Significance
The urinary bladder, which was previously believed to be a sterile environment, is actually a habitat for commensal microbes. However, the presence of the bladder microbiome and its functional relationship with the underlying uroepithelial cells have remained a mystery. In this work, we have shown that Lactobacilli, the most abundant commensal bacteria in human bladders, promote type I interferon production in bladder epithelial cells, resulting in mature phagolysosomes with enhanced cathepsin D and acidification. Functionally, Lactobacilli create a microenviroment that protects the host from infection by uropathogenic Escherichia coli. These findings provide compelling evidence that Lactobacilli can modulate innate immunity in the urinary bladder and exert therapeutic effects against urinary tract infection.
Keywords: type I interferon, urinary tract infection, commensal bacteria, Lactobacilli
Abstract
Many urinary tract infections (UTIs) are recurrent because uropathogens persist within the bladder epithelial cells (BECs) for extended periods between bouts of infection. Because persistent uropathogens are intracellular, they are often refractive to antibiotic treatment. The recent discovery of endogenous Lactobacillus spp. in the bladders of healthy humans raised the question of whether these endogenous bacteria directly or indirectly impact intracellular bacterial burden in the bladder. Here, we report that in contrast to healthy women, female patients experiencing recurrent UTIs have a bladder population of Lactobacilli that is markedly reduced. Exposing infected human BECs to L. crispatus in vitro markedly reduced the intracellular uropathogenic Escherichia coli (UPEC) load. The adherence of Lactobacilli to BECs was found to result in increased type I interferon (IFN) production, which in turn enhanced the expression of cathepsin D within lysosomes harboring UPECs. This lysosomal cathepsin D–mediated UPEC killing was diminished in germ-free mice and type I IFN receptor–deficient mice. Secreted metabolites of L. crispatus seemed to be responsible for the increased expression of type I IFN in human BECs. Intravesicular administration of Lactobacilli into UPEC-infected murine bladders markedly reduced their intracellular bacterial load suggesting that components of the endogenous microflora can have therapeutic effects against UTIs.
The advent of 16S ribosomal RNA (rRNA) sequencing has revealed the existence of a steady-state microbiome in the human bladder, which can be readily monitored by examining healthy urine collected by transurethral catheterization or suprapubic aspiration (1, 2). Recent studies of urine from healthy women have revealed that the bladder microbiome is remarkably similar to the vaginal microbiome, albeit of lower biomass (2, 3). The dominant genera are primarily Lactobacillus, Gardnerella, and Streptococcus (4). Among Lactobacillus (L.) species, four are predominant in the bladder microbiome (2): L. crispatus, L. gasseri, L. inners, and L. jensenii.
Currently, there is no clear consensus regarding the physiological role of the bladder microbiome (4). Some have suggested that overabundance of L. gasseri in the bladder microbiome might contribute to the pathogenesis of certain bladder disorders such as interstitial cystitis/bladder pain syndrome and overactive bladder (3–5). Other members of the bladder microflora, such as S. anginosus and G. vaginalis, have been linked to the symptoms of urinary incontinence (3). However, to this day, no direct evidence exists on the actual contribution of the microflora to bladder pathology.
Besides, a small but growing body of data has suggested that the endogenous bladder microflora, especially Lactobacillus species (spp.), is essential for bladder homeostatic maintenance. Few studies have suggested that bladder microflora directly or indirectly play a protective role by inhibiting the colonization of uropathogenic Escherichia coli (UPEC) (6). Women harboring high levels of Lactobacillus spp. were protected from postoperative or postinstrumentation urinary tract infections (UTIs) (2). In contrast, perturbation of the Lactobacilli bacterial barrier led to an increased incidence of UTIs (7). Despite the scarce supporting data, Lactobacillus spp. has been suggested to serve as a physical barrier to the UPEC colonization of the bladder (8). In addition, Lactobacillus spp. in other mucosal sites such as the gut, airway, and reproductive tract were reported to promote the maturation of epithelial cells inducing the secretion of mucus and antimicrobial peptides (9). However, the functional role of the interaction between Lactobacilli and bladder epithelial cells (BECs) and their underlying mechanism needs further investigation.
Of note, UTI is ranked the second most frequent infection in humans with more than 150 million cases per year worldwide (10). UPEC accounts for the vast majority of UTIs. These bacteria gain access into the bladder from the gut through the progressive colonization of the urethra (11). Upon reaching the bladder, UPECs adhere tightly to the superficial BECs via their fimbriae (12, 13). This binding triggers the rapid entry of UPECs into Rab27b+ fusiform vesicles of BECs that serve to supply the extramembrane required for bladder expansion (14). Upon entry into BECs, UPECs multiply initially in vesicles and later within the cytosol, forming intracellular bacteria communities that sustain bladder infections (15). In their intracellular location, UPECs are protected not only from antibiotics but also from antimicrobial actions of host phagocytic cells and antibacterial peptides. Studies in mouse models of UTIs have revealed that UPECs can persist for months in the bladder without accompanying inflammation (16), suggesting that these intracellular bacteria could be the critical nidus for recurrent UTIs.
Given their potential for recurrence, there is great interest in identifying and developing novel strategies to eliminate reservoirs of UPECs from the bladder. Here, we investigated the role of endogenous microbial flora in the bladder of healthy and diseased individuals. We found that Lactobacilli controlled host innate immunity of the bladder via type I interferon (IFN) responses, which help the maturation of phagolysosomes containing intracellular UPEC in BECs. We also offer new insights into the role of endogenous microflora in the homeostasis of the bladder and provide the basis for the development of a microflora-based therapeutic modality as a potentially viable alternative to antibiotic therapy.
Results
Dominant Presence of Lactobacillus spp. in Urine from Healthy Women but Not from Patients with UTIs.
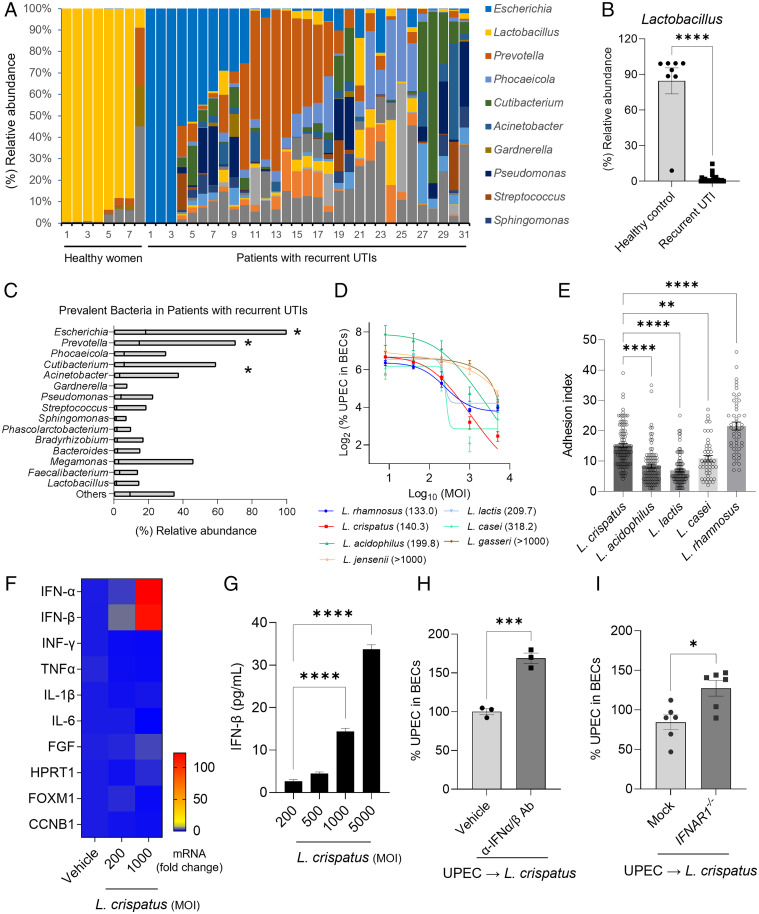
In view of the potential role of Lactobacillus spp. in impacting bladder infections, we investigated their prevalence in the endogenous microflora of the bladder of 31 female patients with recurrent UTIs and eight healthy control female subjects (SI Appendix, Table S1). By using urethral catheterization, we obtained urine from all subjects ensuring that any bacteria detected in these samples would have most likely originated from the bladder. By using next-generation sequencing of 16S rRNA in urine pellets, we could discern the bacterial profiles in each of the two groups. We specifically found that whereas the majority (∼90%) of bladder microflora in healthy women were Lactobacillus spp., the bladder microflora of patients with recurrent UTIs was much more heterogeneous (Fig. 1A). Notably, we observed a sharp reduction in the abundance of Lactobacillus spp., which was found to be less than 20% (Fig. 1B). The most abundant bacteria identified in patients with recurrent UTIs was E. coli. Other prominent bacterial species found in these subjects were, in decreasing order of prevalence, Prevotella, Phocaeicolla, Cutibacterium, and Acinetobacter species (Fig. 1C). We found, conclusively, that the overwhelming majority of microflora in healthy bladders are Lactobacillus spp. However, a drastic reduction in the Lactobacilli population was observed in patients with recurrent UTIs, accompanied by the emergence of other microflora species, including potentially pathogenic E. coli. This finding raised the notion that the presence of Lactobacilli deters the bladder colonization by E. coli and other potential pathogens, and any perturbation in this bladder flora might predispose to UPEC infections.
Fig. 1.
Lactobacilli, prevalent in healthy women, decreased intracellular UPEC load by triggering the expression of type I IFN in BECs. (A) Distribution of bacterial genera in the urine of healthy women or patients with recurrent UTIs. Color-coded columns indicate bacterial genera. (B) Comparison of the relative population of Lactobacilli in healthy women (n = 8) or patients with recurrent UTIs (n = 31). Each data point represents one person. (C) Floating bar graphs indicate relative bacterial abundance in the urine of patients with recurrent UTIs. The line in each bar represents the mean value. Except for Prevotella, Escherichia has a statistically higher abundance. Prevotella outnumbers all other bacteria except Phocaeicola and Cutibacterium. (D) Intracellular UPEC killing by Lactobacilli. UPEC CI5 strain infected human 5637 BECs (MOI 200). After 6 h, infected cells were treated with Lactobacilli at four different MOI (5000, 1000, 200, and 40). The intracellular number of UPECs in treated BECs was counted after overnight incubation. The numbers in parenthesis are the MOI values for 50% reduction (IC50) which were interpolated from the curves. (E) Adhesion of five Lactobacilli strains (adhered Lactobacillus/BEC) was compared by treating 5636 BECs with Lactobacilli at MOI 1000. After 2 h of incubation, cells were thoroughly washed and stained using a Gram staining solution. (F) Human 5637 BECs were treated with L. crispatus at 200 or 1000 MOI for 6 h. qRT-PCR was used to assess changes in messenger RNA (mRNA) expression. (G) Human IFN-β secreted by human 5637 BECs in response to L. crispatus exposure was assessed by ELISA. (H) Neutralization of type I IFN following treatment with specific antibodies. UPEC-infected cells were pretreated with anti-IFN-α/β antibodies before exposure to L. crispatus. The intracellular numbers of UPEC in pretreated BECs was assessed after 16 h. (I) Increased UPEC load on IFNAR1−/−. The IFNAR gene of human 5637 BECs was deleted by using the CRISPR/Cas9 system (pX459). IFNAR1−/− or mock control BECs (empty pX459) were infected with UPEC CI5 strain (MOI 200) for 6 h, and then treated with L. crispatus (MOI 500). The intracellular UPEC number was counted overnight. Error bars in each bar graph show mean ± SEM. Each experiment (D–I) was independently repeated two to three times with similar results. Data were analyzed by unpaired two-tailed Student t test (B, H, I) or an ordinary one-way ANOVA (C, E, G) with Tukey’s multiple comparison posttest. *P < .05; **P < .01; ***P < .001; ****P < .0001. Ab, antibody.
Exposure of Infected BECs to Lactobacillus spp. in vitro Markedly Reduces the Intracellular Load of UPEC.
The apparent inverse correlation observed between the presence of Lactobacilli and the absence of uropathogens in the microflora of human bladders suggested that the former might possess intrinsic properties impeding bladder colonization by uropathogens. One way to demonstrate the putative UPEC-impeding activity of Lactobacilli is to investigate whether addition of Lactobacillus spp. to UPEC-infected BECs would result in a reduced intracellular UPEC burden in BECs. Studies of UPEC infection of BECs have previously revealed that after bacteria adhere to the cell surface, a significant number of these bacteria become internalized into distinct compartments bearing Rab27b markers (14). Whereas, some of the invading UPEC are expelled back into the extracellular medium during the early stages of bacterial infection (14), a considerable number of intracellular UPEC try to avoid this fate by escaping Rab27b+ vesicle compartments to enter the BEC cytosol. However, these UPEC are also soon snared by autophagosomes and driven into lysosomes (17). A few of these cytosolic bacteria presumably escape these intracellular antibacterial actions in BECs to form intracellular bacterial colonies (17), representing the nidus for subsequent bladder colonization by UPEC. To investigate whether Lactobacilli reduced the intracellular UPEC burden in infected BECs, we infected human 5637 BECs with an ampicillin-resistant UPEC CI5 strain and exposed these cells to gentamicin to selectively kill all extracellular UPEC. After that, the antibiotic was removed, and the infected BECs were exposed to various strains of Lactobacilli for 16 to 18 h to investigate the impact of these bacteria to the numbers of intracellular UPEC. We examined the effects of seven different Lactobacilli strains (L. rhamnosus, L. crispatus, L. lactis, L. acidophilus, L. casei, L. jensenii, and L. gasseri) by exposing them to UPEC-infected BECs at different multiplicities of infection (MOI). We observed that all Lactobacillus spp. tested led to a dose-dependent reduction in the intracellular loads of UPEC in BECs, albeit to different degrees of efficiency (Fig. 1D). We found that whereas L. rhamnosus, L. acidophilus, and L. crispatus exhibited a clear dose-dependent effect in reducing the bacterial load with decreasing MOI (Fig. 1D), L. casei, L. lactis, L. jensenii, and L. gasseri exhibited a partial dose dependency (Fig. 1D). We also calculated the half-maximal inhibitory concentration (IC50) and determined that L. rhamnosus and L. crispatus exhibited the lowest IC50 values compared with those of other species (Fig. 1D, numbers in parentheses).
This Lactobacillus spp.–mediated reduction in the intracellular UPEC load could be attributed to either an increased Rab27b+ vesicle–dependent bacterial expulsion (14) or increased intracellular killing within BEC lysosomes (18). To distinguish between these two possibilities, we examined the intracellular bacterial burden at 6 h postexposure of UPEC-infected BECs to L. crispatus, which is the time of the likely maximal Rab27b+ vesicle–dependent bacterial expulsion (18). Because we did not notice any reduction in bacterial numbers (SI Appendix, Fig. S1A) at this time point, we considered it unlikely that the enhanced expulsion of UPEC was responsible for the Lactobacillus-induced reduction in the UPEC burden. SI Appendix, Fig. S1B reveals that L. crispatus reduced the intracellular load of UPEC in BECs in a time-dependent manner. If the intracellular killing of UPEC was responsible for the drop in the UPEC numbers in infected BECs, then treating UPEC-infected BECs with N-acetyl-L-cysteine, which inhibits production of reactive oxygen species, or ammonium chloride, which neutralizes lysosome pH, should impede intracellular killing. Since only ammonium chloride treatment was effective (SI Appendix, Fig. S2 A and B), it seems that L. crispatus–mediated reduction in intracellular UPEC numbers was attributable to the acidification of bacteria-containing vesicles in BECs.
Capacity of Various Lactobacillus Species to Adhere to BECs.
The promotion of intracellular bacterial killing of infected BECs by Lactobacillus spp. would presumably require their adherence to BEC surfaces. We examined the capacity of various Lactobacillus spp. to bind to BECs by using a standard adhesion assay and found that they exhibited different degrees of adherence capabilities (19). More specifically, we noticed that L. rhamnosus and L. crispatus, which were the most efficacious species at reducing the bacterial burden in infected BECs, exhibited the most robust adhesion with adhesion indices exceeding 15, whereas the remaining species exhibited adhesion indices between 7 and 10 (Fig. 1E). Other Lactobacillus spp. were significantly less effective in BEC adhesion. Representative microscopic images showing the adherence of each species to BECs are shown in SI Appendix, Fig. S3A. In addition, SI Appendix, Fig. S3B shows the adherence of L. crispatus to UPEC-infected BECs. We found that the adherence of L. crispatus to BECs and UPEC-infected BECs was comparable (SI Appendix, Fig. S3B), indicating that previous exposure of BECs to UPEC did not impact the subsequent binding of Lactobacilli. Thus, the ability of various Lactobacillus spp. to reduce the UPEC burden in infected BECs could be related to their capacity to adhere to BEC surfaces.
L. crispatus Induces the Expression of Type I IFN in UPEC-Infected BECs Reducing Intracellular Bacterial Burden.
Next, we sought to investigate the underlying mechanisms for the Lactobacilli-mediated reduction of the UPEC load in infected BECs. The adherence of Lactobacilli on BECs implied that adherent Lactobacilli can activate BECs, promoting enhanced intracellular killing. Because the enhanced intracellular killing of UPEC by BECs is reminiscent of enhanced antiviral activities typically induced by IFNs, we examined the production of IFN by BECs after exposure to L. crispatus (2). By using qRT-PCR, we observed that the production of type I IFNs (IFN-α or IFN-β) (Fig. 1F) but not type II IFN (IFN-γ), proinflammatory cytokines (interleukin-1β [IL-1β]), IL-6, tumor necrosis factor-α [TNF-α]), or other cellular factors (Fig. 1F and SI Appendix, Fig. S4) was explicitly induced in BECs upon exposure to L. crispatus. A dose-dependent increase in IFN-β secretion by BECs in response to L. crispatus exposure was confirmed using enzyme-linked immunosorbent assay (ELISA) (Fig. 1G). The functional significance of IFN-β was demonstrated with type I IFN-neutralizing antibody, which impaired L. crispatus–induced intracellular killing of UPEC (Fig. 1H). By binding the IFN-α receptor (IFNAR), type I IFN is known to typically mediate the potent antiviral activity of infected cells, triggering the secretion of mediators that abrogate viral replication (20). To examine the functional contribution of type I IFN in reducing the UPEC burden in BECs, we generated IFNAR1−/− (type I IFN-α receptor) BECs, which fail to recognize type I IFN, by using the CRISPR/Cas9 system. We compared the intracellular UPEC burden in control and IFNAR1−/− BECs at 16 h after infection and found that whereas the bacterial burden in control BECs was reduced, the numbers in mutant BECs were increased over the numbers observed at the time of infection, reflecting a lack of intracellular killing and even intracellular growth (Fig. 1I and SI Appendix, Fig. S5). Taken together, we found that the L. crispatus–triggered production of type I IFN from BECs contributed to the intracellular killing of UPEC in infected BECs. Interestingly, we observed that the L. crispatus–induced intracellular production of type I IFN cytokines in BECs was not accompanied by the production of proinflammatory cytokines such as TNF-α, IL-6, or IL-1β.
Intracellular Bactericidal Effect of Type I IFN Involved Lysosomal Maturation.
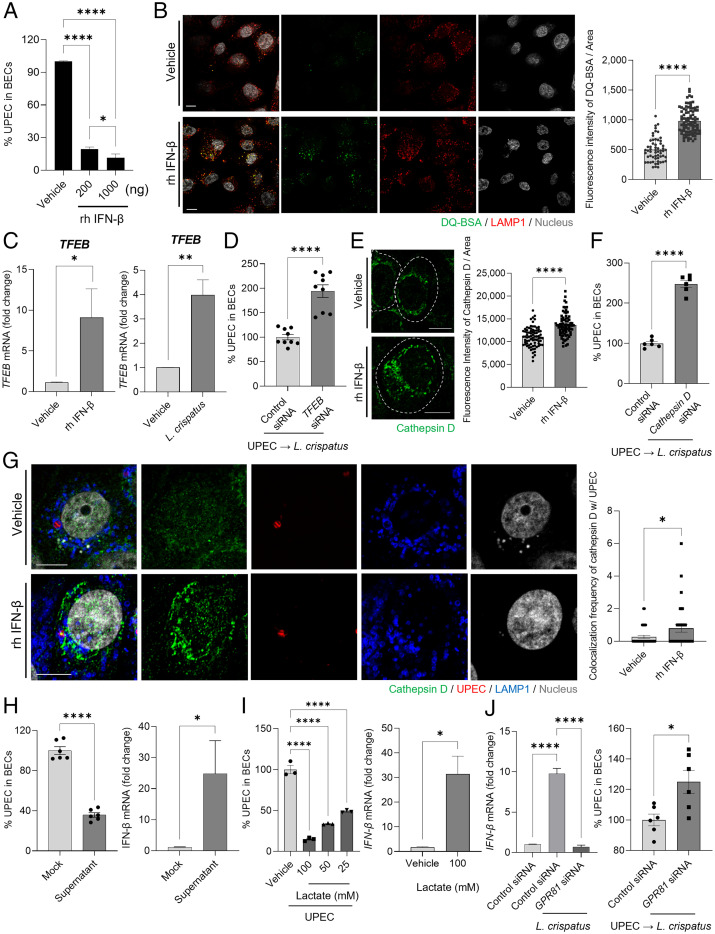
Next, we sought to investigate the mechanism by which the enhanced production of type I IFN by BECs contributes to the reduction in intracellular UPEC numbers. We directly applied different amounts of recombinant human IFN-β (200 or 1000 ng) to UPEC-infected 5637 BECs to see if we could mimic the L. crispatus–mediated effects. We accordingly observed a dose-dependent drop in intracellular UPEC numbers in BECs after exposure to IFN-β (Fig. 2A). This finding suggested that IFN-β induced bacterial killing in BECs by promoting the bactericidal activity of bacteria-harboring lysosomes. After invasion of BECs, UPEC are found in various intracellular vesicles, with most of them eventually ending up in LAMP1+ lysosomes where they are either expelled (17) or killed by reactive oxygen species and lysosomal enzymes (21). To account for the enhanced bactericidal activity of UPEC-infected BECs exposed to IFN-β, we hypothesized that type I IFNs promoted the bactericidal actions of LAMP1+ lysosomal vesicles. To test this notion, we examined the degradative activity of lysosomes in IFN-β–treated BECs by monitoring the fluorescence of the DQ-BSA reporter compound, the digestion of which evokes strong fluorescence signals. We observed that LAMP1+ lysosomes contained brightly fluorescent DQ-BSA following treatment with IFN-β (Fig. 2B). Quantitative analysis of the DQ-BSA signal, which is an indicator of lysosomal degradative activity, revealed its significant enhancement in IFN-β–treated BECs over vehicle-treated BECs (Fig. 2B, Right). We also noticed that IFN-β markedly enhanced the maturation of endocytic vesicles by increasing its acidity, as measured using a lysotracker (SI Appendix, Fig. S6). Because the transcription factor EB (TFEB) has been reported to be a master regulator of lysosomal function by regulating the expression of lysosomal enzymes (22), we examined whether the expression of TFEB in BECs was enhanced by treatment with IFN-β. Accordingly, we found the significantly enhanced expression of TFEB (Fig. 2C) in IFN-β–treated or L. crispatus–treated BECs. To ascertain the effect of TFEB in lowering UPEC load in BECs, we compared intracellular UPEC numbers in control or TFEB short interfering RNA (siRNA)–transfected BECs 16 h after infection (Fig. 2D and SI Appendix, Fig. S7A). The increased bacterial burden in BECs transfected with TFEB siRNA suggested that lysosomal enzymes played a key role in regulating intracellular numbers of UPEC. Treatment with IFN-β also enhanced the expression of cathepsin D, a lysosomal aspartic proteinase known to enhance the degradative activity in lysosomes (Fig. 2E) in BECs treated with IFN-β. Quantitative analysis of the expression of cathepsin D revealed its significant enhancement in IFN-β-treated cells compared with vehicle-treated BECs (Fig. 2E, Right). Cathepsin D is known to manifest its antibacterial activity through the cleavage of bacterial virulence factors and inhibition of bacterial replication within lysosomes (23). We found that silencing cathepsin D expression in UPEC-infected BECs markedly diminished intracellular killing of UPEC, indicating that cathepsin D was a critical contributor to the bactericidal actions of BECs (Fig. 2F and SI Appendix, Fig. S7B). To demonstrate the association between the enhanced expression of cathepsin D and intracellular bacterial killing, we probed the lysosomes of UPEC-infected BECs for cathepsin D 12 h after treatment with IFN-β or vehicle. We found that treatment with IFN-β significantly enhanced the expression of cathepsin D in all LAMP1+ lysosomes, including those harboring UPEC (Fig. 2G). Quantitative analysis revealed a significant increase in the colocalization of cathepsin D in UPEC-containing LAMP1+ vesicles after treatment with IFN-β (Fig. 2G, Right). Thus, the enhanced bactericidal activity in UPEC-infected BECs by adherent L. crispatus was linked to the enhanced production of type I IFN, which in turn promoted the enhanced production of cathepsin D in lysosomes, including those harboring UPEC.
Fig. 2.
Intracellular bactericidal action of type I IFN is mediated by lysosomal maturation. (A) Intracellular UPEC killing by type I IFN. Human 5637 BECs were treated with L. crispatus at various MOI (200). After 6 h of infection, the bacterial burden was measured 20 h after treatment with recombinant human (rh) IFN-β (200 or 1000 ng). (B) Lysosomal maturation by IFN-β. Human 5637 BECs were treated with rh IFN-β (500 ng). After 24 h of treatment, cells were exposed to DQ-BSA (green) for 6 h and then immunostained using an anti-LAMP1 (red) antibody for confocal microscopic imaging. (B, Right) The mean fluorescence intensity of DQ-BSA was measured in random fields. (C) After treatment with rh IFN-β (500 ng) for 6 h, the expression of TFEB mRNA in cells treated with IFN-β or L. crispatus was compared to that in vehicle-treated control using qPCR. (D) TFEB mediated L. crispatus-induced intracellular UPEC killing in BECs. After human BECs were transfected with control or TFEB siRNA, UPEC-infected BECs were exposed to L. crispatus. The intracellular number of UPEC in treated BECs was assessed after 16 h. (E) Enhanced cathepsin D expression was assessed by treating BECs with rh IFN-β (500 ng) for 24 h followed by anti-cathepsin D (green) immunostaining for confocal imaging. The dotted line indicates the periphery of BECs. (E, Right) The fluorescence intensity of cathepsin D was measured in random fields. (F) Cathepsin D regulates the size of intracellular UPEC population. After human 5637 BECs were transfected with control or cathepsin D siRNA, UPEC-infected BECs were exposed to L. crispatus. The intracellular number of UPEC in treated BECs was assessed after 16 h. (G) UPEC-infected BECs were treated for 12 h with IFN-β (500 ng) and then immunostained for cathepsin D (green), UPEC (red), and LAMP1 (blue). (G, Right) The frequency of colocalization between cathepsin D and UPEC was measured in random fields. (H) Effects of CFS of L. crispatus on human BECs. (H, Left) UPEC-infected human BECs were CFS or mock treated for 16 h, and the bacterial load was determined. (H, Right) The relative change of IFN-β mRNA expression by human BECs was analyzed using qPCR after 6 h of exposure to CFS of L. crispatus. (I) Effects of lactate on human BECs. (I, Left) UPEC-infected human BECs were treated with lactate in a dose-dependent manner or mock treated for 16 h; thereafter, the intracellular bacterial load was determined. (I, Right) The relative change in expression levels of IFN-β mRNA in human BECs was assessed using qPCR after 6 h of treatment with lactate (100 mM). (J) The lactate effect on BECs is mediated by GPR81. After human 5637 BECs were transfected with control or GPR81 siRNA, (J, Left) the relative change in IFN-β mRNA expression levels was assessed using qPCR after 6 h of treatment with L. crispatus. (J, Right) UPEC-infected BECs were exposed to L. crispatus (MOI 1000), and the intracellular number of UPECs in treated BECs was assessed after 16 h. Error bars in each bar graph show mean ± SEM. Each experiment (A–J) was independently repeated two to three times with similar results. Data were analyzed by unpaired two-tailed Student t test (B–J) or an ordinary one-way ANOVA (A and I) with Tukey’s multiple comparison posttest. *P < .05; **P < .01; ****P < .0001. Scale bars: 10 μm.
Lactobacillus-Derived Lactate Promotes Intracellular Bactericidal Actions of BECs.
Next, we sought to identify the factor(s) produced by L. crispatus responsible for promoting bactericidal actions of BECs. We started by examining the L. crispatus culture supernatant and found that exposing UPEC-infected BECs to the culture supernatant was sufficient for promoting bactericidal actions of BECs (Fig. 2H). Since Lactobacillus spp. are known to produce bacteriocins, which are proteinaceous antimicrobial compounds (24), we sought to distinguish the intracellular antibacterial activity we have described from any possible effects of bacteriocins. When we exposed L. crispatus culture supernatants to various proteases such as trypsin, proteinase K, or α-amylase (25) followed by heat treatment (95 °C for 30 min), we did not observe any significant loss in the capacity of the culture supernatant to promote bactericidal actions of BECs (SI Appendix, Fig. S8). This result points to the active component in the cell-free Lactobacillus spp. supernatant as being a non-proteinaceous agent.
It is noteworthy that exposing infected BECs to L. crispatus culture supernatant significantly increased IFN-β expression, which was the critical determinant of intracellular bacterial killing by BECs (Fig. 2H). Since enhanced cellular IFN-β expression has previously been ascribed to the presence of extracellular lactate, a major metabolic product of growing Lactobacillus spp (26), we examined the possibility that the L. crispatus product responsible for promoting BEC bactericidal activity is lactate. Indeed, exposure of UPEC-bearing BECs to increasing amounts of lactate resulted in a dose-dependent decrease in intracellular bacterial load in infected BECs (Fig. 2I). In addition, exposure to purified lactate (100 mM) significantly enhanced IFN-β expression in BECs (Fig. 2I). Previous studies have described G protein–coupled receptor 81 (GPR81) as a specific cell surface receptor for lactate (27, 28). To determine whether GPR81 was relevant to IFN-β expression and intracellular UPEC killing by BECs, we silenced the expression of GPR81 in human BECs by using GPR81 siRNA. Knockdown of GPR81 failed to enhance IFN-β expression and was not able to reduce intracellular UPEC numbers in human BECs after exposure to L. crispatus (Fig. 2J and SI Appendix, Fig. S9). Taken together, our findings suggests that the L. crispatus factor responsible for promoting bactericidal actions of BECs is lactate, a major metabolite of this bacterium.
Endogenous Microbiome Upregulates the Expression of Cathepsin D in the Bladder Epithelium, Protecting Against Infection.
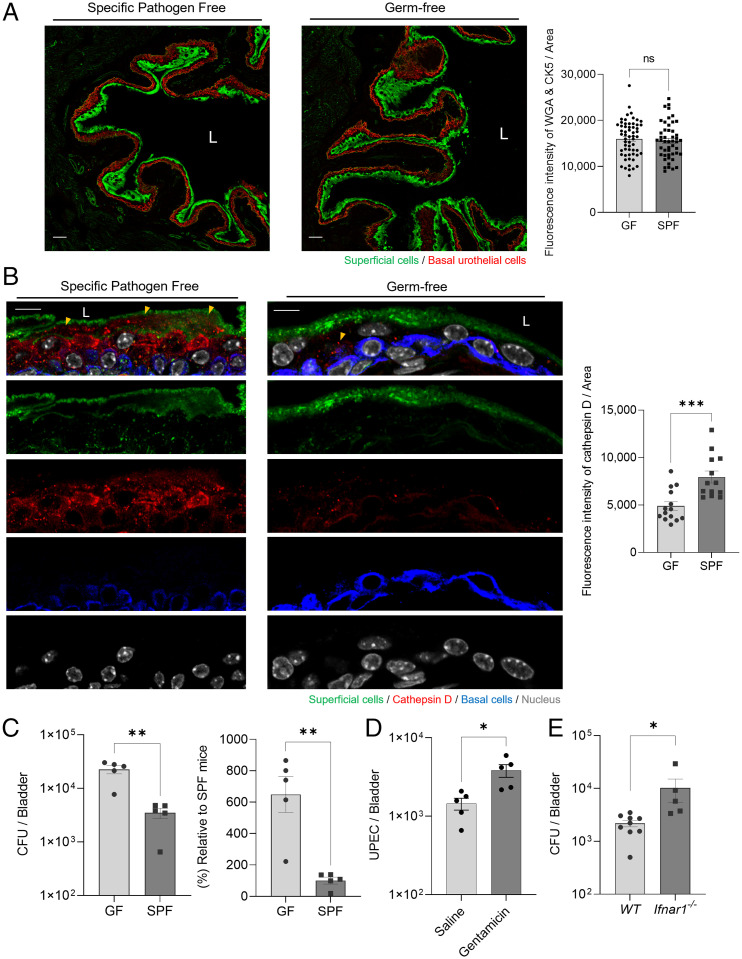
Even though the endogenous microflora of the mouse bladder have yet to be characterized, we sought to investigate their impact on UPEC infections of the mouse bladder. For these studies, we compared C57BL/6J germ-free (GF) mice against specific-pathogen-free (SPF) mice, which presumably harbor endogenous microbes in their bladders. When we probed cross-sections of the murine bladders’ antibodies directed at the epithelium, no morphologic differences were apparent (Fig. 3A). Quantitative analysis of gene expression in BECs from GF or SPF mice revealed no statistical differences (Fig. 3A, Right). However, when we probed for the expression of cathepsin D, we found that numerous cathepsin D+ vesicles were harbored in the bladder epithelium of SPF mice, but very few were evident in GF mice (Fig. 3B). Quantitative analysis revealed a significantly lowered expression of cathepsin D in GF BECs compared with that in SPF BECs (Fig. 3B, Right). These results suggested that the endogenous microflora might contribute to the enhanced expression of cathepsin D in the murine bladder epithelium.
Fig. 3.
Host protective role of endogenous microbiome mediated by upregulation of cathepsin D expression. (A) Comparison of the expression of BEC markers in C57BL/6J GF SPF mice. Immunostaining of superficial epithelial cells (green; wheat-germ-agglutinin-fluorescein isothiocyanate [WGA-FITC]) and basal urothelial cells (red; cytokeratin 5 [CK5]) in bladders of C57BL/6J GF or SPF mice. Scale bars: 50 μm. “L” denotes bladder lumen. (A, Right) The fluorescence intensity of WGA and CK5 was measured in random fields. (B) Bladders sectioned from GF or SPF mice were stained using anti-cathepsin D, CK5 antibodies, or WGA-FITC. Yellow arrowheads indicate cathepsin D expression in BECs. Representative images were chosen. (B, Right) The fluorescence intensity of cathepsin D was measured in random fields. Scale bars: 10 μm. (C) Intravesical infection of GF or SPF mice with UPEC CI5 (1 × 108 CFU). Bladders from each group were harvested after 24 h to count the bacterial burden. (D) Host protective role of gentamicin-sensitive endogenous microbiome in the bladder. WT C57BL/6J mice were administered gentamicin (10 mg/kg) intraperitoneally and (1 mg/kg) intravesically three times one day apart. Three days after the last treatment, the bacterial load of UPEC-infected mouse bladders was examined. (E) Intravesical infection of WT or Ifnar1−/− mice with UPEC CI5 (1 × 108 CFU). Bladders from each group were harvested after 24 h to count the bacterial burden. Each data point represents one mouse. Error bars in each bar graph show mean ± SEM. Each experiment (A–E) was independently repeated, and representative results were presented. Data were analyzed by unpaired two-tailed Student t test (A–E). *P < .05; **P < .01; ***P < .001. ns, not significant.
Because our in vitro studies revealed that the enhanced expression of cathepsin D in BECs correlated with the enhanced clearance of UPEC, we examined whether the endogenous microflora in the murine bladder was protective against UPEC infections. To address this question, we challenged the bladders of both groups of mice with 1 × 108 colony-forming units (CFU) of UPEC strain CI5 and assessed the bacterial load in the bladder of each mouse 24 h later. We found that in contrast to the bladders of GF mice, the bladders of SPF mice harbored numbers of UPECs that were 6 times lower (Fig. 3C). Since antibiotic treatment can be effectively used to deplete the microbiota from various mucosal sites of the mouse urinary tract (29), we pretreated SPF C57BL/6J mice with gentamicin, an antibiotic known to clear endogenous microbiota (29), and then compared the capacity of UPEC CI5 to infect the urinary tract of these mice compared with saline-treated SPF mice. We found that mice pretreated with gentamicin harbored a higher load of UPEC compared with mice treated with saline (Fig. 3D), which indicates that the endogenous microbiota in the urinary tract were a deterrent to UPEC infection. To determine whether the reduced UPEC load in SPF mice that were not treated with gentamicin was mediated by the type I IFN response, we examined the bacterial burden in murine bladders using Ifnar1−/− mice. We compared the UPEC burden in SPF wild-type (WT) and Ifnar1−/− mice for 24 h after infection and found that the bacterial burden was significantly higher in mutant than in WT mice, indicating a lack of intracellular death mediated by the type I IFN response (Fig. 3E). Thus, the capacity of SPF mice to resist UPEC bladder infections seemed to be correlated with the expression of cathepsin D in the bladder epithelium.
Administration of L. crispatus into UPEC-Infected Bladders Reduces Bacterial Burden.
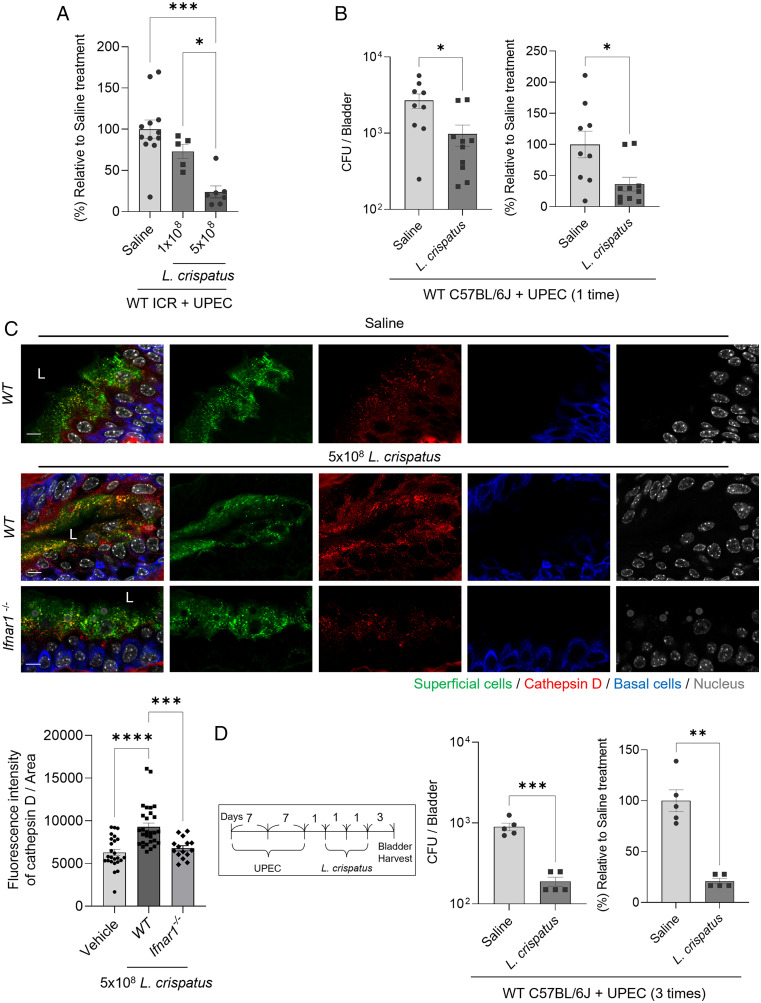
Our in vitro and in vivo results implied that the endogenous bladder flora could be protective against UPEC infections. To test this notion, we administered L. crispatus into the bladders of WT ICR mice infected 24 h previously with 1 × 108 CFU from a UPEC CI5 strain. We considered that UPEC numbers would be stabilized in the bladder of mice by then (30). Briefly, L. crispatus (1 × 108 CFU) was administered into the infected bladders by using a catheter similar to that used for administering UPEC. Compared with the UPEC load in saline-challenged control mice, the UPEC load in L. crispatus–challenged mice was significantly reduced (30%) (Fig. 4A). When we increased the L. crispatus challenge dose to fivefold higher amounts (5 × 108 CFU), we noticed that the reduction in the UPEC burden was as high as 75% (Fig. 4A). To exclude the possibility of mouse strain–specific effects, we substituted ICR mice with C57BL/6J mice and found similar results (Fig. 4B). When we examined whether L. crispatus influenced the expression of cathepsin D, we found a significantly increased number of cathepsin D+ vesicles in the bladder superficial cells of L. crispatus-treated mice (Fig. 4C). However, far fewer numbers of cathepsin D+ vesicles were seen in Ifnar1−/− mice compared with WT mice even though both were treated with L. crispatus (Fig. 4C). Finally, we infected C57BL/6J mice multiple times to mimic the recurrent UTIs in humans and then examined the effect of the Lactobacillus challenge (Fig. 4D, Left). We observed that three consecutive intravesical instillations at 1 d apart significantly decreased the UPEC load in infected bladders (Fig. 4D). Thus, applying Lactobacillus spp. to UPEC-infected mouse bladders significantly reduced the UPEC burden in these animals.
Fig. 4.
Reduced bacterial burden in UPEC-infected bladders mediated by treatment with L. crispatus. (A) Intravesical infection of ICR female mice with UPEC CI5 strain (1 × 108 CFU). After 24 h, infected mice were intravesically inoculated with L. crispatus (1 × 108 or 5 × 108 CFU). The bladders fromeach group of mice were harvested after 24 h. (B) Intravesical infection of C57BL/6J mice with UPEC CI5 strain (1 × 108 CFU). After 24 h, infected mice were intravesically inoculated with L. crispatus (5 × 108 CFU). The bladders from each group of mice were harvested after 24 h. (C) Intravesical inoculation of L. crispatus (5 × 108 CFU) or saline into C57BL/6J mice or Ifnar1−/− mice. After 24 h, WGA (superficial epithelial cell), anti-cathepsin D, or anti-cytokeratin 5 (basal urothelial cell) antibodies were used to stain each bladder, and representative images were chosen. (D) The UPEC CI5 strain (1 × 108 CFU) was used to intravesically infect C57BL/6J mice three times over a period of 7 d. L. crispatus (5 × 108 CFU) was intravesically instilled three times after 24 h from the last infection. The bladders from each group of mice were harvested to assess the bacterial burden. Each data point represents one mouse. Error bars in each bar graph show mean ± SEM. Each experiment (A–D) was independently repeated two to three times with similar results. Data were analyzed by an ordinary one-way ANOVA (A and C) with Tukey’s multiple comparison posttest or unpaired two-tailed Student t test (B and D). *P < .05; **P < .01; ***P < .001; ****P < .0001.
Discussion
To this day, few studies have examined the endogenous microflora in the bladders of healthy subjects because this organ had long been assumed to be sterile. Since the recent discovery that the healthy bladder harbors an endogenous population of microorganisms (2, 3), there has been keen interest in exploring whether this microflora (many members of which are Lactobacillus spp.) confers any benefits to the host. Our findings from the 16S rRNA sequencing screening of urine samples obtained via catheters from patients with recurrent UTIs and healthy controls revealed that Lactobacillus spp. predominate in the bladder microflora of healthy controls, whereas Lactobacillus constitutes only a small fraction of the microflora in patients with recurrent UTIs. Since the predominant bacterial species in patients with UTIs was the uropathogen, we infer that the predominance of Lactobacillus in the bladder of healthy subjects was protective, because it seemed to prevent the predominance of uropathogens. Recent studies that used various next-generation sequencing techniques to identify the bladder microbiome have similarly revealed a correlation between dysbiosis and UTIs (31); however, the role of the bladder microbiome continues to remain unclear (32). Previous studies of vaginal and urethra microflora have portrayed the protective role of Lactobacillus species by occupying entry sites into the urinary tract and preventing prospective pathogens from establishing a foothold. In addition, Lactobacilli can process glycogen, and its breakdown products can lower the vaginal pH of ≤4.5, which is not conducive to the survival of most uropathogens.
In view of the protective role of Lactobacilli in the genitourinary tract, we were keen on evaluating the potential therapeutic value of administering these bacteria into the UPEC-infected bladders. We initially undertook a series of in vitro studies that examined the effects of exposing human UPEC-infected BECs to various species of Lactobacillus. We found that Lactobacilli from at least 3 species (L. rhamnosus, L. acidophilus, and L. crispatus) were capable of markedly reducing the UPEC burden in infected BECs in a dose-dependent fashion. This ability seemed to be related to the capacity of these bacterial species to adhere to the surface of BECs. However, unlike the known effects of adherent Lactobacillus species in inhibiting extracellular E. coli and other prospective pathogens from colonizing the vaginal epithelial surface, the inhibitory effect in the bladder seemed to be targeted at UPEC harbored within BECs. Indeed, we found that adherent Lactobacillus enhanced the intracellular killing of UPEC because the addition of known agents that blocked lysosomal killing by host cells, such as ammonium chloride, inhibited this Lactobacillus-mediated effect. Interestingly, enhanced killing of intracellular UPEC by BECs specifically paralleled their production of type I IFNs (IFN-α or IFN-β) but not of type II IFNs (e.g., IFN-γ) or proinflammatory cytokines such as IL-1β, IL-6, and TNF-α. The critical role of type I IFN in mediating the intracellular killing of UPEC was confirmed by the findings that L. crispatus failed to reduce the intracellular UPEC burden in infected IFNAR1−/− BECs and that treating UPEC-infected human BECs with recombinant human IFN-β was sufficient to recapitulate the Lactobacillus-mediated effect. Our studies further revealed a molecular determinant that promoted the IFN-β–induced enhanced acidity and degradative capabilities of lysosomes. This agent was found to be cathepsin D, an aspartic protease previously implicated in binding to and degrading various bacteria within lysosomes (23). We observed that upon their exposure to IFN-β, infected BECs could greatly enhance the expression of cathepsin D in all their lysosomes, including those harboring intracellular UPEC.
Further support that endogenous bladder microflora promoted the production of cathepsin D in BECs came from our observations that the bladder epithelium of SPF mice contained numerous cathepsin D+ vesicles whereas they were greatly reduced in GF mice. Although the endogenous microflora in the murine bladder have yet to be identified, we found that they conceivably promoted “maturation,” including the production of cathepsin D in murine BECs, similar to our observation of Lactobacillus species that induce the IFN-β–dependent production of cathepsin D in human BECs. Commensal bacteria are known to promote the cellular development and function of epithelial cells and sometimes even the maturation of the immune system (9). The signals emanating from the endogenous flora in the gut are often soluble microbial byproducts such as microbial cell wall components, short-chain fatty acids (acetate, butyrate, and propionate), taurine, and polyamides (33). Our studies revealed that lactate, a Lactobacilli metabolic product, is responsible for promoting the intracellular bactericidal activity of BECs. Indeed, lactate was observed to specifically induce IFN-β expression; furthermore, knocking down of GPR81, the putative lactate receptor on host cells (27, 34), was shown to reduce the bactericidal actions of Lactobacillus. This finding is reminiscent of earlier findings that show gut commensal bacteria promoting maturation and differentiation of intestinal cells via lactate secretion (35). Since any cell maturation cues secreted by bladder commensal bacteria are likely to be quickly removed during voiding, the capacity of certain Lactobacilli spp. to bind BECs may be a critical factor in determining their protective efficacy, as indicated in our study.
We also found that direct administration of Lactobacillus spp. into the bladder of UPEC mice had a therapeutic advantage. We chose to administer Lactobacillus 24 h after UPEC infection, which was sufficient time for the stabilization of UPEC numbers in the bladders of mice (30). We found that administering Lactobacilli had a dose-dependent impact in reducing the UPEC load in the bladders of mice. Indeed, when we administered doses as high as 5 × 108 CFU of Lactobacilli, we noticed a reduction in the UPEC load in the bladders of up to 75%, which is remarkable considering that this was achieved by a single administration of Lactobacilli. The therapeutic effect of Lactobacillus was evident even in mice that had experienced multiple UTIs, which is reminiscent of the situation in patients with recurrent UTIs. Presumably, the addition of Lactobacillus induced a burst in the lysosomal expression of cathepsin D resulting in massive loss of viability of intracellular UPEC. It is noteworthy that during acute UTIs, the bladder is highly inflamed with a large number of proinflammatory mediators released by BECs and recruited immune cells. Because the administration of Lactobacilli failed to provoke the release of proinflammatory cytokines such as IL-1β, IL-6, and TNF-α, administration of Lactobacilli could potentially temper the harmful bladder inflammation while also reducing the UPEC burden. At this time, it remains unknown whether additional treatments with Lactobacilli would be beneficial for the complete clearance of UPEC and what impact, if any, these repeated challenges with endogenous microflora would have on the immune system in the bladder.
Our study revealed the potential of using L. crispatus as a probiotic therapy for UTIs. In view of the protective role of commensal bacteria located at different body sites in preventing the colonization and infection of these sites by prospective pathogens, a few clinical trials have already examined the use of urogenital commensal bacteria as a treatment for recurrent UTIs (7). Even though some of these studies showed promise for this mode of therapy, the underlying mechanisms involved remain unclear (36–38). Our study reveals a novel mechanism by which endogenous microflora such as Lactobacillus protect against recurrent UTIs. However, additional studies are warranted to expand on this mode of treatment as a potentially viable alternative to antibiotic therapy.
Methods
Bacterial Strains and BEC Lines.
Uropathogenic E. coli CI5 strain or CI5 strain transformed with pWSK29 (ampicillin resistance gene–containing plasmid) was used. UPECs were statically grown overnight in lysogeny broth (BD). Lactobacilli (L. rhamnosus, L. crispatus, L. lactis, L. acidophilus, L. casei) were purchased from Korean Collection for Type Cultures. L. jensenii and L. gasseri are clinical isolates from Dr. Choi Laboratory (Korea University). Lactobacilli were cultured in De Man, Rogosa and Sharpe broth (BD) at 37 °C with 5% CO2 overnight. The human BEC line 5637 (American Type Culture Collection, HTB-9) was grown in RPMI 1640 (Gibco) containing 10% fetal bovine serum (HyClone) and incubated at 37 °C with 5% CO2. Recombinant human IFN-β (BioLegend) was used to examine the phenotype. To delete IFNAR1 in the 5637 human BEC line, we used a genome-editing approach targeting the endogenous IFNAR1 gene through clustered regularly interspaced short palindromic repeats (CRISPR) and the endonuclease Cas9. Three CRISPR single-guide RNA (sgRNA) candidates targeting exons 7 and 8 of the IFNAR1 gene were designed by using the CRISPR design database and were cloned into the pSpCas9(BB)-2A-Puro (pX459) plasmid (Addgene). We first examined the efficacy of three sgRNAs and selected the most efficient one among the three candidates (SI Appendix, Fig. S5). The selected oligonucleotide sequences of sgRNAs are forward: 5′- CAC CGA TCG GTG CTC CAA AAC AGT C -3′ and reverse: 5′- AAA CGA CTG TTT TGG AGC ACC GAT C -3′. The pX459 with sgRNA targeting IFNAR1 was transfected with transfection reagent (ScreenFect-A-plus) into the 5637 BEC line for 24 h, followed by the puromycin (5 μg/mL) selection for 3 d. Deletion of IFNAR1 gene expression was confirmed by using immunoblotting with an anti-IFNAR1 antibody (Santa Cruz Biotechnology). To knock down the expression of specific genes (SI Appendix, Table S2), 100 pmol of siRNA targeting the desired host molecules or nonspecific siRNA duplexes (GenePharma) with ScreenFect-A-plus (FUJIFILM Wako Pure Chemical Corporation) were used to transfect human 5637 BECs. After 24 h, transfected 5637 BECs were used for assays of interest. The knockdown was confirmed via western blot analysis using antibodies against the target molecule.
Mice and Human Urine Samples.
Eight-week-old female mice were used for in vivo experiments. WT ICR strain, WT C57BL/6J mice, and Ifnar1−/− mice were used in this study. Ifnar1−/− mice were kindly provided by Nam-Hyuk Cho (Seoul National University College of Medicine). GF inbred C57BL/6J mice were kindly provided by the POSTECH Biotech Center. The experimental procedures were performed with the approval of the Korea University Institutional Animal Care & Use Committee. The human urine sample study was approved by the Institutional Review Board at Soonshunhyang University Bucheon Hospital. The study protocol adheres to the ethical guidelines outlined in the World Medical Association’s Declaration of Helsinki. All human subjects provided informed consent before participating in this study. Patients who were pregnant or who had prolonged indwelling catheters, kidney stones, or anatomical abnormalities were excluded from this study. This study enrolled women (patients with recurrent cystitis, 55.2 ± 13.3 years old; healthy controls, 45.7 ± 12.9 years old) (SI Appendix, Table S1). Patients with recurrent cystitis had medical records that included pyuria and bacteriuria twice in the last six months or three times a year. In addition, these patients exhibited one or more of the following symptoms: suprapubic discomfort, dysuria, frequency, or urgency.
Analysis of Bacterial Burden.
The human 5637 BECs were seeded onto 24-well plates at a density of 2.5 × 105 cells per well and were infected with UPEC CI5 strain (AmpR) at MOI 200. After 1 h of infection, the extracellular bacteria were killed by gentamicin for an additional hour. The infected cells were then rinsed thoroughly, replaced with growth media, and incubated for 6 h at 37 °C with 5% CO2. When necessary, a human type I IFN neutralizing antibody mixture (PBL Assay Science) was used. Lactobacillus spp. were added at different MOI and incubated overnight at 37 °C. After removing media, BECs were treated with 0.1% Triton X-100 in phosphate-buffered saline (PBS) to lyse the cells and plated on MacConkey agar plates (BD) with carbenicillin.
qPCR.
RNA isolation and qRT-PCR. The BECs were lysed, and total RNA was isolated by using RNeasy Mini Kit (QIAGEN), as indicated in the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized with an M-MLV cDNA synthesis kit (Enzynomics). TB Green Premix Ex Taq (Takara Bio) and a CFX384 Touch Real-Time PCR Detection System (Bristol Myers Squibb) were used for qRT-PCR. All target gene RNA expressions were normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression. Data were analyzed manually by the ΔΔCT method and were statistically demonstrated by using GraphPad Prism v.9.2.1. The primer sequences for qPCR are listed in SI Appendix, Table S3.
DNA Extraction, 16S rRNA Sequencing, and Bioinformatic Analysis.
The 30-mL urine samples were collected and centrifuged to obtain urine pellets. After being resuspended in PBS, the DNA from urine pellets was extracted by using a chemagic DNA Stool Kit (PerkinElmer) with a modified pretreatment bead-beating step. The 16S library was constructed on the basis of the prepared DNA samples and analyzed by using the NEXTFLEX 16S V4 Amplicon-Seq Library Prep Kit (Bioo Scientific). Paired-end sequencing was performed with the MiSeq Reagent Kit v2 Nano and the Illumina MiSEq 2000 high-throughput DNA sequencing service in April 2013. Cluster generation, sequencing, and analysis were all performed according to the manufacturer’s instructions. QIIME 2 microbiome analysis package was used to perform the bioinformatic analysis. Taxonomy classification used naïve Bayesian classifiers trained with the Reference Sequence databases.
Immunofluorescent Staining and Microscopy.
The bladder tissue was fixed in 4% paraformaldehyde and frozen in optimal cutting temperature compound. To section the tissues, we used a Leica Biosystems CM1850 cryostat (Leica Biosystems Inc). The sliced tissues were blocked and permeabilized in the buffer, which contained 0.3% Triton and 2.5% normal goat serum (Gibco) in 1% bovine serum albumin (BSA)-PBS. Primary antibodies against E. coli (BioRad), cytokeratin 5 (Abcam), or cathepsin D (Santa Cruz Biotechnology) were added overnight. To visualize with a confocal microscope, fluorescently conjugated wheat germ agglutinin (Molecular Probes) or fluorescently conjugated secondary antibodies (Jackson ImmunoResearch) was used, and samples were mounted with Prolong Diamond Antifade Reagent (Invitrogen). The images were captured via a channel-series approach using a Zeiss LSM700 or LSM800 upright confocal microscope.
Human 5637 BECs were grown on a round glass coverslip and DQ-BSA (Thermo Fisher Scientific) was added to assess lysosomal protease activity. To examine the acidity of lysosomes, 100 nM Lysotracker (Invitrogen) was added to culture media. After 4% perfluoroalkoxy fixation, the samples were permeabilized and blocked with 0.1% saponin in 1% BSA-PBS solution. The primary antibody was anti-LAMP1 (Abcam), and anti-cathepsin D antibodies (Santa Cruz Biotechnology) were also used. The secondary antibodies were fluorescently conjugated antibodies (Jackson ImmunoResearch).
ELISA.
Human 5637 BECs were dose-dependently exposed to L. crispatus. After 9 h of treatment, the cell-free supernatant was analyzed by using the human IFN-β Quantikine ELISA Kit (R&D Systems).
Gram Staining of Adhered Lactobacillus on BECs.
Human 5637 BECs were plated on glass coverslips and treated with Lactobacilli at MOI 1000. After 4 h, the coverslip was washed with PBS. The methanol-fixed cells were stained with Gram solution (Sigma). All the images were taken with a light microscope at ×1000 magnification. Lactobacilli were Gram positive colored rod-shaped bacteria. Lactobacillus adhesion was assessed by using the adhesion index, which indicates adhering bacteria per number of cells. The adherent Lactobacilli were counted as a total quantity of bacteria adherent to 50 randomly chosen intact BECs.
Immunoblots.
The samples were lysed in radioimmunoprecipitation assay buffer and vortexed three times for 30 s each. After centrifugation at 13,000g, the lysates were mixed with sample buffer and separated on 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels. Proteins on SDS-PAGE gels were transferred to PVDF membranes (Immobilon) and then blocked for 1 h in 3% BSA in Tris-buffer saline with Tween-20. Membranes were treated with a human anti-IFN-α/βRα mouse monoclonal antibody, anti-TFEB antibody, anti-cathepsin D antibody (Santa Cruz Biotechnology), and anti-GPR81 antibody (Novus Biological) overnight at 4 °C. Secondary antibodies were anti-mouse immunoglobulin G (IgG) or anti-rabbit IgG antibodies (BioRad) coupled with horseradish peroxidase.
Antimicrobial Activity of Cell-Free Supernatants.
The agar well diffusion method was used to determine the antibacterial activity of cell-free supernatants (CFSs) against UPEC. Briefly, MRS plates were overlaid with a soft 0.5% agar solution premixed with 108 CFU of the UPEC CI5 strain. Then, wells with a diameter of 7 mm were sterilely prepared and filled with 100 μL of CFS from L. crispatus or CFS mixed with α-amylase, trypsin, or proteinase K (Sigma). The bacterial inhibition zone was determined 48 h after incubation at 37 °C.
Agar Overlay Method.
L. crispatus (1 × 108 CFU in 20 µL) was inoculated and dried at five different spots on MRS agar plates. Then, the MRS agar plates with Lactobacilli in spots were overlaid with a soft 0.5% agar solution premixed with 108 CFU of the UPEC CI5 strain. After solidifying the overlaid agar medium, the plates were incubated at 37 °C for 24 h. The experiment switching L. crispatus to UPEC CI5 strain was performed, in which cultured UPECs were inoculated on L. crispatus–cultured agar plates.
Quantification and Statistical Analysis.
Prism (GraphPad Software v.9.2.1) was used to analyze the data and produce graphs. Microsoft Excel was used to produce Fig. 1A. Dots displayed in each graph (in vitro experiments) represent samples obtained from two to three independent experiments. Dots in each graph (mouse experiments) refers to the number of mice from which bladders were obtained. A two-tailed unpaired Student t test was used for comparisons between two groups, and an ordinary one-way ANOVA with Tukey’s multiple comparisons test was used to calculate statistical significance among more than two groups. Each experiment was repeated independently two to three times with similar results. Data are presented as mean ± SEM. Posttest P values were as follows: *P < .05; **P < .01; ***P < .001; ****P < .0001; ns, not significant.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant 2020R1C1C1003257, by Korea University’s internal grants, and by the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Disease Grant K12DK100024 (K12 Urologic Research Career Development Program).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2117904119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Langille M. G., et al. , Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas-White K., et al. , Culturing of female bladder bacteria reveals an interconnected urogenital microbiota. Nat. Commun. 9, 1557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pearce M. M., et al. , The female urinary microbiome: A comparison of women with and without urgency urinary incontinence. MBio 5, e01283-e14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolfe A. J., Brubaker L., Urobiome updates: Advances in urinary microbiome research. Nat. Rev. Urol. 16, 73–74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nickel J. C., et al. ; MAPP Research Network, A culture-independent analysis of the microbiota of female interstitial cystitis/bladder pain syndrome participants in the MAPP Research Network. J. Clin. Med. 8, 415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Whiteside S. A., Razvi H., Dave S., Reid G., Burton J. P., The microbiome of the urinary tract—A role beyond infection. Nat. Rev. Urol. 12, 81–90 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Stapleton A. E., et al. , Randomized, placebo-controlled phase 2 trial of a Lactobacillus crispatus probiotic given intravaginally for prevention of recurrent urinary tract infection. Clin. Infect. Dis. 52, 1212–1217 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan F., Polk D. B., Probiotics and immune health. Curr. Opin. Gastroenterol. 27, 496–501 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khosravi A., et al. , Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15, 374–381 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamm W. E., Norrby S. R., Urinary tract infections: Disease panorama and challenges. J. Infect. Dis. 183 (suppl. 1), S1–S4 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Svanborg C., Godaly G., Bacterial virulence in urinary tract infection. Infect. Dis. Clin. North Am. 11, 513–529 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Kuehn M. J., Heuser J., Normark S., Hultgren S. J., P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature 356, 252–255 (1992). [DOI] [PubMed] [Google Scholar]
- 13.Abraham S. N., Sun D., Dale J. B., Beachey E. H., Conservation of the D-mannose-adhesion protein among type 1 fimbriated members of the family Enterobacteriaceae. Nature 336, 682–684 (1988). [DOI] [PubMed] [Google Scholar]
- 14.Bishop B. L., et al. , Cyclic AMP-regulated exocytosis of Escherichia coli from infected bladder epithelial cells. Nat. Med. 13, 625–630 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Mulvey M. A., Schilling J. D., Hultgren S. J., Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 69, 4572–4579 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz D. J., Conover M. S., Hannan T. J., Hultgren S. J., Uropathogenic Escherichia coli superinfection enhances the severity of mouse bladder infection. PLoS Pathog. 11, e1004599 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miao Y., Li G., Zhang X., Xu H., Abraham S. N., A TRP channel senses lysosome neutralization by pathogens to trigger their expulsion. Cell 161, 1306–1319 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sachdeva K., Sundaramurthy V., The interplay of host lysosomes and intracellular pathogens. Front. Cell. Infect. Microbiol. 10, 595502 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y., et al. , Evaluation of the inhibitory effects of Lactobacillus gasseri and Lactobacillus crispatus on the Adhesion of seven common lower genital tract infection-causing pathogens to vaginal epithelial cells. Front. Med. (Lausanne) 7, 284 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Isaacs A., Lindenmann J., Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147, 258–267 (1957). [PubMed] [Google Scholar]
- 21.Nathan C., Shiloh M. U., Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. U.S.A. 97, 8841–8848 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sardiello M., et al. , A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 (2009). [DOI] [PubMed] [Google Scholar]
- 23.Carrasco-Marín E., et al. , The innate immunity role of cathepsin-D is linked to Trp-491 and Trp-492 residues of listeriolysin O. Mol. Microbiol. 72, 668–682 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Vieco-Saiz N., et al. , Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 10, 57 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewus C. B., Sun S., Montville T. J., Production of an amylase-sensitive bacteriocin by an atypical Leuconostoc paramesenteroides strain. Appl. Environ. Microbiol. 58, 143–149 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kandler O., Carbohydrate metabolism in lactic acid bacteria. Antonie van Leeuwenhoek 49, 209–224 (1983). [DOI] [PubMed] [Google Scholar]
- 27.Cai T. Q., et al. , Role of GPR81 in lactate-mediated reduction of adipose lipolysis. Biochem. Biophys. Res. Commun. 377, 987–991 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Ge H., et al. , Elucidation of signaling and functional activities of an orphan GPCR, GPR81. J. Lipid Res. 49, 797–803 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Kennedy E. A., King K. Y., Baldridge M. T., Mouse microbiota models: Comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 9, 1534 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choi H. W., et al. , Loss of bladder epithelium induced by cytolytic mast cell granules. Immunity 45, 1258–1269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perez-Carrasco V., Soriano-Lerma A., Soriano M., Gutiérrez-Fernández J., Garcia-Salcedo J. A., Urinary microbiome: Yin and yang of the urinary tract. Front. Cell. Infect. Microbiol. 11, 617002 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gasiorek M., Hsieh M. H., Forster C. S., Utility of DNA next-generation sequencing and expanded quantitative urine culture in diagnosis and management of chronic or persistent lower urinary tract symptoms. J. Clin. Microbiol. 58, e00204–e00219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Solis A. G., Klapholz M., Zhao J., Levy M., The bidirectional nature of microbiome-epithelial cell interactions. Curr. Opin. Microbiol. 56, 45–51 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu C., et al. , Lactate inhibits lipolysis in fat cells through activation of an orphan G-protein-coupled receptor, GPR81. J. Biol. Chem. 284, 2811–2822 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Lee Y. S., et al. , Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host Microbe 24, 833–846.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Reid G., et al. , Oral probiotics can resolve urogenital infections. FEMS Immunol. Med. Microbiol. 30, 49–52 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Akgül T., Karakan T., The role of probiotics in women with recurrent urinary tract infections. Turk. J. Urol. 44, 377–383 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mueller E. R., Wolfe A. J., Brubaker L., Female urinary microbiota. Curr. Opin. Urol. 27, 282–286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.