Significance
The neuropeptide system encompasses the most diverse family of neurotransmitters, but their expression, cellular localization, and functional role in the human brain have received limited attention. Here, we study human postmortem samples from prefrontal cortex (PFC), a key brain region, and employ RNA sequencing and RNAscope methods integrated with published single-cell data. Our aim is to characterize the distribution of peptides and their receptors in 17 PFC subregions and to explore their role in chemical signaling. The results suggest that the well-established anatomical and functional heterogeneity of human PFC is also reflected in the expression pattern of the neuropeptides. Our findings support ongoing efforts from academia and pharmaceutical companies to explore the potential of neuropeptide receptors as targets for drug development.
Keywords: anterior cingulate cortex, in situ hybridization, RNA-seq, classic neurotransmitter coexistence
Abstract
Human prefrontal cortex (hPFC) is a complex brain region involved in cognitive and emotional processes and several psychiatric disorders. Here, we present an overview of the distribution of the peptidergic systems in 17 subregions of hPFC and three reference cortices obtained by microdissection and based on RNA sequencing and RNAscope methods integrated with published single-cell transcriptomics data. We detected expression of 60 neuropeptides and 60 neuropeptide receptors in at least one of the hPFC subregions. The results reveal that the peptidergic landscape in PFC consists of closely located and functionally different subregions with unique peptide/transmitter–related profiles. Neuropeptide-rich PFC subregions were identified, encompassing regions from anterior cingulate cortex/orbitofrontal gyrus. Furthermore, marked differences in gene expression exist between different PFC regions (>5-fold; cocaine and amphetamine–regulated transcript peptide) as well as between PFC regions and reference regions, for example, for somatostatin and several receptors. We suggest that the present approach allows definition of, still hypothetical, microcircuits exemplified by glutamatergic neurons expressing a peptide cotransmitter either as an agonist (hypocretin/orexin) or antagonist (galanin). Specific neuropeptide receptors have been identified as possible targets for neuronal afferents and, interestingly, peripheral blood-borne peptide hormones (leptin, adiponectin, gastric inhibitory peptide, glucagon-like peptides, and peptide YY). Together with other recent publications, our results support the view that neuropeptide systems may play an important role in hPFC and underpin the concept that neuropeptide signaling helps stabilize circuit connectivity and fine-tune/modulate PFC functions executed during health and disease.
Prefrontal cortex (PFC) represents the most rostral part of the frontal lobe and is highly developed in the human brain. Functionally, human PFC (hPFC) is a region of high complexity involved in multiple functions, including cognitive processes such as working memory and decision making as well as generation and regulation of emotions (1–3), and likely in many psychiatric disorders. Thus, monitoring regional cerebral blood flow, a “hypofrontal” pattern was early on reported in hPFC of “deteriorated schizophrenics” (4), and later confirmed and solidified (5) and complemented with aspects of chemical signaling (6). PFC is also a key node of the circuitry related to, for example, stress and depression (3, 7–9), attention disorders (10), drug abuse (11, 12), and pain (13, 14).
Chemical signaling in PFC has been thoroughly reviewed (1), but knowledge of the expression and distribution patterns of neuropeptides (NPs) (15), the most diverse of all neurotransmitter families (>100 members) (16, 17), is still comparatively limited. However, the field of NPs and NP receptors (NPRs) in human cerebral cortex has been investigated (18, 19), both with histochemical (20–24) and biochemical (25, 26) methods. Moreover, there are reports dealing with NP systems specifically in hPFC as recently reviewed, often in relation to disease (27, 28). For example, in human dorsolateral prefrontal cortex (DLPFC), somatostatin (SST) (29) and somatostatin receptor 2 (SSTR2) (30) messenger RNA (mRNA) levels are decreased in schizophrenia, and transcripts for several peptides are changed in this disease during development (31). In the same brain region of subjects who committed suicide/suffered from major depression disorder (MDD), neuropeptide Y receptor Y2 (NPY2R) transcript levels are elevated (32), and SST mRNA levels are reduced (33). Also, in human subgenual anterior cingulate cortex (sgACC), both SST mRNA and the precursor protein are markedly reduced in a subset of GABAergic neurons, female transcript levels more so than male ones (34), in agreement with the well-known sexual dimorphism in mood disorders (35–37). Another example is significant differences in the galanin system between MDD subjects and controls (38, 39). These and other findings indicate a therapeutic potential by targeting modulatory NP systems in hPFC (28).
NPs have an early evolutionary origin (40) and are widely expressed and stored in large dense core vesicles; they are auxiliary, modulatory transmitters, fine-tuning networks and exerting their actions via seven-transmembrane, G protein–coupled receptors (GPCRs); in contrast with classic transmitters, peptides do not have a reuptake mechanism (transporter) but have to be replaced by new protein synthesis (41–44). These messenger molecules are preferentially released extrasynaptically in response to intense/burst firing (41, 45–48). More recently, NPs have been proposed to contribute to stabilizing circuit connectivity (49). They distinctly contrast with fast transmitters like the amino acids GABA and glutamate that are stored in classic synaptic vesicles, mostly are released at synapses, exert effects at the millisecond level, and have a reuptake mechanism. The NPs coexist with classic transmitters, for example, monoamines (50), as well as with GABA in cortical inhibitory interneurons (51, 52).
Here, we present an extensive analysis of the transcriptomic landscape for 17 subregions of hPFC and three reference cortical regions (RefCorts) (frontal, parietal, and temporal cortex), encompassing a large number of microdissected samples from both male and female donors, including both left and right hemispheres. We have chosen to work with “bulk” samples to include all cell types and detected low-abundance transcripts in each microdissected sample enabling comparison between regions of PFC. This is especially relevant for the detection of transcripts for NPs and their receptors that in some cases are limited to a small population of cells. In addition to the expression patterns of the peptidergic systems, we include notes on molecules related to classic and some other neurotransmitters. The RNA sequencing (RNA-seq) results are complemented with spatial RNAscope of selected NPs and NPRs. To classify the peptidergic systems in PFC, we also map the expression of NPs and NPRs to cell type–specific expression profiles of GABAergic, glutamatergic, and non-neuronal cells.
Results
Transcriptomic Analysis of Microdissected Human Brain Cortical Subregions.
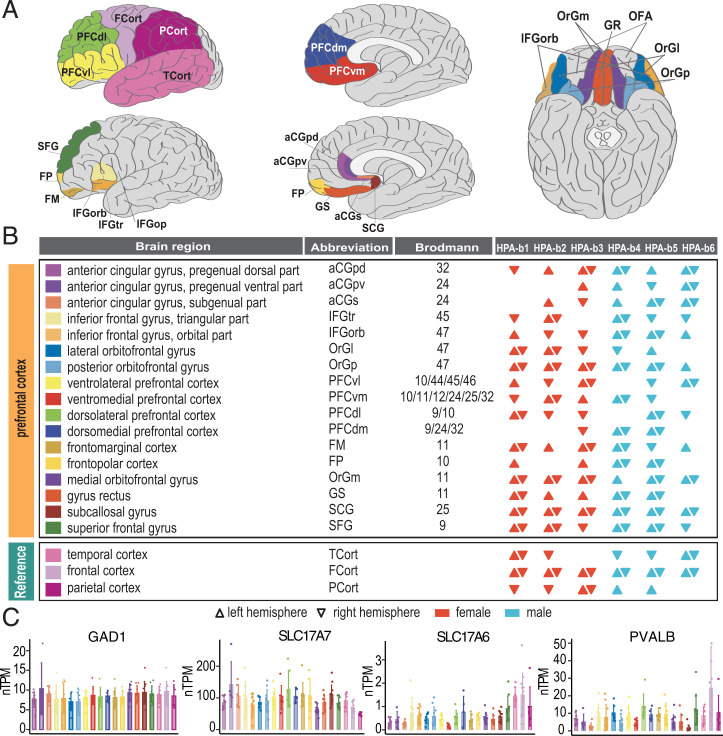
Seventeen hPFC subregions and three RefCorts, depicted schematically in Fig. 1A, were microdissected from both left and right hemispheres of three male and three female donor brains between the ages of 61 and 94 y (Fig. 1B and Dataset S1A). Expression maps for all individual human protein-coding genes (n = 19,670) across the 20 brain cortical subregions are presented in the Human Protein Atlas (HPA) Brain Atlas (https://v20.proteinatlas.org/humanproteome/brain) (53) as an open-access resource (more details are in SI Appendix, Materials and Methods and Fig. S1). As examples, bar plots of four important markers for cortical cell subpopulations are shown in Fig. 1C. Glutamic acid decarboxylase 1 (GAD1) is highly expressed and present in most cortical interneurons, as is vesicular glutamate transporter 1 (VGLUT1) (also known as solute carrier family 17 member 7; SLC17A7) in pyramidal neurons. Both are evenly distributed in all regions. In contrast, markedly different but much lower levels (up to 17-fold) and regional variations were observed for vesicular glutamate transporter 2 (VGLUT2; also called SLC17A6) with highest expression levels in RefCorts. Also, the levels of parvalbumin (PVALB), a calcium-binding protein (CaBP), and a marker for subsets of GABAergic interneurons varied markedly (14-fold) between regions.
Fig. 1.
Transcriptomic analysis of the 17 microdissected PFC regions and 3 cortical reference regions. (A) Schematic overview of the 20 cortical subregions and the associated Broca areas. (B) A summary of the 165 included human brain samples (for technical details, see SI Appendix, Fig. S1). (C) Examples of expression profiles for three genes in 20 cortical subregions. Shown are bar plots for transcripts for GAD1, the GABA-synthesizing enzyme, the vesicular glutamate transporter VGLUT1 (SLC17A7) and VGLUT2 (SLC17A6), as well as the calcium-binding protein parvalbumin (PVALB) (note difference in levels, see http://v20.proteinatlas.org/humanproteome/brain for details regarding all 19,670 human protein-coding genes and their expression profiles in these subregions of PFC). Error bars represent mean ± SD.
Neuropeptides and Their Receptors.
The complete list of NPs was based on Burbach (16) with the addition of two pituitary hormones, prolactin (PRL) and growth hormone (GH). In total, 78 NPs, including 7 precursors, and 83 corresponding NPRs were analyzed (Dataset S3). Among them, 60 (77%) NPs and 60 (72%) NPRs were detected in at least one hPFC subregion (Fig. 2A and B). Information on general aspects of NPs and NPRs and their relation to disease is provided in SI Appendix, Notes I and II.
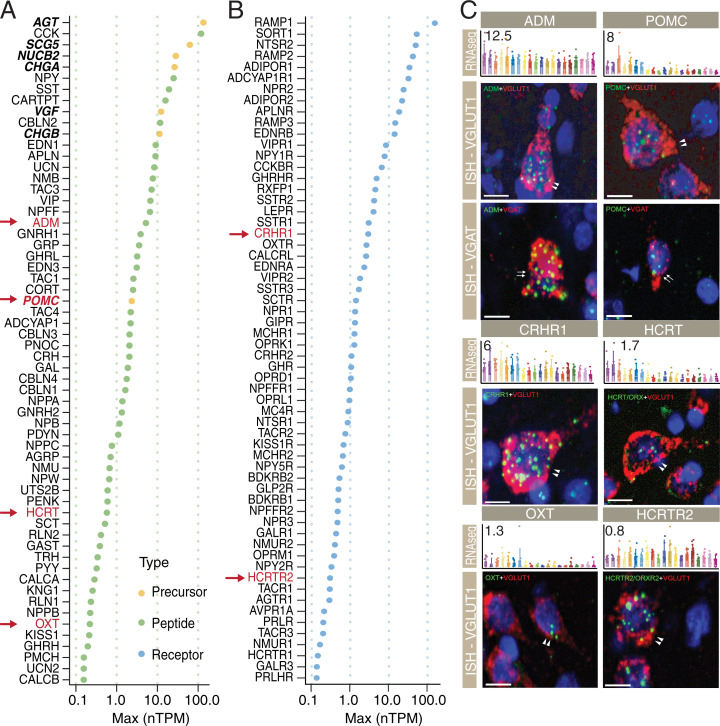
Fig. 2.
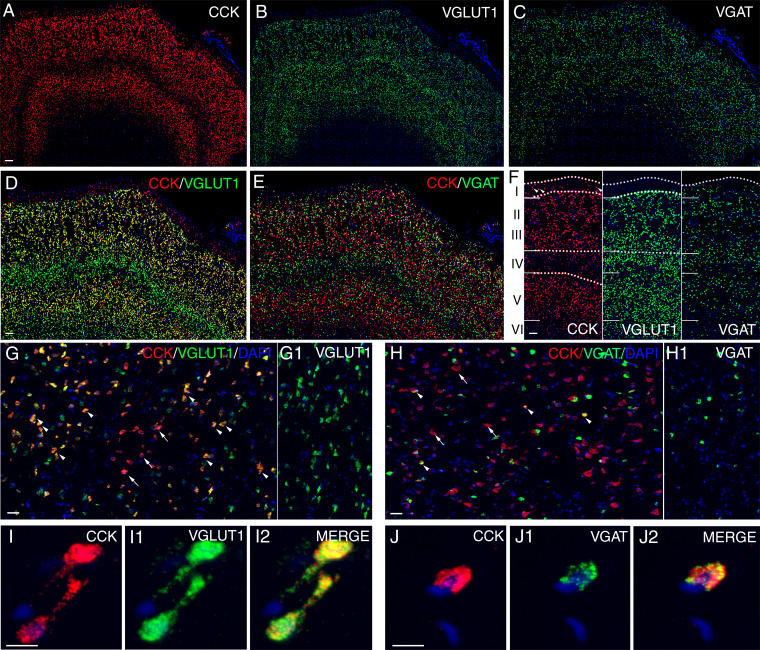
Expression levels of NPs and NPRs in hPFC. (A) Maximum expression levels of detected NPs (n = 60). (B) Maximum expression levels of detected NPRs (n = 60). Genes with nTPM >0.1 were defined as detected genes. The color code indicates the type of genes (orange, precursor; green, NP; blue, NPR). Red arrows in A and B point to the transcripts studied with ISH in C. (C) Examples of expression profiles and in situ RNA hybridization data for adrenomedullin (ADM), proopiomelanocortin (POMC), corticotropin-releasing hormone receptor 1 (CRHR1), hypocretin NP/orexin precursor (HCRT), oxytocin (OXT), and HCRT/ORX receptor 2 (HCRTR2). The y axis represents the average nTPM values for each gene. The x axis represents the 20 human cortical subregions. Error bars represent mean ± SD. The color codes are the same as in Fig. 1B. Bar plots are examples of ISH results for each gene in the corresponding cell types. Color code: cell type analyzed, red; gene of interest, green. Arrowheads depict positive cells for VGLUT1 and arrows indicate VGAT-positive cells. (Scale bars, 5 μm.) The expression levels of exemplified NPs and NPRs with in situ RNA hybridization data are outlined in A and B with red arrows.
The distribution of expression levels of precursors, NPs, and NPRs in hPFC is shown in SI Appendix, Fig. S2A. The levels markedly vary both for peptide and receptor transcripts. The highest levels were found for precursor molecules, which generate several bioactive NPs after tissue- and region-specific posttranslational processing by prohormone convertases (54). They include angiotensinogen (AGT), chromogranin A (CHGA), chromogranin B (CHGB), nucleobindin 2 (NUCB2), proopiomelanocortin (POMC), secretagogin V (SCG5), and neurotrophic growth factor–inducible (VGF). AGT is part of the renin–angiotensin system (RAS) (55). However, we detected only angiotensin I converting enzyme (ACE) (normalized transcripts per million [nTPM] 1.0 to 3.2), ACE2 (∼10-fold lower levels), angiotensin receptor 1 (AGTR1), and MAS1 protooncogene (MAS1) but, importantly, no renin (REN) (see gene-specific pages in the HPA Brain Atlas), possibly indicating that classic angiotensin peptides are not produced in PFC. Cerebellins (CBLNs) 1 to 4 are special, earlier considered precursors, with the highest transcript levels for CBLN2 > CBLN3 > CBLN4 > CBLN1. They are secreted glycoproteins with signal peptides and act as synaptic organizers associated with neurexins and glutamate receptors GluD1 and -D2 for CBLN1 and 2 (56, 57) (SI Appendix, Fig. S2F) and, in fact, control spinogenesis also in hPFC (58).
Regarding NPs, the expression level of cholecystokinin (CCK) is the highest (Fig. 2A). Early histochemical studies reported the presence of CCK only in interneurons, but later in situ hybridization (ISH) showed transcripts also in many pyramidal neurons (59), giving the very high cortical CCK NP levels a reasonable explanation (see below). The NPR levels were, in general, somewhat lower than those for precursors and NPs (SI Appendix, Fig. S2A).
GPCR transcripts for “classic” interneuron NPs like adenylate cycles activating polypeptide 1 (ADCYAP1; also known as PACAP) (ADCYAP1R1), neuropeptide Y (NPY; NPY1R), CCK (CCKB), and SST (SSTR1 and -R2) are all highly expressed in hPFC. However, the highest levels in hPFC are seen for RAMPs (receptor activity–modifying proteins) (Fig. 2B). RAMPs were first described for calcitonin gene–related peptide (CGRP; CALCA and -B), and represent auxiliary proteins important for forming a functional complex with the GPCR (60). Also, the transcript for sortilin-related receptor 1 (SORT1) is very highly expressed (Fig. 2B). SORT1 was originally thought to be a neurotensin receptor, but has multiple functions (sorting, vesicle transportation, and activation of nascent receptor molecules) (61). The likely main receptor for neurotensin is the also very highly expressed neurotensin receptor 2 (NTSR2). In situ RNA hybridization was further performed and confirmed the expression of selected medium- and low-abundance NPs/NPRs on the single-cell level (Fig. 2C). In addition, a multifactor ANOVA for all NPs and corresponding NPRs was conducted to investigate the effects from sex, hemisphere, and PFC subregions on gene expression levels (Dataset S4). A ternary plot shows that most variability of the genes can be observed between regions (SI Appendix, Fig. S2B).
PFC Regions Rich in Peptides and Peptide Receptors.
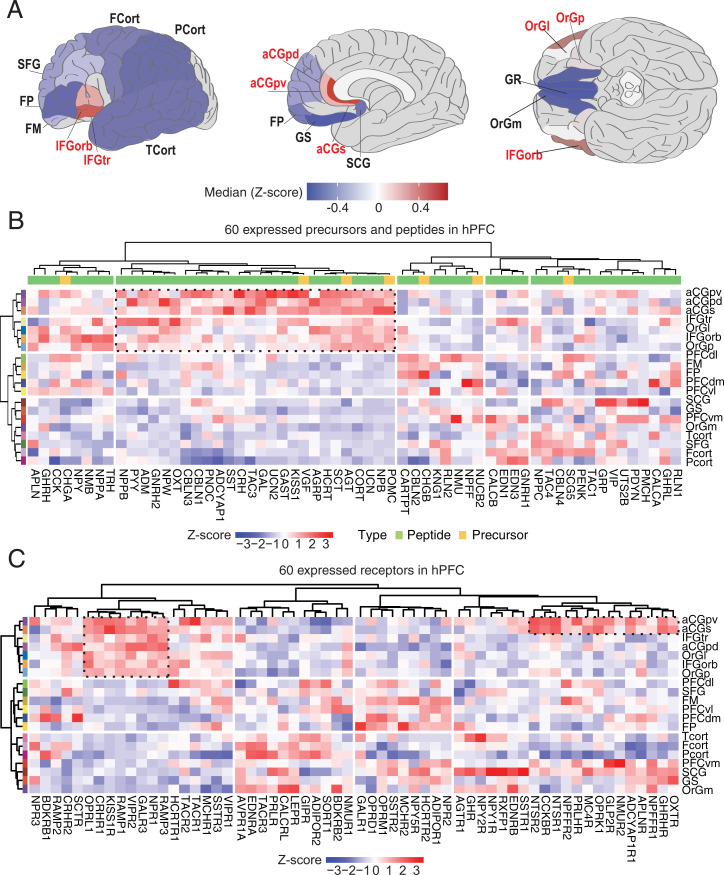
By calculating the z score, we compared the relative expression levels of the NPs in hPFC and RefCorts. Interestingly, NPs were found to be enriched in seven “ACC-ORB” subregions, including ACC (subgenual, pregenual dorsal and ventral; BA24 and BA32), ORB (lateral and posterior orbitofrontal gyrus; BA47), and inferior frontal gyrus (triangular part, Broca and orbital part; BA45 and BA47), from here on referred to as “NP-rich” regions (Fig. 3A and SI Appendix, Fig. S2C). Several coexpression clusters could be identified but, of particular interest, one encompassed 26/34 NPs expressed at high levels in ACC-ORB (Fig. 3B and Dataset S5A). Pearson correlation analysis also revealed the overall similarity of these NP-rich regions as compared with the other cortical subregions based on the expression profiles of NPs (SI Appendix, Fig. S2D). Nevertheless, and importantly, each peptide has a unique profile based on subregion localization and expression levels, suggesting that an NP may differentially affect/modulate neurocircuits in cortical areas that form hPFC.
Fig. 3.
Regional variation of NPs and NPRs in hPFC. (A) Levels of NP transcripts in different cortical subregions as well as the localization of NP-rich regions (labeled in red). The color code indicates the median z score of all detected NPs (SI Appendix, Fig. S2B shows the distribution of z scores of NPs). (B) A heatmap presenting the expression patterns of 60 detected NP transcripts across the 20 cortical subregions. (C) A heatmap presenting the expression patterns of 60 detected NPR transcripts across the 20 cortical subregions.
Relative expression levels of NPRs across the 20 cortical regions are shown in Fig. 3C, SI Appendix, Fig. S2E, and Dataset S5B, and several cases of pairs of NPs and corresponding NPRs (proopiomelanocortin [POMC]/melanocortin-4 receptor [MC4R]; oxytocin [OXT]/OXTR; galanin [GAL]/GAL3; kisspeptin1 [KISS1]/KISS1R; corticotropin-releasing hormone [CRH]/CRHR1; secretin [SCT]/SCTR) were highly coexpressed in certain regions, evidencing putative local circuitries as shown by Smith et al. (62). The subregional variation for NPR transcripts is less pronounced than for NP transcripts but, for example, CRHR1, KISS1R, opioid-related neuropeptide (OPN) (also known as nociceptin [NOC] and orphanin [FQ]/OPRL1), as well as RAMP1 and -3 are more abundant in the NP-rich cluster, whereas TAC3 (also known as neurokinin B) (TAC3)/TACR3, leptin (LEP)/LEPR, and adiponectin (ADIPOR)/ADIPOR2 are higher in the RefCorts. Fifteen examples were selected to show the regional variation of peptides and receptors (SI Appendix, Fig. S2F).
To further explore the differences between the expression levels of NPs and NPRs in hPFC and RefCorts, we conducted a differential expression analysis using ANOVA. A total of 9 NPs and 19 NPRs were identified with significantly higher levels in hPFC (SI Appendix, Fig. S3A and Dataset S5), including KISS1 (nTPM 0.2 in PFC vs. 0.03 in RefCorts) and the corresponding NPR, KISS1R (nTPM 0.67 vs. 0.06) (SI Appendix, Fig. S3B), thus representing interneuron candidates.
Classification of Peptides and Receptors.
To gain insights into neuronal cell types expressing NPs and NPRs in NP-rich regions, we mapped the expression of NPs and NPRs to published cell taxonomy profiles for GABAergic, glutamatergic, and non-neuronal cells from single-nucleus RNA-seq (snRNA), with focus on ACC, reported by Khrameeva et al. (63) (Fig. 4 and SI Appendix, Fig. S4). A large fraction of NPs (n = 31, 52%) and NPRs (n = 40, 67%) was also detected in at least one of the cell types (GABAergic, glutamatergic, and non-neuronal cells) (Dataset S6). In addition, we observed the expression of 29 NPs and 20 NPRs in ACC not detected by snRNA, indicating a higher detection sensitivity of bulk RNA-seq.
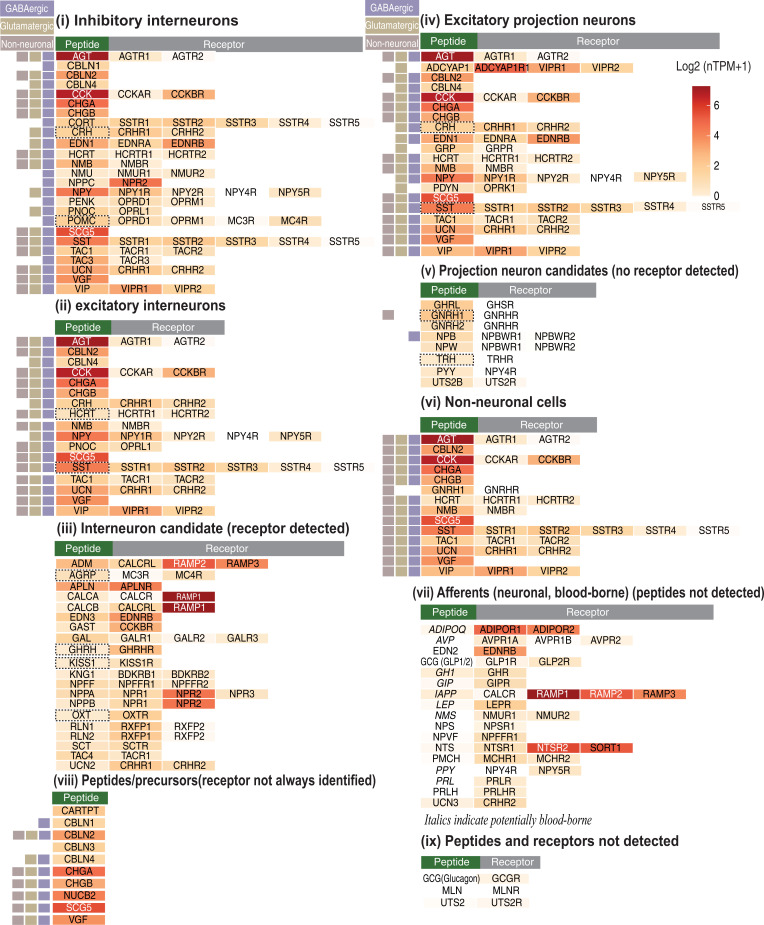
Fig. 4.
Categorization of peptidergic cell types in ACC based on expression profiles of NPs and their NPRs. Overview of the gene expression of 78 NPs and their corresponding NPRs in ACC. The three boxes (Left) indicate the existence of the NPs in GABAergic, glutamatergic, and nonneuronal cells which were inferred from a published snRNA dataset of the hACC region from Khrameeva et al. (63). The color code of the heatmap indicates the expression levels of the NPs/NPRs in ACC. The hypothalamic NPs are outlined with dashed boxes.
The 78 NPs and 83 of their corresponding NPRs were thus further classified based on their expression in ACC and in three basic cell types (GABAergic, glutamatergic, and non-neuronal cells) from the same region. Eight peptidergic cell classes based on the expression of NPs and NPRs are proposed: 1) inhibitory interneurons (NP and its NPR[s] expressed in inhibitory neurons, n = 25); 2) excitatory interneurons (NP and its NPR[s] expressed in excitatory neurons, n = 17); 3) interneuron candidates (both NP and its NPR[s] expressed, n = 20); 4) excitatory projection neurons (NP expressed in excitatory neurons, n = 20); 5) projection neuron candidates (NP expressed, but not its NPR, n = 8); 6) non-neuronal cells (NP expressed in non-neuronal cells, n = 14); 7) afferent input, either neuronal from other brain regions or blood-borne peptides (NPR expressed, but not NP, n = 17); and 8) precursors/NPs (no receptor known, n = 10); some NPs and NPRs could not be detected (Fig. 4 and Dataset S7). Note that several NPs are listed in more than one class.
Interneurons in cortex expressing a NP represent an important group, as shown in animal studies (19, 62, 64) and human studies (64). A total of 25 NPs were listed as expressed in inhibitory interneurons in hPFC, including, for example, CCK, NPY, SST, vasoactive intestinal polypeptide (VIP), CRH, hypocretin/orexin (HCRT/ORX), TAC1 (neurokinin A, substance P, and more), and TAC3 (neurokinin B), as well as 17 NPs that are expressed in excitatory interneurons (“near-projecting”), including, for example, prepronociceptin (PNOC) and urocortin (UCN) (Fig. 4). In addition, we suggest that 20 NPs are expressed in candidate interneurons based on local presence of both the NP and the corresponding NPR(s). They include, for example, adrenomedullin (ADM), apelin (APLN), gastrin (GAST), OXT, and SCT.
Projection (pyramidal) neurons represent the largest cell group in cortex. They usually release glutamate and NPs in other cortical regions, thalamus, or more distal subcortical regions, and often lack local NPRs for their NPs. These NPs include, for example, gastrin-releasing peptide (GRP), neuromedin B (NMB), prodynorphin (PDYN), and CCK. Since receptors for ghrelin (GHRL), gonadotropin-releasing hormone 1/2 (GNRH1/2), neuropeptide B (NPB), neuropeptide W (NPW), thyrotropin-releasing hormone (TRH), peptide YY (PYY), and urotensin 2B (UTS2B) were not detected, these NPs are most likely released and active outside hPFC in regions innervated by neurons located in hPFC (Fig. 4). Cocaine and amphetamine–regulated transcript peptide (CARTPT) represents a special case: Even though discovered already in 1995, still no receptor has been identified (65). It is robustly expressed with >5-fold differences between subregions, and the lowest levels are found in ACC-ORB. Interestingly, pyramidal neurons projecting to extracortical regions often do not synthesize NPs themselves but do express NPRs (64). Thus, cortical NPs released from inhibitory interneurons may indirectly influence extracortical regions via exciting or inhibiting such NP-sensing projection neurons.
A few of the NPs and NPRs were also detected in non-neuronal cells (Fig. 4). Of these, GNRH1 and three of the NPRs, APLNR, NTSR2, and RAMP3, are so far only detected in non-neuronal cells.
In Situ RNA Hybridization of Peptidergic Systems in ACC.
To extend the RNA-seq results to the cellular level, we have further employed the RNAscope method for a selected number of NPs and NPRs focusing on ACC, the NP-rich region. Altogether, we have analyzed 11 transcripts and throughout used double labeling for VGAT and VGLUT transcripts, namely in an attempt to decide if the NPs/NPRs are expressed, principally, in GABAergic interneurons and/or glutamatergic projection neurons. We have chosen both abundant and rare transcripts and succeeded in obtaining a signal for 9 transcripts (Fig. 3 and SI Appendix, Fig. S4). CCK mRNA was found to be expressed in both GABA and glutamatergic neurons and in all the cortical layers, except layer 4 (Fig. 5). Layer 1 apparently expressed CCK alone, and further phenotypic characterization is needed. In addition, layer 4 showed very few CCK-positive cells. We can confirm the high expression of CCK both in GABA and glutamate neurons, but we also found transcripts that were not phenotyped in the single-cell analysis (63): ADM, POMC, and the receptors for CRHR1 and growth hormone–releasing hormone (GHRHR). ADM (high levels) and POMC (low levels) were expressed both in GABA and glutamate neurons, and OXT only at low levels in a few cells and only in glutamate neurons. The receptors CRHR1 (strong signal) and GHRHR1 (weak signal) were expressed both in GABA and glutamate neurons. AGTR1 was only found in a few, undefined cells.
Fig. 5.
Distribution of CCK, VGLUT1, and VGAT mRNA-positive cells in ACC. ISH was performed with RNAscope probes targeting CCK (red), VGLUT1 (green), and VGAT (green). Nuclei were counterstained with DAPI (blue) (A–C). CCK mRNA was coexpressed in glutamatergic (D) as well as GABAergic (E) cells. Layer-specific expression of CCK, VGLUT1, and VGAT mRNA (F). Note the lack of CCK transcripts in lamina IV. Examples of coexpression of CCK with VGLUT1 (G) and VGAT (H) are indicated by arrowheads, and arrows represent CCK-only cells. CCK coexists much more frequently with VGLUT1 than with VGAT, and VGLUT1-positive cells (G1) are much more abundant than VGAT-positive cells (H1). High-magnification images at the single-cell level for CCK, VGLUT1, and VGAT (I–J2). (Scale bars, 100 μm [A–F], 50 μm [G and H], and 5 μm [I–J2].) See Figshare for a higher-quality version of Fig. 5 (116).
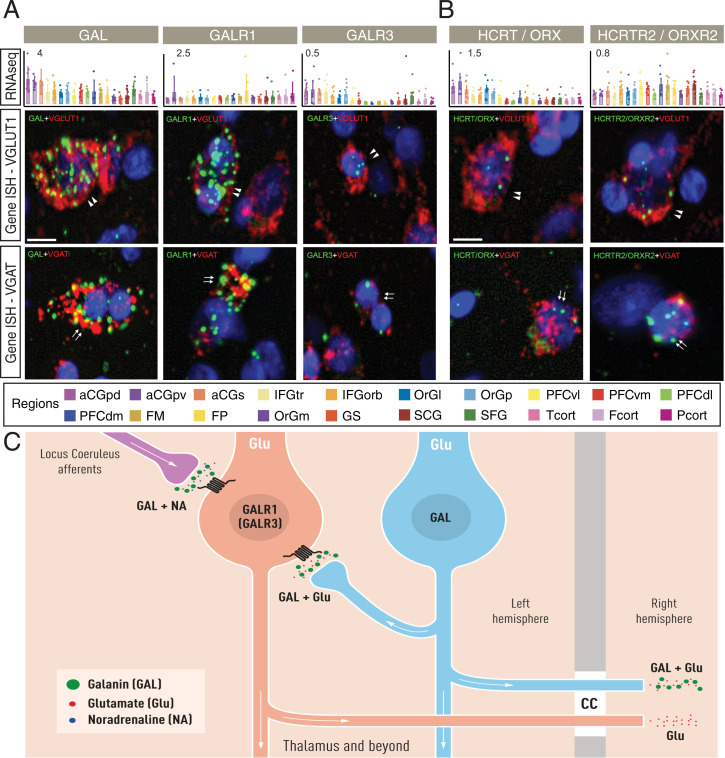
We have paid special attention to the galanin (GAL and GALR1 to R3) and the HCRT/ORX and HCRTR2/ORXR2 systems. Thus, the signals for both GAL and GALR1 are strong versus weak for GALR3, but all three were found both in glutamate and GABAergic neurons. GALR2 could not be detected, in agreement with the RNA-seq results. A weak to moderately strong signal for both HCRT/ORX and HCRTR2/ORX2 was seen in glutamate and GABAergic neurons. An analysis of the expression profiles of these two peptidergic systems in hPFC will be described more in depth below.
Local Circuits of “Hypothalamic” Neuropeptides in PFC.
Some NPs detected in hPFC have long since been associated with hypothalamus and its functions (indicated as dashed boxes in Fig. 4). Most of their corresponding NPRs were robustly expressed in hPFC, supporting the existence of additional local circuits (Fig. 4). Exceptions were TRH and GNRH1. They may influence intracortical and subcortical signaling processes, through actions via GABAergic interneurons and glutamatergic projection neurons, possibly also forming feedback loops for blood-borne hormones.
An interesting example is OXT, which is expressed in hypothalamic magnocellular neurons and released from dendrites and nerve endings in posterior pituitary (66, 67). OXT is involved in the milk-ejection reflex and uterine contractility and is also a stress hormone (68) with projections to many brain areas including, in the mouse, PFC (69). In addition to intraparenchymal/extracellular volume transmission, OXT may reach hPFC after passing the blood–brain barrier (BBB) via RAGE (also known as AGER; advanced glycosylation end product–specific receptor) (70), which is expressed at low levels in all hPFC regions and RefCorts (see gene-specific pages in the HPA Brain Atlas). The present results suggest a fourth, minor local OXT source in PFC: OXT-positive glutamatergic neurons (Fig. 2C and SI Appendix, Fig. S2E). Low levels of OXT-positive, glutamatergic neurons have also been observed in nonpathological PFC samples by Mathys et al. (71).
These results are interesting in view of OXT’s pleiotropic actions, involving stress, social behavior anxiety (67), and involvement in cortical development (72), all functions potentially related to PFC. In contrast, OXTRs are expressed in all 20 cortical subregions in glutamatergic projection neurons (SI Appendix, Fig. S2E). Thus, OXT signaling may also influence other cortical and subcortical regions, in addition to forming an intrinsic oxytocinergic hPFC circuitry. CD38, a transmembrane glycoprotein important for OXT release and social behavior (73), has recently been found across the human brain (74), and as shown here also in all hPFC regions (see gene-specific pages in the HPA Brain Atlas).
Also, four hypothalamic “releasing” hormones, discovered by Guillemin’s, Schally’s, and Wylie Vale’s teams, reviewed by Guillemin (75), were expressed in PFC with current investigation—TRH, GNRH1, CRH, and GHRH—likely having cortical functions and complementing another hypothalamic hormone, SST, an inhibitor of GH release, long since known as a cotransmitter in inhibitory GABAergic interneurons (51, 52).
POMC and agouti-related peptide (AgRP), often referred to as the melanocortin system (76), were detected in hPFC and are associated with hypothalamic arcuate nucleus and, in particular, with feeding. These two NPs act, among others, via MC4R, expressed in all hPFC regions. As mentioned, both HCRT/ORX and KISS1 and their corresponding NPRs (HCRTR2 and KISS1R) are expressed in the ACC-ORB region, suggesting local circuits. MC4R and HCRTR2 in PFC may also be activated by the respective NP released from nerve endings originating from hypothalamic parent cell bodies. In addition, receptors of two pituitary hormones, PRL and GH, were detected in hPFC, indicating that these two hormones may reach and influence cortical activities (SI Appendix, Note II).
Integrative Analysis of Peptidergic Systems.
Mapping chemical circuits at the cellular level in the human brain represents a major challenge. This contrasts with the situation in rodents, where novel methods introduced during the last decade(s) have offered unique opportunities (77). However, based on results presented in this study, by integrating RNA-seq and ISH data, we can suggest some hypothetical transmitter-identified microcircuitries in hPFC as a resource for further dissection of NP effects via their GPCRs. As an example, an integrative analysis of the galanin system was conducted. Both GAL and GALR3 are highest in ACC, whereas GALR1 is evenly distributed in all 20 regions (Fig. 6A). Using histochemical and cellular analysis of the ACC region with RNAscope, we observed a distinct signal from three of the four GAL system transcripts in glutamatergic neurons, GALR3 being very low (Fig. 6A). In addition, GAL and GALR1 were also observed in some VGAT cells. Overall, this is in agreement with the single-cell data by Mathys et al. (71) showing much higher GAL and higher GALR1 transcript levels in excitatory than in inhibitory neurons, whereas GALR3 is detected (at low levels) only in excitatory neurons. This contrasts with rodents, where the cortical galanin system is very discrete (78, 79). This may point to a distinct species difference and indicate a more profound role of the cortical galanin system in humans. A hypothetical overview of this system in hACC is outlined in Fig. 6C and SI Appendix, Fig. S6.
Fig. 6.
Expression of the GAL system in ACC. (A) Bar plots showing the expression profiles of GAL and two of its receptors, GAL1R and GAL3R (the latter just above detectable threshold, nTPM >0.1, and only in ACC), in hPFC and reference cortical subregions. The y axis represents the consensus nTPM values for each gene. The x axis represents the 20 human cortical subregions (color code; Bottom). Error bars represent mean ± SD. Bar plots are ISH results for each gene in glutamatergic (VGLUT1) and GABAergic (VGAT) cells. Color code: cell type analyzed, red; gene of interest, green. (Scale bar, 5 μm.) (B) Bar plots showing the expression profiles of transcripts for HCRT/ORX and HCRTR2/ORXR2 in hPFC and reference cortical subregions. Color code: cell type analyzed, red; gene of interest, green. Arrowheads depict positive cells for VGLUT1 and arrows for VGAT-positive cells. (Scale bar, 5 μm.) (C) Schematic illustration of the GAL system (hypothesis). We speculate that GAL, released together with noradrenaline (NA) from afferents originating in locus coeruleus [established in rat experiments (97)], could act on glutamatergic, pyramidal neurons expressing GALR1 (or GALR3, expressed at very low levels). These receptors can also be targeted by GAL released from glutamatergic, pyramidal neurons. The potential projections of the pyramidal neurons described in this figure have been defined in some detail in SI Appendix, Fig. S6. The present cartoon is principally also valid for the HCRT/ORX system. However, its afferents originate in lateral hypothalamus and are glutamatergic [established in rat experiments (115)]. Both HCRT/ORX as well as the receptors are also expressed in glutamatergic, pyramidal cells (a detailed hypothesis is in SI Appendix, Fig. S7). Both GAL and HCRT/ORX peptides and receptors are in addition expressed in GABAergic interneurons, but are not included in this cartoon.
The HCRT/ORX system was independently discovered by two groups and found, in rodents, only in a discrete, bilateral region in lateral hypothalamus (80, 81). Here, we find that the peptide is expressed in both glutamatergic projection neurons and GABAergic interneurons (63) (Fig. 6B), and likely in hypothalamic afferents. The type 2 receptor was detected in glutamatergic neurons (Fig. 6B). These findings are in agreement with single-cell results (71), but here no significant differences in transcript levels were observed between excitatory neurons and interneurons. Fig. 6C is applicable also for the cellular localization of the HCRT/ORX system in hPFC. For functional implications, see Discussion (GAL) and SI Appendix, Note II (HCRT/ORX).
PFC May Sense Peptidergic Inputs from Afferents.
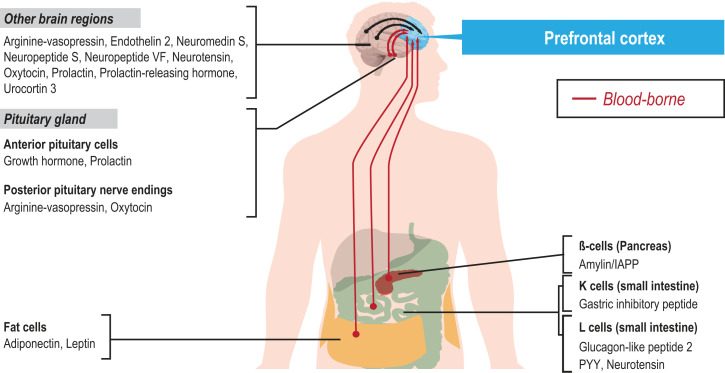
The presence of afferent peptidergic inputs to the 17 hPFC subregions is indicated by the expression of the NPR but apparent lack of NP. In total, 17 such cases were identified in hPFC, including receptors, for example, for arginine-vasopressin (AVP), endothelin 2/B (EDN2/B), NTS, neuropeptide FF (NPVF), neuropeptide S (NPS), prolactin-releasing hormone (PRLH), neuromedin S (NMS), and urocortin 3 (UCN3) (Fig. 4). Those NPs are thus released from afferents originating in other brain regions, or arrive via volume transmission (82). The tissue or brain region origins of the afferent NPs are shown in SI Appendix, Fig. S8, based on the integrative analysis of the tissue elevated genes from the HPA Tissue Atlas (83), brain regionally expressed genes from the HPA Brain Atlas (84), and blood secreted proteins from the human secretome (85). As an example, NTS is expressed in many other brain regions but cannot be detected in PFC. A summary of the origins of the afferent NPs is also outlined in Fig. 7.
Fig. 7.
Peptides from diverse afferent sources. Schematic overview of the possible origins of afferent peptidergic inputs to hPFC. There are 16 NPs that cannot be detected in hPFC, contrasting with the presence of, often robust, receptor expression levels. These receptors are likely activated by peptides released from neurons outside hPFC (released from neuronal afferents or via volume transmission), or alternatively by blood-borne hormones from endocrine glands or fat cells.
Gut and Fat Hormones May Participate in hPFC Signaling.
Four “gastrointestinal” (GI) peptides were detected in PFC: SCT, GAST, PYY, and GHRL. The receptors of both secretin (SCTR) and gastrin (together with CCK) have been found in hPFC. However, no receptor could be detected for PYY (NPYR4) or ghrelin/obestatin (GHRL). Thus, PYY and GHRL are putatively expressed in, among others, projection neurons.
There are receptors in hPFC that may be activated by peripheral peptides produced in fat cells (adiponectin [ADIPOQ], leptin [LEP]), pancreatic β-cells (islet amyloid peptide [IAPP]/amylin), K cells (gastric inhibitory peptide [GIP]; also known as glucose-dependent insulinotropic polypeptide), or L cells (PYY, NTS, and glucagon-like peptide 2 [GLP2]). K and L cells are located in the GI tract (Fig. 7). These peptides may access the brain via penetration of the BBB or via regions with a “leaky” BBB.
LEP is a protein important for feeding control but with many other functions (86), even including synaptogenesis (87). Its transport across endothelial cells may be mediated by robustly expressed LEP receptors (88). LEP receptors are found in glutamatergic and GABAergic neurons as well as in endothelial cells and astrocytes (64), the latter two cell types being part of the BBB. Also, ADIPOQ is assumed to reach the brain, even if only at 0.1% of blood levels (89).
Nonpeptidergic Systems Including Monoamine and Cholinergic Receptors, Growth Factors, Nitric Oxide Synthases, and Amino Acid Systems.
SI Appendix, Note III and Fig. S9 include details.
Discussion
We here present the neuropeptide system in hPFC based on gene expression in microdissected (bulk) samples from 17 subregions and 3 RefCorts. The results revealed that NP and NPR genes are expressed with a unique subregional profile, often at distinctly higher levels in hPFC than in RefCorts, and especially in the ACC-ORB subregions—the NP-rich region. ACC is known to be part of the affective network and to be activated by reward and emotions (2, 3, 9, 90). Sub- and pregenual ACC are related to, respectively, autonomic processes and subjective emotional feelings. The former receives inputs from medial orbitofrontal cortex, insula, amygdala, and hypothalamus, and also from the lower brainstem (periaqueductal gray, parabrachial nucleus), the latter in addition to dorsomedial and rostromedial PFC, as well as the subgenual region (2, 8, 9). For example, the latter shows structural abnormalities (reduction in volume) and reduced metabolic activity in depressed patients (3). Interestingly, depressed patients receiving electromagnetic stimulation at cortical sites, especially DLPFC, show a marked improvement in global depression scores when connected with subgenual ACC (91–93), even if this effect is indirect and relayed via an intrinsic circuit (94).
By mapping the results to published cell type–specific expression profiles, we managed to identify several NPs that are expressed by interneurons and some NPs that are markers for efferent projection (pyramidal) neurons. In addition, certain NPRs are identified as targets for extraregional/extracortical peptidergic neuronal afferents/diffusion by volume transmission or for blood-borne hormones that thus may influence signaling in hPFC. Several NPs, firmly associated with hypothalamic circuitries, fat cells, or GI endocrine cells, are also expressed in hPFC. This may appear surprising, but the name of the NP often just reflects the tissue/organ from which an NP was first isolated, and/or the very first function associated with the NP. NPs are cogwheels in a large machinery, and the same NP is produced and active in many parts of the nervous system, like classic transmitters. Their effects then depend on the type of GPCRs and G proteins involved. The varying sources of the peptides, including being blood-borne, may influence PFC physiology and pathophysiology.
We further explored the GAL system described above (cf. Fig. 6C) by integrating the results from a single-cell dataset from the Allen Brain Atlas (64, 95), in which 120 cell types have been identified based on samples from multiple cortex regions. A hypothesis on circuitry has been generated: Galanin released bilaterally from nerve endings of GAL-positive, intratelencephalic-projecting pyramidal neurons act on GALR1 expressed in glutamatergic neurons projecting to thalamus (SI Appendix, Fig. S6). Since GALR1 and GALR3 mostly are postsynaptic and cause inhibition by opening potassium channels (96), we suggest that the role of galanin is, for example, to prevent excessive glutamate release, namely to eventually protect against excitotoxicity. Based on animal experiments, this effect could be enhanced by galanin released from noradrenergic afferent nerves costoring galanin (97), acting on GALR1/-3 postsynaptic receptors (98). Taken together, GAL may surveil and suppress excitability in the ACC circuitry. Since GAL from pyramidal neurons is coreleased with the excitatory transmitter glutamate, this could be an example of cotransmitter antagonism. We have created a similar scenario for the excitatory NP HCRT/ORX system, but this is an example of cotransmitter agonism, since HCRT/ORX is an excitatory peptide (SI Appendix, Note II, Hypocretin/Orexin and Fig. S7).
The GAL system has previously been associated with, among others, depression and resilience against depression via mechanisms intrinsic to pontine noradrenergic locus coeruleus (LC) (44, 99–101). The present results show that the GAL system is also expressed in hPFC, mainly in glutamatergic neurons. In agreement, both genetic (38) and neurochemical (39) analyses suggest association between the GAL system and major depression disorder, possibly also involving glutamatergic neurons, in addition to the pontine LC system. This is interesting in view of the proposed “reconceptualization of the biology of depression,” that is, a shift from the monoamine hypothesis to amino acids (102). One reason is breakthrough research showing that the glutamate receptor antagonist ketamine has a rapid effect on treatment-resistant depression (102, 103).
Most of the NPs, often expressed in cortex/PFC, have been discussed in relation to multiple mental and/or neurological disorders, primarily based on animal experiments (27). Several NPs (NPY, SST, CRF, and DYN) have in clinical studies been shown to be up- or down-regulated in disorders such as schizophrenia, bipolar disorder, depression, and alcohol use disorder. Moreover, it has been reported that ADIPOQ, APLN, and NOC are associated with depression, and ADIPOQ, ADM, APLN, bradykinin, CARTPT, EDNs, relaxin, GAL, GIP, GLP1, GLP2, and VIP are associated with Alzheimer’s disease (AD). Even more NPs are related to stress.
The large number of NPRs represents potential targets for drug development (28,104–107). In fact, today nearly 50 drugs acting via GPCRs have been approved by the Food and Drug Administration for various therapeutic interventions (106), some acting via neuropeptide systems: a substance P antagonist for treatment of chemotherapy-induced nausea (108), orexin antagonists for treatment of insomnia (109), and CGRP antibodies/antagonists for treatment of migraine (110). Finally, GLPR1 agonists are not only useful for treatment of metabolic syndrome (111) but together with GIPR and GLPR2 agonists they have in animal experiments/models (e.g., AD and Parkinson’s disease) been shown to reduce chronic inflammation and mitochondrial damage, improve memory, and reduce plaque load (112, 113). In fact, recently a clinical trial has been initiated by Novo Nordisk, the Danish pharmaceutical company, with an agonist (Rybelsus) at the peptide receptor GLP1R for treatment of AD.
In summary, we present a peptidergic transcriptomic PFC landscape, where closely located and functionally different subregions are heterogeneous with unique peptide/transmitter–related transcript profiles, including microcircuitries, involving local glutamatergic neurons coexpressing HCRT/ORX, GAL, or OXT and respective receptors. Here we report such molecules in this critically important part of the human brain. Here, the peptide may represent a cotransmitter agonist (HCRT/ORX) or antagonist (GAL). Specific hPFC receptors have been identified as targets for neuronal afferents, like NTS, and for peripheral blood-borne peptide hormones, like LEP, ADIPOQ, and GIP, as well as GH and PRL. Importantly, NP-rich regions (the seven ACC-ORB subregions) were identified with abundant NPs and NPRs. Also, some markers for catecholaminergic, cholinergic, histaminergic (HRH3), and nitrergic signaling (NOS3) are enriched there. These insights into the global gene expression of PFC reveal a spectrum of peptidergic systems that may fine-tune/modulate functions executed during health and disease. Moreover, the sustained actions over longer periods characteristic of NPs may contribute to stabilizing circuit connectivity. The results will contribute to the basic understanding of PFC and will facilitate efforts to identify novel therapeutic strategies of relevance for human brain disorders.
Limitations of the Study.
A limitation is that the expression landscape presented here is based on transcriptomics. Apart from the uncertainty of translation into protein, transcript levels may be dependent on peptide release (reflecting neuronal activity) and may thus vary with the state of the brain at the moment of death. Therefore, the fact should be noted that the brains studied were from subjects who died from stroke, although all regions affected by stroke were far from PFC. Another limitation is that an arbitrary and low cutoff for detection of genes has been used (nTPM > 0.1), resulting in a large number of genes detected in the subregions. The fact that we have been able to visualize, for example, GALR3 with RNAscope in ACC, where the nTPM values for the transcript are 0.12 to 0.14, together with some further examples of this type, supports that the low cutoff is valid. Nevertheless, results close to the cutoff, like GALR3 and OXT, need to be confirmed in future studies, and the decision on cutoff will always remain a source of uncertainty. The probe-based RNAscope analysis only encompassed a limited number of transcripts, and not all experiments resulted in detectable signals expected based on RNA-seq results. This is not unexpected, since we, in an earlier radioactive, autoradiographic ISH study experienced that, for example, the GALR3 transcript could only be detected in human brains with very short postmortem time and long exposures (114). Here, we used a separate brain that had been especially processed for RNAscope analysis. The high-abundance neuropeptide transcripts, such as CCK, were easily demonstrated, and we are satisfied that also a number of other important neuropeptide system transcripts could be visualized.
Materials and Methods
Full details regarding the human brain sample collection, processing, RNA extraction, RNA-seq, RNAscope analysis, data quality control, and statistical analysis are provided in SI Appendix. All brain samples were collected by the Human Brain Tissue Bank (HBTB; Semmelweis University) and the Human Brain Research Laboratory (ELKH Institute of Experimental Medicine). The activity of the HBTB has been authorized by the Committee of Science and Research Ethics of the Ministry of Health Hungary (ETT TUKEB: 189/KO/02.6008/2002/ETT) and the Semmelweis University Regional Committee of Science and Research Ethics (32/1992/TUKEB) to remove human brain tissue samples, collect, store, and use them for research. Obtaining human samples by the Human Brain Research Laboratory was approved by the ethics committee at the Regional and Institutional Committee of Science and Research Ethics of the Scientific Council of Health (ETT TUKEB15032/2019/EKU).
Supplementary Material
Acknowledgments
We thank Drs. S. Paabo (Ref. 63), E. Lein (Ref. 64) and L.H. Tsai (Ref. 71) for allowing us to use their results—they have been essential for our study. We thank Mattias Karlén for two excellent drawings (Figs. 6 and 7). We appreciate valuable advice and input from Dr. Huda Zoghbi, as well as from Drs. Rainer Landgraf, Mike Ludwig, Ming Zhao, Zhi-Qing David Xu, Tibor Harkany, and Nolan Williams. We thank Magdolna Toronyay-Kasztner for managing the HBTB database and acknowledge support from the National Genomics Infrastructure in Stockholm funded by the Science for Life Laboratory, Knut and Alice Wallenberg Foundation and the Swedish National Infrastructure for Computing (SNIC)/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. We thank the Knut and Alice Wallenberg Foundation, Swedish Research Council (2018-02753 and 2020-02956), SciLifeLab and Wallenberg Data Driven Life Science Program (Grant KAW 2020.0239), and Hungarian Brain Research Program (2017-1.2.1-NKP-2017-00002) for financial support.
Footnotes
Reviewers: J.D., Consejo Superior de Investigaciones Científicas and Universidad Politécnica de Madrid; I.M., University of Maryland School of Medicine; N.S., Yale University School of Medicine; and E.S., Centre for Addiction and Mental Health.
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2123146119/-/DCSupplemental.
Data Availability
The transcriptomics data reported in this article for the 17 human prefrontal cortex subregions and 3 reference cortical regions (frontal, parietal, and temporal cortex), gene information, and normalized TPM values are available, open-access, and downloadable from the Human Protein Atlas database (https://www.proteinatlas.org/about/download) (53).
All data are included in the article and/or supporting information.
References
- 1.Fuster J., The Prefrontal Cortex (Academic Press, 2015). [Google Scholar]
- 2.Dixon M. L., Thiruchselvam R., Todd R., Christoff K., Emotion and the prefrontal cortex: An integrative review. Psychol. Bull. 143, 1033–1081 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Price J. L., Drevets W. C., Neurocircuitry of mood disorders. Neuropsychopharmacology 35, 192–216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingvar D. H., Franzén G., Abnormalities of cerebral blood flow distribution in patients with chronic schizophrenia. Acta Psychiatr. Scand. 50, 425–462 (1974). [DOI] [PubMed] [Google Scholar]
- 5.Berman K. F., Weinberger D. R., The prefrontal cortex in schizophrenia and other neuropsychiatric diseases: In vivo physiological correlates of cognitive deficits. Prog. Brain Res. 85, 521–536, discussion 536–537 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Goldman-Rakic P. S., The physiological approach: Functional architecture of working memory and disordered cognition in schizophrenia. Biol. Psychiatry 46, 650–661 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Belleau E. L., Treadway M. T., Pizzagalli D. A., The impact of stress and major depressive disorder on hippocampal and medial prefrontal cortex morphology. Biol. Psychiatry 85, 443–453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vogt B., Cingulate Neurobiology and Disease (Oxford University Press, 2009). [Google Scholar]
- 9.Rolls E. T., The orbitofrontal cortex and emotion in health and disease, including depression. Neuropsychologia 128, 14–43 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Robbins T. W., Arnsten A. F., The neuropsychopharmacology of fronto-executive function: Monoaminergic modulation. Annu. Rev. Neurosci. 32, 267–287 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everitt B. J., Robbins T. W., Neural systems of reinforcement for drug addiction: From actions to habits to compulsion. Nat. Neurosci. 8, 1481–1489 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Volkow N. D., Fowler J. S., Wang G. J., The addicted human brain viewed in the light of imaging studies: Brain circuits and treatment strategies. Neuropharmacology 47 (suppl. 1), 3–13 (2004). [DOI] [PubMed] [Google Scholar]
- 13.Osborne N. R., et al. , Abnormal subgenual anterior cingulate circuitry is unique to women but not men with chronic pain. Pain 162, 97–108 (2021). [DOI] [PubMed] [Google Scholar]
- 14.Vogt B. A., Sikes R. W., “Cingulate nociceptive circuitry and roles in pain processing: The cingulate premotor pain model” in Cingulate Neurobiology and Disease, B. A. Vogt, Ed. (Oxford University Press, 2009), pp. 312–339. [Google Scholar]
- 15.de Wied D., The neuropeptide concept. Prog. Brain Res. 72, 93–108 (1987). [PubMed] [Google Scholar]
- 16.Burbach J. P. H., Neuropeptides from concept to online database www.neuropeptides.nl. Eur. J. Pharmacol. 626, 27–48 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Kastin A., Handbook of Biologically Active Peptides (Academic Press, 2013). [Google Scholar]
- 18.DeFelipe J., Neocortical neuronal diversity: Chemical heterogeneity revealed by colocalization studies of classic neurotransmitters, neuropeptides, calcium-binding proteins, and cell surface molecules. Cereb. Cortex 3, 273–289 (1993). [DOI] [PubMed] [Google Scholar]
- 19.DeFelipe J., et al. , New insights into the classification and nomenclature of cortical GABAergic interneurons. Nat. Rev. Neurosci. 14, 202–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouras C., Magistretti P. J., Morrison J. H., An immunohistochemical study of six biologically active peptides in the human brain. Hum. Neurobiol. 5, 213–226 (1986). [PubMed] [Google Scholar]
- 21.Braak E., Braak H., Weindl A., Somatostatin-like immunoreactivity in non-pyramidal neurons of the human isocortex. Anat. Embryol. (Berl.) 173, 237–246 (1985). [DOI] [PubMed] [Google Scholar]
- 22.Chan-Palay V., Allen Y. S., Lang W., Haesler U., Polak J. M., Cytology and distribution in normal human cerebral cortex of neurons immunoreactive with antisera against neuropeptide Y. J. Comp. Neurol. 238, 382–389 (1985). [DOI] [PubMed] [Google Scholar]
- 23.Vincent S. R., et al. , Neuropeptide coexistence in human cortical neurones. Nature 298, 65–67 (1982). [DOI] [PubMed] [Google Scholar]
- 24.Hornung J. P., De Tribolet N., Törk I., Morphology and distribution of neuropeptide-containing neurons in human cerebral cortex. Neuroscience 51, 363–375 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Sagar S. M., Beal M. F., Marshall P. E., Landis D. M., Martin J. B., Implications of neuropeptides in neurological diseases. Peptides 5 (suppl. 1), 255–262 (1984). [DOI] [PubMed] [Google Scholar]
- 26.Taquet H., et al. , Biochemical mapping of cholecystokinin-, substance P-, [Met]enkephalin-, [Leu]enkephalin- and dynorphin A (1-8)-like immunoreactivities in the human cerebral cortex. Neuroscience 27, 871–883 (1988). [DOI] [PubMed] [Google Scholar]
- 27.Brockway D. F., Crowley N. A., Turning the ’tides on neuropsychiatric diseases: The role of peptides in the prefrontal cortex. Front. Behav. Neurosci. 14, 588400 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fee C., Banasr M., Sibille E., Somatostatin-positive gamma-aminobutyric acid interneuron deficits in depression: Cortical microcircuit and therapeutic perspectives. Biol. Psychiatry 82, 549–559 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morris H. M., Hashimoto T., Lewis D. A., Alterations in somatostatin mRNA expression in the dorsolateral prefrontal cortex of subjects with schizophrenia or schizoaffective disorder. Cereb. Cortex 18, 1575–1587 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beneyto M., Morris H. M., Rovensky K. C., Lewis D. A., Lamina- and cell-specific alterations in cortical somatostatin receptor 2 mRNA expression in schizophrenia. Neuropharmacology 62, 1598–1605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fung S. J., et al. , Expression of interneuron markers in the dorsolateral prefrontal cortex of the developing human and in schizophrenia. Am. J. Psychiatry 167, 1479–1488 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Caberlotto L., Hurd Y. L., Neuropeptide Y Y(1) and Y(2) receptor mRNA expression in the prefrontal cortex of psychiatric subjects. Relationship of Y(2) subtype to suicidal behavior. Neuropsychopharmacology 25, 91–97 (2001). [DOI] [PubMed] [Google Scholar]
- 33.Tripp A., Kota R. S., Lewis D. A., Sibille E., Reduced somatostatin in subgenual anterior cingulate cortex in major depression. Neurobiol. Dis. 42, 116–124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sibille E., Morris H. M., Kota R. S., Lewis D. A., GABA-related transcripts in the dorsolateral prefrontal cortex in mood disorders. Int. J. Neuropsychopharmacol. 14, 721–734 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kessler R. C., Epidemiology of women and depression. J. Affect. Disord. 74, 5–13 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Seligowski A. V., Harnett N. G., Merker J. B., Ressler K. J., Nervous and endocrine system dysfunction in posttraumatic stress disorder: An overview and consideration of sex as a biological variable. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 5, 381–391 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seney M. L., Glausier J., Sibille E., Large-scale transcriptomics studies provide insight into sex differences in depression. Biol. Psychiatry 91, 14–24 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Juhasz G., et al. , Brain galanin system genes interact with life stresses in depression-related phenotypes. Proc. Natl. Acad. Sci. U.S.A. 111, E1666–E1673 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barde S., et al. , Alterations in the neuropeptide galanin system in major depressive disorder involve levels of transcripts, methylation, and peptide. Proc. Natl. Acad. Sci. U.S.A. 113, E8472–E8481 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grimmelikhuijzen C. J., Hauser F., Mini-review: The evolution of neuropeptide signaling. Regul. Pept. 177, S6–S9 (2012). [DOI] [PubMed] [Google Scholar]
- 41.van den Pol A. N., Neuropeptide transmission in brain circuits. Neuron 76, 98–115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nusbaum M. P., Blitz D. M., Marder E., Functional consequences of neuropeptide and small-molecule co-transmission. Nat. Rev. Neurosci. 18, 389–403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bargmann C. I., Beyond the connectome: How neuromodulators shape neural circuits. BioEssays 34, 458–465 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Hökfelt T., et al. , Neuropeptide and small transmitter coexistence: Fundamental studies and relevance to mental illness. Front. Neural Circuits 12, 106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lundberg J. M., Hökfelt T., Coexistence of peptides and classical neurotransmitters. Trends Neurosci. 6, 325–333 (1983). [Google Scholar]
- 46.Bartfai T., Iverfeldt K., Fisone G., Serfözö P., Regulation of the release of coexisting neurotransmitters. Annu. Rev. Pharmacol. Toxicol. 28, 285–310 (1988). [DOI] [PubMed] [Google Scholar]
- 47.Bondy C. A., Gainer H., Russell J. T., Effects of stimulus frequency and potassium channel blockade on the secretion of vasopressin and oxytocin from the neurohypophysis. Neuroendocrinology 46, 258–267 (1987). [DOI] [PubMed] [Google Scholar]
- 48.Verhage M., Ghijsen W. E., Lopes da Silva F. H., Presynaptic plasticity: The regulation of Ca(2+)-dependent transmitter release. Prog. Neurobiol. 42, 539–574 (1994). [DOI] [PubMed] [Google Scholar]
- 49.Guillaumin M. C. C., Burdakov D., Neuropeptides as primary mediators of brain circuit connectivity. Front. Neurosci. 15, 644313 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hökfelt T., Johansson O., Ljungdahl A., Lundberg J. M., Schultzberg M., Peptidergic neurones. Nature 284, 515–521 (1980). [DOI] [PubMed] [Google Scholar]
- 51.Hendry S. H., et al. , Neuropeptide-containing neurons of the cerebral cortex are also GABAergic. Proc. Natl. Acad. Sci. U.S.A. 81, 6526–6530 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Somogyi P., et al. , Different populations of GABAergic neurons in the visual cortex and hippocampus of cat contain somatostatin- or cholecystokinin-immunoreactive material. J. Neurosci. 4, 2590–2603 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.The Brain Atlas. The Human Protein Atlas. https://v20.proteinatlas.org/humanproteome/brain. Accessed 26 July 2022. [Google Scholar]
- 54.Seidah N. G., Chrétien M., Proprotein and prohormone convertases: A family of subtilases generating diverse bioactive polypeptides. Brain Res. 848, 45–62 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Jackson L., Eldahshan W., Fagan S. C., Ergul A., Within the brain: The renin angiotensin system. Int. J. Mol. Sci. 19, 876 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dai J., Patzke C., Liakath-Ali K., Seigneur E., Südhof T. C., GluD1 is a signal transduction device disguised as an ionotropic receptor. Nature 595, 261–265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsuda K., et al. , Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science 328, 363–368 (2010). [DOI] [PubMed] [Google Scholar]
- 58.Shibata M., et al. , Hominini-specific regulation of CBLN2 increases prefrontal spinogenesis. Nature 598, 489–494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Burgunder J. M., Young W. S. III, The distribution of thalamic projection neurons containing cholecystokinin messenger RNA, using in situ hybridization histochemistry and retrograde labeling. Brain Res. 464, 179–189 (1988). [DOI] [PubMed] [Google Scholar]
- 60.McLatchie L. M., et al. , RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature 393, 333–339 (1998). [DOI] [PubMed] [Google Scholar]
- 61.Carlo A. S., Nykjaer A., Willnow T. E., Sorting receptor sortilin—A culprit in cardiovascular and neurological diseases. J. Mol. Med. (Berl.) 92, 905–911 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Smith S. J., et al. , Single-cell transcriptomic evidence for dense intracortical neuropeptide networks. eLife 8, e47889 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Khrameeva E., et al. , Single-cell-resolution transcriptome map of human, chimpanzee, bonobo, and macaque brains. Genome Res. 30, 776–789 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hodge R. D., et al. , Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kuhar M. J., Jaworski J. N., Hubert G. W., Philpot K. B., Dominguez G., Cocaine- and amphetamine-regulated transcript peptides play a role in drug abuse and are potential therapeutic targets. AAPS J. 7, E259–E265 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ludwig M., Apps D., Menzies J., Patel J. C., Rice M. E., Dendritic release of neurotransmitters. Compr. Physiol. 7, 235–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grinevich V., Neumann I. D., Brain oxytocin: How puzzle stones from animal studies translate into psychiatry. Mol. Psychiatry 26, 265–279 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Engelmann M., Landgraf R., Wotjak C. T., The hypothalamic-neurohypophysial system regulates the hypothalamic-pituitary-adrenal axis under stress: An old concept revisited. Front. Neuroendocrinol. 25, 132–149 (2004). [DOI] [PubMed] [Google Scholar]
- 69.Knobloch H. S., et al. , Evoked axonal oxytocin release in the central amygdala attenuates fear response. Neuron 73, 553–566 (2012). [DOI] [PubMed] [Google Scholar]
- 70.Yamamoto Y., et al. , Vascular RAGE transports oxytocin into the brain to elicit its maternal bonding behaviour in mice. Commun. Biol. 2, 76 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mathys H., et al. , Single-cell transcriptomic analysis of Alzheimer’s disease. Nature 570, 332–337 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng J. J., et al. , Oxytocin mediates early experience-dependent cross-modal plasticity in the sensory cortices. Nat. Neurosci. 17, 391–399 (2014). [DOI] [PubMed] [Google Scholar]
- 73.Jin D., et al. , CD38 is critical for social behaviour by regulating oxytocin secretion. Nature 446, 41–45 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Quintana D. S., et al. , Oxytocin pathway gene networks in the human brain. Nat. Commun. 10, 668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guillemin R., Hypothalamic hormones a.k.a. hypothalamic releasing factors. J. Endocrinol. 184, 11–28 (2005). [DOI] [PubMed] [Google Scholar]
- 76.Mountjoy K. G., Pro-opiomelanocortin (POMC) neurones, POMC-derived peptides, melanocortin receptors and obesity: How understanding of this system has changed over the last decade. J. Neuroendocrinol. 27, 406–418 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Abbott A., How the world’s biggest brain maps could transform neuroscience. Nature 598, 22–25 (2021). [DOI] [PubMed] [Google Scholar]
- 78.Merchenthaler I., López F. J., Negro-Vilar A., Anatomy and physiology of central galanin-containing pathways. Prog. Neurobiol. 40, 711–769 (1993). [DOI] [PubMed] [Google Scholar]
- 79.O’Donnell D., et al. , “Chapter IV Localization of galanin receptor subtypes in the rat CNS” in Handbook of Chemical Neuroanatomy, R. Quirion, A. Björklund, T. Hökfelt, Eds. (Elsevier, 2002), vol. 20, 195–244. [Google Scholar]
- 80.de Lecea L., et al. , The hypocretins: Hypothalamus-specific peptides with neuroexcitatory activity. Proc. Natl. Acad. Sci. U.S.A. 95, 322–327 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sakurai T., et al. , Orexins and orexin receptors: A family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell 92, 573–585 (1998). [DOI] [PubMed] [Google Scholar]
- 82.Fuxe K., et al. , The discovery of central monoamine neurons gave volume transmission to the wired brain. Prog. Neurobiol. 90, 82–100 (2010). [DOI] [PubMed] [Google Scholar]
- 83.Uhlén M., et al. , Proteomics. Tissue-based map of the human proteome. Science 347, 1260419 (2015). [DOI] [PubMed] [Google Scholar]
- 84.Sjöstedt E., et al. , An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367, eaay5947 (2020). [DOI] [PubMed] [Google Scholar]
- 85.Uhlén M., et al. , The human secretome. Sci. Signal. 12, eaaz0274 (2019). [DOI] [PubMed] [Google Scholar]
- 86.Friedman J., 20 years of leptin: Leptin at 20: An overview. J. Endocrinol. 223, T1–T8 (2014). [DOI] [PubMed] [Google Scholar]
- 87.Sahin G. S., et al. , Leptin increases GABAergic synaptogenesis through the Rho guanine exchange factor β-PIX in developing hippocampal neurons. Sci. Signal. 14, eabe4111 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Di Spiezio A., et al. , The LepR-mediated leptin transport across brain barriers controls food reward. Mol. Metab. 8, 13–22 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bloemer J., et al. , Role of adiponectin in central nervous system disorders. Neural Plast. 2018, 4593530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Beckmann M., Johansen-Berg H., Rushworth M. F., Connectivity-based parcellation of human cingulate cortex and its relation to functional specialization. J. Neurosci. 29, 1175–1190 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fox M. D., Buckner R. L., White M. P., Greicius M. D., Pascual-Leone A., Efficacy of transcranial magnetic stimulation targets for depression is related to intrinsic functional connectivity with the subgenual cingulate. Biol. Psychiatry 72, 595–603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cash R. F. H., et al. , Subgenual functional connectivity predicts antidepressant treatment response to transcranial magnetic stimulation: Independent validation and evaluation of personalization. Biol. Psychiatry 86, e5–e7 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Cole E. J., et al. , Stanford accelerated intelligent neuromodulation therapy for treatment-resistant depression. Am. J. Psychiatry 177, 716–726 (2020). [DOI] [PubMed] [Google Scholar]
- 94.Oathes D. J., et al. , Resting fMRI-guided TMS results in subcortical and brain network modulation indexed by interleaved TMS/fMRI. Exp. Brain Res. 239, 1165–1178 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Allen Institute for Brain Science, Allen Cell Types Database. https://portal.brain-map.org/atlases-and-data/rnaseq/human-multiple-cortical-areas-smart-seq. Accessed 1 October 2019.
- 96.Smith K. E., et al. , Cloned human and rat galanin GALR3 receptors. Pharmacology and activation of G-protein inwardly rectifying K+ channels. J. Biol. Chem. 273, 23321–23326 (1998). [DOI] [PubMed] [Google Scholar]
- 97.Xu Z. Q., Shi T. J., Hökfelt T., Galanin/GMAP- and NPY-like immunoreactivities in locus coeruleus and noradrenergic nerve terminals in the hippocampal formation and cortex with notes on the galanin-R1 and -R2 receptors. J. Comp. Neurol. 392, 227–251 (1998). [DOI] [PubMed] [Google Scholar]
- 98.Ma X., et al. , Effects of galanin receptor agonists on locus coeruleus neurons. Brain Res. 919, 169–174 (2001). [DOI] [PubMed] [Google Scholar]
- 99.Weinshenker D., Holmes P. V., Regulation of neurological and neuropsychiatric phenotypes by locus coeruleus-derived galanin. Brain Res. 1641, 320–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuteeva E., Hökfelt T., Wardi T., Ogren S. O., Galanin, galanin receptor subtypes and depression-like behaviour. Exp. Suppl. 102, 163–181 (2010). [DOI] [PubMed] [Google Scholar]
- 101.Mitsukawa K., Lu X., Bartfai T., Galanin, galanin receptors, and drug targets. Exp. Suppl. 102, 7–23 (2010). [DOI] [PubMed] [Google Scholar]
- 102.Krystal J. H., Abdallah C. G., Sanacora G., Charney D. S., Duman R. S., Ketamine: A paradigm shift for depression research and treatment. Neuron 101, 774–778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kadriu B., et al. , Glutamatergic neurotransmission: Pathway to developing novel rapid-acting antidepressant treatments. Int. J. Neuropsychopharmacol. 22, 119–135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hoyer D., Bartfai T., Neuropeptides and neuropeptide receptors: Drug targets, and peptide and non-peptide ligands: A tribute to Prof. Dieter Seebach. Chem. Biodivers. 9, 2367–2387 (2012). [DOI] [PubMed] [Google Scholar]
- 105.Hökfelt T., Bartfai T., Bloom F., Neuropeptides: Opportunities for drug discovery. Lancet Neurol. 2, 463–472 (2003). [DOI] [PubMed] [Google Scholar]
- 106.Davenport A. P., Scully C. C. G., de Graaf C., Brown A. J. H., Maguire J. J., Advances in therapeutic peptides targeting G protein-coupled receptors. Nat. Rev. Drug Discov. 19, 389–413 (2020). [DOI] [PubMed] [Google Scholar]
- 107.Muttenthaler M., King G. F., Adams D. J., Alewood P. F., Trends in peptide drug discovery. Nat. Rev. Drug Discov. 20, 309–325 (2021). [DOI] [PubMed] [Google Scholar]
- 108.Hargreaves R., et al. , Development of aprepitant, the first neurokinin-1 receptor antagonist for the prevention of chemotherapy-induced nausea and vomiting. Ann. N. Y. Acad. Sci. 1222, 40–48 (2011). [DOI] [PubMed] [Google Scholar]
- 109.Coleman P. J., Gotter A. L., Herring W. J., Winrow C. J., Renger J. J., The discovery of suvorexant, the first orexin receptor drug for insomnia. Annu. Rev. Pharmacol. Toxicol. 57, 509–533 (2017). [DOI] [PubMed] [Google Scholar]
- 110.Edvinsson L., Goadsby P. J., Discovery of CGRP in relation to migraine. Cephalalgia 39, 331–332 (2019). [DOI] [PubMed] [Google Scholar]
- 111.Müller T. D., et al. , Glucagon-like peptide 1 (GLP-1). Mol. Metab. 30, 72–130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang Z. Q., Hölscher C., GIP has neuroprotective effects in Alzheimer and Parkinson’s disease models. Peptides 125, 170184 (2020). [DOI] [PubMed] [Google Scholar]
- 113.Su Y., et al. , A GLP-2 analogue protects SH-SY5Y and neuro-2a cells against mitochondrial damage, autophagy impairments and apoptosis in a Parkinson model. Drug Res. (Stuttg.) 71, 43–50 (2021). [DOI] [PubMed] [Google Scholar]
- 114.Le Mâtre E., Barde S. S., Palkovits M., Diaz-Heijtz R., Hökfelt T. G., Distinct features of neurotransmitter systems in the human brain with focus on the galanin system in locus coeruleus and dorsal raphe. Proc. Natl. Acad. Sci. U.S.A. 110, E536–E545 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Peyron C., et al. , Neurons containing hypocretin (orexin) project to multiple neuronal systems. J. Neurosci. 18, 9996–10015 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhong W., et al. , “The neuropeptide landscape of human prefrontal cortex.” Figshare. 10.6084/m9.figshare.20442714. Deposited 5 August 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptomics data reported in this article for the 17 human prefrontal cortex subregions and 3 reference cortical regions (frontal, parietal, and temporal cortex), gene information, and normalized TPM values are available, open-access, and downloadable from the Human Protein Atlas database (https://www.proteinatlas.org/about/download) (53).
All data are included in the article and/or supporting information.