The type III secretion system (T3SS) encoded in the Salmonella pathogenicity island 1 (SPI-1) is a primary virulence factor for Salmonella, required for initiating the inflammatory diarrhea that is the hallmark of salmonellosis and for invading the intestinal epithelium, leading to potentially lethal systemic infection. Expression of the SPI-1 T3SS is controlled by a complex regulatory circuit. One important aspect of expression is that it is bimodal; under most conditions, only a subset of cells induces the SPI-1 system. In PNAS, Figueroa-Bossi et al. (1) have uncovered an interesting functional interaction among the transcription terminator Rho, the xenogenic silencer H-NS (histone-like nucleoid-structuring protein), and the SPI-1 regulator HilD that contributes to a shift in bimodal SPI-1 expression. This work reveals a deeper role for Rho-mediated transcriptional termination and spurious transcription (or transcriptional noise) in regulation.
The 35-kilobase SPI-1 “island” is a classic example of a horizontally acquired virulence locus. Indeed, its acquisition, some 140 Mya, created the Salmonella species (2). Characteristic of pathogenicity islands, the locus has a low Guanine-Cytosine (GC) content (45%) relative to the ancestral chromosome (52% GC). Also typical of horizontally acquired DNA, transcription of SPI-1 is generally silenced by the nucleoid-associated protein, H-NS, which binds to Adenine-Thymine (AT)-rich nucleation sites and polymerizes on DNA or interacts and bridges binding sites (3, 4). Thus, H-NS blocks binding of transcriptional activators and/or RNA polymerase. Pathogenicity islands are AT rich precisely because they could be silenced by H-NS. Horizontally acquired GC-rich DNA would presumably be expressed in an unregulated fashion and be selected against. Once acquired and silenced by H-NS, the bacterium can evolve mechanisms to appropriately regulate the island genes to the benefit of the cell.
SPI-1 expression is regulated by three AraC-like regulators, HilD, HilC, and RtsA, which act in a complex feed-forward loop to activate expression of hilA, encoding the transcriptional activator of the T3SS structural genes (5). The system has evolved to be highly sensitive to the environment and physiological status of the cell, responding to a number of global regulatory systems. The vast majority of regulatory signals are integrated through HilD, whose expression and activity are controlled at multiple levels: transcription initiation, messenger RNA (mRNA) translation, mRNA stability, and HilD protein activity (6). At the protein level, HilE directly binds to HilD to prevent DNA binding. HilE is produced at a relatively low level and acts as one of the factors setting the threshold for HilD activity required to induce the system (7). HilC and RtsA act as amplifiers of any inducing signals.
A simple model would suggest that H-NS coats the entire SPI-1 locus and that the transcriptional activators act primarily by displacing H-NS, allowing access to the transcriptional machinery. In reality, H-NS action seems to be concentrated at the hilD promoter region (8). The current model suggests that H-NS nucleates upstream of hilD and coats the region asymmetrically for several kilobases. HilD does indeed act primarily to displace H-NS at the hilD promoter to activate its own expression. In contrast, HilD acts as a true activator at the hilA promoter, largely independent of H-NS. Positive signals feeding into the system must induce enough HilD activity to overcome both inhibition by HilE and promoter repression by H-NS. Once this threshold is met, HilD autoactivation gives rise to a switch-like phenomenon, conferring the bimodal characteristic of SPI-1 activation.
H-NS, acting at the hilD promoter, is sensitive to perturbation, lowering the threshold of HilD required to throw the switch. For example, deletions as far as 3.5 kilobases downstream of the hilD promoter cause activation (8). The iron-responsive regulatory protein Fur also activates SPI-1 by binding upstream of the hilD promoter and partially disrupting H-NS, making it easier for HilD to activate (9, 10).
Mutations in rho were originally identified as suppressors of polar mutations in the lacZ gene, allowing expression of the downstream lacY gene (11, 12). However, the transcriptional terminator Rho did not evolve to confound bacterial geneticists. Rather, Rho is responsible for terminating transcription in the ∼20% of chromosomal transcripts that lack intrinsic terminator sequences. More recently, it has been realized that there is an extensive amount of spurious transcription initiation in the cell (13). Rho is required to terminate these transcripts before they can interfere with normal gene expression by, for example, clashing with RNA polymerases that are transcribing true mRNAs or creating antisense transcripts that interfere with translation of these mRNAs.
Interestingly, there is significant overlap between H-NS binding and transcripts most affected by Rho inhibition (13). Because of the AT richness of the pathogenicity islands, spurious promoters should be more common in these regions. However, this also means that Rho utilization sites (rut sites), which are C-rich regions in the transcript, are less common. The data suggest that NusG, required for some but not all Rho-mediated termination, is particularly important in these regions, compensating for the lack of rut sites (13). Thus, there is cooperation and genetic interaction between Rho, NusG, and H-NS. One concept is that H-NS slows RNA polymerase, allowing Rho, acting through NusG, to terminate transcription in these AT-rich regions.
To further study these relationships, Bossi et al. (14) created a Salmonella strain that can be largely depleted of NusG. Their initial results were consistent with the above model, showing that both NusG depletion and certain mutations in rho lead to increased transcription, primarily in H-NS–silenced regions. Mutant Rho proteins defective in rut site binding had little effect in these experiments, consistent with the proposed role of NusG.
In PNAS, Figueroa-Bossi et al. (1) expand on these results. Most of the increased transcription observed upon depletion of NusG is in the SPI-1 locus. Moreover, it was not spurious transcripts that most increased, but rather the SPI-1 mRNAs. This also resulted in a significant shift in the percentage of cells that induced the SPI-1 T3SS. In other words, the cells depleted for NusG were much more likely to switch to the “SPI-1-on” state. How does failure to block spurious transcription lead to induction of SPI-1? The authors provide data suggesting that in a strain depleted of NusG, spurious transcription partially disrupts H-NS binding in the hilD promoter region, allowing HilD autoactivation of SPI-1 and induction of the system. Inserting a foreign promoter outside the hilD promoter region also led to activation of the system. Interestingly, the spurious transcripts that lead to this phenomenon can initiate more than 600 nucleotides and perhaps, more than 1,400 nucleotides from the hilD promoter. Does transcription from the spurious promoters have to proceed all the way through the region coated by H-NS to have an effect? The fact that deletions in the SPI-1 locus 3 kilobases from the hilD promoter can cause a similar activation of hilD expression suggests that the answer is no (8).
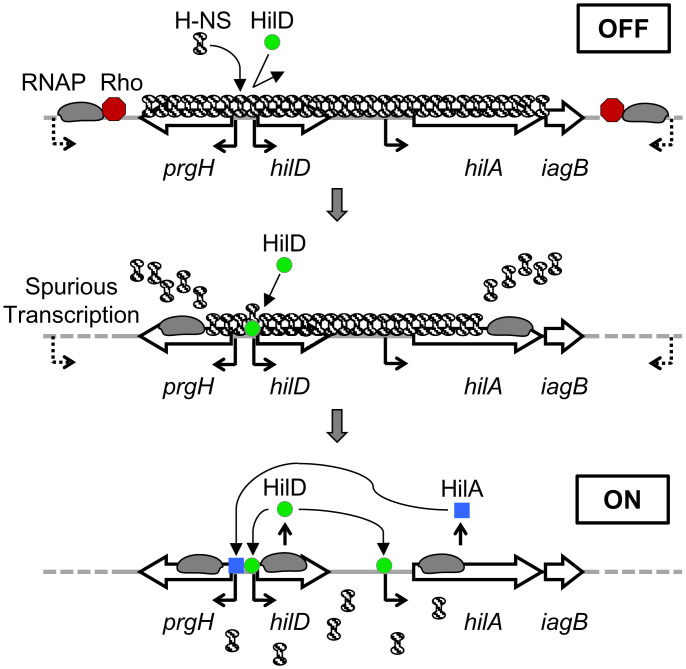
Thus, H-NS inhibits transcription, facilitating Rho-mediated termination. However, the transcription machinery that escapes Rho action is also dislodging H-NS from the DNA. In the case of the autoactivated hilD promoter, this lowers the threshold of HilD required to flip the switch to the “on” state, thus shifting the proportion of cells expressing SPI-1. The effect is amplified by depletion of NusG, but one would expect that some transcripts elongate under normal circumstances. It follows that subtle changes in overall Rho function in the cell would normally affect triggering of the SPI-1 locus (Fig. 1).
Fig. 1.
Regulation of the SPI-1 T3SS. In the “off” state, H-NS silences the hilD promoter region, and there is not enough HilD activity to overcome this repression. Rho, working through NusG, is blocking spurious transcripts initiated outside this region by RNA polymerase (RNAP). If these transcripts escape Rho action, H-NS is dislodged from the DNA by the transcription complex. This lowers the threshold of HilD activity required to autoactivate, leading to the “on” state.
The apparent intimate relationship between Rho and H-NS brings into question the evolutionary functions of these proteins. The function of H-NS as a xenogenic silencer is clear, but perhaps this is just happenstance, and its real role is to work with Rho to block spurious transcription of any sort. Likewise, we have come to think of Rho as simply one of the two mechanisms of transcription termination, and it certainly plays that role for some normal transcripts. Rho is also increasingly recognized as a regulatory factor, affecting expression at the level of transcription elongation, its activity controlled by, for example, small RNAs or riboswitches (15). Are these functions the primary evolutionary role of Rho? Does H-NS have a normal role in these Rho functions? This relationship between the two proteins and the effects on overall cell physiology and gene regulation are exciting areas for further investigation.
Acknowledgments
The research is funded by National Institute of Allergy and Infectious Diseases of the NIH Grants AI163687 and AI166495.
Footnotes
The author declares no competing interest.
See companion article, “Pervasive transcription enhances the accessibility of H-NS–silenced promoters and generates bistability in Salmonella virulence gene expression,” 10.1073/pnas.2203011119.
References
- 1.Figueroa-Bossi N., et al. , Pervasive transcription enhances the accessibility of H-NS–silenced promoters and generates bistability in Salmonella virulence gene expression. Proc. Natl. Acad. Sci. U.S.A., 10.1073/pnas.2203011119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Desai P. T., et al. , Evolutionary genomics of Salmonella enterica subspecies. MBio 4, e00579-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lucchini S., et al. , H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2, e81 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navarre W. W., et al. , Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313, 236–238 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Ellermeier C. D., Ellermeier J. R., Slauch J. M., Hil D., Hil C., HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57, 691–705 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Golubeva Y. A., Sadik A. Y., Ellermeier J. R., Slauch J. M., Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190, 79–90 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grenz J. R., Cott Chubiz J. E., Thaprawat P., Slauch J. M., HilE regulates HilD by blocking DNA binding in Salmonella enterica serovar Typhimurium. J. Bacteriol. 200, e00750-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalafatis M., Slauch J. M., Long-distance effects of H-NS binding in the control of hilD expression in the Salmonella SPI1 locus. J. Bacteriol. 203, e0030821 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellermeier J. R., Slauch J. M., Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J. Bacteriol. 190, 476–486 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teixidó L., Carrasco B., Alonso J. C., Barbé J., Campoy S., Fur activates the expression of Salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro. PLoS One 6, e19711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beckwith J., Restoration of operon activity by suppressors. Biochim. Biophys. Acta 76, 162–164 (1963). [PubMed] [Google Scholar]
- 12.Ratner D., Evidence that mutations in the suA polarity suppressing gene directly affect termination factor rho. Nature 259, 151–153 (1976). [DOI] [PubMed] [Google Scholar]
- 13.Peters J. M., et al. , Rho and NusG suppress pervasive antisense transcription in Escherichia coli. Genes Dev. 26, 2621–2633 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bossi L., et al. , NusG prevents transcriptional invasion of H-NS-silenced genes. PLoS Genet. 15, e1008425 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bossi L., Figueroa-Bossi N., Bouloc P., Boudvillain M., Regulatory interplay between small RNAs and transcription termination factor Rho. Biochim. Biophys. Acta. Gene Regul. Mech. 1863, 194546 (2020). [DOI] [PubMed] [Google Scholar]