Significance
The ability to repair mutations in situ is a large unmet need for neurological disorders, such as Rett syndrome, a disorder due to sporadic mutations in the gene encoding Methyl CpG binding protein 2 (MECP2). In this study, we find that a targeted RNA-editing approach effectively repairs a patient mutation by restoring wild-type MeCP2 protein expression and function in the mouse model. As a result, longevity is greatly improved and respiratory dysfunction, a hallmark phenotype among patients, is rescued to wild-type levels. This study represents a proof-of-concept for use of in vivo targeted RNA editing to attenuate disease phenotypes in mouse models for human neurological disorders.
Keywords: neurological disorder, Rett syndrome, targeted RNA editing
Abstract
Rett syndrome is a neurological disease due to loss-of-function mutations in the transcription factor, Methyl CpG binding protein 2 (MECP2). Because overexpression of endogenous MECP2 also causes disease, we have exploited a targeted RNA-editing approach to repair patient mutations where levels of MECP2 protein will never exceed endogenous levels. Here, we have constructed adeno-associated viruses coexpressing a bioengineered wild-type ADAR2 catalytic domain (Editasewt) and either Mecp2-targeting or nontargeting gfp RNA guides. The viruses are introduced systemically into male mice containing a guanosine to adenosine mutation that eliminates MeCP2 protein and causes classic Rett syndrome in humans. We find that in the mutant mice injected with the Mecp2-targeting virus, the brainstem exhibits the highest RNA-editing frequency compared to other brain regions. The efficiency is sufficient to rescue MeCP2 expression and function in the brainstem of mice expressing the Mecp2-targeting virus. Correspondingly, we find that abnormal Rett-like respiratory patterns are alleviated, and survival is prolonged, compared to mice injected with the control gfp guide virus. The levels of RNA editing among most brain regions corresponds to the distribution of guide RNA rather than Editasewt. Our results provide evidence that a targeted RNA-editing approach can alleviate a hallmark symptom in a mouse model of human disease.
Targeted RNA editing is gaining attention as an alternative base-editing approach for improving symptoms in a variety of diseases (1, 2). Our approach, based on the λN Deaminase Domain (DD)-BoxB system (3), exploits the expression of the catalytic domain of a naturally occurring enzyme, adenosine deaminase acting on RNA 2 (ADAR2), one of two active RNA deaminases expressed to high levels in the brain. The catalytic domain deaminates adenosines to inosines (I) (4) that are usually read by the translational machinery as guanosines (G) (5–7). Native ADAR2 recognizes double-stranded RNA in its natural targets, the best characterized of which are ion channels, where editing within the coding sequence dramatically alters functional properties (8). To target ADAR2-mediated editing to disease mutations requires either expression of guide RNA to recruit the endogenous enzyme (9–15) or, alternatively, virally mediated coexpression of guide RNA, antisense to the sequence containing the mutation, and an engineered ADAR2 catalytic domain that recognizes the hybrid guide RNA (3, 16–20). Most viral studies have capitalized on delivering a mutant hyperactive form of the ADAR2 catalytic domain (21) that leads to efficient RNA editing of guide-directed ADAR2 targets. While targeted editing, in either mode, has demonstrated success in improving cellular phenotypes, no studies have shown improvement of disease symptoms in vivo.
We have been testing the potential of targeted RNA editing to repair patient transcript Guanosine-to-Adenosine (G > A) mutations in mouse models of Rett syndrome, a major genetic cause of intellectual disability in females (22). Rett syndrome is caused by de novo mutations in the gene encoding the X-linked repressor, Methyl CpG binding protein 2 (MECP2) (23). After achieving early developmental milestones, affected females lose speech and purposeful hand motions, and acquire other hallmark neurological abnormalities, such as respiratory irregularities with apneas and seizures (24). Hemizygous males with MECP2 mutations are affected more severely, usually not surviving past the age of 2 y (25). Approximately 45% of Rett syndrome-causing mutations in RNA that are amenable to repair by targeted RNA editing are G > A missense and amber nonsense mutations, or Cytosine > Uracil opal mutations causing stop codons (26). C-terminal truncations resulting from premature stop codons are also potentially amenable to this approach.
Mice expressing Rett syndrome patient mutations exhibit pronounced motor phenotypes reminiscent of features seen in patients, including hallmark respiratory dysfunction, general hypoactivity, and seizures (27, 28). As in human patients, males are affected more severely than females and die prematurely. Although MeCP2 is a repressor (29, 30), its loss-of-function has not revealed a specific druggable target thus far (28). For this reason, and the potential for MeCP2 overexpression toxicity by gene replacement therapy, we previously performed a proof-of-concept study using targeted RNA editing (31). In this study, we introduced adeno-associated virus (AAV), coexpressing an engineered hyperactive catalytic ADAR2 domain (21) and Mecp2 guide optimized for this enzyme, directly into the hippocampus of a mouse model of Rett syndrome. These mice express a rare patient G > A mutation that causes classic Rett syndrome due to diminished binding to chromatin (31). Expression of the editing components resulted in efficient repair of Mecp2 RNA and chromatin association across three different populations of neurons in the hippocampus (17, 31). This study indicated that targeted RNA editing was efficacious in nondividing cells in vivo, extending previous studies illustrating the viability of this approach to repair RNA in cultured nondividing neurons and dividing cells (3, 10, 16–19, 32). However, the study left open the questions of whether targeted RNA editing could be sufficient to mitigate a hallmark Rett symptom in vivo and whether a different Rett syndrome mutation, with a different adenosine context, was amenable to targeted RNA editing with our system.
Here, we have addressed both questions by generating a unique Rett syndrome mouse model expressing the patient mutation, MECP2G311A, which creates a stop codon resulting in lack of MeCP2 protein. Using systemic injection of AAV encoding an engineered wild-type ADAR2 catalytic domain (Editasewt) and Mecp2-targeting guide, we tested whether targeted RNA editing can restore mouse MeCP2 protein and function, and improve Rett-like symptoms in the mutant mice. Control mutant mice were injected with AAV expressing the Editasewt and a nontargeting gfp guide. We also tested in wild-type mice how the expression pattern of the gfp guide RNA under control of the U6 promoter compares to that of Editasewt, and whether wild-type mice injected with the Mecp2-targeting virus exhibit premature lethality or overt abnormal phenotypes compared to noninjected mice.
Results
The MECP2G311A Rett Syndrome Mouse Model.
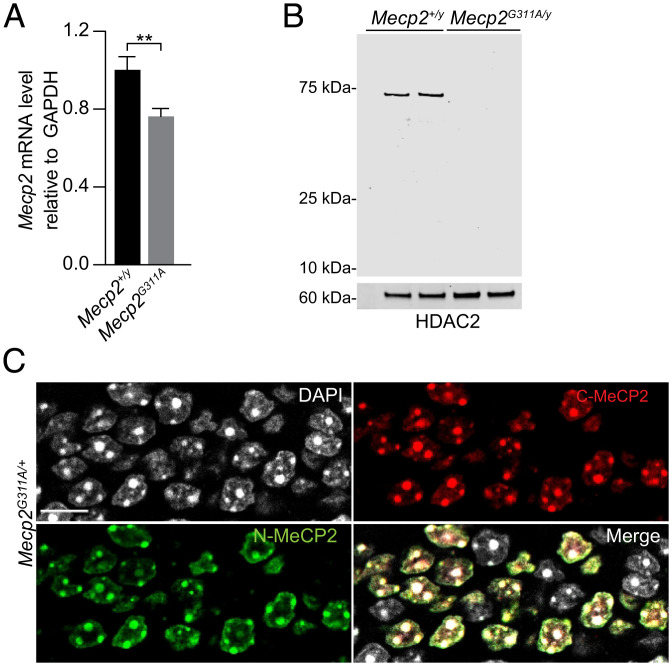
We chose the MECP2G311A mutation, MECP2W104X, because it causes Rett syndrome in humans and the mutant adenosine is in an ideal codon context (UAG) in the RNA for deaminase activity by endogenous ADAR2 (33). The mutation causes a truncated protein lacking the DNA binding and repressor domains. In transfection analyses, we showed previously that targeting the endogenous ADAR2 catalytic domain to cells expressing this mutated Mecp2 RNA resulted in high editing efficiency (76% of the target adenosines edited compared to controls) (17). To generate the mouse model, we used CRISPR/Cas9 methodology (SI Appendix, Fig. S1). Mecp2G311A RNA levels were reduced slightly to 76% of the levels in wild-type mice (Mecp2+/y) (Fig. 1A), indicating sufficient RNA levels for testing RNA-editing efficiency. In contrast, MeCP2 protein was not detected by Western blotting of brain lysates from the mutant mice using an antibody directed against the amino terminus (Fig. 1B). The specificity of N- and C-termini–directed MeCP2 antibodies was validated by immuno-labeling brain sections from female Mecp2G311A/+ mice (Fig. 1C). Both antibodies labeled the same nuclei, in ∼50% of the cells, as predicted by random X chromosome inactivation in the heterozygote.
Fig. 1.
Analysis of Mecp2 RNA and protein in the mutant Mecp2G311A mouse model. (A) Real-time qRT-PCR of Mecp2 RNA in indicated genotypes. n = 3 mice per genotype. Histogram values are mean ± SD, **P < 0.01 using Student’s t test. (B) Western blot of nuclear lysates prepared from brains of postnatal day 56 male mice. n = 2 mice per genotype. The blot was probed with antibodies directed against the N terminus of MeCP2 and HDAC2 (for the loading control). (C) Confocal images acquired from a dentate gryus section of a female Rett syndrome mouse showing mosaic staining for MeCP2 detected by immunolabeling for the N and C termini of MeCP2. (Scale bar, 10 µm.)
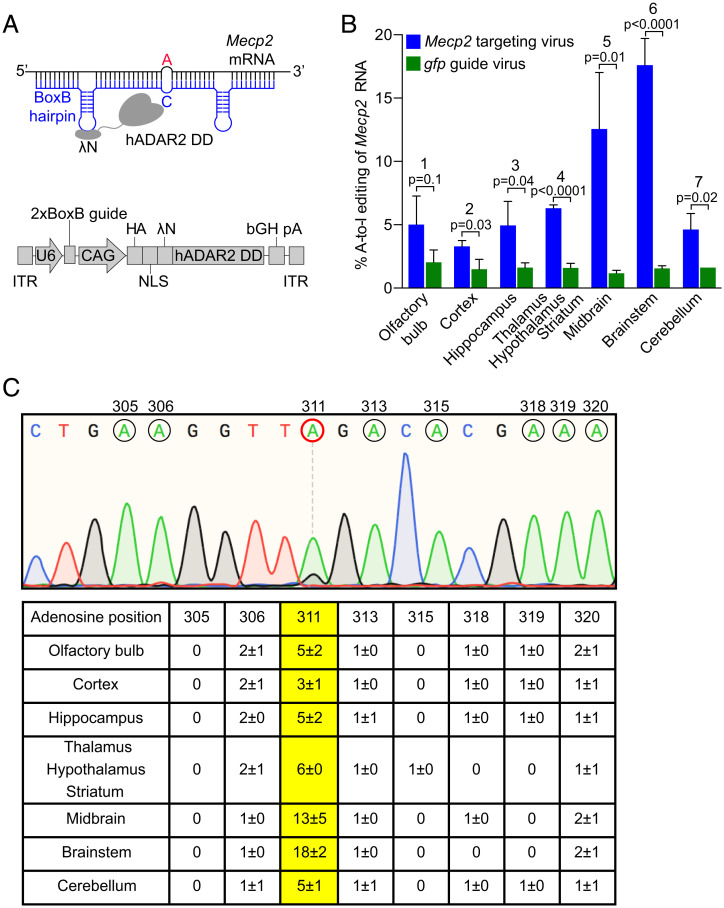
To target RNA editing to the Mecp2G311A mutation in our mouse model, we used an adaptation of the λN DD-BoxB system that we have used previously (17, 31). The engineered editing enzyme, Editasewt, consists of a fusion between the wild-type catalytic subunit of human ADAR2 and the bacteriophage λN peptide (Fig. 2A). We had previously surmised that addition of three copies of a nuclear localization signal (NLS) would be helpful in promoting on-target editing, because ADAR2 is localized normally in the nucleus (17). That the NLS would also limit off-target editing was tested specifically in Vallecillo-Viejo et al. (32), who found similar on-target editing efficiencies with and without the NLS, but dramatically decreased global off-target editing with the NLS addition (32). To identify cells expressing Editasewt in vivo, two copies of an HA epitope were included at the amino terminus. The guide RNAs each contain two copies of the RNA hairpin recognized by the bacteriophage λN peptide (BoxB hairpin) and 46 nucleotides antisense to either Mecp2 or gfp RNA (Fig. 2A). Note that our mice do not contain a gfp transgene. In the Mecp2 guide RNA, a cytosine is placed across from the target adenosine to increase on-target editing efficiency (34). In this system, the hybrid enzymes and guides are both encoded in the same AAV backbone. The enzyme and guide are under control of the cytomegalovirus immediate early enhancer chicken β-actin promoter (CAG) and polymerase III human U6 promoter, respectively. For simplicity, hereafter we refer to the viruses as either Mecp2-targeting virus (encodes both Editasewt and Mecp2 guide RNA) or gfp guide virus (encodes both Editasewt and gfp guide RNA) (Fig. 2A). All editing and MeCP2 functional studies were performed in male mice. This focus avoided the complications of mosaicism in the female model for our molecular analyses of editing efficiencies across brain regions.
Fig. 2.
Essential components of the targeted RNA editing strategy and resulting on-target editing frequencies at Mecp2G311A. (A, Upper) The hybrid Editasewt consists of the bacteriophage λN RNA binding peptide, fused to the native human ADAR2 deaminase domain (hADAR2 DD). A guide RNA (blue) contains 46 nucleotides complementary to Mecp2 mRNA (black) and two copies of the BoxB hairpin from bacteriophage λ, recognized by the λN RNA binding peptide in Editase. As a control for the specificity of editing, a 2xBoxB guide containing sequences complementary to gfp RNA (gfp guide) was substituted for the Mecp2-targeting guide. To increase editing efficiency, opposite of the target A (red), a C is introduced into the guide RNA. (Lower) Schematic of AAV encoded RNA editing components. U6, human U6 promoter; CAG, CMV enhancer, chicken β-actin promoter. Two copies of an HA-epitope tag, and three copies of an NLS, are added to the amino terminus of the Editasewt enzyme. (B) Quantification of on-target RNA-editing percentages based on Sanger sequencing (mean ± SD, n = 3 mice per condition). Bracketed P values above each region represent editing percentages compared between the two viral conditions (blue and green). Statistics are by unpaired two-tailed t test. Numbers above each brain region refer just to the mice injected with Mecp2-targeting virus (blue), and indicate pairwise comparisons of editing percentages across the brain regions. (1, 2; ns), (1, 3; ns), (1, 4; ns), (1, 5; **), (1, 6; ***), (1, 7; ns), (2, 3; ns), (2, 4; ns), (2, 5; ***), (2, 6; ****), (3, 4; ns), (3, 5; ***), (3, 6; ****), (3, 7; ns), (4, 5; *), (4,6; ****), (4, 7; ns), (5, 6; *), (5, 7; **), (6, 7; ****). ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, by repeated-measures one-way ANOVA and Tukey’s multiple comparisons test. (C, Upper) Representative Sanger sequencing chromatogram of cDNA from brainstem of a mouse injected with the Mecp2-targeting virus. Adenosines within the guide region of Mecp2 are indicated by black circles with the base number listed above. Target A is encircled in red. (Lower) Quantification of RNA editing percentages (mean ± SD, n = 3 mice per condition) for all adenosines within the guide region of Mecp2 across each brain region. Editing frequencies of the target A are in yellow.
Targeted RNA Editing Repairs MeCP2 Expression in the Brainstem.
RNA-editing components were delivered systemically by retro-orbital injection of viral vectors into juvenile male mice between postnatal days (P) 28 and 35. We tested for efficacy of our paradigm by determining RNA-editing efficiency in different brain regions by Sanger sequencing, 4 wk postinjection. For all brain regions, on-target RNA-editing efficiencies were significantly higher in the mice injected with the Mecp2-targeting virus compared to mice injected with the gfp virus (Fig. 2B). In the former mice, brainstem exhibited the highest on-target editing efficiency (18 ± 2%), followed by midbrain (13 ± 5%) (Fig. 2 B and C). The high on-target editing efficiencies in these brain regions were not reflected in higher bystander off-target editing percentages within the Mecp2-targeting guide region, which were not significant (Fig. 2C).
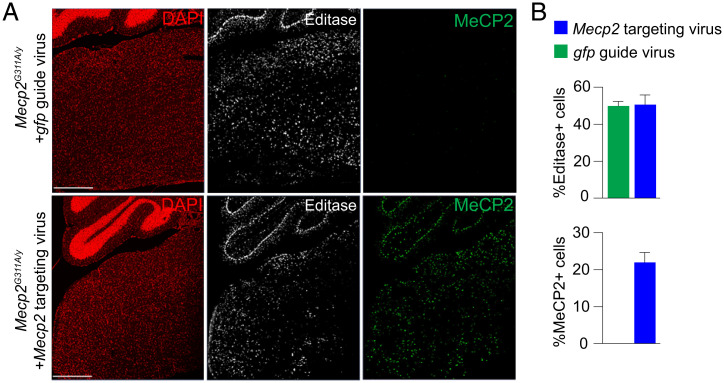
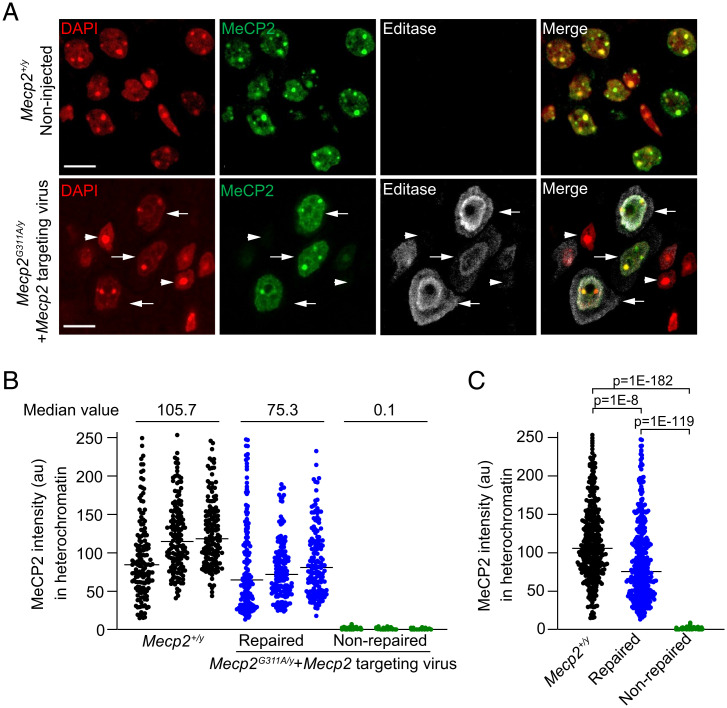
We predicted that RNA repair, in cells expressing the Editasewt and Mecp2-targeting guide, would result in recovery of MeCP2 protein expression in individual cells within the brainstem. In contrast, cells that expressed Editasewt and the gfp guide RNA would be predicted to lack MeCP2 protein. In immunolabeling experiments, using an antibody directed against the HA epitope in the enzyme, we confirmed that Editasewt expression was widespread in cells throughout the brainstem (Fig. 3A) and present in equal numbers of cells in both viral conditions (Fig. 3B). Consistent with the RNA-editing results, within the brainstem, MeCP2 protein was detected only in cells from the mice injected with Mecp2-targeting virus (Fig. 3). As we have done previously (31), we used the association with heterochromatin as a proxy for recovery of the chromatin binding ability of the MeCP2 protein. Heterochromatin is enriched in methylated cytosines in CG dinucleotides, a major site of MeCP2 binding (35–38), which in mouse cells is visualized as discrete nuclear puncta using DAPI staining (Fig. 4A). As expected, in the brainstem of noninjected wild-type mice, MeCP2 immunolabeling was associated with the DAPI puncta in every cell (Fig. 4A). In the mutant mice injected with the Mecp2-targeting virus, one fraction of cells had nuclei showing clear evidence for MeCP2 association with DAPI puncta, and these nuclei were immunolabeled for Editasewt (Fig. 4A). Another fraction showed no evidence of Editasewt expression and lacked MeCP2 expression (Fig. 4A). What is causal to the perimembrane distributions of Editasewt is not known.
Fig. 3.
In vivo Mecp2-targeted RNA editing leads to expression of AAV-encoded Editasewt and rescues endogenous MeCP2 protein expression. (A) Confocal images of sections of brainstem and a portion of cerebellum stained for Editasewt and MeCP2 protein. DAPI staining indicates nuclei. (Upper) Mecp2G311A/y mice injected with nontargeting gfp guide virus. (Lower) Mecp2G311A/y mice injected with Mecp2-targeting virus. Editasewt is expressed in both conditions while MeCP2 immunolabeled nuclei are detected only in the mice injected with Mecp2-targeting virus. (Scale bar, 500 µm.) (B, Upper) Percent Editase immunopositive cells in both viral conditions. (Lower) Percent MeCP2 immunopositive cells in both conditions. Data are relative to total number of DAPI+ cells. n = 3 mice each condition.
Fig. 4.
Mecp2-targeted RNA editing rescues MeCP2 protein expression and function in the brainstem. (A) Confocal images of nuclei in brainstem sections from noninjected wild-type Mecp2+/y male mice (Upper) and mutant Mecp2G311A/y mice injected with the Mecp2-targeting virus (Lower). DAPI staining defines nuclear compartment and heterochromatic foci. Arrows indicate repaired cells expressing MeCP2 and Editasewt; arrowheads indicate nonrepaired cells lacking both MeCP2 and Editasewt expression. Editasewt intensities appear greater near the nuclear membrane and in the cytoplasm. (Scale bar, 10 µm.) (B) Quantification of immuno-labeled MeCP2 associated with heterochromatic puncta for three individual wild-type mice and three mutant mice injected with the Mecp2-targeting virus. Each dot represents the mean of MeCP2 intensities within all heterochromatic foci in an individual nucleus. Median values are indicated for the comparisons; n = 150 cells per mouse. Note that the repaired and nonrepaired nuclei are in the same section of an injected mutant mouse. The criterion for a nonrepaired nucleus is based on estimates from sections from noninjected mutant mice. (C) Histogram showing pooled results for the three mice associated with each condition (n = 450 cells per condition). Horizontal lines represent the median intensities. Statistical comparisons of distributions are by Kruskal–Wallis and Dunn’s multiple comparisons tests with a Bonferroni correction.
We next estimated the amount of functional repair in the brainstem, comparing “repaired” (associated with heterochromatin) and “nonrepaired” (no heterochromatin association) cells in the same brainstem sections. These estimates were also compared to estimates in cells from wild-type mice representing the maximum values for this metric. In each of three mice injected with the Mecp2-targeting virus, the MeCP2 immunofluorescence of nonrepaired cells in the section was at background levels, determined from previous measurements in noninjected null cells (Fig. 4B) (median intensity 0.1). In repaired cells and cells from wild-type mice, we detected a broad distribution of MeCP2 immunofluorescence intensities associated with the heterochromatic puncta that were above background in each case (Fig. 4B) (median intensities 75.3 and 105.7, respectively). The broad distributions are consistent with known cell-type–specific differences in MeCP2 expression in vivo (39, 40). When the immunofluorescence intensities in cells from all three mice were combined, statistical comparisons indicated that repaired cells and cells from wild-type mice were both highly different from nonrepaired cells (Fig. 4C). This analysis also confirmed levels of MeCP2 association with heterochromatin that were intermediate to wild-type and nonrepaired mutant cells. It is notable that the point spreads for wild-type and repaired cells are very similar (Fig. 4C), with some repaired cells showing nearly equivalent MeCP2 intensities in heterochromatin as the top scoring wild-type cells.
Given that repaired MeCP2 protein should be wild-type, we then sought to determine whether the reduced level of MeCP2 immunofluorescence in the heterochromatin in the repaired cells compared to wild-type cells, was due to lower levels of protein, or to weaker association with the heterochromatin. To this end, we measured MeCP2 intensities in the nucleoplasm and within the heterochromatin. In 44 cells measured for each condition, the median nucleoplasm intensities were 23.4 and 42.4 arbitrary units (au) for repaired and wild-type nuclei, respectively; the median heterochromatin intensities were 92.8 and 111.1 au for repaired and wild-type nuclei, respectively. Both compartments were lower in the repaired nuclei, indicating less MeCP2 protein in the repaired nuclei. Thus, our interpretation is that the reduced level of MeCP2 immunofluorescence in heterochromatin of repaired cells reflects less total MeCP2 protein when compared to wild-type cells.
Prolonged Survival and Improved Respiratory Function in Mice Injected with the Mecp2-Targeting Virus Compared to Control Mice.
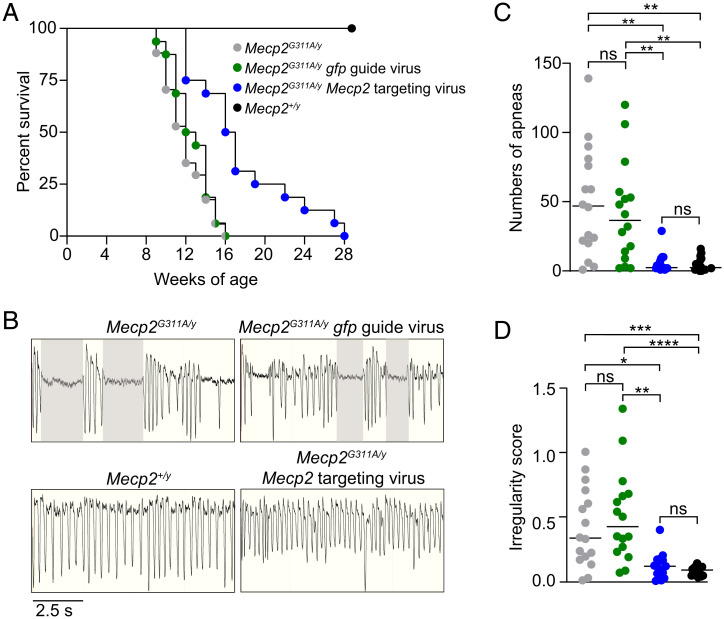
We first compared survival curves among four groups: mutant Mecp2G311A/y mice, mutant mice injected with the Mecp2-targeting guide, mutant mice injected with gfp guide, and wild-type mice. As expected, over a 28-wk time course, none of the wild-type mice died (Fig. 5A). For the other three groups, the survivals differed markedly from wild-type. Specifically, 50% of the noninjected mutant mice, as well as mice injected with the control gfp guide virus, died at 11 and 12 wk, respectively (Fig. 5A), consistent with other null Mecp2 mutant mice (41). In contrast, survival was prolonged in mice injected with the Mecp2-targeting virus. In these mice, 50% survived to 16 wk, with six mice surviving beyond the last living mutant mice and mice injected with the gfp guide virus (Fig. 5A).
Fig. 5.
MeCP2 targeted RNA editing prolongs survival and alleviates abnormal respiratory functions in the Mecp2G311A/y mice. (A) Kaplan–Meier survival curves for male mice with indicated genotypes and guide expression. Noninjected Mecp2G311A/y mice (gray, n = 16), Mecp2G311A/y mice injected with 1E12 viral genomes of the gfp guide virus (green, n = 16), Mecp2G311A/y mice injected with 1E12 of the Mecp2-targeting virus (blue, n = 16), and noninjected Mecp2+/y mice (black, n = 16). (B) Representative whole-body plethysmography traces of mice with indicated genotypes and viral expression condition at 8 wk. Shaded areas represent apneas defined as exceeding 1 s (Materials and Methods). (C) Total number of apneas for the four conditions. (D) Irregularity scores for the four conditions. Colors and n values for genotypes and conditions are the same for A, C, and D. Data were acquired during a 30-min test interval. Horizontal lines represent the median values. ns, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 by Kruskal–Wallis Test and Dunn’s multiple comparisons test.
We next compared respiratory function among the four groups. Respiration was tested specifically because of the high RNA-editing efficiency and MeCP2 functional recovery in the brainstem, a region that is tightly linked to respiration and represents a severe phenotype in female Rett syndrome patients. A high apneic frequency (prolonged pauses) was observed in both mutant mice and mice injected with gfp guide virus (Fig. 5 B and C). The frequencies were not significantly different from one another, but both were significantly higher than the apneic number in mice injected with the Mecp2-targeting virus (Fig. 5 B and C). The mutant mice and mice injected with gfp guide virus equally exhibited irregularities in breathing patterns, expressed as irregularity scores (Fig. 5 B and D). In contrast, the mice injected with Mecp2-targeting virus showed irregularity scores not significantly different from the scores in wild-type mice and were reduced significantly compared to mutant mice and mice injected with the gfp-targeting guide (Fig. 5 B and D).
Taken together, the above results indicated on-target editing efficiency in Mecp2 RNA in the brainstem that manifests in significant rescue of MeCP2 function and alleviation of severe respiration abnormalities. However, on-target editing is usually associated with some degree of off-target editing outside the guide region, which can cause lethality (20). To identify the off-target editing sites in our mice, we performed next-generation sequencing on the brainstem of mutant mice injected with the Mecp2-targeting virus or the gfp guide virus, as well as on the brainstem of noninjected mutant mice (SI Appendix, Fig. S2), using the same RNA that was used in the Sanger sequencing. We also performed exome sequencing to exclude single nucleotide polymorphisms and to exclude endogenous RNA-editing events. The mean on-target editing percentage in mice injected with the Mecp2-targeting virus was ∼15%, corresponding well to the ∼18% on-target editing efficiency from Sanger sequencing, and lending confidence to the next-generation sequencing.
We identified the off-target editing sites in the brainstem of mice injected with the Mecp2-targeting virus by first removing all sites where the percent editing was not statistically different from noninjected mutant mice. This comparison was intended to identify changes caused specifically by the expression of the Mecp2-targeting virus. No off-target editing sites were identified within the Mecp2 transcript. After removing sites that were unchanged between samples from Mecp2 guide-injected and noninjected mice, we identified 350 off-target editing sites (SI Appendix, Fig. S2A). We then asked whether the off-target RNA editing sites in mice injected with the Mecp2 guide occurred at greater frequency compared to samples from mice injected with the gfp guide virus. For this analysis, we performed a pairwise comparison between the off-target sites in mice injected with each virus, after each site was compared to the same site in the mutant mice. The results indicated that the mean editing percentage at each individual off-target site was well-correlated between the two viral conditions (SI Appendix, Fig. S2A) (r = 0.97, P < 0.0001). This result supports previous studies indicating that off-target editing is largely due to naturally occurring double-stranded regions edited by an unguided editing enzyme (18, 31, 32). It is possible that the endogenous ADAR enzymes, ADAR1 and ADAR2, might be contributing to new off-target editing sites in the mice injected with Mecp2-targeting virus. A formal test of this possibility will require repeating the study in Mecp2G311A mice crossed with viable rescued mice deficient in ADAR1 and ADAR2 (42).
To predict the impact of RNA-editing and annotate the sites genome-wide, we used Variant Effect Predictor (SI Appendix, Supplemental Methods). Consistent with activity by endogenous ADAR2, which primarily serves as a fine-tuning mechanism for protein function (43), off-target sites were associated with graded editing activity and ≥70% of the off-target sites were edited to less than 25% and located throughout exonic and intronic regions (SI Appendix, Fig. S2B). To evaluate the biological impact of the off-target editing, we tested for premature lethality that has been reported previously for expression of the hyperactive ADAR2 protein (20). To this end, we injected the Mecp2-targeting virus systemically, by retro-orbital injection, into 4-wk-old wild-type mice. Four weeks postinjection, the mice expressed Editasewt throughout the brain (SI Appendix, Fig. S2C), indicating viral distribution and promoter activity as efficient as in the Mecp2-injected mutant mice. Like the mutant mice injected with the Mecp2-targeting virus, the wild-type mice injected with the same virus also lacked bystander off-target editing (SI Appendix, Fig. S2D). In terms of symptoms, the injected wild-type mice were indistinguishable from noninjected wild-type mice in terms of body weight (SI Appendix, Fig. S2E), as well as in aggregate scores for the observed general health parameters: mobility, gait, general condition, hindlimb clasp, and tremor (SI Appendix, Fig. S2F). The oldest wild-type injected mice were 18-mo old at the time of this submission and these metrics have remained the same as noninjected wild-type mice.
Multiplex BaseScope for In Situ Hybridization and Immunolabeling Reveals High Brainstem Expression of gfp Guide RNA under Control of the U6 Promoter.
We found the brainstem RNA-editing efficiency (18%) (Fig. 2) and a similar percentage of MeCP2+ cells (22%) (Fig. 3) intriguing, given the much higher percentage of Editase+ cells (∼50%) (Fig. 3) expressed in the same virus (44). We hypothesized that the lower levels of RNA editing might be due to differences in the cell-type–specific promoter activities driving the guide and enzyme. To test this idea, we used BaseScope with in situ hybridization (ISH) of guide RNA multiplexed with immunolabeling analysis for Editasewt using the HA epitope to examine the respective distributions across brain regions. Because the question of brain region specificity of the promoters is of general importance, and to mitigate any effects due to the compromised health of the mutant mice injected with the Mecp2-targeting guide, we injected wild-type mice with the gfp guide virus that also expresses Editasewt. Use of the gfp is optimal because an Mecp2 antisense guide will hybridize to the high levels of endogenous Mecp2 RNA and prevent binding of the ISH probe to the guide RNA. However, a limitation of this approach is that ISH cannot distinguish between promoter activity or stabilization of the encoded RNA that is challenging to determine in vivo. If the brain region specificity of the gfp guide matches the brain region specificity of on-target editing programmed by the Mecp2-targeting guide, this would support the idea that U6 promoter activity is driving differences in editing efficiency, but is not conclusive.
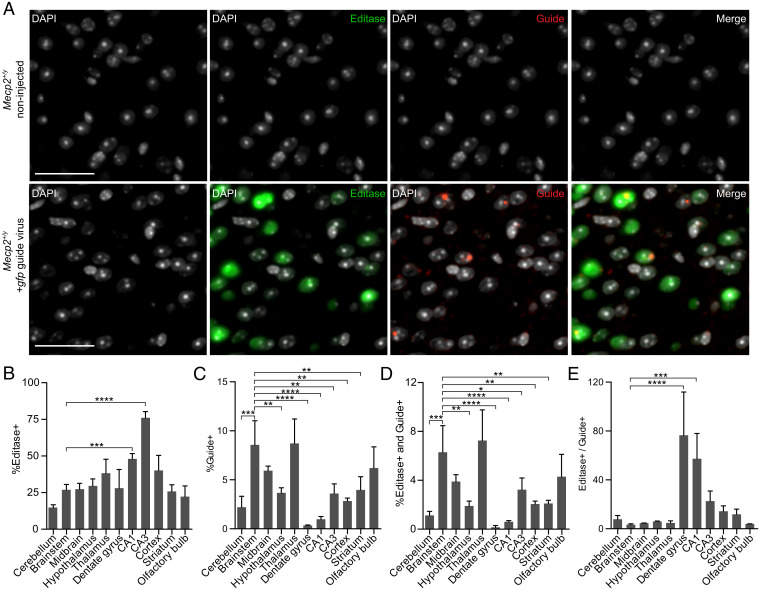
To compare Editasewt and gfp guide RNA distributions, brainstem sections from wild-type mice injected with the gfp guide virus, or sections from noninjected wild-type mice, were analyzed by multiplexed BaseScope 4 wk postinjection (8 wk of age). Representative images from this analysis are shown in Fig. 6A. The noninjected wild-type mice were, as expected, negative for both HA-Editasewt and guide, providing controls for the specificity of the probes (Fig. 6A). Cells in mice injected with gfp guide virus showed cells immunolabeled for both Editasewt and guide (Fig. 6A). However, expression of Editasewt and guide RNA was not uniform, evident by quantitative labeling representing an average over the entire brain regions imaged (Fig. 6B). As might have been predicted from the link between brainstem RNA editing and respiration results, the pattern of cells expressing guide was much more restricted than that of Editasewt expression (Fig. 6C). Specifically, brainstem and midbrain had two of the highest editing percentages, with widespread Editasewt expression throughout the regions, and a high number of guide+ cells. Furthermore, the pattern of cells expressing guide (Fig. 6C), and coexpressing both Editasewt and guide (Fig. 6D), was strikingly similar, with brainstem and midbrain again exhibiting among the highest percentages in each group. To further investigate whether the guide or Editasewt might be limiting overall RNA-editing repair in the brainstem, we computed expression of Editase and guide+ cells in other brain regions as well as the ratio of Editase+ to guide+ cells (Fig. 6E). The result of these comparisons suggested that the high editing efficiencies in brainstem and midbrain were best explained by the widespread guide expression in these regions. The hippocampal sectors had among the lowest editing efficiencies, with high numbers of Editase+ cells but among the lowest number of guide+ cells.
Fig. 6.
Multiplex BaseScope detection of cells expressing Editasewt and guide RNA across brain regions. (A) Representative images from the brainstem of a noninjected male Mecp2+/y mouse (Upper) and an Mecp2+/y mouse injected with the gfp guide virus (Lower). DAPI marks nuclei. (Scale bars, 50 µm.) (B–E) Percentages corresponding to Mecp2+/y mice injected with the gfp guide virus; n = 3 mice. (B) Percentage of cells expressing Editasewt. (C) Percentage of cells expressing gfp guide RNA. (D) Percentage of cells expressing both Editasewt and guide RNA. (E) Ratio of Editase+ to guide+ cells. The percentage (mean ± SD) of cells in each brain region is relative to the total number of DAPI+ cells. Total cells counted in each region are found in SI Appendix, Fig. S4. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. All comparisons lacking asterisks were not significant. Statistics were determined using a one-way ANOVA relative to brainstem values.
Because of the reported ability of PHP.eB to preferentially infect neurons (45), we examined the distribution of guide and Editasewt specifically in neurons. For this purpose, we used BaseScope for guide RNA multiplexed with Editasewt and the neuronal marker protein NeuN for immunolabeling neurons (SI Appendix, Fig. S3A). As might be expected, the fractional representation of neurons in each region (SI Appendix, Fig. S3B) did not correlate directly with cell number (SI Appendix, Fig. S4). For example, the dentate gyrus, CA1, and CA3 sectors in the hippocampus had the lowest overall cell number but the highest percentage of neurons (SI Appendix, Fig. S3B). Brainstem had a much higher total cell number, but the lowest number of neurons. Furthermore, the brain region distributions of total Editase+ cells relative to DAPI (Fig. 6B) were distinct from the distributions of Editase+ cells in neurons (SI Appendix, Fig. S3C). For example, the brainstem had ∼25% Editase+ cells, but only ∼55% of these were NeuN+, suggesting editing in some nonneuronal cell types. In contrast to Editase, the distributions of guide+ cells relative to DAPI (SI Appendix, Fig. S3C) were very similar to guide+ cells in neurons (SI Appendix, Fig. S3D). Additionally, the neuronal guide distribution (SI Appendix, Fig. S3D) was remarkably like the neuronal distribution of cells expressing both Editase and guide (SI Appendix, Fig. S3E). Together, these last two observations again suggest a role for guide expression in setting the upper limit of MeCP2 restoration and repair of the respiration phenotype.
Discussion
There has been accelerating interest in exploiting RNA editing as a therapeutic approach for human disease, but in vivo studies have been relatively few (13, 15, 20, 31, 46, 47). Apart from our previous study (31), all other studies have focused on editing target RNAs outside of the nervous system (13, 15, 20, 46, 47). None have addressed the ability to rescue mouse phenotypes. Here, we generated a Rett syndrome mouse model that expresses a disease-causing patient G > A mutation (Mecp2G311A) causing lack of MeCP2 protein. We used this model to test the ability of targeted RNA-editing components, delivered systemically by AAV PHP.eB, to repair the mutation and alleviate Rett-like phenotypes in the mutant mice.
The central behavioral findings using our approach were prolonged survival and alleviation of respiratory apneas and irregular breathing, the latter features typical of many female Rett syndrome patients (48). With respect to the alleviation of abnormal phenotypes in the mutant mice, the focus on the brainstem was prompted by the opportunity to quantitate phenotypic improvement through measurements of both breathing irregularities and apneas that are under control by the brainstem (48, 49). The 18% editing efficiency, although highest in the brainstem, might underestimate the actual level of per cell repair, as it reflects the composite editing percentages from many different cell types within this brain region. In particular, the editing percentage may be much higher in neurons, based on the higher numbers of cells coexpressing both Editase and guide in NeuN+ cells. In future, it should be possible to determine whether respiration rescue is due to editing repair in the well-defined centers of respiratory control in the brainstem (49), using the recovery of MeCP2 protein in the virally injected mice to guide the experiments. For other brain regions, multiplexed BaseScope analysis, using cell-specific markers other than general neuronal markers, could provide more information on cell-specific RNA editing as the field awaits the development of cell-specific RNA-editing methodology.
We found that the repaired nuclei exhibited a level of MeCP2 protein associated with heterochromatin that was lower than that in wild-type nuclei. However, the repair was improved significantly over that in the null cells. Indeed, some cells showed MeCP2 functional recovery equivalent to the highest measured levels in wild-type cells. The single-cell analysis also indicated that this high level of functional protein recovery for mice injected with Mecp2-targeting virus was higher than might have been predicted by Sanger and next-generation sequencing from the entire brain region. This finding is likely, again, because the sequencing data reflect the average of editing within a heterogeneous brain region. How much MeCP2 is required to achieve normal cell function across brain regions, and in how many cells, remains an important question that could perhaps begin to be addressed by comparing editing results in the Mecp2311G > A mutant using different titers of virus.
To our knowledge, the basis for the premature lethality in male mice expressing Mecp2 mutations is not known. Partial improvement, however, is not unexpected. Indeed, in Matagne et al. (50), after expression of wild-type MeCP2 delivered systemically by AAV, the respiration defect in Mecp2null/y mice was completely rescued with 10 to 20% corrected cells and mean survival time of 43 d. These results closely mirror our mean survival time of 35 d, with a similar percentage of edited cells and the correction of respiration abnormalities. It seems reasonable to propose that a higher editing efficiency per cell, distributed more widely across brain regions, as mentioned above, would improve survival times. Improvement in survival will also necessitate a better understanding of the persistence of editing efficiency throughout the lifetime of the mice, something we could not test in our study of mutant males. Because female Rett mouse models live a normal lifetime, the persistence of editing efficiency, even with the current editing efficiencies, can be resolved, as well as prove informative regarding maintenance of a normal respiratory pattern. Furthermore, the finding that wild-type male mice, injected with the Mecp2-targeting virus, showed no overt health problems or premature death by 18 mo, suggests that the wild-type cells in injected Mecp2G311A/+ heterozygotes will not contribute to toxicity despite off-target editing due to the enzyme. Supporting this idea, we found no evidence for bystander editing activity in the guide region of Mecp2 RNA in the wild-type injected mice.
Using the U6-driven nontargeting guide as a proxy for the Mecp2-targeting guide, our findings suggest individual roles played by guide RNA versus Editasewt expression in setting the levels of MeCP2 restoration. Both enzyme and guide showed uneven distribution among brain regions, but with important distinctions in expression levels. The most important distinction was the consistently high expression levels of both guide and Editasewt distributed throughout brainstem and midbrain, which had similar editing efficiencies. That guide rather than Editase may set the limit of rescued MeCP2 expression was reflected in the striking relationship in cells expressing guide to cells expressing both Editase and guide. This relationship held for both brain-wide cellular quantitation overall and for the smaller subset of NeuN+ neurons. The dependence on guide for setting the levels of MeCP2 restoration is further reflected in the Editase/guide ratio, where a trend was seen with the regions showing the highest numbers of Editase+ and guide+ cells having the lowest Editase/guide ratio and regions with the lowest numbers of Editase+ and guide+ cells tending to have the highest ratio, suggesting a strong dependency on guide for brain-wide differences in cell expression patterns.
To our knowledge, our study examining the distribution of steady-state levels of any RNA programmed by the U6 promoter in the brain is unique. Our study suggests that the U6 promoter is driving the pattern of guide expression, based on the correlation of the gfp guide-expression pattern with the pattern of Mecp2-targeted editing efficiencies in the brainstem and midbrain. We cannot exclude, however, cell-type–specific differences in the gfp and Mecp2 RNA half-lives within the entire brain. Given the technical challenges in measuring RNA half-lives at this level, a more immediate approach to resolving this issue may be to increase expression from the U6 promoter. Such optimization could perhaps be achieved by combining U6 with other polymerase III promoters (51). Increased guide expression would not be expected to markedly change the number of off-target editing events, because as we and others have found, off-target editing is independent of guide and due largely to intrinsic nontargeted editing by the ADAR2 catalytic domain itself (18, 31, 32).
Overall, our study supports the idea that a targeted RNA-editing approach has potential to improve symptoms in mouse models of Rett syndrome, and holds promise for other neurological disease models as well. Our study also raises questions, however, that require further investigation. For example, AAV-mediated targeted RNA editing is subject to the general limitations of therapeutic AAV delivery to the human brain. While AAV-mediated gene expression can endure for years in nonhuman primates (52), the need for development of new AAV serotypes, with improved widespread cell tropisms after systemic or more localized delivery methods into the brain, is still pressing. These issues are already recognized in academic and biotechnology laboratories across the world, so we anticipate the field will continue to advance at a rapid pace.
Materials and Methods
Animal Studies.
All animal procedures were approved by the Institutional Animal Care and Use Committees of Oregon Health and Science University and the University of Connecticut Health Center.
Mecp2311G > A Husbandry and Genotyping.
All mice were housed on a 12:12 light/dark cycle. Initially, female Mecp2G311A/+ mice on the mixed 129/SvEv × C57BL/6J background were mated to pure wild-type C57BL/6J male mice. The Mecp2G311A/+ mice are maintained by crossing to pure wild-type C57BL/6J mice. Genotyping was performed using primers specific for the Mecp2G311A allele. Separately, sex was determined using PCR primers specific for the X and Y chromosomes (SI Appendix, Table S1).
Viral Vector Preparation.
Plasmid DNA was prepared using the Qiagen EndoFree Plasmid Maxi Kit (Qiagen) prior to generating viral vectors. AAV vectors were produced by the University of Pennsylvania Vector Core and titered using digital-droplet PCR. Aliquots were frozen at −80 °C before use.
Viral Injections.
P28 to P35 mice were deeply anesthetized with 4% isofluorane (vol/vol) and placed on a prewarmed surface. For each animal, 100 μL of 1E12 viral genomes was injected into the retro-orbital sinus. Following injections, mice were monitored for pain and distress while recovering on a heated pad prior to being returned to their home cage.
Whole-Body Plethysmography of the Mecp2311G > A Mice.
Mice were back-crossed to C57BL/6J mice for at least seven generations prior to behavioral testing, consistent with previous studies (53, 54). Mice (8-wk old; 4-wk postinjection) were acclimated to the behavioral room for a minimum of 30 min in their home cage. Respiratory parameters were measured using a VivoFlow whole body plethysmography system (SCIReq Corporation) with a ventilation pump flow of 0.5 L/min and were recorded and analyzed using the IOX software package (SCIReq Corporation). The system was calibrated so that the input range was between −1,400 and +1,400 mL/s by injection of known amounts of air into the chamber. Individual nonanesthetized animals were placed in the VivoFlow chamber and their respiration was recorded for a 30-min acclimation period followed by a 30-min test period. The sampling rate was set to 1 kHz, flow threshold was set at 2.5 mL, and breaths occurring at a frequency of greater than 6 Hz were rejected as sniffing behavior. Apneas were defined as a total breath length (TTOT = time of inhalation + time of exhalation) of 1 s or greater. The irregularity score and variance were determined using the formula (TTOT n − TTOT n+1)/(TTOT n+1) where n and n+1 are individual sequential breaths.
RNA Preparation and qRT-PCR Analysis for Mecp2 RNA.
Total RNA was isolated from single hemispheres or individual brain regions using Qiazol reagent (Qiagen, cat# 79306) and the Qiagen miRNeasy Kit reagent (Qiagen, cat# 217004) according to the manufacturer’s instructions. RNA was reverse transcribed using the SuperScript III First-Strand Synthesis System (Invitrogen, cat# 18080051) and was primed using oligo dT. Samples were run in triplicate along with a standard curve using Sybr select master mix reagent (Thermo Fisher Scientific, cat# 4472918) and primers for Mecp2 and GAPDH RNA (SI Appendix, Table S1). The amount of Mecp2 mRNA was calculated relative to GAPDH RNA using the ΔΔ CT method. Primer sequences are listed in SI Appendix, Table S1.
Sanger Sequencing Analysis.
Endogenous Mecp2 cDNA was amplified and analyzed by Sanger sequencing, as previously described (31). Primer sequences are listed in SI Appendix, Table S1.
Immunostaining.
Immunostaining was performed as previously described (31). Briefly, mice were anesthetized using an intraperitoneal injection of 2,2,2-tribromoethanol (Sigma Aldrich, cat# T48402) and killed by transcardial perfusion of PBS pH 7.4, followed by 4% depolymerized paraformaldehyde. Brains were cryoprotected with sucrose, embedded in freezing medium, and stored at −80 °C. Sagittal whole-brain sections were cut at 20 μm using a cryostat and stored at −20 °C until staining. Sections underwent heat mediated antigen retrieval before blocking in PBST (0.1% Triton X-100 in PBS, pH 7.4) and 10% normal donkey serum (RRID: AB_2337258, Jackson Immunoresearch Laboratories) for 30 min at room temperature. Sections were incubated overnight at 4 °C with rabbit anti-MeCP2 (rabbit mab D4F3, RRID: AB_2143849, Cell Signaling 1:500) and mouse anti-HA (mouse mab clone HA.11, RRID: AB_2565336, BioLegend, 1:250) antibodies diluted in blocking buffer. Sections were washed with PBST and incubated for 1 h at room temperature with donkey anti-rabbit IgG CF568 (RRID: AB_1055718, Biotium 1:500) and donkey anti-mouse IgG CF488 (RRID: AB_10561327, Biotium, 1:500) diluted in blocking buffer. Sections were washed and incubated with 300 nM DAPI (Invitrogen, cat# D1306) in PBS for 20 min. After a final wash in PBS, sections were mounted using ProLong Gold antifade reagent (RRID: SCR_01596, Thermo Fisher Scientific).
BaseScope.
ISH and immunostaining.
Mice were anesthetized using an intraperitoneal injection of 2,2,2-tribromoethanol (Sigma Aldrich, cat# T48402) and killed by decapitation. Dissected mouse brain halves were fixed in freshly prepared 4% paraformaldehyde in 1× PBS for 24 h at room temperature and then transferred to 80% ethanol before tissue processing and embedding in paraffin. Sections of the paraffin-embedded brains were cut at a thickness of 5 μM, placed onto glass slides, and deparaffinized. Brain sections were treated with 3% H2O2 for 10 min and then underwent heat-induced epitope retrieval by boiling in ACDBio Target Retrieval Reagent (cat# 322000) for 15 min. The sections were then treated with ACDBio Protease III (cat# 322337; diluted 1:10 in PBS) for 20 min at 40 °C before incubation with the BaseScope probe against gRNA-GFP (cat# 1083211-C1) for 2 h. Probe signal amplification was performed with a custom ACDBio BaseScope Brown kit that allowed tyramide reaction (with Biotium CF568; 92173) to visualize probe staining. The slides were then washed in TBS-T (TBS with 0.1% Tween 20) and incubated with primary antibodies against the HA-tag (Mouse anti-HA; BioLegend 901501), NeuN (rabbit anti-NeuN; Cell Signaling D3S3I) and GFAP (chicken anti-GFAP; Milipore Sigma AB5541) overnight at room temperature. Slides were then washed with TBS-T and incubated with fluorescently tagged secondary antibodies against mouse IgG (ThermoFisher cat# SA5-10171; CF755), rabbit IgG (ThermoFisher cat# A31573; AF647), and chicken IgY (Milipore Sigma cat# SAB46000321; CF488) for 2 h at room temperature. Sections were washed with TBS-T, stained with DAPI, and mounted with Prolong Gold.
Imaging and analysis.
A Zeiss AxioScan Slide Scanner was used to acquire single plane images with the laser strengths for each channel adjusted independently to fall within the nonsaturating range. Images were analyzed using the HALO imaging analysis platform (Indica Labs; HighPlex FL v4.0.4). Individual brain regions were defined manually using a stereotaxic mouse brain atlas as a reference. Cells were defined by the presence of nuclear DAPI. Brain sections from noninjected animals served as a negative control and were used to threshold detection of fluorescence.
Western Blotting.
Whole-brain samples were collected, bisected down the midline with a razor blade, and single hemispheres were snap frozen in liquid nitrogen before storage at −80 °C. The details for preparing the nuclear brain extracts are described in SI Appendix. Protein concentrations from the nuclear extracts were measured using the BCA protein assay kit (Pierce Biotechnology, Thermo Fisher Scientific, cat# 23225) and 20 μg of protein lysate per sample was separated on NuPage 4–12% Bis–Tris gels (Thermo Fisher Scientific cat# NP0335) in NuPage MES-SDS running buffer (Thermo Fisher Scientific, cat# NP0002). Proteins were transferred onto a nitrocellulose membrane (GE Healthcare Life Sciences, cat# 45004001). The membrane was blocked with 5% nonfat milk in 1× TBS-T for 1 h and then incubated with a mouse antibody raised against the N terminus of MeCP2 (RRID: AB_477235, Sigma-Aldrich; 1:1,000) and a rabbit antihistone deacetylase 2 (HDAC2 RRID: AB_2533908, Invitrogen; 1:1,000) antibody overnight at 4 °C with continual rocking. After washing three times with 1× TBS-T, membranes were incubated with anti-rabbit IgG DyLight 680 (RRID: AB_614946, Thermo Fisher Scientific; 1:10,000) and anti-mouse IgG Dylight 800 (RRID: AB_1660927, Rockland; 1:10,000) diluted in 5% nonfat milk in 1× TBS-T for 1 h. After secondary antibody incubation, membranes were washed three times in 1× TBS-T before being imaged using the Odyssey Imaging System (LI-COR Biosciences).
Quantification and Statistical Analysis.
All statistical tests, with exception of the whole transcriptome analysis, were performed using GraphPad v9.0 software (RRID: SCR_002798).
Supplementary Material
Acknowledgments
We thank Gabe Haw and Erin Fonseca for mouse husbandry and pilot studies; the laboratories of Adrian Bird (Wellcome Trust Centre for Cell Biology, University of Edinburgh), Michael E. Greenberg (Harvard Medical School), and Paul Brehm (Oregon Health and Science University), and members of the G.M. laboratory, for helpful discussions and encouragement throughout the study; Paul Brehm for critical reading of the manuscript; and Guanming Wu and Jimmy Kelly for help with statistics. This work was funded by NIH Award NS110868 (to G.M.) and a grant from the Rett Syndrome Research Trust (to J.R.S.).
Footnotes
Reviewers: Y.H., Yale University School of Medicine; and H.Z., Baylor College of Medicine.
Competing interest statement: G.M. and J.R.S are coinventors on a pending patent licensed by Oregon Health and Science University to Vico Therapeutics on the therapeutic use of RNA editing. G.M. is a cofounder and shareholder of Vico Therapeutics. Vico Therapeutics does not stand to financially gain from this work. All other authors declare no conflicts of interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2206053119/-/DCSupplemental.
Data Availability
Whole-exome DNA sequencing and RNA sequencing data have been deposited in the Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA849938) (55). All study data are included in the article and/or SI Appendix.
References
- 1.Reardon S., Step aside CRISPR, RNA editing is taking off. Nature 578, 24–27 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Bhakta S., Tsukahara T., Artificial RNA editing with ADAR for gene therapy. Curr. Gene Ther. 20, 44–54 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Montiel-Gonzalez M. F., Vallecillo-Viejo I., Yudowski G. A., Rosenthal J. J., Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc. Natl. Acad. Sci. U.S.A. 110, 18285–18290 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishikura K., Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basilio C., Wahba A. J., Lengyel P., Speyer J. F., Ochoa S., Synthetic polynucleotides and the amino acid code. V. Proc. Natl. Acad. Sci. U.S.A. 48, 613–616 (1962). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoernes T. P., et al. , Translation of non-standard codon nucleotides reveals minimal requirements for codon-anticodon interactions. Nat. Commun. 9, 4865 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Licht K., et al. , Inosine induces context-dependent recoding and translational stalling. Nucleic Acids Res. 47, 3–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishikura K., A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Woolf T. M., Chase J. M., Stinchcomb D. T., Toward the therapeutic editing of mutated RNA sequences. Proc. Natl. Acad. Sci. U.S.A. 92, 8298–8302 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wettengel J., Reautschnig P., Geisler S., Kahle P. J., Stafforst T., Harnessing human ADAR2 for RNA repair—Recoding a PINK1 mutation rescues mitophagy. Nucleic Acids Res. 45, 2797–2808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukuda M., et al. , Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-to-I RNA editing. Sci. Rep. 7, 41478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merkle T., et al. , Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 37, 133–138 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Reautschnig P., et al. , CLUSTER guide RNAs enable precise and efficient RNA editing with endogenous ADAR enzymes in vivo. Nat. Biotechnol. 40, 759–768 (2022). [DOI] [PubMed] [Google Scholar]
- 14.Qu L., et al. , Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 37, 1059–1069 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Yi Z., et al. , Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022). [DOI] [PubMed] [Google Scholar]
- 16.Montiel-González M. F., Vallecillo-Viejo I. C., Rosenthal J. J., An efficient system for selectively altering genetic information within mRNAs. Nucleic Acids Res. 44, e157 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sinnamon J. R., et al. , Site-directed RNA repair of endogenous Mecp2 RNA in neurons. Proc. Natl. Acad. Sci. U.S.A. 114, E9395–E9402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox D. B. T., et al. , RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel P., et al. , Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat. Methods 15, 535–538 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katrekar D., et al. , In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat. Methods 16, 239–242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuttan A., Bass B. L., Mechanistic insights into editing-site specificity of ADARs. Proc. Natl. Acad. Sci. U.S.A. 109, E3295–E3304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kilstrup-Nielsen C., Landsberger N., “Rett syndrome: From the involved gene(s) to treatment” in Neurobiology of Brain Disorders: Biological Basis of Neurological and Psychiatric Disorders, Zigmond M. J., Rowland L. P., Coyle J. T., Eds. (Academic Press, 2015), pp. 98–119. [Google Scholar]
- 23.Amir R. E., et al. , Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 23, 185–188 (1999). [DOI] [PubMed] [Google Scholar]
- 24.Neul J. L., et al. ; RettSearch Consortium, Rett syndrome: Revised diagnostic criteria and nomenclature. Ann. Neurol. 68, 944–950 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kankirawatana P., et al. , Early progressive encephalopathy in boys and MECP2 mutations. Neurology 67, 164–166 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Fyfe S., Cream A., de Klerk N., Christodoulou J., Leonard H., InterRett and RettBASE: International Rett syndrome association databases for Rett syndrome. J. Child Neurol. 18, 709–713 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Tillotson R., Bird A., The molecular basis of MeCP2 function in the brain. J. Mol. Biol. 432, 1602–1623 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Sandweiss A. J., Brandt V. L., Zoghbi H. Y., Advances in understanding of Rett syndrome and MECP2 duplication syndrome: Prospects for future therapies. Lancet Neurol. 19, 689–698 (2020). [DOI] [PubMed] [Google Scholar]
- 29.Nan X., et al. , Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393, 386–389 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Meehan R. R., Lewis J. D., Bird A. P., Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 20, 5085–5092 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinnamon J. R., et al. , In vivo repair of a protein underlying a neurological disorder by programmable RNA editing. Cell Rep. 32, 107878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallecillo-Viejo I. C., Liscovitch-Brauer N., Montiel-Gonzalez M. F., Eisenberg E., Rosenthal J. J. C., Abundant off-target edits from site-directed RNA editing can be reduced by nuclear localization of the editing enzyme. RNA Biol. 15, 104–114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eggington J. M., Greene T., Bass B. L., Predicting sites of ADAR editing in double-stranded RNA. Nat. Commun. 2, 319 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong S. K., Sato S., Lazinski D. W., Substrate recognition by ADAR1 and ADAR2. RNA 7, 846–858 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nan X., Campoy F. J., Bird A., MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell 88, 471–481 (1997). [DOI] [PubMed] [Google Scholar]
- 36.Lagger S., et al. , MeCP2 recognizes cytosine methylated tri-nucleotide and di-nucleotide sequences to tune transcription in the mammalian brain. PLoS Genet. 13, e1006793 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinde B., Wu D. Y., Greenberg M. E., Gabel H. W., DNA methylation in the gene body influences MeCP2-mediated gene repression. Proc. Natl. Acad. Sci. U.S.A. 113, 15114–15119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skene P. J., et al. , Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol. Cell 37, 457–468 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shahbazian M. D., Antalffy B., Armstrong D. L., Zoghbi H. Y., Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum. Mol. Genet. 11, 115–124 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Lioy D. T., et al. , A role for glia in the progression of Rett’s syndrome. Nature 475, 497–500 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guy J., Hendrich B., Holmes M., Martin J. E., Bird A., A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat. Genet. 27, 322–326 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Chalk A. M., Taylor S., Heraud-Farlow J. E., Walkley C. R., The majority of A-to-I RNA editing is not required for mammalian homeostasis. Genome Biol. 20, 268 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park E., Jiang Y., Hao L., Hui J., Xing Y., Genetic variation and microRNA targeting of A-to-I RNA editing fine tune human tissue transcriptomes. Genome Biol. 22, 77 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan K. Y., et al. , Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mathiesen S. N., Lock J. L., Schoderboeck L., Abraham W. C., Hughes S. M., CNS transduction benefits of AAV-PHP.eB over AAV9 are dependent on administration route and mouse strain. Mol. Ther. Methods Clin. Dev. 19, 447–458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katrekar D., et al. , Efficient in vitro and in vivo RNA editing via recruitment of endogenous ADARs using circular guide RNAs. Nat. Biotechnol. 40, 938–945 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monian P., et al. , Endogenous ADAR-mediated RNA editing in non-human primates using stereopure chemically modified oligonucleotides. Nat. Biotechnol., 10.1038/s41587-022-01225-1 (2022). [DOI] [PubMed] [Google Scholar]
- 48.Ramirez J. M., Ward C. S., Neul J. L., Breathing challenges in Rett syndrome: Lessons learned from humans and animal models. Respir. Physiol. Neurobiol. 189, 280–287 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feldman J. L., Del Negro C. A., Gray P. A., Understanding the rhythm of breathing: So near, yet so far. Annu. Rev. Physiol. 75, 423–452 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matagne V., et al. , A codon-optimized Mecp2 transgene corrects breathing deficits and improves survival in a mouse model of Rett syndrome. Neurobiol. Dis. 99, 1–11 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Goguen R. P., et al. , Efficacy, accumulation, and transcriptional profile of anti-HIV shRNAs expressed from human U6, 7SK, and H1 promoters. Mol. Ther. Nucleic Acids 23, 1020–1034 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rivera V. M., et al. , Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer. Blood 105, 1424–1430 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Brown K., et al. , The molecular basis of variable phenotypic severity among common missense mutations causing Rett syndrome. Hum. Mol. Genet. 25, 558–570 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tillotson R., et al. , Radically truncated MeCP2 rescues Rett syndrome-like neurological defects. Nature 550, 398–401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinnamon J. R., et al. , Targeted RNA editing in brainstem alleviates respiratory dysfunction in a mouse model of Rett syndrome. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA849938. Deposited 1 August 2021. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-exome DNA sequencing and RNA sequencing data have been deposited in the Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra (accession no. PRJNA849938) (55). All study data are included in the article and/or SI Appendix.