Significance
The genetic architecture of chronic kidney disease (CKD) remains incompletely understood. Variants in the kidney-specific gene UMOD cause autosomal dominant tubulointerstitial kidney disease (ADTKD) and are associated with kidney function and the risk of CKD in the general population. We identified an intermediate-effect variant, p.Thr62Pro, detected in ∼1/1,000 individuals of European ancestry that showed a high genetic load in familial clusters of CKD and was associated with an odds ratio (OR) of approximately four for kidney failure in the 100,000 Genomes Project and the UK Biobank. Compared with canonical ADTKD mutations, p.Thr62Pro carriers displayed reduced disease severity and an intermediate trafficking defect. These findings complete the spectrum of UMOD-associated kidney diseases and provide a paradigm for the genetic contribution to CKD.
Keywords: genetic architecture, uromodulin, Autosomal Dominant Tubulointerstitial Kidney Disease, rare disease, ER stress
Abstract
The kidney-specific gene UMOD encodes for uromodulin, the most abundant protein excreted in normal urine. Rare large-effect variants in UMOD cause autosomal dominant tubulointerstitial kidney disease (ADTKD), while common low-impact variants strongly associate with kidney function and the risk of chronic kidney disease (CKD) in the general population. It is unknown whether intermediate-effect variants in UMOD contribute to CKD. Here, candidate intermediate-effect UMOD variants were identified using large-population and ADTKD cohorts. Biological and phenotypical effects were investigated using cell models, in silico simulations, patient samples, and international databases and biobanks. Eight UMOD missense variants reported in ADTKD are present in the Genome Aggregation Database (gnomAD), with minor allele frequency (MAF) ranging from 10−5 to 10−3. Among them, the missense variant p.Thr62Pro is detected in ∼1/1,000 individuals of European ancestry, shows incomplete penetrance but a high genetic load in familial clusters of CKD, and is associated with kidney failure in the 100,000 Genomes Project (odds ratio [OR] = 3.99 [1.84 to 8.98]) and the UK Biobank (OR = 4.12 [1.32 to 12.85). Compared with canonical ADTKD mutations, the p.Thr62Pro carriers displayed reduced disease severity, with slower progression of CKD and an intermediate reduction of urinary uromodulin levels, in line with an intermediate trafficking defect in vitro and modest induction of endoplasmic reticulum (ER) stress. Identification of an intermediate-effect UMOD variant completes the spectrum of UMOD-associated kidney diseases and provides insights into the mechanisms of ADTKD and the genetic architecture of CKD.
Chronic kidney disease (CKD) has an estimated global prevalence above 10%, with high burden at both the individual and societal levels and limited therapeutic options (1). There is a strong genetic predisposition to CKD, as the heritability of the CKD-defining trait estimated glomerular filtration rate (eGFR) is between 30 and 50% (2, 3). The largest and most recent genome-wide association study (GWAS) for eGFR identified >250 genetic loci and explained nearly 20% of that heritability. Among those, variants in the UMOD locus display the largest effect size on eGFR and CKD (4). This effect, thought to be associated with increased UMOD expression (5), is consistent across different ethnic groups and is also observed for longitudinal traits, including rapid decline of eGFR (6).
The biological relevance of the UMOD locus is immediately clear, as it is a kidney-specific gene coding for uromodulin (UMOD), the most abundant protein excreted in the normal urine (2, 7). UMOD, which is essentially produced by the thick ascending limb (TAL) of the loop of Henle, plays multiple roles in blood pressure control and protection against kidney stones and urinary tract infections (7, 8). In TAL cells, UMOD enters the secretory pathway to undergo a complex maturation involving the formation of 24 intramolecular disulphide bonds, a rate-limiting step for its proper apical sorting and release into the tubule lumen (9). Ultrarare (minor allele frequency [MAF] < 10−4) mutations in UMOD cause autosomal dominant tubulointerstitial kidney disease (ADTKD; ADTKD-UMOD; Mendelian Inheritance in Man [MIM] #162000), characterized by tubular damage and interstitial fibrosis (10). More than 95% of the UMOD mutations associated with ADTKD are missense, often targeting cysteine residues and leading to the formation of UMOD aggregates within the endoplasmic reticulum (ER; gain-of-toxic function) with a sharp decrease of its excretion in urine. These UMOD mutations invariably lead to kidney failure in adulthood with a penetrance of 100% (10, 11). Over 100 mutations in UMOD have been associated with ADTKD-UMOD with an overall prevalence of ∼2% in patients with kidney failure, representing one of the most common monogenic kidney diseases (12). UMOD is thus implicated at both extremes of the genetic disease spectrum: ultrarare variants with large-effect size (ADTKD-UMOD) and common (MAF > 0.05) low-impact variants associated with reduced eGFR and risk of CKD in the general population (13).
The availability of large genomic datasets suggests that the dichotomy of rare high-effect variants causing Mendelian disorders and frequent low-effect variants involved in complex diseases must be completed by variants with intermediate-effect sizes. These intermediate-effect variants lie on a spectrum based on allelic frequency (AF), biological consequence, and phenotypical readout. They can manifest as a nonfully penetrant Mendelian disease or an oligo-/polygenic model modifying disease expressivity (14, 15). The absence of ultrarare and predicted pathogenic variants in the majority of CKD patients (16) and the missing heritability when studying polymorphisms (17) suggest that intermediate-effect variants, not captured by GWAS but amenable to sequencing approaches, may be part of a continuum underlying CKD. Because UMOD is involved at both ends of the genetic spectrum, it is a plausible candidate gene for intermediate-effect variants modulating the risk of CKD.
Here, we identified and characterized intermediate-effect variants in UMOD contributing to CKD by crossing general population datasets with curated variants reported in ADTKD, analyzing biological and phenotypical effect sizes using in silico modeling, cell systems, databases, and biobanks; and validating the impact on kidney failure in the 100,000 Genomes Project and the UK Biobank (SI Appendix, Fig. S1).
Results
Identification of Candidate Intermediate-Effect Variants in UMOD.
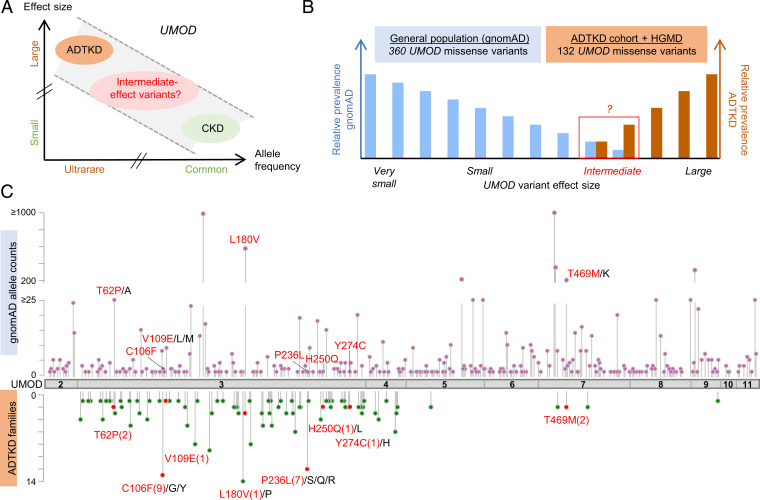
The UMOD gene is implicated at both extremes of a CKD risk continuum (Fig. 1A); however, bona fide high-effect variants cannot be more prevalent than the disease they cause. We first assessed the missense variants in UMOD reported in the Genome Aggregation Database (gnomAD; “controls”) (18) as well as in the International ADTKD Cohort (11) and the Human Gene Mutation Database (HGMD; “cases”). Shared UMOD variants should be either benign (falsely associated with disease) or intermediate-effect variants associated with nonfully penetrant or milder late-onset disease (Fig. 1B and SI Appendix). The majority of UMOD variants listed in gnomAD (360/1,000; 36%) were missense variants (SI Appendix, Fig. S2), with an AF ranging from 2.0 × 10−2 to 3.6 × 10−6 and only 4 variants with an AF exceeding 10−3 (SI Appendix, Table S1). Most of these missense variants in gnomAD were rare (258 variants with allele counts three or less), and 8% among them (30/360) had strong in silico pathogenic scores (rare exome variant ensemble learner [REVEL] scores of >0.75) (19) (SI Appendix, Table S2). A total of 132 UMOD missense variants associated with ADTKD families were included in the International ADTKD Cohort and the HGMD (SI Appendix, Table S3). As expected, these variants associated with ADTKD were generally ultrarare (MAF < 10−4 in 129/132; 98%), with most of them (124/132; 94%) being absent from gnomAD. However, eight missense variants (seven in exon 3 and one in exon 7) were detected in both populations (Fig. 1C and Table 1). These eight variants were identified in 879 individuals in gnomAD, with three being detected relatively frequently (AF of 3.5 × 10−4 to 3.3 × 10−3), whereas the five others were considerably rarer (4.1 × 10−6 to 1.4 × 10−5) (Table 1). Since in silico predictions and prevalence of these five ultrarare variants remain compatible with high-effect penetrant variants (Table 1 and SI Appendix, Table S2), we focused our investigations of potential intermediate-effect variants on the three more frequent UMOD missense variants p.Thr62Pro, p.Leu180Val, and p.Thr469Met (Table 1). Ages of p.Thr62Pro and p.Thr469Met carriers are consistent with the overall age distribution in gnomAD, suggesting that bias due to recruitment before clinical presentation of ADTKD-UMOD is not a concern.
Fig. 1.
Identification of intermediate-effect UMOD variants. (A) Genetic architecture of UMOD-associated diseases showing the continuum of CKD risk associated with ultrarare (AF < 10−4) high-effect to common (AF > 0.05) low-effect UMOD variants. Adapted from ref. 17. (B) Schematic representation of the study design; 360 distinct missense UMOD variants are reported in the gnomAD with an individual AF of 2.0 × 10−2 to 3.6 × 10−6, and 132 distinct missense UMOD variants reported in ADTKD-UMOD have been extracted from the International ADTKD Cohort (726 patients from 585 families) and from the HGMD. The theoretical maximum credible AF of high-effect UMOD variants (pathogenic) in the overall population considering the disease prevalence, allelic and genetic heterogeneity, and a penetrance of 100% is estimated to be 1 × 10−7 (46) (SI Appendix), excluding those (in theory) from gnomAD. Plotted here are the expected relative prevalences of very small– to large–effect size UMOD variants in gnomAD and reported in ADTKD-UMOD patients. Conferring nonfully penetrant ADTKD or reduced disease expressivity, intermediate-effect UMOD variants might be found overlapping at the lower prevalence spectrum in both gnomAD and reported variants in ADTKD-UMOD. (C) The UMOD protein (NM_003361) is in gray with exon boundaries marked by dotted lines. UMOD missense variants reported in the gnomAD consortium are plotted on top of the protein with respect to the amino acid position. The y axis represents gnomAD allele counts. UMOD missense variants reported in the International ADTKD Cohort and in HGMD are plotted below the protein. The y axis represents the number of reported families. Variants that are reported both in gnomAD and in at least one ADTKD family are marked in red (with family numbers in parentheses). Protein and variant visualization used ProteinPaint (47) and cBioPortal MutationMapper (48, 49).
Table 1.
UMOD variants detected in gnomAD and ADTKD
| Position (GRCh38) | rs identification | HGVS coding | HGVS protein | gnomAD AC (AF) | TOPMed AC (AF) | ALFA AC (AF) | ADTKD families | AA change | REVEL | ClinVar |
|---|---|---|---|---|---|---|---|---|---|---|
| 16:20349117:T:G | rs143248111 | c.184A > C | p.(Thr62Pro) | 99/282,046/0 (3.5 × 10−4) | 51/125,568 (4.1 × 10−4) | 47/94,922 (5.0 × 10−4) | 2 | neu, pol > neu, nonpol | 0.54 | Conflicting (B, VUS) |
| 16:20348984:C:A | rs398123697 | c.317G > T | p.(Cys106Phe)* | 2/184,308/0 (1.1 × 10−5) | 1/125,568 (8.0 × 10−6) | 1/11,176 (9.0 × 10−5) | 9 | neu, nonpol > neu, nonpol | 0.91 | Conflicting (VUS, 4× LP) |
| 16:20348975:A:T | rs780462125 | c.326T > A | p.(Val109Glu)† | 3/215,296/0 (1.4 × 10−5) | 5/125,568 (4.0 × 10−5) | 1/2,188 (4.6 × 10−4) | 1 | neu, nonpol > neg, pol | 0.76 | LP |
| 16:20348763:G:C | rs187555378 | c.538C > G | p.(Leu180Val) | 578/174,180/8 (3.3 × 10−3) | 1,277/12,5568 (1.0 × 10−2) | 8/11,174 (7.2 × 10−4) | 1 | neu, nonpol > neu, nonpol | 0.51 | B |
| 16:20348594:G:A | rs1447458978 | c.707C > T | p.(Pro236Leu)‡ | 1/207,972/0 (4.8 × 10−6) | / | / | 7 | neu, nonpol > neu, nonpol | 0.97 | P/LP |
| 16:20348551:G:T | rs1475702916 | c.750C > A | p.(His250Gln)§ | 1/223,344/0 (4.5 × 10−6) | / | / | 1 | pos, pol > neu, pol | 0.80 | / |
| 16:20348480:T:C | rs375848177 | c.821A > G | p.(Tyr274Cys)¶ | 1/244,864/0 (4.1 × 10−6) | 1/125,568 (8.0 × 10−6) | / | 1 | neu, pol > neu, nonpol | 0.96 | / |
| 16:20341262:G:A | rs143583842 | c.1406C > T | p.(Thr469Met) | 203/282,716/1 (7.2 × 10−4) | 81/125,568 (6.5 × 10−4) | 82/88,416 (9.3 × 10−4) | 2 | neu, pol > neu, nonpol | 0.56 | Conflicting (LB, 2× VUS) |
Overlapping UMOD variants with frequencies in the general population, the number of ADTKD families, and in silico pathogenicity scores are shown. The 3 most frequent UMOD missense variants further studied are indicated in bold. GnomAD (18) allele counts (ACs) are reported as ACs/total allele number/number of homozygotes (with AF in parentheses). The theoretical maximum tolerated AC for high-effect UMOD variants (pathogenic [P]) in gnomAD is estimated to be one (46) (SI Appendix has more details). Annotation and in silico analyses were performed using the VarSome platform (51) and Ensembl VEP considering ENST00000302509.8 as the reference transcript. For REVEL (19), a score >0.5 corresponds to a sensitivity of 0.75 and a specificity of 0.89 for P variants in the training dataset. ClinVar was last accessed April 2021. GRCh38, Genome Reference Consortium Human Build 38; rs, Reference SNP; HGVS, Human Genome Variation Society; AA, amino acid; ClinVar, Clinical Variations database; VEP, Variant Effect Predictor; neu, neutral; nonpol, non polar; pol, polar; neg, negative; pos, positive; ALFA, National Center for Biotechnology Information (NCBI) Allele Frequency Aggregator (https://www.ncbi.nlm.nih.gov/snp/docs/gsr/alfa/); B, benign; LB, likely benign; LP, likely pathogenic; TOPMed, Trans-Omics for Precision Medicine Program (50).
*Using CRISPR-Cas9 gene editing, a mouse model with the homologous mutation was created that showed intracellular and secreted UMOD aggregates, but characterization of ER stress or kidney function was not shown (52). In vitro trafficking of this UMOD isoform was similar to the wild type (in vitro score of one) (20).
†In the reported ADTKD family, the index patient displayed kidney failure at 20 to 24 y, and intracellular UMOD accumulation was described in the kidney biopsy. However, two first-degree relatives were heterozygous for p.Val109Glu without presenting hyperuricemia or CKD. In vitro analysis indicated ER retention of the UMOD isoform (43, 53).
‡An in vitro score of three indicates considerable ER retention in stably transfected MDCK cells (20).
§The 45- to 49-y-old index patient had classical presentation (CKD, gout with low fractional excretion of uric acid (FEUA), small kidneys, and reduced urinary UMOD levels). However, the family history was negative (mutation status in parents was not known), and her clinically unaffected child carried the variant but was only 10 to 14 y old at the time of investigation (54).
¶The index patient was described with features not classically associated with ADTKD: early-onset CKD, microhematuria, mental retardation, and hearing impairment. The kidney biopsy showed glomerulosclerosis and tubular atrophy. Two siblings and a grandparent have been reported as affected (44).
UMOD p.Thr62Pro Shows an Intermediate Intracellular Trafficking Defect.
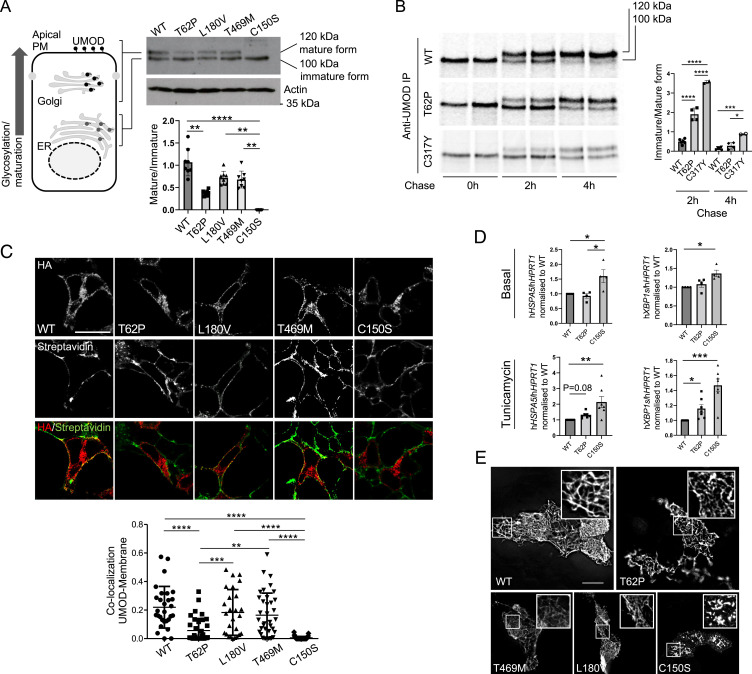
The functional consequences of the p.Thr62Pro, p.Thr469Met, and p.Leu180Val UMOD variants were assessed by monitoring UMOD processing in transiently transfected HEK293 cells. In control conditions, both the immature (100 kDa, “ER glycosylated”) and mature (120 kDa, “Golgi glycosylated”) UMOD bands can be detected (Fig. 2A). While no difference is noted between wild-type, p.Leu180Val, and p.Thr469Met UMOD isoforms, p.Thr62Pro UMOD shows an approximately twofold decrease of the mature/immature ratio compared with wild-type UMOD (Fig. 2A). In this setting, a typical ADTKD-UMOD mutant as p.Cys150Ser is fully retained in the ER, as evidenced by the absence of the mature form, thus demonstrating an intermediate phenotype for the p.Thr62Pro isoform between wild-type UMOD and ADTKD mutants. Of note, the direct comparison of p.Thr62Pro and p.Cys150Ser (unpaired Student’s t test with Welch’s correction) shows a statistically significant difference (P < 0.0001). The effect of p.Thr62Pro on protein maturation was confirmed using pulse chase experiments in stably transfected HEK293 cells (used because of their kidney origin and high capacity for protein synthesis) close to the TAL cells producing UMOD. The p.Thr62Pro UMOD showed an intermediate cellular phenotype between wild-type and p.Cys317Tyr UMOD [a reference ADTKD mutant (20)], with an increased amount of immature UMOD at 2 h after chase (Fig. 2B). Consistent with an intermediate phenotype, colocalization of UMOD with a plasma membrane marker is decreased for p.Thr62Pro compared with wild-type UMOD and with the two variants p.Leu180Val and p.Thr469Met, while it is increased compared with p.Cys150Ser (Fig. 2C). Of interest, p.Thr62Pro expression has no evident effect on ER homeostasis at baseline, while the ADTKD mutant p.Cys150Ser induces ER stress, as evidenced by the increased expression level of the ER chaperone glucose-regulated protein 78 kDa (GRP78; HSPA5) and of the spliced isoform of the unfolded protein response (UPR) effector Xbp1 (hXBP1s). However, upon treatment with a low dose of tunicamycin, affecting ER protein folding, cells expressing p.Thr62Pro show increased GRP78 and hXBP1s expression at an intermediate level between the wild type and p.Cys150Ser, suggesting increased ER stress susceptibility (Fig. 2D). Yet, p.Thr62Pro, p.Leu180Val, and p.Thr469Met UMOD isoforms are all able to form extracellular polymeric filaments similar to the ones observed for the wild-type protein, contrasting with pathological aggregates observed for p.Cys150Ser in Madin-Darby canine kidney (MDCK) cells (Fig. 2E). These data reveal a unique UMOD isoform p.Thr62Pro with an intermediate defect in processing from the ER to Golgi, contrasting with wild-type and other low-frequency variants p.Leu180Val and p.Thr469Met and with canonical ultrarare ADTKD-UMOD mutations. Moreover, under mild stress conditions, p.Thr62Pro induces ER stress at a level that is intermediate between the wild type and a typical ADTKD mutant.
Fig. 2.
p.Thr62Pro substitution induces intermediate UMOD trafficking and maturation defect. (A) Western blot analysis of UMOD expression in HEK293 cells 6 h after transfection of the indicated UMOD isoforms. Actin is shown as a loading control. The graph represents the ratio of Golgi glycosylated form (mature) to ER glycosylated form (immature). Bars indicate mean ± SD. n = 8 independent experiments. **P < 0.01 using the Kruskal–Wallis test (P < 0.0001) followed by Dunn’s multiple comparison test; ****P < 0.0001 using the Kruskal–Wallis test (P < 0.0001) followed by Dunn’s multiple comparison test. (B) Pulse chase experiments performed in HEK293 cells stably expressing the indicated UMOD isoform. The maturation to fully glycosylated protein is delayed for the p.Thr62Pro isoform compared with the wild-type (WT) one, but it is more efficient than the one observed for the p.Cys317Tyr mutant. Bars indicate the mean ± SD of the ratio of immature to mature form. n = 6, 4, and 2 for WT, p.Thr62Pro, and p.Cys317Tyr, respectively. *P < 0.05 using one-way ANOVA (P < 0.0001) followed by Bonferroni’s multiple comparison test; ***P < 0.001 using one-way ANOVA (P < 0.0001) followed by Bonferroni’s multiple comparison test; ****P < 0.0001 using one-way ANOVA (P < 0.0001) followed by Bonferroni’s multiple comparison test. IP, immunoprecipitation (C) Immunofluorescence analysis of HEK293 cells 10 h after transfection with the indicated UMOD isoform. UMOD is seen in red (stained with HA), and the plasma membrane (PM) is in green (stained with streptavidin-FITC after biotinylation) in the merged picture. The graph reports the Mander’s 2 coefficient as a readout for UMOD at the PM. The mean ± SD is indicated. (Scale bar, 10 µm.) **P < 0.01 using one-way ANOVA (P < 0.0001) followed by Bonferroni’s multiple comparison test; ***P < 0.001 using one-way ANOVA (P < 0.0001) followed by Bonferroni’s multiple comparison test; ****P < 0.0001 using one-way ANOVA (P < 0.0001) followed by Bonferroni’s multiple comparison test. HA, hemagglutinin; FITC, fluorescein isothiocyanate. (D) GRP78 (HSPA5) and spliced XBP1 (XBP1s) expression assessed by real-time qPCR in HEK293 cells expressing the indicated UMOD isoform in the basal condition (Upper) or after 12 h of treatment with a low dose of tunicamycin (Lower). Expression is normalized to HPRT1, and bars indicate mean ± SEM. *P < 0.05 using the Kruskal–Wallis test (baseline: P = 0.009 HSPA5, P = 0.003 XBP1s; tunicamycin: P = 0.0009 HSPA5, P < 0.0001 XBP1s) followed by Dunn’s multiple comparison test (n = 4 to 7 independent experiments); **P < 0.01 using the Kruskal–Wallis test (baseline: P = 0.009 HSPA5, P = 0.003 XBP1s; tunicamycin: P = 0.0009 HSPA5, P < 0.0001 XBP1s) followed by Dunn’s multiple comparison test (n = 4 to 7 independent experiments); ***P < 0.001 using the Kruskal–Wallis test (baseline: P = 0.009 HSPA5, P = 0.003 XBP1s; tunicamycin: P = 0.0009 HSPA5, P < 0.0001 XBP1s) followed by Dunn’s multiple comparison test (n = 4 to 7 independent experiments). (E) Immunofluorescence analysis of unpermeabilized MDCK cells stably showing the indicated UMOD isoform at the PM. Insets show UMOD polymers at higher magnification. All the analyzed variants form polymers on the PM, while the pathogenic ADTKD mutant p.Cys150Ser forms aggregates. (Scale bar, 10 µm.)
Modeling of the UMOD p.Thr62Pro Effect on Protein Folding.
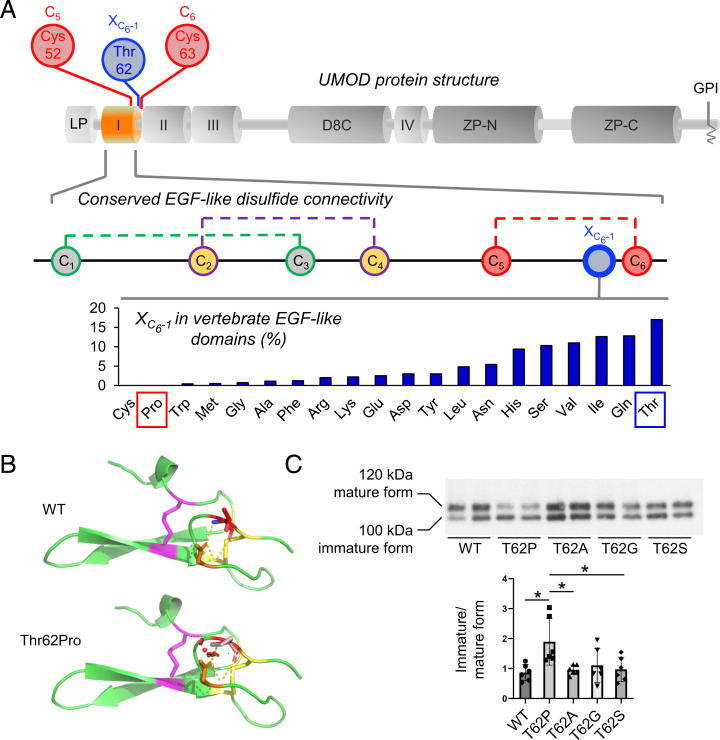
UMOD contains 48 conserved cysteine residues, which are all engaged in intramolecular disulphide bonds (21) (SI Appendix, Fig. S3). There is a major enrichment of cysteine substitutions in patients with ADTKD-UMOD (∼55% of all missense variants) (11), such that most cysteine positions in UMOD (35/48; 73%) have been substituted in patients with ADTKD-UMOD (SI Appendix, Table S3) vs. only 10% (5/48) in gnomAD. In contrast, all the other amino acids, apart from tryptophan, show more genetic variability in gnomAD compared with the ADTKD population, supporting that cysteine residues in UMOD are intolerant to genetic variation (SI Appendix, Fig. S4A). In fact, Thr62 is directly adjacent to Cys63, which is predicted to form a disulphide bridge with Cys52 (from Uniprot; PROSITE-ProRule:PRU00076) (22) (Fig. 3A, Top and Middle), and substitutions of Cys52 and Cys63 are identified in multiple ADTKD-UMOD families (SI Appendix, Table S3). We, therefore, postulated that p.Thr62Pro might affect UMOD maturation by impacting neighboring disulphide bridge formation. Proline has a cyclic pyrrolidine side chain determining high conformational rigidity and potential high impact on protein stability (23). Thus, missense variants replacing an amino acid with a proline are poorly tolerated when adjacent to a cysteine bond (24). Four different cysteine-adjacent positions are substituted with proline in ADTKD-UMOD families, more than for any other amino acid in UMOD (SI Appendix, Fig. S4B). In line, when aligning >8,000 vertebrate epidermal growth factor (EGF)-like domain sequences, we observed that proline is never present at the position corresponding to Thr62 (i.e., preceding the sixth cysteine of the domain [XC6-1]), while all the other amino acids except cysteine can be found (Fig. 3A, Bottom), further supporting a damaging effect of proline at this position.
Fig. 3.
Modeling of the p.Thr62Pro impact on protein structure. (A, Top) Schematic representation of the UMOD domain structure. The EGF-like domain 1 is shown in orange; the positions of Cys52 and Cys63, predicted to form a disulphide bond, and of neighboring Thr62 are shown. D8C, domain with eight cysteines; GPI, glycosylphosphatidylinositol anchoring site; I to IV, EGF-like domains; LP, leader peptide; ZP, bipartite zona pellucida domain. (A, Middle) Schematic representation of the EGF-like domain showing the conserved disulphide bond connectivity (C1 to C3, C2 to C4, C5 to C6; data from PROSITE documentation PDOC00021). The position corresponding to Thr62 in UMOD is indicated in blue (one amino acid before the sixth cysteine, XC6-1). (A, Bottom) Vertebrate EGF-like domain sequences from protein families database (Pfam) family PF12947 (8,511 sequences) were aligned, and the frequency of residues at the position preceding C6 (XC6-1) was calculated. Pro and Cys are the only amino acids that are never found, strongly suggesting a damaging effect of Pro at this position. Note that Thr is the most frequently encountered amino acid at position XC6-1. (B) Modeling of the UMOD p.Thr62Pro isoform. Polar contacts are indicated by yellow dashed lines. Clashes in the structure are represented by red dots and are visible only in the p.Thr62Pro isoform. The figure was made with PyMOL (Schrödinger LLC). (C) Western blot analysis for UMOD in cell lysates from HEK293 cells transiently transfected with the indicated UMOD isoforms. The immature and mature forms of UMOD proteins are indicated on the left. The ratios of the intensities of immature to mature forms are indicated below. Bars indicate mean ± SD. Note that only the proline substitution increases the ratio of immature to mature forms, indicating defective trafficking. n = 6 independent experiments. WT, wild type. *P < 0.05 using one-way ANOVA (P = 0.0077) followed by Bonferroni’s multiple comparison test.
To analyze the effect of the substitution, we constructed a model of the first EGF of UMOD (amino acids 28 to 65) using the software Phyre2 and introduced the threonine to proline change in PyMOL. The presence of structural clashes is detected when proline is inserted (Fig. 3B). In contrast, nonproline substitutions at Thr62 in UMOD appear to be tolerated (SI Appendix, Figs. S4C and S5). These results were confirmed by using the prediction programs Missense3D (25) and SDM (SI Appendix, Fig. S4C) (26), which identified proline as the only substitution affecting protein structure and as the most destabilizing. Furthermore, only Thr62Pro, but not Thr62Ala, Thr62Gly, or Thr62Ser, led to a trafficking defect in HEK293 cells (Fig. 3C). Of note, consistent with cellular studies, the two variants p.Leu180Val and p.Thr469Met have no predicted effect on UMOD structure (SI Appendix, Fig. S6). Collectively, these results strongly suggest a unique effect of p.Thr62Pro on protein maturation, likely via an impact on the conserved neighboring disulphide bridge within the canonical EGF-like domain.
High Genetic Load of the UMOD p.Thr62Pro Variant in Familial Tubulointerstitial CKD Clusters.
The UMOD p.Thr62Pro variant was initially detected in proband III.1 of family CH1, presenting with CKD, hyperuricemia with gout, small multicystic kidneys, and normal urinalysis (negative for other ADTKD genes) (SI Appendix, Fig. S7 and Table S4). However, the p.Thr62Pro variant did not fully segregate with disease in this family, being detected in apparently unaffected relatives. The variant was also reported in a German family (GE1) as a variant of unknown significance (VUS) segregating with ADTKD. Kidney biopsy revealed interstitial fibrosis with tubular atrophy (27). Carriers of p.Thr62Pro were identified in 13 additional ADTKD families (SI Appendix, Fig. S7) and in five unrelated probands with kidney disease (i.e., CKD, gout, kidney cysts) in the UK Genomics England 100,000 Genomes Project (SI Appendix, Fig. S7 and Table S4). We first tested for pathogenic variants in MUC1 [causing ADTKD-MUC1 and missed by massively parallel sequencing (27, 28)] in all available DNA samples (n = 19 individuals from 14 families, all negative). To exclude variants in other nephrogenes in the 20 families, whole-genome sequencing was performed in 5 families (Genomics England 100,000 Genomes Project), whole-exome sequencing was performed in another 5 families, and gene panel testing was performed in 4 other families (SI Appendix, Tables S4 and S5). The five families recruited in Genomics England are labeled unsolved after review of the clinical and genetic data. Similarly, no alternative genetic explanations for CKD were detected in the other CKD families with UMOD p.Thr62Pro (SI Appendix, Table S5). Segregation analysis confirmed incomplete penetrance for the p.Thr62Pro variant, also detected in unaffected carriers (SI Appendix, Fig. S7, CH1: II.2, III.2; UK2: IV3; and GEL4: I.1). Incomplete penetrance (or late-onset disease) is also evident in the 100,000 Genomes Project, where p.Thr62Pro was detected in 52 unaffected relatives and 57 rare disease patients without a primary kidney diagnosis and with an average age of 41.5 y (SI Appendix, Tables S6 and S7). Thus, the p.Thr62Pro variant is detected in a significant number of families with tubulointerstitial CKD and evidence for dominant inheritance. Combining the ADTKD-UMOD families reported thus far (SI Appendix, Table S3) and including the families from this study, p.Thr62Pro is detected in ∼8% (20/247) of them. This corresponds to an AF for p.Thr62Pro of ∼0.041 (20/494) among ADTKD family index patients, >100× increased compared with the AF for p.Thr62Pro in gnomAD (∼0.00035).
Milder Phenotype in UMOD p.Thr62Pro Carriers with Kidney Disease.
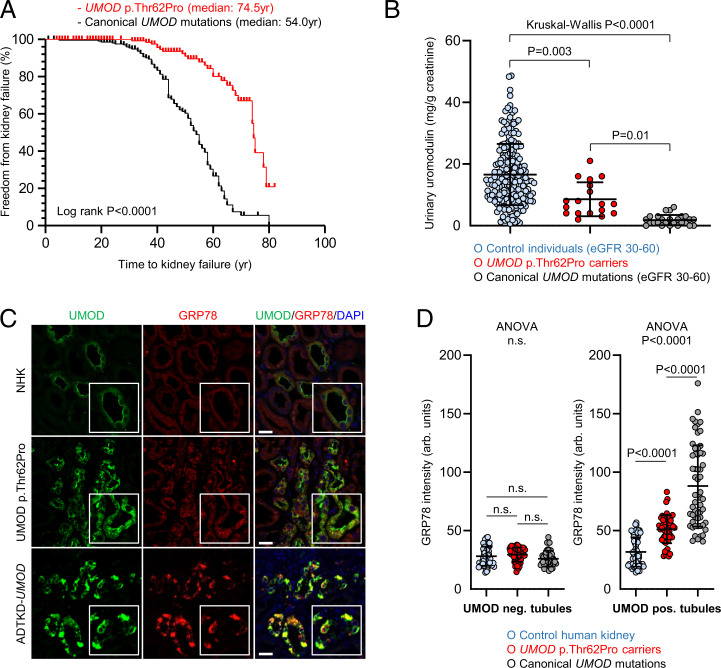
We next tested whether UMOD p.Thr62Pro was associated with an intermediate phenotype, consistent with its biological effect. Analysis of 157 p.Thr62Pro carriers from our CKD families (SI Appendix, Table S4) and from the Genomics England 100,000 Genomes Project compared with ADTKD-UMOD patients carrying canonical UMOD mutations indicated that p.Thr62Pro was associated with 20-y-longer kidney survival (median age of kidney failure: 74.5 vs. 54.0 y; log-rank P < 0.0001) (Fig. 4A). Among those patients who did reach kidney failure, mean age at onset of kidney failure was later in patients carrying p.Thr62Pro compared with typical ADTKD-UMOD patients (58.6 ± 14.6 vs. 47.5 ± 12.4 y, respectively; P = 0.0002).
Fig. 4.
Mild kidney disease phenotype and intermediate UMOD trafficking defect and ER stress in p.Thr62Pro carriers. (A) Kaplan–Meier curves of renal survival in all p.Thr62Pro carriers identified in Genomics England 100,000 Genomes Project data and in p.Thr62Pro-positive families/cases with CKD (n = 157) compared with ADTKD-UMOD patients from the International ADTKD Cohort (n = 225). Only patients with confirmed p.Thr62Pro status have been included in the analysis. Median age at kidney failure was 74.5 y in UMOD p.Thr62Pro carriers and 54.0 y in ADTKD-UMOD patients with canonical UMOD mutations. (B) Urinary UMOD levels normalized to urinary creatinine are depicted for control individuals matched for eGFR (30 to 60 mL/min per 1.73 m2), UMOD p.Thr62Pro carriers, and ADTKD-UMOD patients with canonical UMOD mutations matched for eGFR (30 to 60 mL/min per 1.73 m2). Outlier removal was performed using Robust regression and Outlier removal (ROUT) method (Q = 1%). The graph depicts individual values, and bars indicate mean ± SD. The Kruskal–Wallis test was used with Dunn's multiple comparisons test. (C) Immunofluorescence staining for UMOD (green) and GRP78 (red) in normal human kidney (NHK; from a tumor nephrectomy), a UMOD p.Thr62Pro human kidney biopsy, and kidney tissue from an ADTKD-UMOD patient with a canonical UMOD mutation (p.Arg185Ser). Insets show UMOD-expressing tubules at higher magnification. DAPI, 4′,6-diamidino-2-phenylindole. (Scale bars, 25 μm). (D) Quantification of the fluorescent signal intensity for ER stress–associated GRP78 in UMOD-negative and UMOD-positive tubules from three NHK samples (tumor nephrectomy), three p.Thr62Pro kidney biopsies, and three ADTKD-UMOD kidney samples. Further details can be found in SI Appendix, Table S9. The graph depicts tubule values in arbitrary units, and bars indicate mean ± SD. One-way ANOVA was followed by Tukey’s multiple comparison test. ns, not significant.
Markedly decreased levels of uromodulin in the urine (uUMOD) are a hallmark of ADTKD-UMOD, reflecting mutant UMOD retention in the kidney (11). Compared with uUMOD levels in control individuals (17 ± 10 mg/g creatinine) and ADTKD-UMOD patients (2 ± 2 mg/g creatinine), intermediate levels were observed in p.Thr62Pro carriers (9 ± 6 mg/g creatinine; Kruskal–Wallis P < 0.0001). The latter never showed the very low uUMOD concentrations (<1 µg/mL) often detected in ADTKD-UMOD samples. Conversely, lower levels of uUMOD were also seen in the few p.Thr62Pro carriers with preserved eGFR (Fig. 4B and SI Appendix, Table S8).
Comparative immunofluorescence studies in three human kidney biopsies from p.Thr62Pro carriers revealed increased intracellular UMOD staining as compared with strongly predominant apical staining in normal kidneys and exclusive intracellular staining in patients harboring canonical ADTKD-UMOD mutations (p.Arg185Ser, p.Tyr274Cys) (Fig. 4C and SI Appendix, Figure S8 and Table S9). Quantification of ER stress marker GRP78 signal intensity revealed a moderate increase in p.Thr62Pro kidneys, intermediate between signals observed in control and ADTKD-UMOD kidneys and specifically in UMOD-positive tubules (Fig. 4D and SI Appendix, Table S9). These data indicate that the fraction of UMOD p.Thr62Pro carriers who presents with CKD shows a milder kidney disease, with progression to kidney failure delayed by ∼20 y compared with classical ADTKD-UMOD, paralleled by intermediate levels of uUMOD reduction reflecting a milder trafficking defect with intracellular UMOD accumulation and intermediate ER stress levels.
Wondering whether the UMOD locus haplotype, modulating UMOD expression levels (2, 5), affects p.Thr62Pro disease penetrance, we assessed genotypes at two different haplotype-tagging single-nucleotide polymorphisms (SNPs) among carriers of rare UMOD variants. While the global Genomics England population displayed the expected allelic distribution for European populations (∼70% homozygous for major and 3% homozygous for minor alleles; associated with higher and lower UMOD expression, respectively) (5), we detected a striking shift toward minor alleles in carriers of UMOD p.Thr62Pro but not in carriers of UMOD p.Thr469Met and p.Leu180Val. In fact, none of the 121 individuals with UMOD p.Thr62Pro were homozygous for the major SNP alleles. In line with these results, haplotype estimations phased UMOD p.Thr62Pro with the minor UMOD haplotype (SI Appendix, Figs. S9 and S10). These results show that UMOD p.Thr62Pro (in contrast to p.Thr469Met and p.Leu180Val) is linked with the minor UMOD haplotype, associated with lower UMOD expression levels (5). This configuration could potentially contribute to the reduced penetrance observed for UMOD p.Thr62Pro.
UMOD p.Thr62Pro Is Enriched in Patients with Kidney Failure.
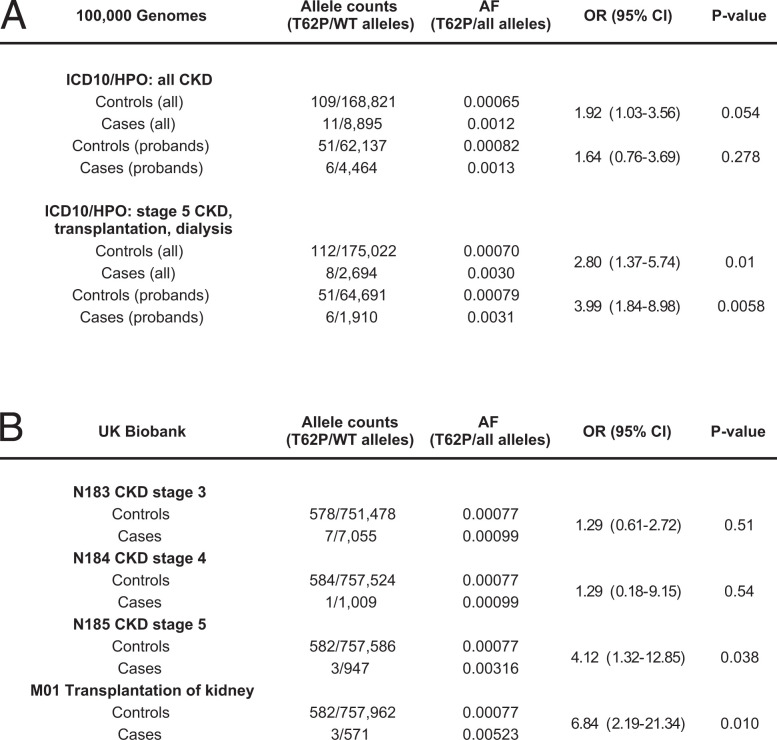
To investigate whether intermediate-effect UMOD variants contribute to risk of CKD at the population level, we performed a single–variant load analysis in the Genomics England 100,000 Genomes Project. Among the top 14 low-frequency (gnomAD AF > 10−3) or rare (gnomAD 10−3 > AF > 10−4) UMOD missense variants (SI Appendix, Table S1), p.Thr62Pro is the only variant with higher prevalence in the three tested modalities (1] 100,000 Genomes kidney disease categories, 2] Human Phenotype Ontology [HPO] term CKD, and 3] HPO term CKD stage 5) compared with gnomAD and the 100,000 Genomes control population (SI Appendix, Figs. S11 and S12). We also found enrichment for p.Thr62Pro (but not for p.Thr469Met or p.Leu180Val) among probands recruited with “Unexplained kidney failure” when compared with probands recruited to all other rare disease groups in the 100,000 Genomes Project (SI Appendix, Fig. S13). Next, we selected 1,351 cases (958 probands) in the 100,000 Genomes Project with stage 5 CKD and/or treated by kidney transplantation and/or dialysis as well as 87,567 controls (32,371 probands) in which none of these diagnoses were present and tested for the prevalence of p.Thr62Pro (SI Appendix, Table S7). A significant enrichment for p.Thr62Pro was detected when considering the whole cohort OR = 2.80 [1.4 to 5.7], P = 0.01) and when considering only probands (OR = 3.99 [1.8 to 9.0], P = 0.006) (Fig. 5A). As the percentage of individuals with White ethnicity was similar between cases and controls, it is unlikely that stratification based on genetic ancestry was driving this association with p.Thr62Pro, which is enriched in Europeans (SI Appendix, Table S7). When considering only individuals reported with a White ethnic background in Genomics England, the risk conferred by UMOD p.Thr62Pro for kidney failure was confirmed (OR = 4.2 [1.9 to 8.6], P = 0.002) (SI Appendix, Table S10). Using the same pipeline in individuals with a reported Black ethnic background, we detected comparable risk for kidney failure associated with the presence of two risk alleles at APOL1 (29) compared with the wild type (OR = 4.3 [2.6 to 7.1]) or with one risk allele (OR = 4.0 [2.3 to 6.8]) (SI Appendix, Table S10). Similar results were observed in ancestry-matched individuals from the UK Biobank, where UMOD p.Thr62Pro was enriched in probands with CKD stage 5 (OR = 4.12 [1.3 to 12.9], P = 0.038) and probands with kidney transplantation (OR = 6.84 [2.2 to 21.3], P = 0.010) (Fig. 5B). In both databases, no significant enrichment for p.Thr62Pro was detected for milder stages of CKD. UMOD variants p.Leu180Val and p.Thr469Met were not significantly enriched in probands with kidney disease (SI Appendix, Figs. S13 and S14), which together with the negative in vitro and in silico data, makes their contribution to human disease unlikely.
Fig. 5.
UMOD p.Thr62Pro confers risk for kidney disease in UK cohorts. (A) Prevalence of p.Thr62Pro alleles in all individuals and in probands from the Genomics England 100,000 Genomes Project with diagnoses of CKD (ICD10: N18 and/or HPO HP0012622 diagnoses, including mapped and descendant concepts) or kidney failure (ICD10: N185, Z940, Z491, and Y841 and/or HPO HP0003774 diagnoses, including mapped and descendant concepts) and in all control individuals or family probands in which these diagnoses are absent. P values were computed using Fisher’s exact test. (B) Prevalence of p.Thr62Pro alleles in unrelated White British controls and individuals with indicated kidney phenotypes in the UK Biobank SNP array data. P values were computed using Fisher’s exact test. WT, wild type. ICD10, International Classification of Diseases, Tenth Revision.
Finally, we analyzed clinical indications for 3,122 unrelated patient whole-exome datasets obtained in Western Europe (Eurofins Biomnis). UMOD p.Thr62Pro was detected in 2 (FR1 and IRL1) of 508 exomes (0.39%) performed for cystic kidney disease, tubulointerstitial kidney disease, or CKD of unknown etiology, but it was absent from the remaining 2,614 exomes performed for glomerular or vascular kidney disease or nonnephrological indications—in line with enrichment of p.Thr62Pro in ADTKD-like disease (SI Appendix, Table S11). Altogether, we detected 27 individuals with kidney disease and UMOD p.Thr62Pro in the UK Biobank, the 100,000 Genomes Project, and the Eurofins exomes (SI Appendix, Table S11).
Discussion
The UMOD gene is of particular interest because it encodes the most abundant protein excreted in the normal urine, is associated with an autosomal dominant kidney disease (ADTKD-UMOD), and is the top GWAS signal for eGFR and CKD in the general population (4). By intersecting missense variants associated with ADTKD-UMOD or present in gnomAD, we detected a unique UMOD variant p.Thr62Pro with intermediate-sized biological effects associated with kidney failure and enriched in CKD families with a milder clinical course compared with ADTKD-UMOD. The identification of an intermediate-sized effect variant completes the spectrum of UMOD-associated kidney diseases and provides insights into the genetic architecture of CKD.
Using large-scale reference datasets, we confirm that UMOD variants associated with ADTKD are generally ultrarare and absent from gnomAD, as expected for bona fide high-effect variants causing a dominant Mendelian disease (11). The eight UMOD variants detected both in ADTKD patients and gnomAD showed a prevalence of ∼1/160 in gnomAD, similar to the prevalence (∼1/165) of purported pathogenic variants in PKD1 reported in the Exome Aggregation Consortium (ExAC) database (30). These observations could be due to either annotation errors or altered penetrance and expressivity of purported pathogenic variants in control datasets. The MAF of p.Thr62Pro in the general population (between 0.035 and 0.05%) and the fact that it is among the most frequent UMOD missense variants listed in gnomAD are not compatible with a high-effect size variant for ADTKD-UMOD, whose prevalence is estimated between 1/50,000 and 1/100,000 (10). However, p.Thr62Pro is enriched in kidney disease populations, with an MAF of ∼0.4% among UK patients with kidney failure and up to 4% among ADTKD family index patients. These epidemiological data are compatible with UMOD p.Thr62Pro occupying an intermediate position in terms of AFs between common and ultrarare variants and also compatible with an intermediate-effect size.
What could be the basis for the increased risk of CKD associated with the UMOD p.Thr62Pro variant? While ADTKD-UMOD is caused by gain-of-toxic function mutations altering ER homeostasis (10, 31, 32), the risk of CKD associated with common GWAS UMOD variants is thought to be related to increased UMOD expression, which may, over time, stress the ER protein folding capacity and lead to tubulointerstitial damage (5). Our data suggest that the same cellular pathways, involving UMOD ER homeostasis and maturation, may sustain the CKD risk conferred by the p.Thr62Pro UMOD variant. The variant shows intermediate trafficking defect and induction of ER stress between wild-type and canonical ADTKD mutant isoforms. Interestingly, induction of ER stress is only observed in HEK293 cells under mild tunicamycin treatment, an experimental setting that reflects the high production and secretion of UMOD by TAL cells and mimics the reduction of chaperoning systems in aging cells (33). Consistently, patients with the p.Thr62Pro variant show intermediate urinary UMOD levels, paralleled by mild intracellular accumulation of UMOD and modest ER stress as quantified on patient tissue. Such significant but mild biological effects match the intermediate clinical effect sizes for p.Thr62Pro, as evidenced by significantly longer kidney survival and older age at kidney failure compared with those observed for the canonical ADTKD-UMOD mutations. The in vitro data also suggest that additional stressors may be required to induce cell toxicity, possibly reflected by the incomplete penetrance and later onset of disease in pThr62Pro carriers. These findings corroborate the recent evidence that, in ADTKD-UMOD, the severity of the mutant UMOD trafficking defect correlates with disease severity (20). Further in vitro and in silico evidence substantiate the specific effect of the proline substitution on UMOD folding and ER exit, possibly by impacting the neighboring disulphide bond, in line with mutagenesis datasets showing missense proline insertions being associated with the highest predicted damage (34). Whether and how p.Thr62Pro could have additional deleterious effects on the function of UMOD remain to be investigated. Our findings implicate ER proteostasis and different levels of UPR in a spectrum of UMOD-associated kidney disease risk. Strategies to rescue protein misfolding and UPR using, for instance, chemical chaperones might, therefore, be viable therapeutic options that should be tested in CKD patients with intermediate, potentially more rescuable, or high-impact UMOD variants. A recent high-content screen identified the small molecule BRD4780 as a promising lead to treat ADTKD-MUC1, and it has been proposed to induce mutant UMOD degradation in vitro (35).
While the majority of UMOD p.Thr62Pro carriers do not develop overt kidney disease, the variant appears to segregate with CKD in some families. The basis for such heterogeneity remains speculative. No other plausible potential pathogenic variants in nephrogenes were detected in these families using whole-exome or -genome sequencing or gene panel analysis and after analysis of the MUC1 gene. One could speculate that other genetic factors, possibly combined with environmental factors, could modulate the threshold for disease expression and CKD (15, 36). Such modifiers could involve the ER–proteostasis pathway, in line with induction of ER stress in p.Thr62Pro-expressing cells and on patient biopsies, or genes that determine the inflammatory response (10). Arguably, the co-occurrence of functional nephrogene variants with UMOD p.Thr62Pro would not invalidate the hypothesis of p.Thr62Pro being an intermediate-effect variant under an oligogenic disease model. Our data indicate that CKD progression is significantly delayed in p.Thr62Pro carriers compared to carriers of classical ADTKD mutations and that disease might be subclinical in some younger adults. In line, we saw that p.Thr62Pro carriers that were labeled with CKD in the 100,000 Genomes Dataset were notably older than carriers in which no CKD was described (SI Appendix, Table S7). In addition to its intrinsic milder effect on trafficking and ER stress, the intriguing observation that the p.Thr62Pro variant appears to be linked with the minor UMOD locus haplotype could contribute to reduced penetrance. Indeed, the minor “CKD-protective” UMOD haplotype is associated with lower UMOD expression levels (2, 5) and possibly, delayed ER accumulation of mutant UMOD p.Thr62Pro. The true lifelong risk for developing CKD in aging p.Thr62Pro carriers remains to be determined.
Thus far, only a few examples of genes showcasing a spectrum of effect sizes have been documented. Reported pathogenic variants in the prion protein gene PRNP, causing autosomal dominant prion disease due to gain-of-function prion protein misfolding, are at least 30× more common in the population than expected based on disease incidence (37). The authors suggested that some of these variants confer intermediate amounts of lifetime risk rather than being fully penetrant. About 1% of individuals from the general population carry bona fide pathogenic variants in Mendelian diabetes mellitus genes yet remain euglycemic through middle age (38). HNF1A has been implicated in a spectrum of phenotypes ranging from multifactorial polygenic diabetes risk to autosomal dominant early-onset diabetes (39). Whole-exome sequencing of Latino populations has revealed a single low-frequency missense variant (p.Glu508Lys) in HNF1A associated with an intermediate functional effect and a fivefold increase in type 2 diabetes prevalence. Of note, only one variant in HNF1A and three variants in PRNP have been associated with intermediate-effect sizes, compatible with the single variant described here (37, 40). In addition, previous studies investigating intermediate-effect variants in type 2 diabetes mellitus and Alzheimer’s disease reported low numbers of cases carrying these alleles, similar to our low numbers of stage 5 CKD patients with UMOD p.Thr62Pro (40, 41). The identification of additional rare variants with intermediate-effect sizes contributing to diverse human phenotypes and of larger patient numbers may require even better-powered studies.
The identification of a UMOD variant with incomplete penetrance has implications for genetic testing and counseling. The implementation of massively parallel sequencing in routine care means that more VUSs in UMOD will be identified, requiring careful characterization and mapping into the disease spectrum. Previous analyses reported an absence of positive family history in ∼15% of cases as well as incomplete penetrance in ADTKD-UMOD (42–44), suggesting the possibility of further atypical UMOD variants with intermediate-effect sizes. Several atypical UMOD variants were identified here (Table 1) but in contrast to p.Thr62Pro, with very low population frequencies (MAF ∼ 10−5). Taking into account a possible oligogenic disease model, we would encourage continuation of diagnostic workup even in individuals carrying UMOD p.Thr62Pro. The fact that numerous ultrarare and predicted pathogenic UMOD variants are present in gnomAD is noteworthy (SI Appendix, Table S2). If the 45 individuals harboring such ultrarare variants with an REVEL score of >0.75 [corresponding to a specificity of ∼0.95 for pathogenic variants (19) and excluding the probable benign variant p.Gly131Asp] would develop mild or late-onset CKD, then the prevalence of UMOD-associated disease would rise close to 1 in 3,000 (45 carriers in ∼140,000 gnomAD subjects).
This study is a systematic investigation of intermediate-sized effects for UMOD variants based on analysis of large databases; detailed characterization of the effect in silico, in vitro, and in patient samples; and validation in large patient and population-based cohorts. While this study is limited to UMOD, an important gene for individual and population burden of kidney disease, the approach and concept here are potentially applicable to other genes, organs, and traits. Limitations of the study include using previously reported ADTKD-UMOD variants rather than resequencing patients, thereby limiting the discovery stage. Most of the patient and control dataset is of European ancestry, thus neglecting potential variants operating in other backgrounds. For instance, the p.Leu180Val UMOD variant, ultrarare in UK databases, is principally detected in Africans. Approximately 1 in 1,000 people of European ancestry carry p.Thr62Pro; based on our findings, these individuals have a fourfold increase in risk for kidney failure, comparable with the risk we detected for Black individuals with two APOL1 risk alleles for all-cause kidney failure in the Genomics England database. Better-powered studies conducted in Black individuals detected higher ORs associated with two APOL1 risk variants for various defined kidney diseases predominantly through a recessive model (OR ∼ 7 to 89) (29, 45). The fact that p.Thr62Pro is significantly rarer than APOL1 risk variants impedes for the moment similar risk stratification into better-defined disease ontologies. Longitudinal follow-up studies are required to determine the clinical significance of the p.Thr62Pro carrier status in the general population, its positive predictive value for CKD, and whether UMOD p.Thr62Pro could also act as a modifier in other types of kidney diseases.
In conclusion, we identified a single rare missense variant (p.Thr62Pro) in UMOD, characterized by an intermediate biological effect in vitro and in vivo, associated with a milder course of ADTKD and conferring an increased risk for kidney failure in UK cohorts. These findings extend the understanding of UMOD-associated kidney diseases and of the genetic architecture of CKD.
Materials and Methods
A detailed description of the materials and methods (patient and population cohorts, in vitro and in silico analyses, statistics) is presented in SI Appendix. Informed written consent was obtained from all participants, and individual-level data were deidentified. All methods were carried out in accordance with approved guidelines and the Declaration of Helsinki. Participating cohorts/centers and ethics committees as well as local institutional review boards are detailed in SI Appendix, Table S12.
Supplementary Material
Acknowledgments
We thank all participating patients and families. The Cohorte Lausannoise is acknowledged for providing reference urine samples and eGFR information. We thank the Broad Institute of Massachusetts Institute of Technology (MIT) and Harvard; the First Faculty of Medicine, Charles University (LM2018132); and the Friedrich-Alexander Universität Erlangen-Nürnberg in Germany for providing MUC1 testing. We also thank Emily Cornec-Le Gall and Marie-Pierre Audrézet (Centre Hospitalier Régional et Universitaire [CHRU] Brest, France) for providing gene panel testing in CKD families, Anne Kipp for her help in generating preliminary results, Valeria Berno from the San Raffaele Advanced Light and Electron Microscopy BioImaging Center for helpful discussions, and Nadine Nägele for technical assistance. E.O. is supported by Postdoc Mobility-Stipendien of the Swiss National Science Foundation Grants P2ZHP3_195181 and P500PB_206851, Kidney Research UK Grant Paed_RP_001_20180925, Fonds National de la Recherche Luxembourg Grant 6903109, and the University Research Priority Program “Integrative Human Physiology, Zurich Center for Integrative Human Physiology (ZIHP)” of the University of Zurich. K.K., S.K., and A.J.B. were supported by Ministry of Health of the Czech Republic Grant NU21-07-00033. E.A.E.E. is funded by the Royal College of Surgeons in Ireland Blackrock Clinic Strategic Academic Recruitment (StAR) Doctor of Medicine (MD). A.D.K. was supported by University of Zurich Grant Forschungskredit FK-14-035. L.R. is supported by Italian Ministry of Health Grants RF‐2010‐2319394 and RF‐2016‐02362623. The work of A.K. was supported by German Research Foundation (DFG) Grant KO 3598/5-1, DFG Project-ID 431984000 SFB 1453. M.W. is supported by DFG Projektnummer 387509280 Grant SFB 1350, TP C4. J.A.S. is supported by Kidney Research UK and the Northern Counties Kidney Research Fund. O.D. is supported by the European Union’s Horizon 2020 Research and Innovation Program under Marie Skłodowska-Curie Grant 860977, European Reference Network for Rare Kidney Diseases Project 739532, the Swiss National Science Foundation’s National Center of Competence in Research Kidney Control of Homeostasis program, Swiss National Science Foundation Grant 310030-189044, the Gebert-Rüf Foundation for research on ADTKD-UMOD, and the University Research Priority Program Innovative Therapies in Rare Diseases (ITINERARE) at the University of Zurich. This research was made possible through access to the data and findings generated by the 100,000 Genomes Project. This research has been conducted using data from UK Biobank (Project ID 43879), a major biomedical database with open access to researchers (https://www.ukbiobank.ac.uk/).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2114734119/-/DCSupplemental.
Contributor Information
Collaborators: J. C. Ambrose, P. Arumugam, R. Bevers, M. Bleda, F. Boardman-Pretty, C. R. Boustred, H. Brittain, M.A. Brown, M. J. Caulfield, G. C. Chan, A. Giess, J. N. Griffin, A. Hamblin, S. Henderson, T. J. P. Hubbard, R. Jackson, L. J. Jones, D. Kasperaviciute, M. Kayikci, A. Kousathanas, L. Lahnstein, A. Lakey, S. E. A. Leigh, I. U. S. Leong, F. J. Lopez, F. Maleady-Crowe, M. McEntagart, F. Minneci, J. Mitchell, L. Moutsianas, M. Mueller, N. Murugaesu, A. C. Need, P. O’Donovan, C. A. Odhams, C. Patch, D. Perez-Gil, M. B. Pereira, J. Pullinger, T. Rahim, A. Rendon, T. Rogers, K. Savage, K. Sawant, R. H. Scott, A. Siddiq, A. Sieghart, S. C. Smith, A. Sosinsky, A. Stuckey, M. Tanguy, A. L. Taylor Tavares, E. R. A. Thomas, S. R. Thompson, A. Tucci, M. J. Welland, E. Williams, K. Witkowska, S. M. Wood, and M. Zarowiecki
Data Availability
All study data are included in the article and/or SI Appendix. Previously published data were also used for this work. Data presented here were derived from the following resources available in the public domain: the gnomAD version 2.1.1 (https://gnomad.broadinstitute.org/) and Ensembl (release 100; https://useast.ensembl.org/index.html). The International ADTKD Cohort and the UMOD in vitro scoring have also been published (11, 20).
References
- 1.Eckardt K.-U., et al. , Evolving importance of kidney disease: From subspecialty to global health burden. Lancet 382, 158–169 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Devuyst O., Pattaro C., The UMOD locus: Insights into the pathogenesis and prognosis of kidney disease. J. Am. Soc. Nephrol. 29, 713–726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tin A., Köttgen A., Genome-wide association studies of CKD and related traits. Clin. J. Am. Soc. Nephrol. 15, 1643–1656 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wuttke M., et al. ; Lifelines Cohort Study; V. A. Million Veteran Program, A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat. Genet. 51, 957–972 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trudu M., et al. ; SKIPOGH team, Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat. Med. 19, 1655–1660 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gorski M., et al. , Meta-analysis uncovers genome-wide significant variants for rapid kidney function decline. Kidney Int. 99, 926–939 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devuyst O., Olinger E., Rampoldi L., Uromodulin: From physiology to rare and complex kidney disorders. Nat. Rev. Nephrol. 13, 525–544 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Schaeffer C., Devuyst O., Rampoldi L., Uromodulin: Roles in health and disease. Annu. Rev. Physiol. 83, 477–501 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Malagolini N., Cavallone D., Serafini-Cessi F., Intracellular transport, cell-surface exposure and release of recombinant Tamm-Horsfall glycoprotein. Kidney Int. 52, 1340–1350 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Devuyst O., et al. , Autosomal dominant tubulointerstitial kidney disease. Nat. Rev. Dis. Primers 5, 60 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Olinger E., et al. , Clinical and genetic spectra of autosomal dominant tubulointerstitial kidney disease due to mutations in UMOD and MUC1. Kidney Int. 98, 717–731 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Gast C., et al. , Autosomal dominant tubulointerstitial kidney disease-UMOD is the most frequent non polycystic genetic kidney disease. BMC Nephrol. 19, 301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wuttke M., Köttgen A., Insights into kidney diseases from genome-wide association studies. Nat. Rev. Nephrol. 12, 549–562 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Walsh R., Tadros R., Bezzina C. R., When genetic burden reaches threshold. Eur. Heart J. 41, 3849–3855 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Katsanis N., The continuum of causality in human genetic disorders. Genome Biol. 17, 233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Groopman E. E., et al. , Diagnostic utility of exome sequencing for kidney disease. N. Engl. J. Med. 380, 142–151 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manolio T. A., et al. , Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karczewski K. J., et al. ; Genome Aggregation Database Consortium, The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581, 434–443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ioannidis N. M., et al. , REVEL: An ensemble method for predicting the pathogenicity of rare missense variants. Am. J. Hum. Genet. 99, 877–885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kidd K., et al. , Genetic and clinical predictors of age of ESKD in individuals with autosomal dominant tubulointerstitial kidney disease due to UMOD mutations. Kidney Int. Rep. 5, 1472–1485 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fletcher A. P., Neuberger A., Ratcliffe W. A., Tamm-Horsfall urinary glycoprotein. The chemical composition. Biochem. J. 120, 417–424 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selander-Sunnerhagen M., et al. , How an epidermal growth factor (EGF)-like domain binds calcium. High resolution NMR structure of the calcium form of the NH2-terminal EGF-like domain in coagulation factor X. J. Biol. Chem. 267, 19642–19649 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Bajaj K., et al. , Stereochemical criteria for prediction of the effects of proline mutations on protein stability. PLoS Comput. Biol. 3, e241 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carlier E., et al. , Disulfide bridge reorganization induced by proline mutations in maurotoxin. FEBS Lett. 489, 202–207 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Ittisoponpisan S., et al. , Can predicted protein 3D structures provide reliable insights into whether missense variants are disease associated? J. Mol. Biol. 431, 2197–2212 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandurangan A. P., Ochoa-Montaño B., Ascher D. B., Blundell T. L., SDM: A server for predicting effects of mutations on protein stability. Nucleic Acids Res. 45 (W1), W229–W235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ekici A. B., et al. , Renal fibrosis is the common feature of autosomal dominant tubulointerstitial kidney diseases caused by mutations in mucin 1 or uromodulin. Kidney Int. 86, 589–599 (2014). [DOI] [PubMed] [Google Scholar]
- 28.Kirby A., et al. , Mutations causing medullary cystic kidney disease type 1 lie in a large VNTR in MUC1 missed by massively parallel sequencing. Nat. Genet. 45, 299–303 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Genovese G., et al. , Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329, 841–845 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mallawaarachchi A. C., Furlong T. J., Shine J., Harris P. C., Cowley M. J., Population data improves variant interpretation in autosomal dominant polycystic kidney disease. Genet. Med. 21, 1425–1434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piret S. E., et al. , A mouse model for inherited renal fibrosis associated with endoplasmic reticulum stress. Dis. Model. Mech. 10, 773–786 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaeffer C., Merella S., Pasqualetto E., Lazarevic D., Rampoldi L., Mutant uromodulin expression leads to altered homeostasis of the endoplasmic reticulum and activates the unfolded protein response. PLoS One 12, e0175970 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown M. K., Naidoo N., The endoplasmic reticulum stress response in aging and age-related diseases. Front. Physiol. 3, 263 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gray V. E., Hause R. J., Luebeck J., Shendure J., Fowler D. M., Quantitative missense variant effect prediction using large-scale mutagenesis data. Cell Syst. 6, 116–124.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dvela-Levitt M., et al. , Small molecule targets TMED9 and promotes lysosomal degradation to reverse proteinopathy. Cell 178, 521–535.e23 (2019). [DOI] [PubMed] [Google Scholar]
- 36.Gibson G., Rare and common variants: Twenty arguments. Nat. Rev. Genet. 13, 135–145 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minikel E. V., et al. ; Exome Aggregation Consortium (ExAC), Quantifying prion disease penetrance using large population control cohorts. Sci. Transl. Med. 8, 322ra9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Flannick J., et al. , Assessing the phenotypic effects in the general population of rare variants in genes for a dominant Mendelian form of diabetes. Nat. Genet. 45, 1380–1385 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Althari S., et al. , Unsupervised clustering of missense variants in HNF1A using multidimensional functional data aids clinical interpretation. Am. J. Hum. Genet. 107, 670–682 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Estrada K., et al. ; SIGMA Type 2 Diabetes Consortium, Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA 311, 2305–2314 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cruchaga C., et al. ; Alzheimer’s Research UK (ARUK) Consortium, Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature 505, 550–554 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bollée G., et al. , Phenotype and outcome in hereditary tubulointerstitial nephritis secondary to UMOD mutations. Clin. J. Am. Soc. Nephrol. 6, 2429–2438 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gong K., et al. , Autosomal dominant tubulointerstitial kidney disease genotype and phenotype correlation in a Chinese cohort. Sci. Rep. 11, 3615 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zaucke F., et al. , Uromodulin is expressed in renal primary cilia and UMOD mutations result in decreased ciliary uromodulin expression. Hum. Mol. Genet. 19, 1985–1997 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Friedman D. J., Pollak M. R., APOL1 nephropathy: From genetics to clinical applications. Clin. J. Am. Soc. Nephrol. 16, 294–303 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whiffin N., et al. , Using high-resolution variant frequencies to empower clinical genome interpretation. Genet. Med. 19, 1151–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou X., et al. , Exploring genomic alteration in pediatric cancer using ProteinPaint. Nat. Genet. 48, 4–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao J., et al. , Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cerami E., et al. , The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taliun D., et al. ; NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium, Sequencing of 53,831 diverse genomes from the NHLBI TOPMed program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kopanos C., et al. , VarSome: The human genomic variant search engine. Bioinformatics 35, 1978–1980 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Plotkin M., et al. , A uromodulin mutation drives autoimmunity and kidney mononuclear phagocyte endoplasmic reticulum stress. Am. J. Pathol. 190, 2436–2452 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu M., et al. , Novel UMOD mutations in familial juvenile hyperuricemic nephropathy lead to abnormal uromodulin intracellular trafficking. Gene 531, 363–369 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Raffler G., Zitt E., Sprenger-Mähr H., Nagel M., Lhotta K., Autosomal dominant tubulointerstitial kidney disease caused by uromodulin mutations: Seek and you will find. Wien. Klin. Wochenschr. 128, 291–294 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix. Previously published data were also used for this work. Data presented here were derived from the following resources available in the public domain: the gnomAD version 2.1.1 (https://gnomad.broadinstitute.org/) and Ensembl (release 100; https://useast.ensembl.org/index.html). The International ADTKD Cohort and the UMOD in vitro scoring have also been published (11, 20).