxin PTX3 is expressed in a diverse range of tissues including the respiratory tract, muscles, and female reproductive tissues, which reflect its role in pathogen recognition (2), tissue repair (3), and as a structural component of the extracellular matrix (ECM) (4). Structures of short pentraxins C-reactive protein (CRP) and serum amyloid P component (SAP) exhibit a pentamer of single pentraxin domain (PTX) monomers (5, 6). However, there are no complete high-resolution structures of any long pentraxin, which comprise a C-terminal PTX domain, common to all pentraxins, and an unconserved N-terminal region with a sequence that is unique to each long pentraxin (1) (Fig. 1A).
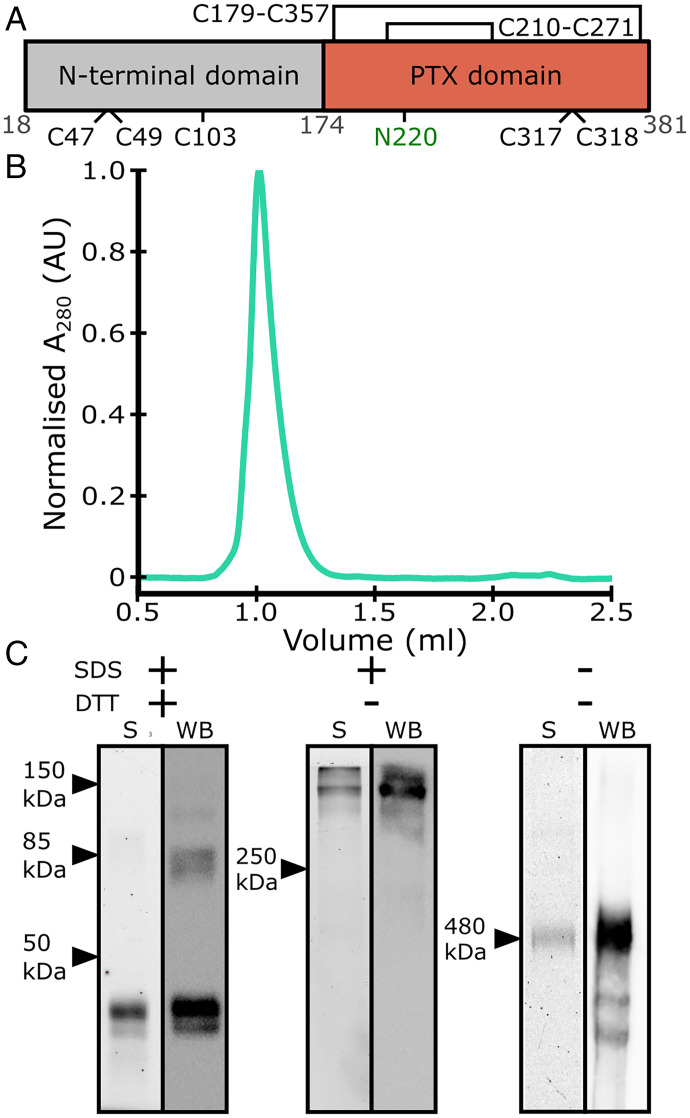
Fig. 1.
Purification of recombinant human PTX3. (A) Schematic of the PTX3 monomer, indicating sites of glycosylation (N220) and disulfide bonds. Intramonomeric disulfide bonds are shown as closed loops. Numbering of residues is shown in gray. (B) SEC elution profile of PTX3 after Ni-NTA affinity and SEC purification. (C) Gel electrophoretic profiles based on TCE staining (S) and Western blots (WB) of PTX3 after purification under in the presence of SDS and/or DTT. Arrows indicate standard protein markers of the indicated molecular weight.
Previous biochemical analysis has revealed that PTX3 forms higher-order oligomers held together by an extensive disulfide bond network and the N-terminal region (7, 8). Additionally, small-angle X-ray scattering (SAXs) revealed a low-resolution molecular envelope, suggesting PTX3 forms an asymmetrical octamer (8), all of which makes PTX3 structurally distinct from CRP and SAP. PTX3 is also a glycoprotein, with an N-linked glycan at N220 that is crucial to regulate interactions with the human immune system (9, 10). When sialylated, this glycan has been shown to be critical for potent antiviral properties of PTX3, and most recently PTX3 has been linked to COVID-19 severity in human patients (11, 12), and been shown to protect against acute lung injury in mouse models of coronavirus infection (13).
Here, we use cryoelectron microscopy (cryo-EM), mass spectrometry (MS), and AlphaFold-based structure prediction to build a complete model of PTX3. Cryo-EM yielded a 2.5-Å map of the C-terminal PTX domains, which form an octameric complex exhibiting a complex disulphide bond network and surface glycosylation. Flexibility in the N-terminal region inhibited full resolution of the protein complex. To overcome this, the artificial intelligence protein prediction tool, AlphaFold (14), was used to model the N termini and complete the full octameric model of PTX3, which was then validated using cryo-EM class averaging. The purified protein was able to bind C1q and activate complement, and this high-resolution data allowed the mapping of the putative C1q binding sites and other ligand-interaction regions. This representation, in which the complete structure of a long pentraxin has been resolved, is unique and may provide a platform whereby other members of the long-pentraxin family can be studied.
Results and Discussion
Biochemical Characterization of Recombinant Human PTX3.
His-tagged recombinant human PTX3 was produced by secretion from mammalian cells, and eluted as a single peak in size-exclusion chromatography (SEC) experiments after Ni-NTA and SEC purification (Fig. 1B). Contrary to this, PAGE and Western blotting showed heterogenous species (Fig. 1C). Under both denaturing and reducing conditions (+/+), two prominent species between 40 and 50 kDa were present, close to the calculated mass of monomeric PTX3 (expected mass = 42 kDa). This presence of multiple species corresponding to monomeric PTX3 is indicative of heterogeneity via variable glycosylation at N220 (Fig. 1A) (9, 15). Western blotting of the same sample revealed the presence of higher-order oligomers of PTX3 that corresponded to dimers or tetramers that persisted even at higher DTT concentrations (Fig. 1C and SI Appendix, Fig. S1). Under denaturing but not reducing conditions (Fig. 1C) (+/−), the presence of higher molecular weight species were detected with masses >250 kDa, using both gel electrophoresis, stained with 2,2,2-trichloroethanol (TCE), and Western blots showing one prominent band consistent with higher-order oligomers of PTX3. Finally, under native conditions (−/−), SDS/PAGE gave similar results as under nonreducing conditions (+/−), with one prominent band corresponding to higher-order oligomers of PTX3. However, Western blot revealed the presence of a smear and several other species of PTX3, consistent with the presence of several glycosylation states or different oligomeric states, which were also seen on SDS/PAGE at higher protein concentrations (SI Appendix, Fig. S1).
These data indicate that the quaternary structure of PTX3 is regulated via disulfide bonds, as has previously been reported (7). Furthermore, this disulfide bond network appears to be partially resistant to concentrations of DTT up to 250 mM (SI Appendix, Fig. S1). PTX3 contains nine cysteine residues, of which five are involved in intermonomer oligomerization (Fig. 1A) (7). Within the higher-order PTX3 oligomer, there are therefore multiple disulphide bonds mediating oligomerization, and this extensive disulphide bond network apparently bestows PTX3 resistance to reduction. Together with the various oligomeric states, glycosylation likely contributes to the variable masses seen under both native and nonreducing conditions. In addition, smearing on gels is characteristic of glycosylation due to the heterogenous nature of the glycans, which can also affect interactions with SDS in the gel (16).
Purified PTX3 was functionally active and able to activate complement via the classic pathway. ELISA of PTX3 in the presence of normal human serum showed binding of C1q and deposition of C4b (SI Appendix, Fig. S1). Bio-layer inferometry confirmed binding of C1 to PTX3, with a KD of 17.4 nM (±1.04 nM) (SI Appendix, Fig. S1 and Table S1), comparable to previously reported values (17, 18).
Cryo-EM Structure Determination.
To investigate the three-dimensional (3D) structure of PTX3, cryo-EM was used to image the purified complex via single-particle analysis (SPA). In line with previous cryo-EM studies of the short pentraxin CRP, PTX3 exhibited aggregation and adhesiveness to both the grid support film and the air–water interface (5), with the latter being ameliorated with the addition of Tween20 detergent at 0.05% (wt/vol). Furthermore, Quantifoil holey carbon grids coated with a layer of graphene oxide were required to obtain an optimal particle distribution and concentration. While PTX3 still exhibited slight preferential orientation (SI Appendix, Fig. S2), this was compensated for by collecting a large number of micrographs on a 300-kV Titan Krios microscope (SI Appendix, Table S2). Particles displayed four to eight rigid domains assembled in a tetrameric or octameric pattern, and subsequent two-dimensional (2D) class averaging of these particles yielded images that exhibited a range of views of the C-terminal PTX domain, with some classes containing poorly defined density protruding from either end (Fig. 2A). Subsequent 3D refinement resulted in the reconstruction of two maps, one without symmetry applied (C1) and another with D4 symmetry, both resulting in high-resolution (<3 Å) reconstructions (SI Appendix, Fig. S2). The D4 symmetric map reached the highest resolution at 2.5 Å, which clearly showed a dice-like octameric complex with dimensions of 7 × 7 nm (Fig. 2B). Also visible in certain 2D classes was a lower-resolution extension corresponding to part of the N-terminal region (Fig. 2A). Noisy density was visible in this region of the 2.5-Å map, so instead a low-pass filter with a frequency cutoff at 5 Å was applied to the map, which rendered that region of the map more tractable for model building (Fig. 2C).
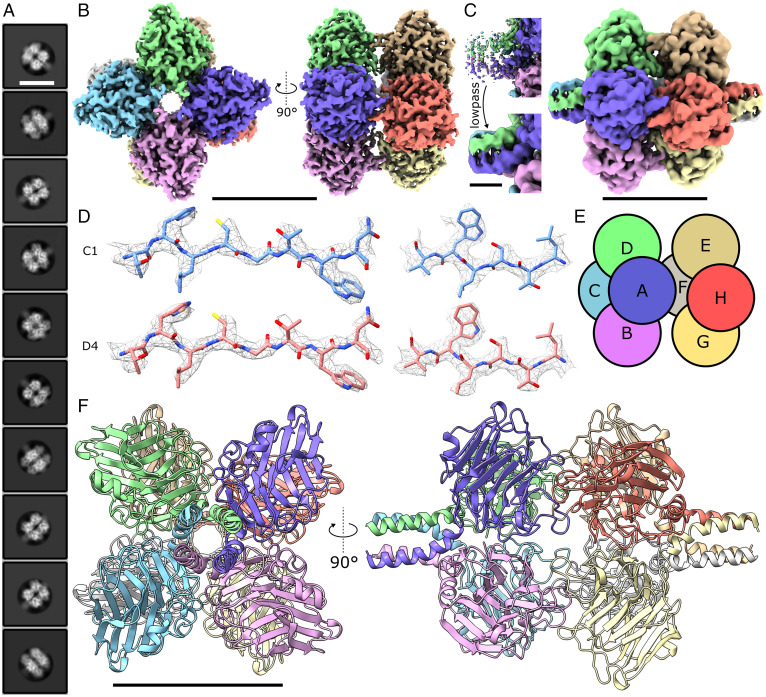
Fig. 2.
Cryo-EM structure of human PTX3. (A) Class averages show octameric structures in solution with less well-defined density protruding from the octameric core. (Scale bar, 110 Å.) (B) Cryo-EM map of the octameric PTX domains of PTX3 resolved with D4 symmetry applied. Each monomer is colored differently. (Scale bar, 50 Å.) (C) Noisy density was visible at the N-terminal regions at lower isosurface thresholds, which a low-pass filter reveals to be structured density protruding from the PTX domain core. (Scale bar, 50 Å, Right, and 10 Å, Left.) (D) The C1 (no applied symmetry, Upper) and D4 symmetrized (Lower) maps both provided sufficient density to model side chain rotamers. (E) Subunit schematic showing PTX domains arranged in a dice-like structure. (F) Model of the PTX domains and partial N-terminal coiled-coil region. (Scale bar, 50 Å.)
The 2.5-Å D4-symmetric map obtained allowed a complete model of the PTX domains and a short section of the N-terminal domain to be built. Both the C1 and D4 symmetrical maps displayed clear side-chain density, which allowed unambiguous modeling of many rotamer states (Fig. 2D and SI Appendix, Table S2). The C1 map fitted into the D4 symmetrized map with a high cross-correlation coefficient of 0.9605 (SI Appendix, Table S2). Together with an improvement in resolution upon map symmetrization, this strongly indicates that the octameric PTX domains form a symmetrical complex in solution.
PTX Domain Architecture of PTX3.
The model showed PTX3 to contain a pentraxin signature sequence (H-X-C-S/T-Y-X-S), present in all PTX domains (SI Appendix, Table S3), that formed a β-strand in a similar fashion to all other resolved human pentraxin domains (SI Appendix, Fig. S3A). The model also revealed an octameric complex (Fig. 2E), with each identical PTX3 monomer comprising a canonical PTX domain (Fig. 2F), as found in all known structures of pentraxins (SI Appendix, Fig. S3B). Although the PTX domain of PTX3 only shares ∼28% sequence identity with CRP, SAP, and the PTX domain from neuronal pentraxin 1 (NPTX1), as determined by BLAST, these structures aligned to PTX3 with a backbone root mean square deviation of 0.9 to 3.5 Å, showing a high degree of structural similarity. The PTX domains form a dimer of tetramers (A–D and E–H, respectively, in Fig. 2E), with each tetramer forming a planar ring held together with noncovalent interactions. Both CRP and SAP also form planar rings (although of five monomers, not four as in PTX3), however the intersubunit interfaces only minimally overlap with those of PTX3 (SI Appendix, Fig. S3C). PTX3 therefore represents a unique architecture for human PTX domain oligomerization.
Density at the N-terminal portion of the model extended beyond the PTX domains, and was clearly modeled as the start of a tetrameric coiled coil comprising four parallel α-helices. Only seven residues of each α-helix were resolved in the 2.5-Å map, but filtering the map to 5 Å enabled extension of the helices to 19 residues (Fig. 2C), which formed a tetrameric coiled coil with a radius of ∼7.4 Å, as determined by TWISTER (19). This region has previously been shown to be important in tetramerization of PTX3 (7), which here is revealed to be mediated by two tetrameric coiled coils that extend from opposing ends of the core PTX octamer (Fig. 2 C and F).
In PTX3, 379 ordered water molecules were identified (SI Appendix, Fig. S4A), and PTX3 was also measured to possess high thermostability observed in thermal shift assays compared to the other classic complement pathway activators CRP and IgG1 (SI Appendix, Fig. S4B). Both the high number of ordered water molecules and disulfide bonds (Fig. 1A and SI Appendix, Fig. S4A), as well as the novel pentraxin oligomerization (SI Appendix, Fig. S3C), may be a consequence of, or contributing factors to, this high thermostability.
Of the 379 waters identified in PTX3, 9 were conserved across the human pentraxins, defined as within 2.5 Å of each other in all four structures (SI Appendix, Fig. S4C). Two of these conserved water molecules were found proximal to the conserved histidine of the pentraxin motif (H269) (SI Appendix, Fig. S4D), hydrogen-bonded to the imidazole nitrogen atoms, as previously highlighted for CRP (5). Intriguingly, although PTX3 is the only pentraxin without a metal-binding site—which is occupied by Ca2+ in CRP, SAP, and NPTX1 but replaced by an extended loop in PTX3—there is also a conserved water proximal to the modified metal-binding region in PTX3 and all human pentraxin structures solved to date (SI Appendix, Fig. S4C).
Disulphide Bond Network of the PTX Domain.
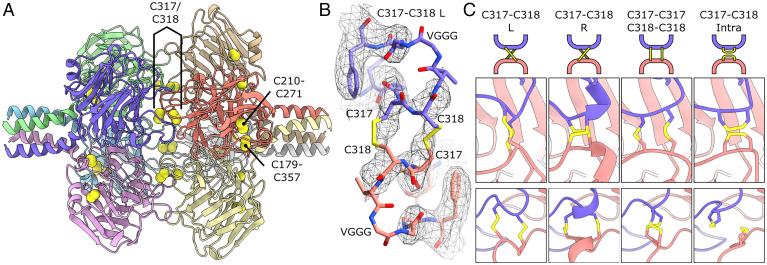
The PTX domain of PTX3 contains six cysteine residues (Fig. 1A), two of which, C317 and C318, are required for octamerization (7). Indeed, in our model each pair of cysteine residues on each monomer subunit face the same two residues on an opposing subunit (e.g., subunits A–H), forming a plane of eight disulfide bonds between the two tetrameric PTX domain rings (Fig. 3A). The map exhibited lower resolution in this region, with the fit best explained by an intermonomer disulfide bond between C317 and C318 with a left-handed twist (C317–C318 L in Fig. 3 B and C). In this configuration, all residues except two glycine residues and a valine of a VGGG linker, fitted well within the map (Fig. 3B). However, biochemical data has indicated some heterogeneity in this region, where both inter- and intramonomer disulfide bonds can form (7). In keeping with this, density in the D4 symmetrical map at high isosurface thresholds displayed heterogeneity in this region (SI Appendix, Fig. S5A), which may indicate a superposition of backbone configurations and differential disulfide bonding. In line with this, the unsymmetrized map showed that one of the four C317/C318 regions displayed heterogeneity, potentially corresponding to a different disulfide bond geometry (Fig. 3C and SI Appendix, Fig. S5B). Extensive 3D classification failed to improve these geometries, and other configurations of this disulfide patch are therefore not precluded from this model (Fig. 3C). The large number of C317/C318 disulfide bonds regulating octamerization, eight per octamer, and their potential to form a mixture of both intra- and intermolecular disulfide bonds likely contributes to the resistance of PTX3 to reduction by DTT (Fig. 1C and SI Appendix, Fig. S1).
Fig. 3.
Cysteine bond network of the PTX domain of PTX3. (A) Octameric arrangement of the PTX domains with cysteine sulfur atoms shown as yellow spheres and disulfide bonds labeled. (B) C317/C318 intertetramer disulfide bond with density mesh showing fit for all atoms except VGG of the glycine linker. The map was low-pass–filtered to 5 Å to aid model building. (C) Reduced resolution in this region does not exclude other potential configurations of the C317/C318 bond network. Shown are a schematic (Top), and two views of the disulfide patch rotated 90° (Middle and Bottom).
Within the PTX domain, there are two more intrasubunit disulfide bonds present; C210–C271 and C179–C357 (Fig. 3A). Homologs of C210–C271 are found in all pentraxins resolved to date, and forms part of the aforementioned PTX signature motif (SI Appendix, Fig. S3A). C179–C357, in contrast, is not found in short pentraxins but a similar motif is present in the long neuronal pentraxin NPTX1. In PTX3, this bond staples the N-terminal α-helix in place, limiting the range of movement of the tetrameric coiled coil in PTX3. The absence of a structure for the NPTX1 N-terminal region inhibits structural analysis of whether this disulfide bond has a similar role for NPTX1 oligomers (20).
PTX3 does not contain a Ca2+ binding site, unlike CRP, SAP, and NPTX1 (5, 6, 20). In CRP and SAP, the Ca2+ binding site is required for lipid binding, where it chelates charged phospholipid headgroups (21). The cognate regions in PTX3 are situated close to both the glycosylation site (N220) and intertetramer cysteine residues (C317 and C318) (SI Appendix, Fig. S5C). Sequence alignment of the calcium binding loop of CRP and the loop containing the C317/C318 disulfide patch in PTX3 reveals a large degree of conservation (SI Appendix, Fig. S5 D and E and Table S3). This loop therefore mediates pentraxin binding: to lipids for CRP and to another monomer for PTX3. Furthermore, the PTX3 glycans regulate binding to various ligands (2, 9, 10). Consequently, the PTX3 glycan and C317/C318 cysteines replace the Ca2+ binding site in CRP and SAP, and as such represents a functionally homologous, but not structurally homologous, region.
Analysis of N-Linked Glycosylation of PTX3.
The PTX domain of PTX3 contains a single glycosylation site at N220 (9, 15), which was hypothesized to be a source of heterogeneity in the electrophoretic profiles of PTX3 (Fig. 1C). The presence of glycosylated structures was confirmed biochemically, as the two bands corresponding to monomers previously seen under reducing conditions (Fig. 1C) were converted into one band upon treatment with PNGase F (Fig. 4A). The presence of sialic acid was confirmed in a similar manner; treatment with neuraminidase resulted in the formation of an intermediate species, with the higher molecular-weight band reducing in mass (Fig. 4A). However, two bands were still detected, unlike with PNGase F treatment, suggesting additional heterogeneity in N-glycosylation beyond the degree of sialylation.
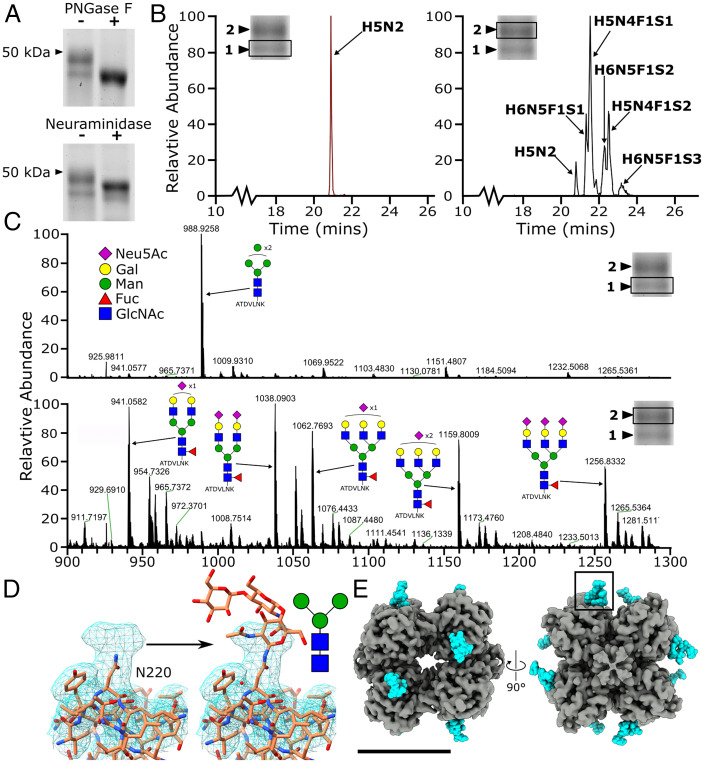
Fig. 4.
Heterogenous N-linked glycosylation of PTX3 at N220. (A) Treatment of monomeric PTX3 with PNGase F (Upper) and neuraminidase (Lower). (B) MS analysis of different glycoforms of the PTX3 tryptic peptide ATDVLNK containing the N-glycosylation site (N220, underlined) in the lower and higher molecular-weight band 1 (Left) and band 2 (Right), respectively. (C) Summed mass spectra (20 to 24 min) from the LC-MS analysis of tryptic peptides from PTX3, corresponding to the lower and higher molecular weight bands (band 1, Upper; and band 2, Lower) shown in A. Predicted species are shown in schematic form. (D) D4 symmetrical map lowpass filtered to 5 Å showing density not accounted for by the protein, with a schematic of the core glycan and detailed view of the boxed region in E. (E) Location of glycans and N220 (cyan) on the low-pass–filtered (4 Å) D4 symmetrical map of the PTX octameric complex. (Scale bar, 50 Å.) The key for the monosaccharides species in C is applicable to D (sialic acid, galactose, mannose, fucose, and N-acetylglucosamine are represented by Neu5Ac, Gal, Man, Fuc, and GlcNAc respectively).
To explore this further, both bands relating to recombinantly produced monomeric PTX3 proteoforms under reducing conditions (without treatment with glycosidases) were excised and investigated via glycoproteomic analysis using MS. Byonic searches (SI Appendix, Table S4) showed that the lower molecular-weight band (band 1 in Fig. 4 B and C) primarily contained high mannose-type glycosylation, as exemplified by the major structure corresponding to the tryptic peptide ATDVLNK carrying two N-acetylhexosamines (N) and five hexoses (H) (m/z 988.926 [M+2H]2+) (Fig. 4C). In contrast, the higher molecular-weight band (band 2, in Fig. 4 B and C) exhibited primarily more complex sugars, with fucosylation (F) and multiple sialylated (S) glycans present on the same tryptic peptide. In addition to this, the higher molecular-weight band (band 2) in Fig. 1C (+/+) was also more intense, indicating that the majority of recombinantly expressed human PTX3 in this study contains these complex glycans. This mixture of glycosylation states supports the notion that the smear and different molecular-weight bands seen during reducing, nonreducing, and native gel electrophoresis were indeed differentially glycosylated PTX3 protomers (Fig. 1C). In our map, density was present that was not accommodated by the protein backbone or sidechains near N220, corroborating the presence of carbohydrate moieties (Fig. 4D). The core glycan observed in the MS data (Fig. 4), comprising the standard two N-acetylglucosamines (GlcNAc) and three mannoses (Man), was built into the map at a low isosurface threshold (Fig. 4D and SI Appendix, Fig. S6), resulting in two carbohydrate moieties per octamer face (Fig. 4E).
AlphaFold Predicted N-Terminal Region of PTX3.
The N-terminal region of PTX3 has no known homologous structure, yet is critical for the structure and biological functions of PTX3 (7). In our map, only the base of the N-terminal region closest to the PTX domains could be resolved, which revealed a parallel tetrameric coiled coil (Fig. 2 C and F). Sequence-based oligomeric state prediction of coiled-coil domains using Logicoil (22) also predicted the N terminus would adopt a near-continuous tetrameric coiled coil from residue F60 to the PTX domain at S173, with a stutter in the heptad repeat around residue 140 (SI Appendix, Table S5).
Extensive class averaging and 3D refinements with extended box, background and mask sizes were attempted to resolve the N-terminal region, but all presented a poorly defined extension, indicating flexibility in this region (SI Appendix, Fig. S7 A and B). To explore the structure of the N-terminal region further, negative-stain electron tomography was utilized to image individual complete PTX3 octamers at low resolution (SI Appendix, Fig. S7 C and D). Slices 15-nm thick through the tomogram revealed a highly heterogenous structure, with individual subtomogram regions showing structures comprising an electron-dense core, corresponding to the octameric PTX domains, with two long, flexible tails protruding from opposing sides, in keeping with the model of extended tetrameric coiled coils.
This flexibility in the N-terminal region hindered conventional SPA reconstruction beyond the PTX domain. To complete the model of PTX3, we turned to AlphaFold to use artificial intelligence to predict the remaining protein structure (SI Appendix, Fig. S6) (14). AlphaFold was unable to predict the oligomeric state, nor overall architecture of octameric or tetrameric PTX3, from sequence data alone. Instead, guided by our high-resolution cryo-EM data, we were able to seed AlphaFold using structures of known tetrameric coiled coils fused in-register C-terminal of the experimentally derived PTX3 coiled coil (SI Appendix, Table S5). AlphaFold consistently predicted a flexible filamentous structure comprising a 175-Å-long tetrameric coiled-coil tail with two proline-rich hinge regions protruding from either end of the octameric PTX core and capped with highly disordered N-terminal regions (Fig. 5A and SI Appendix, Fig. S6 and Table S6). Two cysteine residues per monomer, C47 and C49, are present in this proline-rich disordered section (Fig. 5 A and D). There were no obvious partners to form a disulfide bound for either cysteine, and due to the disordered nature of this area they could presumably participate in intra- or intersubunit disulfide bonds (Fig. 5D). Concurrent mutation of C47 and C49 is associated with the formation of lower-order oligomers, such as trimers and hexamers under denaturing conditions, whereas mutation of only one of the two is markedly less effective (7), suggesting these positions have a degree of redundancy.
Fig. 5.
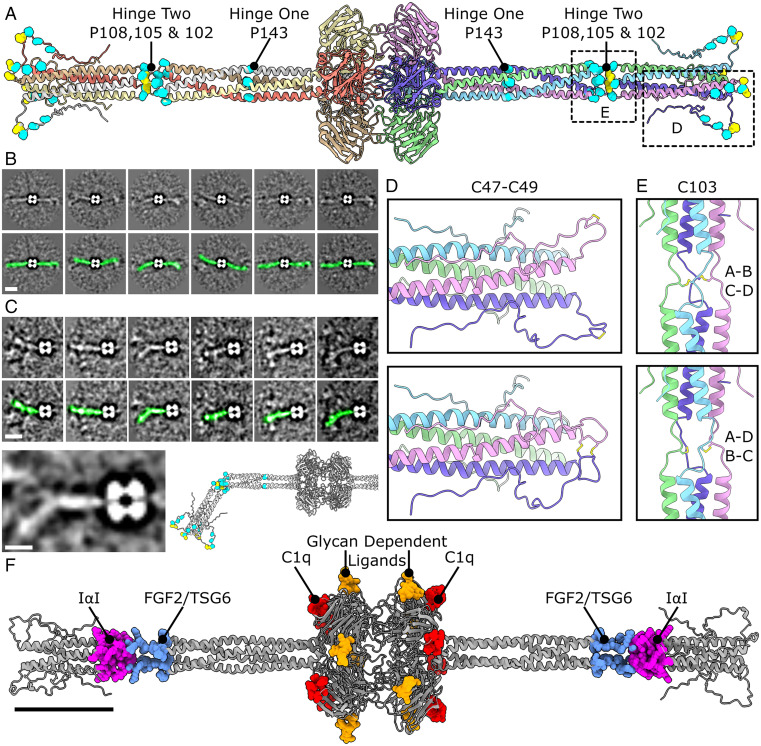
AlphaFold-based structure prediction and validation of the flexible N-terminal region. (A) Structural data from AlphaFold was used to model the entire tetrameric coiled coil N-terminal tail. Cysteine and proline residues in the N-terminal region are represented by yellow and cyan spheres, respectively. (B) Variability analysis of cryo-EM images with masking either side of the pentraxin domain used to visualize the N-terminal region. (Scale bar, 100 nm.) (C) Focused 2D variability analysis with selective masking of one N-terminal region. The atomic model of the N-terminal region could then be modeled onto the 2D classes. Green density in the Lower panels in B and C highlight the N-terminal domain in the 2D classes. (Scale bar in upper panels, 100 nm. Scale bar in lower panel, 50 nm.) (D) Enlarged view of the disordered region showing two possible configurations of C47 and C49 disulfide bonding. (E) Enlarged views of the second hinge region with the two possible configurations of C103-mediated disulfide bonding shown. (F) Binding sites for the ligands shown mapped onto the full-length PTX3 structure. (Scale bar, 50 nm.)
Hinge 1, closest to the PTX domain, is the location of the stutter in the heptad repeat (SI Appendix, Table S5), where proline residue P143 is located. Prolines disrupt the hydrogen bonding network of α-helices and coiled coils, resulting in flexibility and the formation of a hinge. This is also proximal to where the EM density ends in our cryo-EM map, and illustrates why conventional SPA is not able to resolve the protein past this point.
To analyze the range of motion within the N-terminal domain, the original cryo-EM particles were subjected to 2D variability analysis within EMAN2 (23) (Fig. 5 B and C and SI Appendix, Fig. S7 E and F), which produced 2D classes consistent with a model of a central PTX domain core, with two flexible N-terminal regions protruding from either side consisting of rigid units of coiled-coil domains separated by flexible hinge regions. The second hinge contained three proline residues in each helix (Fig. 5A), and indeed appeared to be the location of most of the flexibility observed in both the 2D classes and the tomograms (Fig. 5 B and C and SI Appendix, Fig. S7 C and E). This hinge was also the location of C103, which were oriented on each monomer toward neighboring C103 residues on opposing subunits. As for C47 and C49, there was no obvious partner for each cysteine residue, and presumably this is also a region of heterogeneity, as C103 from chain A can form a disulfide with either cysteine from chain B or D (Fig. 5E). Mutation of C103 was associated with the formation of lower-order oligomers and monomers under denaturing conditions (7) and in keeping with this, C103 mediates covalent dimerization of α-helices within the tetrameric coiled coil in the model presented here. The second hinge region contains the putative binding sites for fibroblast growth factor-2 (FGF2), tumor necrosis factor (TNF)-α–stimulated gene-6 (TSG6), inter-α-trypsin inhibitor (IαI) and the monoclonal antibody MNB4 (24–26) (Fig. 5F and SI Appendix, Table S7), although whether structural changes occur upon ligand association is currently unknown.
Previous biochemical data support the configuration of PTX3 presented here; a dimer of tetramers composed of PTX domains, with each tetramer coupled to a tetrameric coiled-coil N-terminal region. First, mutation of the three N-terminal disulfide bonds (C47, C49, and C103) is still associated with octameric PTX3 under native PAGE (7), indicating that the noncovalent interactions between the PTX domain tetramers and α-helices within the tetrameric coiled coil are sufficient for both tetramerization and octamerization to occur. Furthermore, analytical ultracentrifugation showed that recombinant N-terminal domains are over 95% tetrameric under native conditions (8).
The presence of coiled-coil N-terminal regions has also been predicted in the neuronal long pentraxins NPTX1 and NPTX2, which also contain three N-terminal cysteine residues per monomer involved in the oligomerization of distinct subunits (27). The other human long pentraxins, NPTXR and PTX4, are also predicted to have coiled-coil domains in their N-terminal regions (28), with AlphaFold concurrently predicting a large proportion of α-helical secondary structure at this region in both proteins (14). Our model of PTX3 now strongly implies that PTX4, NPTX1, NPTX2, and NPTXR also utilize their N-terminal coiled-coil domains to form novel pentraxin oligomerization states.
Structural Implications for Ligand Binding.
The complete structure of PTX3, formed by our hybrid cryo-EM/AlphaFold approach, offers insights into ligand binding and structure–function analysis. The structural complexity of PTX3 may explain the vast range of sometimes conflicting functions that have been reported (SI Appendix, Table S7). These include complement activation for immune defense (29, 30), which is mediated via binding to the initiating complex of the classic complement cascade; the C1 complex. Analysis of the atomic model of PTX3 presented here allowed mapping of a putative C1 binding site by identifying homologous residues to those shown to be crucial in C1 binding and complement activation in CRP (31, 32) (SI Appendix, Fig. S8A and Table S3). These residues comprise two clusters, one on the α-helix of the PTX domain and another on a loop region beneath this helix, homologs of which are both present in PTX3 (SI Appendix, Fig. S8B). This creates four binding sites on the faces of the octamer proximal to and facing toward the coiled-coil domain (Fig. 5F), therefore occupying different faces of the protein compared to the glycan moieties (Figs. 4E and 5F). The putative binding site contains predominately acidic residues in both pentraxins (SI Appendix, Fig. S8 A–C; sequences from SI Appendix, Table S3 give combined pIs of 4.09 and 4.03 for PTX3 and CRP, respectively), which is contrasted with the global preponderance of basic residues on the globular head domain of C1q (SI Appendix, Fig. S8C) (gC1q; pIs of 7.91, 9.27 and 8.49 for gC1qA, B and C, respectively), as well as the basic nature of specific residues on gC1q known to be important in binding PTX3 from mutagenesis studies (18, 33).
The PTX3 used in this study bound C1q and caused C4b deposition (SI Appendix, Fig. S1), which combined with our new cryo-EM data showing both structural and chemical conservation in PTX3 of the motifs known to be crucial in C1 binding in CRP, indicates that these same areas in PTX3 form at least part of a putative C1 binding site. However, it is not clear if and how the octameric pentraxin domain architecture of PTX3 relates to the hexameric C1 binding and activation platform of IgG and IgM antibody platforms (34, 35). Previous studies have reported the detection of higher-order structures of PTX3 via SEC, containing 16 protomers (8) or having a mass of ∼900 kDa (29). However, these higher-order oligomers were not detected in any 2D or 3D classes (Fig. 2A and SI Appendix, Fig. S2), which may suggest that the size and flexibility of the N-terminal regions results in an octamer occupying a higher hydrodynamic volume than expected for the given molecular weight. Indeed, in the SEC trace presented here (Fig. 1B), molecular standards of 669 kDa eluted at a similar volume. Alternatively, higher-order oligomers could be transient or weakly bound and require ligand binding to form stable clusters, much like monomeric IgG1 forms hexamers when bound to antigens on lipid membranes (35), and may therefore hint at formation of C1 binding platforms only in the presence of ligands.
Binding of C1q to PTX3 inhibited subsequent binding of C4b-binding protein (C4BP) (36), indicating the same residues or region mediate association of C4BP with PTX3 (SI Appendix, Table S7). Whether PTX3 can bind both C4BP and C1q via different octameric faces, and therefore scaffold inhibitory and activating complement complexes, is unknown. However, such multiface scaffolding could lead to silent clearance of cellular debris to protect against autoimmunity, as has been hypothesized to occur with CRP (37). Similarly, PTX3 can bind the antiinflammatory protein TSG6, which limits neutrophil infiltration and thus damage to host tissues (38, 39). This occurs via the N-terminal region, so concurrent binding of C1 and TSG6 is sterically compatible. TSG6 and the IαI also play crucial roles in ECM organization, especially with regards to female fertility (4), thus these processes could be linked to complement activation and regulation via PTX3. Finally, FGF2 also binds at this N-terminal hinge region, facilitating the inhibition of angiogenesis (40). This suggests PTX3 could regulate the refashioning of damaged or infected regions, such as preventing the creation of neo-vasculature until after a sufficient immune response has been mounted to clear debris or pathogens, after which external cues, such as pH, switch PTX3 to a repair and remodeling mode (3). The arrangement of the PTX domains results in two N220 sites per face of the PTX3 octamer that are distal to the coiled coil regions (Figs. 4E and 5F and SI Appendix, Fig. S8D), with glycans at these positions being observed in both the cryo-EM and MS data (Fig. 4). The carbohydrate moiety is a known modulator of ligand binding (SI Appendix, Table S7) and may form cryptic binding sites on PTX3 via glyco-peptide interactions. Specifically, the presence of terminal sialic acid residues is necessary for some ligands to bind PTX3, but lowers association with the C1 complex (9). Guided by the carbohydrate moieties present in the higher molecular-weight band analyzed by MS (Fig. 4 B and C, band 2), we were able to model a biantennary sialylated glycan comprised of four N-acetylglucosamines, three mannoses, two galactoses, a single fucose, and one terminal sialic acid onto PTX3 using Isolde (41) (SI Appendix, Fig. S8E).
This hypothetical model illustrated that complex carbohydrates could conceivably sterically occlude the putative C1 binding site on the same or neighboring PTX domain monomer (SI Appendix, Fig. S8F), giving a structural basis for biochemical studies reporting the inhibitory effect of sialylation on association with C1q (9). Previous studies have also modeled binding sites of this glycan using molecular dynamics simulations (7), which suggested that the sialylated glycans interacted mostly with polar and charged amino acids to prevent C1 binding, supporting the electrostatic binding interaction between PTX3 and C1q posited above. Whether the stoichiometry of the glycan is tuneable, via the selective glycosylation or deglycosylation of certain faces of PTX3 or N220 residues, is unknown, but could result in the scaffolding of glyco-inhibited with glyco-dependent ligands such the C1 complex (9) and viruses (2).
PTX3 has potent antiviral properties (2, 13), and is correlated with COVID-19 severity in human patients, showing both an increased plasma concentration and expression in lung resident myelomonocytic and endothelial cells (11). Additionally, PTX3 exhibits inhibitory activity against influenza both in vitro and in vivo (2), with the recent finding that PTX3 can bind to the nucleocapsid of SARS-CoV-2 (42). These antiviral properties are associated with different PTX3 glycoforms, with terminal sialic acid sugars forming cryptic binding sites that mimic the same cell surface receptors that viruses, such as influenza, utilize to enter the endocytic system (43). These same glycans are present on our PTX3 structure (Fig. 4), possible configurations of which may represent the antiviral epitopes of PTX3.
Mapping known ligand binding sites onto the full-length structure of PTX3 indicates that PTX3 may be able to bind multiple ligands simultaneously (Fig. 5F). This suggests that PTX3 is a scaffolding protein, being able to link processes, such as complement activation and regulation (29, 30) to antiviral activity (2) or female fertility (7). Consequently, the structural details and functional insights gained from our hybrid cryo-EM/AlphaFold full-length structure of PTX3 may contribute to our understanding of these disorders and processes, and aid the rational design of therapies targeting PTX3.
Methods
Materials and Reagents.
The materials and reagents used are described in detail in SI Appendix.
PTX3 Production.
PTX3 protein was expressed, secreted, and purified from mammalian cells, as described in SI Appendix.
PAGE and Western Blotting.
For denaturing conditions, PTX3 diluted in 2× Laemmli buffer (65.8 mM Tris⋅HCl, 26.3% [wt/vol] glycerol, 2.1% SDS, 0.01% bromophenol blue, pH 6.8), and for reduction DTT was also added at 50 mM. Protein quantities for reducing and nonreducing conditions were 1 and 2 µg, respectively. The sample was subsequently heated to 99 °C for 10 min, briefly centrifuged at 700 × g, and then loaded onto 3% stacking and 8.5%, 8%, or 6% resolving Bis-Tris gels before being run in Tris-glycine-SDS buffer (25 mM Tris, 192 mM glycine, 0.1% SDS) at room temperature for 10 min at 100 V and then 50 min at 200 V. For native conditions, 3 µg of purified recombinant PTX3 was run on 3% stacking and 6% resolving Bis-Tris gels, but without the addition of Laemmli buffer, DTT, or heat, at room temperature for 10 min at 100 V and then 1 h and 50 min at 200 V. Gels contained 1% TCE, which allowed direct protein staining after photoreaction with 500-nm light.
For Western blotting, samples were separated using PAGE as described above, using 2 µg for both reducing and nonreducing denatured conditions and 250 ng for native gels. Samples were transferred to nitrocellulose membranes in Tris-glycine buffer (Tris 12 mM, glycine 96 mM) with 10% methanol. The membranes were then blocked using phosphate buffered saline (PBS) with 0.1% Tween20 and 5% milk (blocking buffer). After washing three times with blocking buffer, proteins were detected with an anti-His6 primary antibody diluted 1:1,000 at 4 °C overnight. The membranes were then washed as before, and subsequently an HRP-linked goat anti-mouse antibody diluted 1:1,000 was added and incubated with the membranes at room temperature for 30 min. The membranes were washed twice in blocking buffer and four times in PBS with 0.1% Tween20. Detection was performed with ECL reagent and imaged on a ChemiDoc gel imaging system (Thermofisher).
Cryo-EM Sample Preparation, Data Collection, and Data Analysis.
PTX3 was prepared for cryo-EM and imaged as described in SI Appendix. For data analysis, motion correction and CTF estimation of the micrographs were performed on-the-fly using WARP (44). Particles were picked from motion-corrected micrographs using the neural network within EMAN2 (23). All subsequent steps were carried out in Relion 3.1 (45) and are described in SI Appendix in detail. Particles underwent several rounds of 2D and 3D classification, as well as CTF estimation and particle polishing to produce the final high-resolution maps with C1 or D4 symmetry applied (SI Appendix, Fig. S2).
To explore variability of the N-terminal domains in 2D, 2× binned 2D classes that contributed to the final map that showed density corresponding to the N-terminal domain were selected. These classes were then separated into three distinct groups representing three clear orientations of the side view of PTX3. Next, particles were reextracted without binning with a box size of 360 pixels. Subsequently, 2D variability analysis was performed using e2motion.py within EMAN2.9 (23). Particles from classes displaying identical orientations of the PTX domain octamer were aligned before masking regions for 2D variability analysis (SI Appendix, Fig. S7). Classes showing clear N-terminal regions were manually identified.
Model Building.
As a starting model, the predicted structure for human PTX3 was downloaded from the AlphaFold Protein Structure Database (14). Residues 1 to 166 were removed, and the resulting monomeric C-terminal PTX domain of PTX3 was placed into the cryo-EM map as a rigid body using University of California San Francisco (UCSF) ChimeraX (46). Disulphides were formed between cysteine residues 179–357 and 210–271 before manual adjustment of the model using ISOLDE (42). The monomer fit was automatically refined using Phenix real-space refine (47) before another monomer was placed into the opposing site (SI Appendix, Fig. S6A). Disulphide bonds were formed between cysteine residues 317 and 318 in four different conformations (Fig. 3C); the best fit was found to be the intermonomer C317–C318 L conformer. This disulphide-linked dimer was again automatically refined using Phenix real-space refine. Next, octameric PTX3 was formed by copying the resulting dimer into the remaining three sites, the interfaces were manually checked using ISOLDE, and another round of refinement was performed using Phenix real-space refine. The core glycan unit comprising two N-acetylglucosamine and three mannose sugars was added to N220 using ISOLDE (SI Appendix, Fig. S6A). Finally, phenix.douse was used to place water molecules in regions of density surrounding the model, before a final round of refinement using Phenix real-space refine was performed (SI Appendix, Fig. S6A).
To model the PTX3 N terminus, the model was extended to include N-terminal residues 153 to 165. The map was low-pass–filtered to 5-Å resolution before residues 153 to 173 were set to be α-helical in ISOLDE and fit into the map as a tetrameric coiled-coil. To seed AlphaFold structure prediction using a tetrameric coiled coil, the CC+ database (48) was searched for proteins containing canonical homotetrameric coiled coils, identifying 1NHL (49) and 1EZJ (50) (PDB codes), which were then submitted to Socket2 (51) to determine the heptad register. Next, the heptad register of N-terminal residues 1 to 173 was determined using Logicoil (22) before the sequences of 1NHL (49) and 1EZJ (50) were fused in-register C-terminal to residue 169 or 166, respectively, yielding the sequences in SI Appendix, Table S5. These were each submitted to the AlphaFold2 server ColabFold (v1.3) (52) as homotetramers. The structures produced are shown in SI Appendix, Fig. S6B. These were all predominantly tetrameric coiled-coil, with long α-helices between residues 55–100 and 107–172, a hinge region formed surrounding cysteine 103, and an unstructured region from residues 1 to 55, containing cysteines 47 and 49. Differences between the models were found primarily in the hinge region and unstructured region (SI Appendix, Fig. S6B). One of the structures was arbitrarily chosen to fuse to the cryo-EM–derived model. Structures derived from AlphaFold and cryo-EM were aligned, and the AlphaFold-derived model was then used to replace residues N-terminal of Glu168 before geometry minimization using Phenix (SI Appendix, Fig. S6A).
Figures were prepared using UCSF ChimeraX (46), as was all structural analysis, such as measuring angles and distances.
In-Gel Trypsin Digestion and MS Analysis.
In-gel trypsin digestion was performed using a Proteineer DP digestion robot (Bruker). Prior to digestion, proteins were first reduced and alkylated using DTT (10 mM) and iodoacetamide (50 mM), respectively. Tryptic peptides were extracted from the gel slices, lyophilized, dissolved in solvent A (95/3/0.1 water/acetonitrile/formic acid [FA] [vol/vol/v]) and subsequently analyzed by on‐line C18 nano-HPLC MS/MS with a system consisting of an Easy nLC 1000 gradient HPLC system (Thermo), and a LUMOS mass spectrometer (Thermo). Samples were injected onto a homemade precolumn (100 μm × 15 mm; Reprosil-Pur C18-AQ 3 μm, Dr. Maisch) and eluted via a homemade analytical nano-HPLC column (15 cm × 50 μm; Reprosil-Pur C18-AQ 3 μm). The gradient was run from 2 to 40% solvent B (20/80/0.1 water/acetonitrile/FA [vol/vol/v]) in 30 min. The nano-HPLC column was drawn to a tip of ∼5 μm, and acted as the electrospray needle of the MS source. The LUMOS mass spectrometer was operated in data-dependent MS/MS (top-20 mode) with a normalized collision energy of 35% and recording of the MS2 spectrum in the Orbitrap. In the master scan (MS1) the resolution was 120,000 with a scan range m/z 300 to 2,000. Dynamic exclusion after n = 1 with exclusion duration of 10 s was applied. For MS/MS, precursors were isolated with the quadrupole with an isolation width of 1.2 Th. The MS2 scan resolution was 30,000. During acquisition a ProductIonTrigger was set on the HexNAc oxonium ion at m/z 204.087. Upon detection of the oxonium ion, additional fragmentation of the same precursor was executed with HCD normalized collision energies of 25%, 32%, and 39%, respectively, using the stepped collision energy mode (i.e., one additional MS/MS scan was recorded). The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (53) partner repository.
MS Data Analysis.
For (glyco)peptide identification, MS/MS spectra were searched against the human database (20,205 sequences, downloaded from Uniprot on April 8, 2020) with Byonic (Protein Metrics, v3.10.10). Precursor mass tolerance was set at 10 ppm and fragment tolerance at 20 ppm. Cleavage C-terminal of K and R was selected (fully specific) and a maximum of two missed cleavages was allowed. As a fixed modification carbamidomethylation of cysteine was selected while acetylation at the protein N-term, and oxidation of methionine were set as variable modifications. For glycopeptide assignment the “N-glycan 309 mammalian no sodium” modification list, as standardly available in Byonic was used. Only glycopeptides with a Byonic score >300 were further selected (SI Appendix, Table S4).
Enzymatic Digestion.
For enzymatic digestion of sugar residues, 5 µg of PTX3 in TBS with SDS 0.1% and 0.05 M DTT were heated to 95 °C for 5 min. Triton X-100 was then added to a final concentration of 1% and the protein was incubated with 0 or 25 units of PNGase F, or separately with 0 or 70 units of α2–3, 6, 8, 9 Neuraminidase for 37 °C for 3 h. The samples were analyzed via SDS/PAGE as described above.
Supplementary Material
Acknowledgments
This work benefited from access to the Netherlands Centre for Electron Nanoscopy at Leiden University, an Instruct-ERIC center with assistance from Ludovic Renault. This research was supported by European Research Council Grant 759517 (to T.H.S.) and The Netherlands Organization for Scientific Research Grants OCENW.KLEIN.291 and VI.Vidi.193.014 (to T.H.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208144119/-/DCSupplemental.
Data Availability
Cryo-EM maps and the associated models of PTX3 are deposited in the Electron Microscopy database (EMDB) and protein database (PDB) (CryoEM-derived PTX3, EMD-14775 (C1) (54), EMD-14774 (D4) (55), and PBD ID code 7ZL1) (56). The full AlphaFold/cryo-EM hybrid model is deposited in PDB-Dev (ID code PDBDEV_00000141) (57). MS data are available via ProteomeXchange (identifier PXD034602) (58). All other study data are included in the main text and SI Appendix.
References
- 1.Garlanda C., Bottazzi B., Bastone A., Mantovani A., Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu. Rev. Immunol. 23, 337–366 (2005). [DOI] [PubMed] [Google Scholar]
- 2.Reading P. C., et al. , Antiviral activity of the long chain pentraxin PTX3 against influenza viruses. J. Immunol. 180, 3391–3398 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Doni A., et al. , An acidic microenvironment sets the humoral pattern recognition molecule PTX3 in a tissue repair mode. J. Exp. Med. 212, 905–925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salustri A., et al. , PTX3 plays a key role in the organization of the cumulus oophorus extracellular matrix and in in vivo fertilization. Development 131, 1577–1586 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Noone D. P., van der Velden T. T., Sharp T. H., Cryo-electron microscopy and biochemical analysis offer insights into the effects of acidic pH, such as occur during acidosis, on the complement binding properties of C-reactive protein. Front Immunol. 12, 757633 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emsley J., et al. , Structure of pentameric human serum amyloid P component. Nature 367, 338–345 (1994). [DOI] [PubMed] [Google Scholar]
- 7.Inforzato A., et al. , Structural characterization of PTX3 disulfide bond network and its multimeric status in cumulus matrix organization. J. Biol. Chem. 283, 10147–10161 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Inforzato A., et al. , The angiogenic inhibitor long pentraxin PTX3 forms an asymmetric octamer with two binding sites for FGF2. J. Biol. Chem. 285, 17681–17692 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inforzato A., et al. , Structure and function of the long pentraxin PTX3 glycosidic moiety: Fine-tuning of the interaction with C1q and complement activation. Biochemistry 45, 11540–11551 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Deban L., et al. , Regulation of leukocyte recruitment by the long pentraxin PTX3. Nat. Immunol. 11, 328–334 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Brunetta E., et al. , Macrophage expression and prognostic significance of the long pentraxin PTX3 in COVID-19. Nat. Immunol. 22, 19–24 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Gutmann C., et al. , SARS-CoV-2 RNAemia and proteomic trajectories inform prognostication in COVID-19 patients admitted to intensive care. Nat. Commun. 12, 3406 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han B., et al. , Protective effects of long pentraxin PTX3 on lung injury in a severe acute respiratory syndrome model in mice. Lab. Invest. 92, 1285–1296 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jumper J., et al. , Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Inforzato A., et al. , The “sweet” side of a long pentraxin: How glycosylation affects PTX3 functions in innate immunity and inflammation. Front. Immunol. 3, 407 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Haan C. A., et al. , The glycosylation status of the murine hepatitis coronavirus M protein affects the interferogenic capacity of the virus in vitro and its ability to replicate in the liver but not the brain. Virology 312, 395–406 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreau C., et al. , Structural and functional characterization of a single-chain form of the recognition domain of complement protein C1q. Front. Immunol. 7, 79 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bally I., et al. , Interaction of C1q with pentraxin 3 and IgM revisited: Mutational studies with recombinant C1q variants. Front. Immunol. 10, 461 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strelkov S. V., Burkhard P., Analysis of alpha-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 137, 54–64 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Suzuki K., et al. , A synthetic synaptic organizer protein restores glutamatergic neuronal circuits. Science 369, eabb4853 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikolajek H., et al. , Structural basis of ligand specificity in the human pentraxins, C-reactive protein and serum amyloid P component. J. Mol. Recognit. 24, 371–377 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Vincent T. L., Green P. J., Woolfson D. N., LOGICOIL—Multi-state prediction of coiled-coil oligomeric state. Bioinformatics 29, 69–76 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Bell J. M., Chen M., Durmaz T., Fluty A. C., Ludtke S. J., New software tools in EMAN2 inspired by EMDatabank map challenge. J. Struct. Biol. 204, 283–290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camozzi M., et al. , Identification of an antiangiogenic FGF2-binding site in the N terminus of the soluble pattern recognition receptor PTX3. J. Biol. Chem. 281, 22605–22613 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Scarchilli L., et al. , PTX3 interacts with inter-alpha-trypsin inhibitor: Implications for hyaluronan organization and cumulus oophorus expansion. J. Biol. Chem. 282, 30161–30170 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Leali D., et al. , Long pentraxin 3/tumor necrosis factor-stimulated gene-6 interaction: A biological rheostat for fibroblast growth factor 2-mediated angiogenesis. Arterioscler. Thromb. Vasc. Biol. 32, 696–703 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu D., et al. , Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron 39, 513–528 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Gómez de San José N., et al. , Neuronal pentraxins as biomarkers of synaptic activity: From physiological functions to pathological changes in neurodegeneration. J. Neural Transm. (Vienna) 129, 207–230 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottazzi B., et al. , Multimer formation and ligand recognition by the long pentraxin PTX3. Similarities and differences with the short pentraxins C-reactive protein and serum amyloid P component. J. Biol. Chem. 272, 32817–32823 (1997). [DOI] [PubMed] [Google Scholar]
- 30.Deban L., et al. , Binding of the long pentraxin PTX3 to factor H: Interacting domains and function in the regulation of complement activation. J. Immunol. 181, 8433–8440 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Bang R., et al. , Analysis of binding sites in human C-reactive protein for FcγRI, FcγRIIA, and C1q by site-directed mutagenesis. J. Biol. Chem. 280, 25095–25102 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Agrawal A., Volanakis J. E., Probing the C1q-binding site on human C-reactive protein by site-directed mutagenesis. J. Immunol. 152, 5404–5410 (1994). [PubMed] [Google Scholar]
- 33.Roumenina L. T., et al. , Interaction of C1q with IgG1, C-reactive protein and pentraxin 3: Mutational studies using recombinant globular head modules of human C1q A, B, and C chains. Biochemistry 45, 4093–4104 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharp T. H., et al. , Insights into IgM-mediated complement activation based on in situ structures of IgM-C1-C4b. Proc. Natl. Acad. Sci. USA 116, 11900–11905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ugurlar D., et al. , Structures of C1-IgG1 provide insights into how danger pattern recognition activates complement. Science 359, 794–797 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Braunschweig A., Józsi M., Human pentraxin 3 binds to the complement regulator c4b-binding protein. PLoS One 6, e23991 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gershov D., Kim S., Brot N., Elkon K. B., C-reactive protein binds to apoptotic cells, protects the cells from assembly of the terminal complement components, and sustains an antiinflammatory innate immune response: Implications for systemic autoimmunity. J. Exp. Med. 192, 1353–1364 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wisniewski H. G., et al. , TNF/IL-1-inducible protein TSG-6 potentiates plasmin inhibition by inter-alpha-inhibitor and exerts a strong anti-inflammatory effect in vivo. J. Immunol. 156, 1609–1615 (1996). [PubMed] [Google Scholar]
- 39.Maina V., et al. , Coregulation in human leukocytes of the long pentraxin PTX3 and TSG-6. J. Leukoc Biol. 86, 123–132 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Presta M., Foglio E., Churruca Schuind A., Ronca R., Long pentraxin-3 modulates the angiogenic activity of fibroblast growth factor-2. Front. Immunol. 9, 2327 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D Struct. Biol. 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stravalaci M., et al. , Recognition and inhibition of SARS-CoV-2 by humoral innate immunity pattern recognition molecules. Nat. Immunol. 23, 275–286 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Imai M., et al. , Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486, 420–428 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tegunov D., Cramer P., Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheres S. H. W., Amyloid structure determination in RELION-3.1. Acta Crystallogr. D Struct. Biol. 76, 94–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pettersen E. F., et al. , UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Testa O. D., Moutevelis E., Woolfson D. N., CC+: A relational database of coiled-coil structures. Nucleic Acids Res. 37, D315–D322 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freedman S. J., Song H. K., Xu Y., Sun Z.-Y. J., Eck M. J., Homotetrameric structure of the SNAP-23 N-terminal coiled-coil domain. J. Biol. Chem. 278, 13462–13467 (2003). [DOI] [PubMed] [Google Scholar]
- 50.Tarbouriech N., Curran J., Ruigrok R. W. H., Burmeister W. P., Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat. Struct. Biol. 7, 777–781 (2000). [DOI] [PubMed] [Google Scholar]
- 51.Kumar P., Woolfson D. N., Socket2: A program for locating, visualising, and analysing coiled-coil interfaces in protein structures. Bioinformatics 37, 4575–4577 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mirdita M., et al. , ColabFold—Making protein folding accessible to all. Nat. Methods. 19, 679–682 (2022). 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Riverol Y., et al. , The PRIDE database resources in 2022: A hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 50 (D1), D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Noone D. P., et al. , Data from “PTX3 structure determination using a hybrid cryoelectron microscopy and AlphaFold approach offers insights into ligand binding and complement activation.” Electron Microscopy Database. http://www.ebi.ac.uk/pdbe-srv/emsearch/atlas/14775_summary.html. Deposited 13 April 2022. [DOI] [PMC free article] [PubMed]
- 55.Noone D. P., et al. , Data from “PTX3 structure determination using a hybrid cryoelectron microscopy and AlphaFold approach offers insights into ligand binding and complement activation.” Electron Microscopy Database. http://www.ebi.ac.uk/pdbe-srv/emsearch/atlas/14774_summary.html. Deposited 23 June 2022. [DOI] [PMC free article] [PubMed]
- 56.Noone D. P., et al. , Data from “PTX3 structure determination using a hybrid cryoelectron microscopy and AlphaFold approach offers insights into ligand binding and complement activation.” Protein Data Bank. 10.2210/pdb7ZL1/pdb. Deposited 21 June 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Noone D. P., et al. , Data from “PTX3 structure determination using a hybrid cryoelectron microscopy and AlphaFold approach offers insights into ligand binding and complement activation.” PDB-Dev. https://pdb-dev.wwpdb.org/entry.html?PDBDEV_00000141. Deposited 29 July 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noone D. P., et al. , Data from “PTX3 structure determination using a hybrid cryoelectron microscopy and AlphaFold approach offers insights into ligand binding and complement activation.” ProteomeXchange. https://www.ebi.ac.uk/pride/archive/projects/PXD034602. Deposited 16 June 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Cryo-EM maps and the associated models of PTX3 are deposited in the Electron Microscopy database (EMDB) and protein database (PDB) (CryoEM-derived PTX3, EMD-14775 (C1) (54), EMD-14774 (D4) (55), and PBD ID code 7ZL1) (56). The full AlphaFold/cryo-EM hybrid model is deposited in PDB-Dev (ID code PDBDEV_00000141) (57). MS data are available via ProteomeXchange (identifier PXD034602) (58). All other study data are included in the main text and SI Appendix.