Abstract
In Lactobacillus casei ATCC 393, the chromosomally encoded lactose operon, lacTEGF, encodes an antiterminator protein (LacT), lactose-specific phosphoenolpyruvate-dependent phosphotransferase system (PTS) elements (LacE and LacF), and a phospho-β-galactosidase. lacT, lacE, and lacF mutant strains were constructed by double crossover. The lacT strain displayed constitutive termination at a ribonucleic antiterminator (RAT) site, whereas lacE and lacF mutants showed an inducer-independent antiterminator activity, as shown analysis of enzyme activity obtained from transcriptional fusions of lac promoter (lacp) and lacpΔRAT with the Escherichia coli gusA gene in the different lac mutants. These results strongly suggest that in vivo under noninducing conditions, the lactose-specific PTS elements negatively modulate LacT activity. Northern blot analysis detected a 100-nucleotide transcript starting at the transcription start site and ending a consensus RAT sequence and terminator region. In a ccpA mutant, transcription initiation was derepressed but no elongation through the terminator was observed in the presence of glucose and the inducing sugar, lactose. Full expression of lacTEGF was found only in a man ccpA double mutant, indicating that PTS elements are involved in the CcpA-independent catabolite repression mechanism probably via LacT.
Lactobacillus casei is a lactic acid bacterium (LAB) found in many food products, such as fermented vegetables, milk, and meat, as well as in the human body and other natural environments. Recently, L. casei has been used in new fermented milk products with original flavors for which certain health benefits are claimed.
During milk fermentation, lactose is fermented by LAB through different pathways that differ in intermediary metabolites and their bioenergetics. However, it is the transport and phosphorylation mechanism that will determine the metabolism of the translocated disaccharide. Three lactose transport mechanisms have been identified in LAB: lactose-galactose antiporters, lactose-H+ symport systems, and the lactose-specific phosphoenolpyruvate-dependent phosphotransferase system (PTS) (19). The lactose-specific PTS (Lac-PTS) is bioenergetically the most efficient one since the sugar is translocated and phosphorylated in a single step. This system has been described only for Streptococcus mutans, Lactococcus lactis, L. casei, and the non-LAB Staphylococcus aureus (1–3, 11, 12, 18, 19, 23, 31, 37).
L. casei ATCC 393 has two lactose assimilation mechanisms, the chromosomal Lac-PTS and a permease/β-galactosidase system encoded by plasmid pLZ15 (13, 21). In L. casei ATCC 393[pLZ15−], the genetic structure and nucleotide sequence of lactose assimilation genes differs from that in S. mutans, L. lactis, and Staphylococcus aureus (22). In L. casei, the lactose genes are transcribed as an operon, where the genes of the tagatose-6-phosphate pathway are not included, as they are in other lac operons described (19). The cluster lacTEGF encodes for a regulatory protein (lacT), lactose-specific PTS proteins (lacE and lacF), and a phospho-β-galactosidase (P-β-Gal) (lacG). The promoter region contains a cre element overlapping the −35 region, which is followed by a highly conserved sequence, the ribonucleic antiterminator (RAT) sequence, and a terminator structure. It has previously been reported (22, 34) that the expression of the lac operon in L. casei ATCC 393[pLZ15−] is subject to dual regulation: carbon catabolite repression (CR) and induction by lactose through transcriptional antitermination. Most CR was shown to be mediated by the general regulatory protein CcpA that regulates lac operon expression, possibly by binding to the cre element at the lactose promoter (lacp). However, an additional CcpA-independent CR effect was observed, which was related to a functional glucose-specific PTS (EIIMan). The second regulatory mechanism involves induction by lactose and is mediated by LacT, a protein that belongs to the BglG family of transcriptional antiterminators (3, 22). This later mechanism is remarkably different from the induction system found in the lac operon in L. lactis, where gene expression is controlled by LacR with tagatose-6-phosphate the likely inducer (18, 19). There is only one other antiterminator protein described in LAB, BglR from L. lactis (10).
Antitermination activity has been extensively studied in homologous proteins, such as BglG from Escherichia coli, which regulates β-glucoside utilization genes (24, 32, 33, 43, 45). However, antiterminators seem to be more frequently found in gram-positive bacteria. In Bacillus subtilis, four different antiterminators have been described: SacT, SacY, LicT, and GlcT, which control the expression of genes related to sucrose, β-glucan, or glucose assimilation (7, 17, 40, 42, 46, 47, 50). The antitermination activity of all of these proteins is dependent on their binding to a RAT sequence, resulting in the unwinding of a neighboring terminator structure in their respective mRNAs (9). BglG from E. coli has been found to be phosphorylated by the β-glucoside PTS transporter, BglF (EIIBgl), which is encoded in the bgl operon. Phosphorylated BglG is monomeric and has no antitermination activity. However, in the presence of β-glucosides, BglG is dephosphorylated, which in turn promotes dimer formation and subsequently full antitermination activity (4–6, 43, 44).
The antiterminator protein SacY controls expression of the sacB gene in B. subtilis; it functions similarly to BglG, and the PTS component involved in this case is SacX (EIIScr) (15, 26). Tortosa et al. (48) found two conserved domains (P1 and P2) common to a number of transcriptional regulators, and through elegant in vitro experiments they demonstrated phosphorylation of SacY by HPr-His-P. Also, SacT and LicT were shown to be activated by phosphorylation by the general components of the PTS (HPr and EI) in B. subtilis (8, 16, 27–29). Recently, Stülke et al. (47) have described the conserved domains common to PTS-controlled transcriptional regulators as the PTS regulation domains (PRDs). They proposed that the PRD closer to the N terminus (PRD-I) is related to the negative control played by the specific sugar permeases, whereas the PRD closer to the C terminus (PRD-II) shows a positive regulation by HPr.
To establish the role of the lac genes in the regulation of the lac operon in L. casei ATCC 393[pLZ15−], different mutants (lacE, lacT, and lacF) were obtained by double crossover and then used to monitor the expression of E. coli β-glucuronidase gene (gusA) as reporter under the control of lacp. Transcriptional analysis was also performed in the three lac mutants, in a man (encoding EIIMan) mutant and in the ccpA man double mutant. These experiments confirmed that the RAT-terminator/LacT interaction is involved in the CcpA-independent CR mechanism and demonstrated that the antiterminator activity of LacT is also negatively regulated by the lactose-specific enzymes, EIILac.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
The L. casei strains and plasmids used in this work are listed in Table 1. L. casei cells were grown in MRS medium (Oxoid) and MRS fermentation broth (Adsa-Micro; Scharlau S.A., Barcelona, Spain) plus 0.5% carbohydrate at 37°C under static conditions. E. coli DH5α was grown with shaking at 37°C in Luria-Bertani (LB) medium. Bacteria were plated on media solidified with 1.5% agar. When required, the concentrations of antibiotics used were 100 μg of ampicillin, 300 μg of erythromycin, or 10 μg of chloramphenicol per ml to select E. coli transformants and 5 μg of erythromycin or 5 μg of chloramphenicol per ml for L. casei.
TABLE 1.
L. casei strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| L. casei strains | ||
| BL23 | ATCC 393[pLZ15−] | B. Chassy (University Illinois, Urbana) |
| BL23D | man | 49 |
| BL71 | ccpA | 34 |
| BL72 | man ccpA | 22 |
| BL153 | BL23 lacE | This work |
| BL154 | BL23 lacT | This work |
| BL155 | BL23 lacF | This work |
| Plasmids | ||
| pUC18 | Amr | Boehringer Mannheim |
| pRV300 | Ermr from pAMβ1 | 30 |
| pBluescriptII SK+ | Amr | Stratagene |
| pT7Blue-T-vector | Amr | Novagen |
| pNZ272 | gusA Cmr | 36 |
| pNZlac | pNZ272 containing the cre element of lacp fused to gusA | 34 |
| pNZRAT | pNZ272 containing the cre element and RAT-terminator of lacp fused to gusA | This work |
| pMJ39 | lacE with 0.965-kb deletion in pRV300 | This work |
| pMJ41 | lacT with a frameshift at PstI site in pRV300 | This work |
| pMJ45 | lacF with a frameshift at SphI site in pRV300 | This work |
| pMJ33 | pBluescript containing a fragment of lacp (Fig. 1) | This work |
| pMJ64 | pBluescript containing a fragment of lacE and lacG (Fig. 1) | This work |
Recombinant DNA procedures.
Genomic DNA from L. casei strains was purified by using a Purogene DNA isolation kit (Gentra Systems, Inc., Minneapolis, Minn.) as described by the manufacturer. Restriction and modifying enzymes were used according to the recommendations of manufacturers. General cloning procedures were performed as described by Sambrook et al. (41).
To obtain plasmid pNZRAT, the promoter lacp was amplified with primers lac11 (5′-TAGCACTGATCATTAAA-3′) and lac33 (5′-TTGCACTGGGAGGGGAT-3′), using L. casei DNA as the template, and the PCR product was cloned into the SmaI site of pUC18. The orientation of the insert was checked by PCR. A clone with the appropriate orientation was digested with EcoRI and PstI, and the resulting fragment was cloned into PstI/EcoRI-digested pNZ272 vector (36). The plasmid obtained, pNZRAT, carries a transcriptional fusion of the L. casei lacTEGF promoter, including the RAT sequence and terminator structure, with the gusA gene of E. coli. pNZlac (34) carries a transcriptional fusion of the lac promoter, lacking the RAT-terminator region, with the gusA gene. L. casei strains were transformed by electroporation with a Gene-Pulser apparatus (Bio-Rad Laboratories, Richmond, Calif.) as described elsewhere (38).
RNA isolation and Northern blot analysis.
L. casei strains were grown in MRS fermentation medium with different sugars to an optical density at 550 nm of 0.8 to 1. Cells from a 10-ml culture were collected by centrifugation, washed with 50 mM EDTA, and resuspended in 1 ml of Trizol (Gibco BRL). One gram of 0.1-mm-diameter glass beads was added, and the cells were broken by shaking in a Fastprep apparatus (Biospec, Bartlesville, Okla.) two times for 45 s. RNA was isolated as described by Gibco BRL, separated by formaldehyde-agarose gel electrophoresis, and transferred to Hybond-N membranes (Amersham).
RNA probes were obtained as follows. Probe Ppt was obtained from plasmid pMJ64. A fragment from nucleotides (nt) −162 to +125 with respect to the transcriptional start site of the lac operon was cloned between the EcoRV and BamHI sites of pBluescriptII SK−. Probe PIIgal was derived from pMJ33 carrying an internal fragment of the lac operon cloned in the EcoRV site of pT7Blue-T-vector (Novagen). Antisense RNAs were synthesized in vitro from linearized plasmids with T3 and T7 RNA polymerase, respectively, using the reagents from the Boehringer digoxigenin-RNA labeling kit as recommended by the manufacturer. Hybridization, washing, and staining were done as described by the supplier.
Enzymatic assays.
P-β-Gal and β-glucuronidase activities were assayed as previously described (14, 36) in permeabilized L. casei cells (49).
Construction of L. casei lac mutants.
Mutations in lacT, lacE, and lacF genes were obtained by Campbell-like recombination using integrative plasmid pRV300 (30).
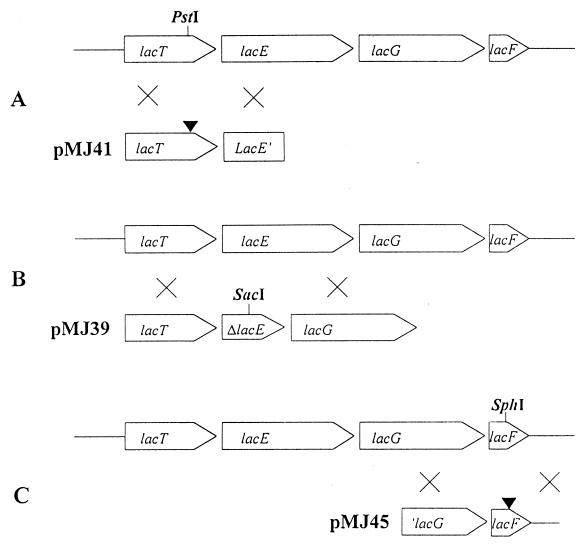
Three plasmids, pMJ41, pMJ39, and pMJ45, were constructed by cloning a fragment of the genes lacT, lacE, and lacF, respectively, in the integration vector. pMJ41 contained a PCR product obtained by using primers lac11 (5′-TAGCACTGATCATTAAA-3′) and lac2 (5′-CAACGATATAAGCGCAGATC-3′). The fragment was made blunt ended and digested with XbaI and then cloned in pRV300 digested with SpeI and EcoRV, and an internal PstI site was made blunt ended with the Klenow fragment of E. coli DNA polymerase, thus generating a frameshift mutation in the cloned fragment. A 1-kb HindIII/BglII fragment from pLZ613 (1) was introduced into pRV300, and the SphI site was treated as was the PstI site described above to give pMJ45. pMJ39 carried a 1.7-kb insert that had a 965-bp deletion in lacE. For its construction, two PCR fragments spanning the regions upstream and downstream of the desired deletion, which also had newly created SacI sites (underlined regions below), were digested with SacI and ligated. The oligonucleotides used as primers in the PCRs were lac11 (5′-TAGCACTGATCATTAAA-3′) and lac25 (5′-CGATATGAGCTCAGATC-3′) for one fragment and lac26 (5′-CAACGAGCTCAACAAAC-3′) and lac6 (5′-CTTGCTGTCTAAATAGCC-3′) for the other fragment. The ligation product was cloned as a KpnI-digested blunt-end fragment into KpnI/EcoRV-digested pRV300. These three plasmids were used to transform L. casei to erythromycin resistance (Ermr), as integration of the DNA fragments into L. casei chromosome occurred through a single crossover by Campbell-like recombination. For each transformation, one Ermr colony was grown for 200 generations without antibiotic. Strains that had undergone a second recombination event due to the excision of the vector could be detected as Erms. The proper first and second recombination events were confirmed by Southern blot hybridization of the chromosomal DNA, and the phenotype of the appropriate mutants was analyzed.
RESULTS
Transcriptional analysis of lac operon in EIIMan and CcpA-deficient mutants.
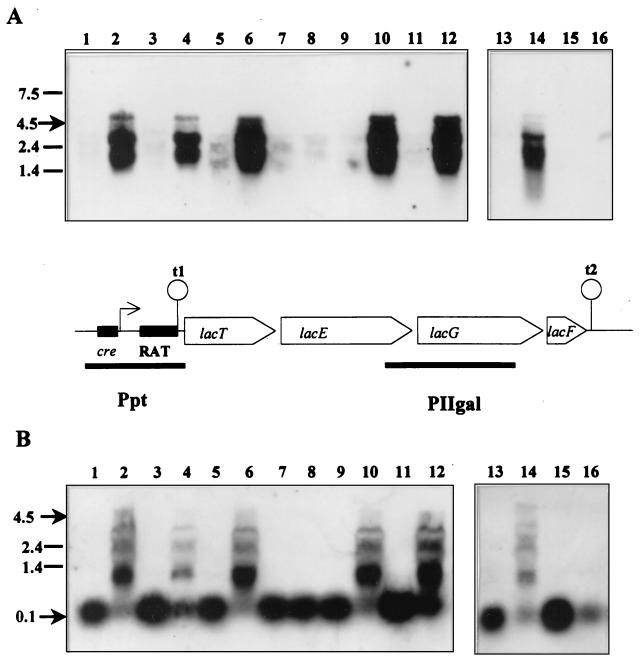
In a previous study (22), P-β-Gal was measured in BL23, BL23D (man), BL71 (ccpA), and BL72 (man ccpA) grown on glucose, lactose, and glucose plus lactose. Only the double mutant, BL72, showed full derepression of P-β-Gal activity when grown on the two latter sugars, whereas BL23D (man) had 24% of the activity found during growth on lactose. To investigate this behavior at the molecular level, Northern blot experiments were performed. With probe PIIgal (Fig. 1A), a signal corresponding to a transcript of 4.5 kb was obtained in all strains when the cells were grown on lactose (Fig. 1A, lanes 2, 6, 10, and 14). This size is in agreement with the expected length (4.8 kb) of a transcript which runs from the transcriptional start site of lac operon to the rho-independent terminator t2 (3, 22). Additional bands corresponding to the 23S and 16S rRNAs were also noticed, which is sometimes the case in Northern blot experiments (25, 35, 39). In the wild-type strain, the 4.5-kb transcript was detected only on lactose-grown cells. When a mixture of glucose and lactose was used to grow all strains, transcription of the complete lac operon occurred only in the man mutant and ccpA man double mutant, although the intensity of the signal was lower in BL23D (Fig. 1A, lanes 4 and 12). However, the 4.5-kb mRNA was never found in glucose- or ribose-grown cells. These results correlate perfectly with the P-β-Gal activities described elsewhere (22).
FIG. 1.
Northern blots prepared with RNA from different L. casei mutants impaired in the catabolite repression signal transduction. The probes used were PIIgal (A) and Ppt (B); the strains used were BL23D (man) (lanes 1 to 4), BL71 (ccpA) (lanes 5 to 8), BL72 (man ccpA) (lanes 9 to 12), and BL23 (wild type) (lanes 13 to 16). Cells were grown on glucose (lanes 1, 5, 9, and 13), lactose (lanes 2, 6, 10, and 14), ribose (lanes 3, 7, 11, and 15) or glucose plus lactose (lanes 4, 8, 12, and 16). The diagram shows the structure of the lac operon and the relative positions of the RNA probes, PIIgal and Ppt, used in this experiment.
RNAs from the same sources were used in another Northern blot with the Ppt probe from the region between nt −162 and +125 with respect to the transcriptional start site of lac operon (Fig. 1). A transcript of about 100 nt was detected in all samples where large mRNA species were not found. This indicates that in the absence of inducer (e.g., ribose-grown cells) transcription stopped at the terminator structure downstream of the promoter (Fig. 1B, lanes 3, 7, 11, and 15). In the wild type, the amount of this 100-nt transcript clearly decreased under repressing conditions (Fig. 1B; compare lanes 13, 15, and 16). However, in BL71 (ccpA), the intensities of the 100-nt RNA obtained under repressing and nonrepressing conditions were nearly identical (Fig. 1B, lanes 5, 7, and 8). This result corroborates previous data (22, 34) and further suggests that CcpA protein mediates catabolite repression at the level of transcription initiation, by binding to the cre site of lacp. However, the absence of a functional CcpA protein is not enough to overcome glucose repression, as full derepression of lac operon was found only in glucose-plus-lactose-grown cells of the BL72 (man ccpA) mutant. Possibly there is an additional CcpA-independent catabolite repression mechanism that involves the transport of glucose by the PTS and also probably the LacT protein.
Construction of L. casei lac mutants and expression studies using the gusA gene as reporter.
The lactose operon, lacTEGF, of L. casei BL23 encodes the regulatory protein LacT, lactose-specific EIIA and EIICB PTS elements (LacF and LacE), and P-β-Gal (LacG) (22). To establish the role of lac gene products in induction of the lacTEGF operon, mutants BL154 (lacT), BL153 (lacE), and BL155 (lacF) were obtained by a double-crossover event (Fig. 2). Although all mutants turned out to be impaired in lactose fermentation, P-β-Gal activity, encoded by lacG, could be used to report the expression of the gene cluster. Consequently, lactose induction of this activity was determined in the lac mutants and in the wild-type strain grown on ribose and ribose plus lactose. In BL23, expression of the lac operon was induced by lactose (9 and 17.9 nmol/min/mg [dry weight], respectively), whereas BL155 (lacF) showed higher P-β-Gal activity under both conditions (27.7 and 29.5 nmol/min/mg [dry weight], respectively). These results indicate that LacF is involved in the induction of the lac operon. No activity was detected in BL154 (lacT), which would be impaired in the antiterminator protein, indicating a lack of induction in the presence of lactose. Surprisingly, no P-β-Gal activity was detected in BL153.
FIG. 2.
Construction of lacT (A), lacE (B), and lacF (C) mutants of L. casei. Plasmids pMJ41, pMJ39, and pMJ45 carrying a frameshift in the PstI site, a 0.965-kb deletion in lacE, and a frameshift in the SphI site, respectively, were used to transform the wild-type strain. The different mutants were selected after double-crossover events.
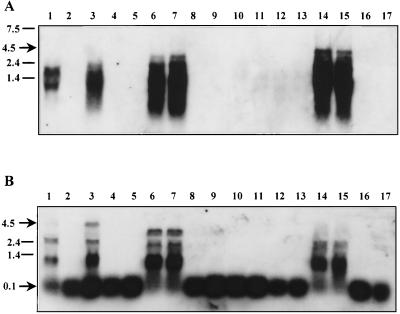
The effect of the mutations in lacE, lacT, and lacF was also studied with the β-glucuronidase reporter system in plasmids pNZlac and pNZRAT (Table 2). In plasmid pNZlac, which lacks the RAT-terminator area but contains the cre element in lacp, β-glucuronidase activity was detected in similar amounts in all strains grown on ribose but was negligible when strains were grown on glucose. Moreover, no induction by lactose was found in the wild type. With pNZRAT, carrying the whole promoter, a remarkable decrease of activity occurred in the lacT mutant with respect to the other strains, consistent with the antiterminator nature of LacT. The fact that β-glucuronidase activity in BL23 grown on ribose was 20-fold higher than that detected in the lacT mutant suggests that antitermination mediated by LacT also occurs to some extent in the absence of lactose and without glucose in the wild-type strain. The highest β-glucuronidase activity was found in BL153 (lacE) and BL155 (lacF) grown under noninducing conditions, indicating a negative effect of EIILac on the antiterminator activity of LacT. No release of glucose repression was observed in the different strains transformed with pNZlac or pNZRAT, but there was a greater repression when the RAT-terminator element was present in the fusion used, except in the lacT strain, suggesting that there is a glucose repression effect mediated by the LacT protein. In the case of strain BL153 (lacE), divergent results were found between the homologous (P-β-Gal) and heterologous (gusA) reporter systems. However, the disruption of lacE might have caused a translational defect in lacG, as the operon seems to be transcribed at a high rate under nonrepression conditions (Fig. 3A, and B, lanes 6 and 7).
TABLE 2.
Expression of different transcriptional fusions in L. casei strains
| Strain | Relevant genotype | Sugar | β-Glucuronidase activitya (nmol/min/mg [dry wt])
|
|
|---|---|---|---|---|
| pNZlac | pNZRAT | |||
| BL23 | Lactose | 101.3 ± 7.2 | 2,318.5 ± 443.5 | |
| Ribose | 139.7 ± 15.6 | 1,025.0 ± 17.5 | ||
| Glucose | 6.3 ± 1.8 | 15.2 ± 2.3 | ||
| BL153 | lacE | Ribose | 144.4 ± 7.9 | 2,964.0 ± 262.0 |
| Glucose | 7.3 ± 1.9 | 10.5 ± 0.6 | ||
| BL154 | lacT | Ribose | 94.7 ± 4.9 | 47.6 ± 13.4 |
| Glucose | 9.5 ± 0.8 | 5.5 ± 2.2 | ||
| BL155 | lacF | Ribose | 120.3 ± 7.5 | 2,860.0 ± 545.0 |
| Glucose | 15.1 ± 5.4 | 8.0 ± 2.4 | ||
Measured with p-nitrophenyl-β-d-glucuronic acid in permeabilized cells grown with 0.5% sugar to an optical density at 550 nm of 0.8. The values (means and standard deviations) are from three independent experiments.
FIG. 3.
Northern blot analysis of RNA from the wild-type strain and different lac mutants. The probes used were PIIgal (A) and Ppt (B); the strains used were BL23 (wild type) (lanes 1 to 5), BL153 (lacE) (lanes 6 to 9), BL154 (lacT) (lanes 10 to 13), and BL155 (lacF) (lanes 14 to 17). Cells were grown on lactose (lane 1), on ribose (lanes 2, 6, 10, and 14), ribose plus lactose (lanes 3, 7, 11, and 15), glucose (lanes 4, 8, 12, and 16), or glucose plus lactose (lanes 5, 9, 13, and 17).
Influence of Lac-PTS elements on transcription of the lactose operon.
We analyzed in vivo the role of lactose-specific proteins by means of Northern blotting using the same RNA probes (PIIgal and Ppt) as before. With the PIIgal probe, strong signals were obtained on ribose- and ribose-plus-lactose-grown cells of BL155 (lacF) (Fig. 3A, lanes 14 and 15), in agreement with the P-β-Gal activity detected (see above). Very intense signals were also observed in BL153 (Fig. 3A, lanes 6 and 7). The remarkable difference between the latter result and that with the wild-type strain (Fig. 3A, lanes 2 and 3) indicates that in lacE and lacF mutants, lactose induction is not required for the antitermination activity. This suggests that in vivo, LacE or LacF interacts with the antiterminator, LacT. Again, there is an excellent correlation between the hybridization patterns obtained with both probes. In the presence of glucose, no large mRNA was detected, while the Ppt probe showed that 100-nt mRNA was observed in all samples, albeit with slight changes in intensity (Fig. 3). A smaller size of the full transcript was noticed in ribose-grown cells of BL153, confirming the deletion generated by recombination (Fig. 3B, lanes 6 and 7). In BL154 (lacT), no 4.5-kb mRNA or any other transcript larger than 100 nt could be detected under any conditions (Fig. 3 and B, lanes 10 to 13), indicating that RNA polymerase always stops at the terminator site when LacT is absent.
DISCUSSION
The lactose operon, lacTEGF, in L. casei ATCC 393 is located on the chromosome and encodes the transcriptional antiterminator LacT, lactose-specific PTS proteins, and P-β-Gal (22). Previous reports suggested that this operon could have a regulatory system different from that described for the lactose operons in L. lactis, S. aureus, and S. mutans (19). In this work, we describe the construction and analysis of lacE, lacF, and lacT mutants showing the involvement of EII elements of the Lac-PTS in modulation of the lac operon of L. casei.
Lactose induction: modulation of LacT activity of EIILac.
Regulation by antitermination has been described for several operons in low-GC, gram-positive bacteria, such as sacPA, sacB, bgl, licTS, and glc of B. subtilis (7, 17, 40, 42, 46, 47, 50). In gram-negative bacteria, this system has been found in the bgl operon of E. coli and the arb operon of Erwinia chrysanthemi (20, 24, 32, 33, 43, 45). Antiterminator proteins of these systems have been assigned to the BglG family on the basis of sequence homology, cross-complementation, and the finding that their DNA targets share a consensus RAT sequence (40). LacT from L. casei shows homology to proteins of this family and has been shown to complement the sacB system of B. subtilis (3). Indeed, the His residues which are potentially phosphorylated in PRD-I (H101 and H159) and PRD-II (H210 and H273) of SacY are conserved in LacT (47). In wild-type L. casei ATCC 393, a transcript corresponding to the whole operon lacTEGF was detectable only in the presence of lactose. Taking into account the evidence shown here and knowledge gained from homologous systems, the general mechanism by which LacT controls transcription of the lac operon would be as follows. LacT binds to the RAT sequence and prevents formation of the terminator in the presence of lactose. In the absence of the inducer, the antiterminator would be inactive, and transcription would start and proceed only until the RNA polymerase reaches the rho-independent terminator. Hence, only a small transcript spanning a region of about 100 nt (from the transcription start site to the terminator) could be detected. As expected, in a lacT mutant constitutive termination (the presence of 100-nt transcript in all conditions) was observed, consistent with the model for LacT.
The antiterminator proteins BglG in E. coli and SacY in B. subtilis are regulated through phosphorylation by sugar-specific PTS components or Hpr (40, 47, 48). We examined the role of LacE (EIICB) and LacF (EIIA) in the regulation of LacT antiterminator activity, by inactivation of the lacE and lacF genes. Transcriptional studies showed an inducer-independent antiterminator activity in the lacE and lacF mutants, indicating that their corresponding proteins in the wild type may be involved in the inactivation of the antiterminator LacT. The model described for B. subtilis antiterminators (40, 47) could apply here, as in the absence of functional IILac elements, the antiterminator LacT was active, possibly because it cannot be phosphorylated at one of the conserved PTS regulation domains (PRD-I) (47). In the wild type, the phosphoryl group would be transferred to the incoming inducing sugar, with the same effect. This is the first report in which sugar-specific PTS elements from a lactic acid bacterium have been conclusively shown to be involved in the induction mechanism via an antiterminator protein.
Involvement of LacT in catabolite repression.
Preliminary reports showed that lactose utilization was repressed by glucose, since L. casei ATCC 393 displayed a likely diauxie (plateau lasting 15 to 20 h) when grown on both sugars (49). It was then shown that transcription of the lac operon was subject to CcpA-mediated CR (22, 34). However, an additional CcpA-independent CR mechanism was suggested when P-β-Gal activity was monitored in mutants lacking CcpA and EIIMan individually and in a ccpA man double mutant (22, 34). This additional CR mechanism was dependent on a functional glucose-PTS transporter. In the bgl and lev operons of B. subtilis, HPr-dependent phosphorylation of the regulator proteins controls a CcpA-independent CR effect (27, 28, 40). However, the experiments with lacp-gusA fusions suggest that the RAT sequence and LacT are involved in the proposed CcpA-independent catabolite repression mechanism. In L. casei, Northern blot analysis showed that transcription initiation of lacTEGF operon is fully derepressed in the ccpA mutant, but that there was no elongation beyond the terminator in the presence of glucose. The double mutant BL72 (man ccpA) exhibits full expression of the lactose operon when grown on glucose plus lactose; therefore, LacT is active. This strain is impaired in the glucose-PTS transporter, which probably links the EIIMan or other PTS elements with the glucose repression mediated by LacT, which was observed in the ccpA mutant.
The model of PTS-mediated control of PRD-containing regulators described by Stülke et al. (47), which also includes the mentioned antiterminators, could explain the regulation of LacT activity. In this model, PRD-I would be phosphorylated by inducer-specific EII components of the PTS in the absence of inducer and PRD-II would be phosphorylated by HPr in absence of glucose (or repressing carbohydrates). Consequently, the antiterminator LacT would exist in three forms: (i) active, when dephosphorylated in PRD-I and phosphorylated in PRD-II; (ii) inactive (noninduced), when phosphorylated in both domains; and (iii) inactive (CR), when PRD-II is dephosphorylated by HPr when grown on glucose. The presence (nonphosphorylated PRD-I) or absence (phosphorylated PRD-I) of the inducer, lactose, would not affect this later form. However, we cannot exclude the possibility that phosphorylation of PRD-II can be carried out by the sugar-specific PTS transporter, EIIMan. Clearly the analysis of defined L. casei mutants lacking the HPr gene (ptsH) or defective ptsH and either lacF or ccpA could lead to a further understanding of this regulation system, as could studies on the phosphorylation of LacT.
ACKNOWLEDGMENTS
We thank B. M. Chassy for providing L. casei ATCC 393[pLZ15−] and F. Morel-Deville for supplying the pRV300 vector.
This work was financed by the EU project BIO4-CT96-0380 and by funds of the Spanish CICyT (Interministerial Commission for Science and Technology) (ref. ALI 95-0038). V.M. was supported by a grant of the Consellería de Educación y Ciencia de la Generalitat Valenciana.
REFERENCES
- 1.Alpert C-A, Chassy B M. Molecular cloning and nucleotide sequence of the factor IIIlac gene of Lactobacillus casei. Gene. 1988;62:277–288. doi: 10.1016/0378-1119(88)90565-3. [DOI] [PubMed] [Google Scholar]
- 2.Alpert C-A, Chassy B M. Molecular cloning and DNA sequence of lacE, the gene encoding the lactose-specific enzyme II of the phosphotransferase system of Lactobacillus casei. J Biol Chem. 1990;265:22561–22568. [PubMed] [Google Scholar]
- 3.Alpert C-A, Siebers U. The lac operon of Lactobacillus casei contains lacT, a gene coding for a protein of BglG family of transcriptional antiterminators. J Bacteriol. 1997;179:1555–1562. doi: 10.1128/jb.179.5.1555-1562.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amster-Choder O, Houman F, Wright A. Protein phosphorylation regulates transcription of the β-glucoside utilization operon in E. coli. Cell. 1989;58:847–855. doi: 10.1016/0092-8674(89)90937-9. [DOI] [PubMed] [Google Scholar]
- 5.Amster-Choder O, Wright A. Regulation of activity of a transcriptional antiterminator in E. coli by phosphorylation. Science. 1990;249:540–542. doi: 10.1126/science.2200123. [DOI] [PubMed] [Google Scholar]
- 6.Amster-Choder O, Wright A. Transcriptional regulation of bgl operon of Escherichia coli involves phosphotransferase system-mediated phosphorylation of a transcriptional antiterminator. J Cell Biochem. 1993;51:83–90. doi: 10.1002/jcb.240510115. [DOI] [PubMed] [Google Scholar]
- 7.Arnaud M, Débarbouillé M, Rapoport G, Saier M H, Jr, Reizer J. In vitro reconstitution of transcriptional antiterminator by the SacT and SacY proteins of Bacillus subtilis. J Biol Chem. 1996;271:18966–18972. doi: 10.1074/jbc.271.31.18966. [DOI] [PubMed] [Google Scholar]
- 8.Arnaud M, Vary P, Zagorec M, Klier A, Débarbouillé M, Postma P, Rapoport G. Regulation of the sacPA operon of Bacillus subtilis: identification of phosphotransferase system components involved in SacT activity. J Bacteriol. 1992;174:3161–3170. doi: 10.1128/jb.174.10.3161-3170.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aymerich S, Steinmetz M. Specificity determinants and structural features in the RNA target of the bacterial antiterminator proteins of the BglG/SacY family. Proc Natl Acad Sci USA. 1992;89:10410–10414. doi: 10.1073/pnas.89.21.10410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bardowski J, Ehrlich S D, Chopin A. BglR protein, which belongs to the BglG family of transcriptional antiterminators, is involved in β-glucoside utilization in Lactococcus lactis. J Bacteriol. 1994;176:5681–5685. doi: 10.1128/jb.176.18.5681-5685.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breidt F J, Hengstenberg W, Finkeldei U, Stewart G C. Identification of genes for lactose-specific components of the phosphotransferase system in lac-operon of Staphylococcus aureus. J Biol Chem. 1987;262:16444–16449. [PubMed] [Google Scholar]
- 12.Breidt F J, Stewart G C. Nucleotide and deduced amino acid sequences of Staphylococcus aureus phospho-β-galactosidase gene. Appl Environ Microbiol. 1987;53:969–973. doi: 10.1128/aem.53.5.969-973.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chassy B M, Gibson E, Giuffrida A. Evidence for extrachromosomal elements in Lactobacillus. J Bacteriol. 1976;127:1576–1578. doi: 10.1128/jb.127.3.1576-1578.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chassy B M, Thompson J. Regulation of lactose-phosphoenolpyruvate-dependent-phosphotransferase system and β-d-phosphogalactosidase galactohydrolase activities in Lactobacillus casei. J Bacteriol. 1983;154:1195–1203. doi: 10.1128/jb.154.3.1195-1203.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crutz A M, Steinmetz M. Transcription of the Bacillus subtilis sacX and sacY genes, encoding regulators of sucrose metabolism, is both inducible by sucrose and controlled by the DegS-DegU signalling system. J Bacteriol. 1992;174:6087–6095. doi: 10.1128/jb.174.19.6087-6095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crutz A M, Steinmetz M, Aymerich S, Richter R, Le Coq D. Induction of levansucrase in Bacillus subtilis: an antitermination mechanism negatively controlled by the phosphotransferase system. J Bacteriol. 1990;172:1043–1050. doi: 10.1128/jb.172.2.1043-1050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Débarbouillé M, Arnaud M, Fouet A, Klier A, Rapoport G. The sacT gene regulating the sacPA operon in Bacillus subtilis shares strong homology with transcriptional antiterminators. J Bacteriol. 1990;172:3966–3973. doi: 10.1128/jb.172.7.3966-3973.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Vos W M, Boerrigter I, Van Rooyen R J, Reiche B, Hengstenberg W. Characterization of lactose-specific enzymes of the phosphotransferase system in Lactococcus lactis. J Biol Chem. 1990;265:22554–22560. [PubMed] [Google Scholar]
- 19.de Vos W M, Vaughan E E. Genetics of lactose utilization in lactic acid bacteria. FEMS Microbiol Rev. 1994;15:217–237. doi: 10.1111/j.1574-6976.1994.tb00136.x. [DOI] [PubMed] [Google Scholar]
- 20.El Hassouni M, Henrissat B, Chippaux M, Barras F. Nucleotide sequences of arb genes, which control β-glucoside utilization in Erwinia chrysanthemi: comparison with the Escherichia coli bgl operon and evidence for a new β-glycohydrolase family including enzymes from eubacteria, archeabacteria, and humans. J Bacteriol. 1992;174:765–777. doi: 10.1128/jb.174.3.765-777.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flickinger J L, Porter E V, Chassy B M. Abstracts of the 86th Annual Meeting of the American Society for Microbiology. Washington, D.C: American Society for Microbiology; 1986. The characterization of a plasmid isolated from Treponema hyodysenteriae and Treponoma innocens, abstr. H-174; p. 156. [Google Scholar]
- 22.Gosalbes M J, Monedero V, Alpert C-A, Pérez-Martínez G. Establishing a model to study the regulation of the lactose operon in Lactobacillus casei. FEMS Microbiol Lett. 1997;148:83–89. doi: 10.1111/j.1574-6968.1997.tb10271.x. [DOI] [PubMed] [Google Scholar]
- 23.Honeyman A L, Curtiss R I. Isolation, characterization and nucleotide sequence of Streptococcus mutans lactose-specific enzyme II (lacE) gene of the PTS and the phospho-β-galactosidase (lacG) gene. J Gen Microbiol. 1993;139:2685–2694. doi: 10.1099/00221287-139-11-2685. [DOI] [PubMed] [Google Scholar]
- 24.Houman F, Diaz-Torres M R, Wright A. Transcriptional antitermination in bgl operon of E. coli is modulated by a specific RNA binding protein. Cell. 1990;62:1153–1163. doi: 10.1016/0092-8674(90)90392-r. [DOI] [PubMed] [Google Scholar]
- 25.Huang F, Coppola G, Calhoun D H. Multiple transcripts encoded by the ilvGMEDA gene cluster of Escherichia coli K-12. J Bacteriol. 1992;174:4871–4877. doi: 10.1128/jb.174.15.4871-4877.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Idelson M, Amster-Choder O. SacY, a transcriptional antiterminator from Bacillus subtilis, is regulated by phosphorylation in vivo. J Bacteriol. 1998;180:660–666. doi: 10.1128/jb.180.3.660-666.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krüger S, Hecker M. Regulation of the putative bglPH operon for aryl-β-glucoside utilization in Bacillus subtilis. J Bacteriol. 1995;177:5590–5597. doi: 10.1128/jb.177.19.5590-5597.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krüger S, Gertz S, Hecker M. Transcriptional analysis of bglPH expression in Bacillus subtilis: evidence for two distinct pathways mediating carbon catabolite repression. J Bacteriol. 1996;178:2637–2644. doi: 10.1128/jb.178.9.2637-2644.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Coq D, Lindner C, Krüger S, Steinmetz M, Stülke J. New β-glucoside (bgl) genes in Bacillus subtilis: the bglP gene product has both transport and regulatory functions similar to those of BglF, its Escherichia coli homolog. J Bacteriol. 1995;177:1527–1535. doi: 10.1128/jb.177.6.1527-1535.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leloup L, Ehrlich S D, Zagorec M, Morel-Deville F. Single crossing-over integration in the Lactobacillus sake chromosome and insertional inactivation of the pts and the lacI genes. Appl Environ Microbiol. 1997;63:2117–2123. doi: 10.1128/aem.63.6.2117-2123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeda S, Gasson M J. Cloning, expression and location of Streptococcus lactis gene for phospho-β-galactosidase. J Gen Microbiol. 1986;132:331–340. doi: 10.1099/00221287-132-2-331. [DOI] [PubMed] [Google Scholar]
- 32.Mahadevan S, Reynolds A E, Wright A. Positive and negative regulation of bgl operon in Escherichia coli. J Bacteriol. 1987;169:2570–2578. doi: 10.1128/jb.169.6.2570-2578.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mahadevan S, Wright A. A bacterial gene involved in transcription antitermination: regulation at a Rho-independent terminator in bgl operon of E. coli. Cell. 1987;50:485–494. doi: 10.1016/0092-8674(87)90502-2. [DOI] [PubMed] [Google Scholar]
- 34.Monedero V, Gosalbes M J, Pérez-Martínez G. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J Bacteriol. 1997;179:6657–6664. doi: 10.1128/jb.179.21.6657-6664.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murakawa G J, Kwan C, Yamashita J, Nierlich D P. Transcription and decay of the lac messenger: role of an intergenic terminator. J Bacteriol. 1991;173:28–36. doi: 10.1128/jb.173.1.28-36.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Platteeuw C, Simons G, de Vos W M. Use of the Escherichia coli β-glucuronidase (gusA) gene as a reporter gene for analyzing promoters in lactic acid bacteria. Appl Environ Microbiol. 1994;60:587–593. doi: 10.1128/aem.60.2.587-593.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porter E V, Chassy B M. Nucleotide sequence of the β-d-phospho-galactosidase gene of Lactobacillus casei: comparison to analogous pbg genes of other Gram-positive organisms. Gene. 1988;62:263–276. doi: 10.1016/0378-1119(88)90564-1. [DOI] [PubMed] [Google Scholar]
- 38.Posno M, Leer R J, van Luijk N, van Giezen M J F, Heuvelmans P T H M, Lokman B C, Pouwels P H. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl Environ Microbiol. 1991;57:1822–1828. doi: 10.1128/aem.57.6.1822-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raya R, Bardowski J, Andersen P S, Ehrlich S D, Chopin A. Multiple transcriptional control of the Lactococcus lactis trp operon. J Bacteriol. 1998;180:3174–3180. doi: 10.1128/jb.180.12.3174-3180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutberg B. Antitermination of transcription of catabolic operons. Mol Microbiol. 1997;23:413–421. doi: 10.1046/j.1365-2958.1997.d01-1867.x. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Schentz K, Stülke J, Gertz S, Krüger S, Krieg M, Hecker M, Rak B. LicT, a Bacillus subtilis transcriptional antiterminator protein of the BglG family. J Bacteriol. 1996;178:1971–1979. doi: 10.1128/jb.178.7.1971-1979.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnetz K, Rak B. Regulation of bgl operon of Escherichia coli by transcriptional antitermination. EMBO J. 1988;7:3271–3277. doi: 10.1002/j.1460-2075.1988.tb03194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schentz K, Rak B. β-Glucoside permease represses the bgl operon of E. coli by phosphorylation of the antiterminator protein and also interacts with glucose-specific enzyme II, the key element in catabolic control. Proc Natl Acad Sci USA. 1990;87:5074–5078. doi: 10.1073/pnas.87.13.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnetz K, Toloczki C, Rak B. β-Glucoside (bgl) operon of Escherichia coli K-12: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J Bacteriol. 1987;169:2579–2590. doi: 10.1128/jb.169.6.2579-2590.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stülke J, Martin-Verstraete I, Zagorec M, Rose M, Klier A, Rapoport G. Induction of Bacillus subtilis ptsGHI operon by glucose is controlled by a novel antiterminator, GlcT. Mol Microbiol. 1997;25:65–78. doi: 10.1046/j.1365-2958.1997.4351797.x. [DOI] [PubMed] [Google Scholar]
- 47.Stülke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD-a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- 48.Tortosa P, Aymerich S, Lindner C, Saier M H, Jr, Reizer J, Le Coq D. Multiple phosphorylation of SacY, a Bacillus subtilis antiterminator negatively controlled by the phosphotransferase system. J Biol Chem. 1997;272:17230–17237. doi: 10.1074/jbc.272.27.17230. [DOI] [PubMed] [Google Scholar]
- 49.Veyrat A, Monedero V, Pérez-Martínez G. Glucose transport by the phosphoenolpyruvate: mannose phosphotransferase system in Lactobacillus casei ATCC 393 and its role in carbon catabolite repression. Microbiology. 1994;140:1141–1149. doi: 10.1099/13500872-140-5-1141. [DOI] [PubMed] [Google Scholar]
- 50.Zukovski M M, Miller L, Cosgwell P, Chen K, Aymerich S, Steinmetz M. Nucleotide sequence of the sacS locus of Bacillus subtilis reveals the presence of two regulatory genes. Gene. 1990;90:153–155. doi: 10.1016/0378-1119(90)90453-x. [DOI] [PubMed] [Google Scholar]