Significance
Because microbiome research generally focuses on a single host or habitat, we know comparatively little about the diversity and distribution of microbiomes at a landscape scale. Our study demonstrates that most of the microbial diversity present within a watershed is maintained within environmental substrates like soil or stream water, and microbiomes of organisms are generally subsets of those that are lower on the food chain. This result challenges the notion that sources of microbial inoculum are likeliest derived from close relatives. By identifying sources of shared microbial diversity within the landscape, we can better understand the origins and assembly processes of symbiotic microbes and how this might abet global conservation, restoration, or bio-engineering goals, such as preserving biodiversity and ecosystem services.
Keywords: landscape microbial ecology, ridge-to-reef connectivity, biogeography, nestedness, watershed microbiome
Abstract
Microbes are found in nearly every habitat and organism on the planet, where they are critical to host health, fitness, and metabolism. In most organisms, few microbes are inherited at birth; instead, acquiring microbiomes generally involves complicated interactions between the environment, hosts, and symbionts. Despite the criticality of microbiome acquisition, we know little about where hosts’ microbes reside when not in or on hosts of interest. Because microbes span a continuum ranging from generalists associating with multiple hosts and habitats to specialists with narrower host ranges, identifying potential sources of microbial diversity that can contribute to the microbiomes of unrelated hosts is a gap in our understanding of microbiome assembly. Microbial dispersal attenuates with distance, so identifying sources and sinks requires data from microbiomes that are contemporary and near enough for potential microbial transmission. Here, we characterize microbiomes across adjacent terrestrial and aquatic hosts and habitats throughout an entire watershed, showing that the most species-poor microbiomes are partial subsets of the most species-rich and that microbiomes of plants and animals are nested within those of their environments. Furthermore, we show that the host and habitat range of a microbe within a single ecosystem predicts its global distribution, a relationship with implications for global microbial assembly processes. Thus, the tendency for microbes to occupy multiple habitats and unrelated hosts enables persistent microbiomes, even when host populations are disjunct. Our whole-watershed census demonstrates how a nested distribution of microbes, following the trophic hierarchies of hosts, can shape microbial acquisition.
Microbial partners metabolize our food, fight off disease, and run the machinery that sustains the air we breathe, water we drink, and soil under our feet. Despite their importance, most host-associated microbes are generally not present at birth and are instead acquired (1). Because microbial symbionts can influence host health and fitness, the processes that determine how different microbiomes assemble within different hosts is a matter of active and urgent inquiry. Microbial ecologists have made great progress in determining how factors such as abiotic conditions (2–4), host evolution (5, 6), and microbial traits (7–9) shape environmental microbiomes, but considerably less is known about how surrounding environments or different guilds of host organisms contribute to host-associated microbiome composition. Longitudinal studies show that microbial richness accumulates and community composition changes over time across a wide diversity of hosts and habitats (1), but we know comparatively little about from where these microbes originate. To better understand microbial transmission and its role in community composition, we propose a framework that relies on theory from foodweb and landscape ecology.
The concept of a foodweb has had a place in the ecological lexicon since at least the time of Elton (1927; (10)), and others such as Lindeman (11) and Odum (12) significantly expanded upon this notion to include how macroorganisms interact within their environments, in addition to their feeding relationships. The units of study for foodwebs are ecosystems, which are spatially explicit and include all organisms along with their abiotic environments and their interactions within its bounds (13). This definition was born from the efforts of the founders of the Hubbard Brook Ecosystem Study (HBES; 1963), who recognized that a watershed naturally delineates the boundaries of an ecosystem, an idea that parallels the Hawaiian ahupuaʻa concept. Since then, the HBES and its framework have led to numerous milestones in our understanding of processes such as the effects of long-term changes in acidification (14) and ecosystem impacts of global warming (15). Here, we adopt the notion of the watershed as an entire discrete ecosystem to better understand the landscape ecology of microbes. Landscape ecology is a means to understand how spatial processes affect biodiversity (16). In classic landscape ecology theory, the structure (heterogeneity) and fragmentation of habitats (or patches) within a matrix of otherwise inhospitable areas affect species’ dispersal ability and establishment. This ultimately shapes species’ abundance and distributions across the landscape (17). Contemporary landscape ecology theory extends this idea to include the concept of a landscape continuum, where continuous environmental variables, as opposed to discrete habitat patches surrounded by a matrix, better describe species’ distributions. Connecting these concepts, foodwebs are embedded in landscapes, and watersheds constitute a useful unit of measure to better understand their interactions.
To expand concepts from foodweb and landscape ecology to be inclusive of microbes, we must first consider the following: a landscape for microbes can be both structural (e.g., different land covers or hydrology) and biotic (e.g., variation in the distribution of host populations). Also, microbes might better fit a continuous landscape model rather than a patch model if their distributions are not governed merely by the presence of a compatible host or habitat, but rather, if they exist among multiple hosts across a gradient of environmental conditions. This requires microbes to be generalists to some degree and/or a matrix that is at least partially hospitable (18). These considerations are important because while microbial transmission among related hosts is one obvious means of microbiome assembly, this model, in and of itself, is insufficient to sustain microbiomes (defined here as communities of bacteria and archaea) across a dynamic landscape. For example, many plants and animals are either sparse, seasonal, or ephemeral, requiring that their symbiotic microbes be capable of residing, at times, in alternate nearby hosts or environments. This potential for a microbe to persist in, and disperse among, hosts of different kingdoms and guilds, or even between liquid and land, is a trait with the potential to add an additional dimension to microbiome assembly theory (19). Where, then, might a host’s microbes reside when not inside that host? In addition, what factors might predict microbiome distributions among potentially interacting hosts and environments?
Variability in matrix suitability and host specialization may result in differing microbial communities reflected in one of three nonmutually exclusive patterns, each of which leaves a diagnostic imprint on microbiome structure. If any host or environment has an equal likelihood of harboring microbes that are present in any other host or environment, we might expect host–microbe interaction networks that are randomly structured. Alternatively, if microbes are more likely to co-occur among related hosts or guilds, we might expect these to contain unique and specific consortia of microbes (modules) that are not found elsewhere in the interaction network. Finally, host–microbe interactions might be best characterized as stratified, resulting in a network topology in which microbial diversity is nested such that taxa-poor microbiomes are subsets of those that are taxa-rich. In this scenario, nonhost environmental matrices (e.g., soil, sediment, water) serve as reservoirs of broad microbial diversity that is subsequently, and hierarchically, partitioned into simpler microbiomes. While this concept is fairly intuitive, there are actually few, if any, studies that demonstrate transmission among environmental microbiomes and multiple hosts at ecosystem scales. Instead, many of the insights gleaned into assembly processes of microbiomes are owed to studies of single hosts, tractable model systems, or global syntheses (20). We address this gap by sampling microbiomes from aquatic, marine, and terrestrial foodwebs within a single watershed to examine the dynamics of sources and sinks of microbial diversity.
Here, we present a microbial census of a model ecosystem metacommunity in which continental-scale environmental heterogeneity is recapitulated within a comparatively small watershed. Because of this, we can surmise the distribution limits of microbiomes across land, stream, and sea, a feat that would not be plausible in most other landscapes of similar size or environmental variability. From ridge to reef, our compact watershed spans a roughly 3.5 m rainfall differential, ∼27 times that encountered along the Mississippi, the largest watershed in continental North America. Also, our model ecosystem is located on the most isolated archipelago on the planet, making exogenous microbial inputs infrequent, if not unlikely. Furthermore, owing to parallels in environmental heterogeneity and foodweb structure across this compact watershed compared to others, our findings are potentially relevant for highly connected ecosystems that span substantially larger geographic areas.
For example, a long-standing question in biogeography is the relationship between organisms’ local distributions and those at larger scales. Many factors influence the distributions of microbes, including their physiology, size, population density, and dispersal abilities (21–23). A common assumption is that niche breadth should also predict the range size of an organism, since the ability to survive in broader environments, and to use a greater array of resources, should indicate the ability to occupy more habitats that occur over greater distances (24, 25). This is an important component of source and sink dynamics, because it suggests that local occupancy should predict global distributions. This relationship is seldom tested empirically, however, because small areas rarely contain, or are sampled for, broad climatic variability and host diversity. In the absence of phenotypic, genomic, or even well-resolved taxonomic information about the majority of the earth’s microbial biodiversity, geographic range is one of the few traits that can be directly inferred from short environmental DNA sequence reads. By examining our ecosystem-wide microbiome census within the context of the global survey of the Earth Microbiome Project (26), we assess the relationship between global and local microbial distributions.
A Model Microbial Mesocosm Containing Continental-Scale Gradients
We characterize microbiomes within Oʻahu’s Waimea watershed, selected for its contiguity, isolation, and environmental heterogeneity. In fewer than 12 km, the main rivers of Waimea Valley plunge from a high elevation bog, through a rainforest, into a protected estuary, and out over sand flats to a coral reef. This short distance spans steep gradients in elevation (0–682 m) and precipitation, (1.1–4.6 m rainfall/y; Fig. 1A). Many abiotic variables within the gradient are highly colinear, such that is impossible to disentangle their individual effects (i.e., rainfall, temperature, solar irradiance) on microbiomes. Instead, the main advantage of the gradient is that conditions diverge rapidly among four Köppen climate types as a function of distance between locations. Furthermore, within this watershed exists a wide diversity of terrestrial, stream, and marine habitats adjacent to each other. In essence, biome diversity at a continental scale is represented within a small spatial area, enabling us to measure whether and how the environment constrains microbial distributions and microbiome structure within foodwebs.
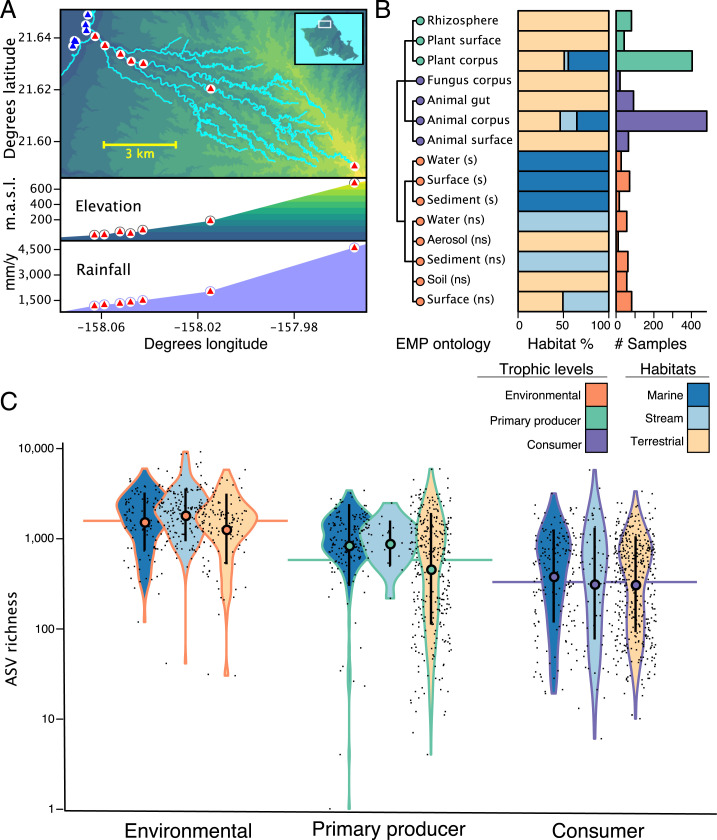
Fig. 1.
Sampling within the Waimea watershed on Oʻahu Island. (A) Terrestrial and stream samples were paired and spanned the entirety of the catchment. Plot positions (n = 21) along elevation and rainfall gradient are indicated with triangles (blue triangles are marine, red triangles are terrestrial/stream). “m.a.s.l.” indicates meters above sea level. (B) Distribution of n = 1,562 samples. Samples are classified at level 3 of the EMP metadata ontology. Stacked barchart colors indicate habitat of origin. Histogram colors indicate environmental/trophic status of sample; “ns” indicates nonsaline, “s” indicates saline. (C) Violin plots indicate distributions of ASV richness organized by trophic level (outline) and habitat (fill). Microbial richness tracks environmental/trophic position of the sample. Circles are median, vertical lines indicate the interquartile range, and horizontal lines indicate the mean. Mean richness of environmental/trophic levels differ significantly (ANOVA, F = 173.9, P < 0.0001).
Along the entirety of the watershed, we assessed microbiome diversity and microbial distributions among habitats. We sampled seven pairs of stream and terrestrial plots (20 m diameter) as well as seven plots in the near-shore sand flats and coral reefs of the bay (21 plots total; Fig. 1A). Within each plot, we collected 113 + 54.5 (SD) biological samples from both host organisms and environmental substrates (Fig. 1B and SI Appendix, Fig. S5 and Dataset S1, Table S1). Sampling among plots was roughly balanced by the number of samples collected, as well as the position of those samples on the environmental/trophic hierarchy (derived from host-independent environmental substrates, primary producers, or consumers).
Samples were circumscribed within an ontology devised by the Earth Microbiome Project (26) that discriminates at its most granular level by sample origin (e.g., saline, nonsaline), substrate type (e.g., soil, water, sediment), host, and location in or on a host (e.g., plant rhizosphere, plant corpus, sediment, aerosol; EMPO (Earth Microbiome Project ontology) level 3; Fig. 1B). We sampled a total of 15 out of the 17 EMPO 3 categories across the entire watershed. Samples were categorized by location, habitat (stream, marine, or terrestrial), and position within their respective foodweb. Within each habitat type, we sampled the same number of replicates within each EMPO level 3 category, although host species identity varied across the watershed within habitats. Unless otherwise specified, sample type refers to EMPO level 3 category.
From these samples, we enumerated microbiomes to test the following four predictions. 1) Independent of whether from land, sea, or stream, environmental samples contain the majority of microbial diversity within the watershed and asymmetrically contribute to the microbiome composition of hosts. 2) Continuous landscape variables such as elevation and precipitation would best predict microbiome compositions among stream and terrestrial habitats. 3) Microbiome composition, regardless of habitat type, would be nested such that environmental microbiomes serve as sources for primary producers followed by consumer microbiomes. We posited that high nestedness values are predictive of a more hospitable matrix where nonhost environments contribute to host patches. In contrast, lower nestedness values and higher modularity would indicate stronger patch effects consistent with high host specificity and a less hospitable matrix. Patch and matrix dynamics are not mutually exclusive, and we predicted that each may act simultaneously to shape microbiome distributions within an entire ecosystem, but their relative contribution to microbiome structure may vary over the landscape, depending upon site-specific environmental conditions. 4) Microbes that inhabit the widest range of habitats, environments, and hosts within our tropical watershed should also be those with the widest geographical distributions globally.
Results and Discussion
Abiotic Substrates as Microbial Diversity Reservoirs.
Within foodwebs, energy is transferred among trophic levels from producers to apex predators in a largely unidirectional and energy-inefficient manner, such that ∼90% of available energy is lost to entropy between any two levels (27). Analogously, and in support of our first prediction, we find that independent of habitat type, microbial richness decreases predictably from environmental substrates up the trophic hierarchy (Fig. 1C). It bears mentioning that microbes are not strictly passed up the food chain via consumption; rather, this is only one of many possible modes of microbial transmission among hosts, including other types of biotic interactions and/or dispersal. Nevertheless, our data demonstrate a strong pattern, tied to foodwebs, that offers a vehicle for understanding microbiome complexity and linkages at the landscape scale.
Richness gradients, however, in and of themselves, are minimally predictive of source sink dynamics since they do not indicate the extent to which microbial composition is shared among sample types or trophic levels. To assess the second portion of our first prediction, that environmental microbiomes contribute to those of hosts, we ranked sample types by their contribution to unique richness of amplicon sequence variants (ASV; an operational taxonomic unit circumscribed at 100% sequence identity), which better indicates their potential as sources for landscape diversity. In other words, this analysis demonstrates which sample types contain the most complete set of ASVs found across all sample types combined. We established a downsampling procedure where each sample type in every habitat was equally represented by the same number of samples and the same number of sequences within each sample. This procedure eliminates artifactual hierarchies resulting from sampling asymmetries. Conglomerate categories of sample type by habitat were then ranked by their contribution to cumulative ASV richness.
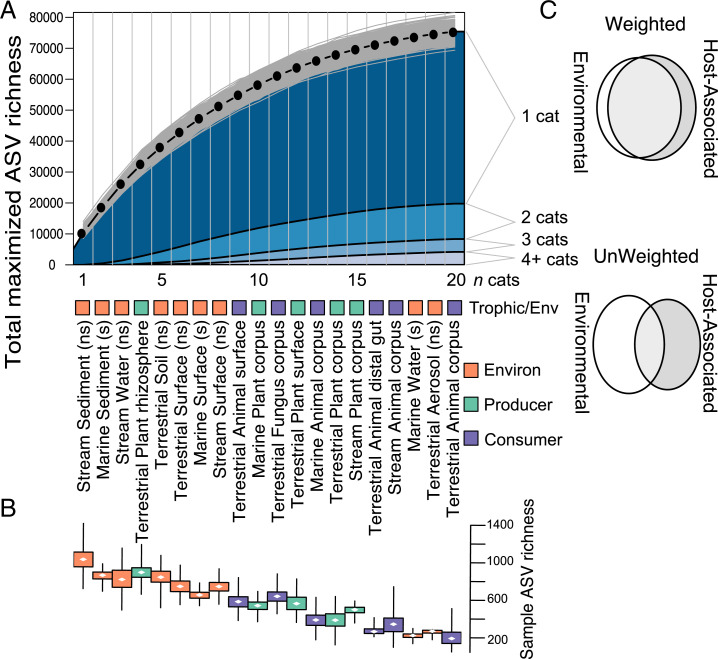
Our hierarchical analysis demonstrates that at the watershed level, nonhost environmental microbiomes ranked the highest in their contribution to ecosystem microbial richness, comprising seven of the top eight sample types (Fig. 2A). Rhizosphere microbiomes, the single high-ranking host-associated category, may themselves be considered partially nonhost since rhizospheres combine components of both the plant-surface and the soil environment (28). Combined, environmental microbiomes contained more than 55% of total watershed ASV richness (Fig. 2 A and B). This suggests that ever-present environments like water or sediments might provide environmental “waiting rooms” for microbes to colonize hosts when available. Microbial richness is particularly concentrated in subsurface samples in each of the three habitats: the combination of marine sediment, stream sediment, and soil contains more than 34% of the overall ASV diversity, indicating their potential as “universal donors” to ecosystem-wide microbial diversity. Host-associated microbiomes, in contrast, are comparatively ASV poor (Fig. 1C), and their contributions to unique microbial richness are generally those microbes that are detected in low abundance. When we account for an ASV’s abundance in the sequence dataset, more than 77% of weighted microbial richness is contained in environmental host-independent samples (Fig. 2C). In other words, environmental samples contain both the highest microbial richness and the fraction of microbes detected most frequently. What is less clear is whether the functional traits of these microbes are similar regardless of where within the ecosystem they reside, as we expect that a microbe’s ecological role is at least partially dependent on context.
Fig. 2.
Environmental samples contribute the most to novel diversity of the watershed microbiome. (A) Accumulation curve of ASV richness maximized for n sample type:habitat categories. For a given n, black dots represent the average ASV richness for the optimized collection of n categories, given 1,000 randomizations (gray lines). Labels indicate the rankings of categories by their contribution to maximized richness. Colored box indicates environment/trophic level. “ns” indicates nonsaline, “s” indicates saline. (B) Boxplots show median, interquartile range, and data extent of ASV richness across randomizations. (C) Euler diagrams that depict the overlap of environmental and host-associated ASV diversity in cases where the ASVs are weighted by their numerical abundance in the dataset and not.
However, encountering a microbial DNA molecule in a host or habitat does not indicate reproductive or metabolic activity there, and we might attribute some of the richness detected among samples to microbes that arrive via dispersal from suitable habitats, but remain quiescent. (24, 25). For example, among both plant and animal samples, surface samples are richer than those collected from inside a host organism, presumably due to transient microbial associates. This dispersal-driven fraction of microbial richness is, nevertheless, a critical component of metacommunity dynamics where less desirable environments provide potential stepping-stones to hosts (19), a phenomenon with relevance for microbial distributions at both large and small scales.
Continuous Environmental Variables Shape Microbiome Composition and Distributions.
Location along the environmental gradient within the watershed was a significant determinant of microbial composition for stream and terrestrial samples (PERMANOVA; R2 = 0.029, P < 0.001), partially supporting our second prediction. However, the interaction between location and sample type was more predictive (R2 = 0.082, P < 0.001). This indicates that the distribution of microbes among patches (i.e., specific hosts or substrates) matters more or less depending on the environmental conditions in which they interact.
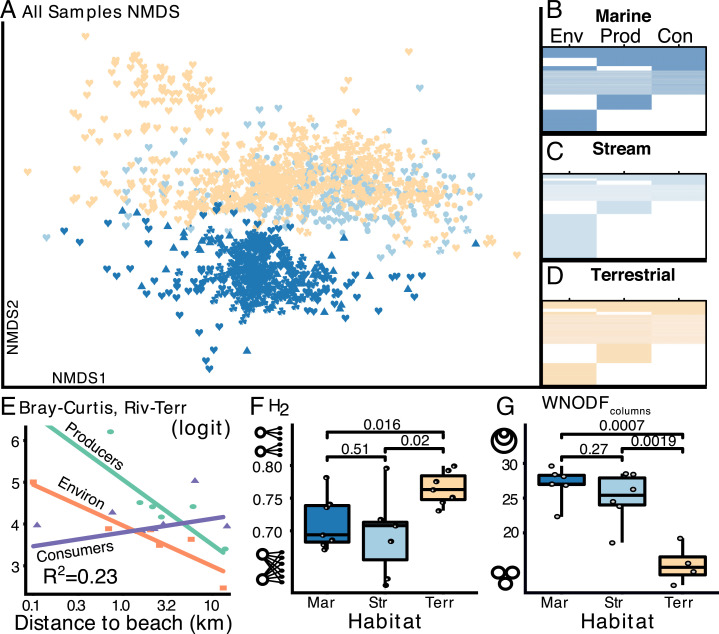
We also found that environmental and primary producer microbiomes differ from those of consumers across the environmental gradient (Fig. 3E). Terrestrial and stream microbiomes of environmental substrates and primary producers, though not consumers, became more similar to each other as a function of distance from the ocean, a cline that corresponds to an increase in elevation and rainfall (Fig. 3E; generalized additive model [GAM], degrees of freedom (DF) = 8,727, R2 = 0.23, P < 0.002). In other words, the wetter, cooler, and higher sites had more similar stream and terrestrial microbiomes. This result indicates that even at the broad circumscription of microbiomes as either environmental, primary producer, or consumer-associated, environmental context affects the degree to which there is transmission between habitats.
Fig. 3.
Microbial community differentiation across habitats and trophic levels. (A) NMDS plots of the reduced dataset containing n = 1,410 samples, colored by habitat; shapes indicate EMPO 2 ontology (♥ animal, ♣ plant,  fungus, ▴ salt water, ● fresh water). (B–D) Nestedness of matrices subsetted to contain equal numbers of samples and sequences. Columns are ordered by richness (in all cases, environmental samples [Env] > primary producers [Prod] > consumers [Con]); rows (ASVs) are ordered by occurrence across the environmental/trophic categories. WNODFc values: B = 44.3, C = 53.04, D = 47.29. (E) Bray-Curtis dissimilarity between paired stream and terrestrial site samples decreased along the transect, but consumer microbiomes did not. Mean Bray-Curtis dissimilarity between stream and terrestrial microbiomes within sites are shown as points, colored by environmental/trophic level. Bray-Curtis values are shown as logit-transformed values. Fit lines are predictions from the GAM used to model these data. (F) Distributions of (n = 21) plot-level bipartite network specificity with samples grouped by habitat. Boxplots are as in Fig. 2. Pairwise P values are shown between habitats (Tukey honestly significant difference). Cartoon diagrams along the y axis demonstrate extremes of network topologies ranging from H2 = 0–1. “Mar” indicates marine, “Str” indicates stream, “Terr” indicates terrestrial. (G) Distributions of plot-level nestedness (WNODFc) grouped by habitat; cartoon Euler diagrams demonstrate extremes of nestedness ranging from 0 to 100.
fungus, ▴ salt water, ● fresh water). (B–D) Nestedness of matrices subsetted to contain equal numbers of samples and sequences. Columns are ordered by richness (in all cases, environmental samples [Env] > primary producers [Prod] > consumers [Con]); rows (ASVs) are ordered by occurrence across the environmental/trophic categories. WNODFc values: B = 44.3, C = 53.04, D = 47.29. (E) Bray-Curtis dissimilarity between paired stream and terrestrial site samples decreased along the transect, but consumer microbiomes did not. Mean Bray-Curtis dissimilarity between stream and terrestrial microbiomes within sites are shown as points, colored by environmental/trophic level. Bray-Curtis values are shown as logit-transformed values. Fit lines are predictions from the GAM used to model these data. (F) Distributions of (n = 21) plot-level bipartite network specificity with samples grouped by habitat. Boxplots are as in Fig. 2. Pairwise P values are shown between habitats (Tukey honestly significant difference). Cartoon diagrams along the y axis demonstrate extremes of network topologies ranging from H2 = 0–1. “Mar” indicates marine, “Str” indicates stream, “Terr” indicates terrestrial. (G) Distributions of plot-level nestedness (WNODFc) grouped by habitat; cartoon Euler diagrams demonstrate extremes of nestedness ranging from 0 to 100.
Rainfall, which comprises a particularly steep gradient (Fig. 1A), might be the mechanism by which stream and terrestrial samples become more similar as a function of distance from the ocean. Rainfall both increases the hydraulic connectivity (e.g., runoff) between stream and terrestrial microbiomes and also increases the water content of absorptive substrates like soil and moss. Elsewhere, headwater streams exhibit increased similarity to the surrounding soils among both biogeochemical parameters (29, 30) and microbial community composition (31–33). This pattern did not hold for consumers, among which there was no correlation between stream/terrestrial microbial compositional overlap and location along the transect (Fig. 3E). The stability of consumer microbiomes might be attributable to animal physiology or behavior, since association with an animal host might provide a greater degree of insulation from external environments. For example, microbes inside rodent gastrointestinal tracts are more influenced by host identities than by diet or geography (34). In addition, some highly mobile animals (such as birds or rats) likely interact with a greater geographic area which could result in more uniform microbiomes across the gradient compared to those of sessile plants.
The habitat origin of a sample—stream, marine, or terrestrial—is a robust predictor of microbiome composition (Fig. 3A), regardless of sample type. Marine microbiomes were compositionally distinct from terrestrial and stream samples (Fig. 3A and SI Appendix, Fig. S2), supporting previous results that found salinity was the second most discriminatory variable of global microbiomes (26). Microbiomes were further discriminated by environmental substrate/trophic level (PERMANOVA [permutational multivariate ANOVA], R2 = 0.036, P ≤ 0.001) and to a greater extent by sample type (PERMANOVA, R2 = 0.13, all P = 0 < 001).
Microbiomes Are Compositionally Nested and Follow Foodweb Hierarchies.
In line with our third prediction, we found that regardless of habitat type, microbiomes were nested such that consumer microbiomes were subsets of primary producers, which were subsets of their environmental microbiomes (Fig. 3 B–D and G). Despite the fact that the majority of ASVs were unique to single sample types (Fig. 2A), this is largely driven by ASVs found in low relative abundance, as is typical (7). Nestedness values also increased as rare microbes were sequentiallyculled from the dataset (SI Appendix, Fig. S8B), indicating stronger overlap among the more abundant fraction of microbial diversity. High-abundance microbes also have the highest co-occurrence values and therefore the greatest probability of transmitting from environment to host and vice versa. Nestedness patterns occur among microbiomes at both large and small scales (26, 35), but our results demonstrate a correlation between this nestedness topology and the organization of foodwebs that is consistent, to some degree, across habitats (Fig. 3 B–D and SI Appendix, Fig. S8).
Despite this robust and constant pattern, the degree of specialization varied among habitats (Fig. 3 F and G). Terrestrial microbiomes were more specialized (Fig. 3F) and less nested than those of stream or marine habitats (Fig. 3G). We suspect that the liquid matrix of aquatic habitats is more hospitable and results in higher rates of dispersal and mixture of microbes, leading to significantly lower specificity and higher nestedness compared to dry land. An alternative, and nonexclusive, hypothesis is that liquid may diminish some of the physicochemical differences among sample types (such as pH or temperature) that would otherwise result in more specific, structured microbiomes within a foodweb. This convergence among aquatic microbiome network properties stands in contrast to the fact that stream microbiomes are more compositionally similar to terrestrial microbiomes (Fig. 3A and SI Appendix, Fig. S2), an indication that differences in network-level microbial specificity between liquid and land might be the result of abiotic forces affecting dispersal in addition to (or instead of) innate microbial traits conferring specialization (33). Measured differences between liquid and land network topologies raise an important point about the extent to which our results might extend to less connected ecosystems (e.g., deserts or glaciers). Understanding these constraints requires higher replication than what a single model watershed might provide.
The largest deviation from a perfectly nested topology is attributable to unique microbial diversity among primary producers, particularly in terrestrial habitats (Fig. 3D). Plant microbiomes are partitioned by their location above- or belowground, and leaves, in particular, demonstrate high variability (although low within-sample richness) compared with other plant parts (35). This, combined with self-inoculation via litter fall (36), vertical transmission (37), and obligate-coevolved symbioses (38), might all contribute to a significant fraction of the plant microbiome that is unique from other hosts and environments. Although consumers may possess some of these same mechanisms, they appear to contain lower modularity overall.
Local Distributions of Microbes Predict Their Global Distributions.
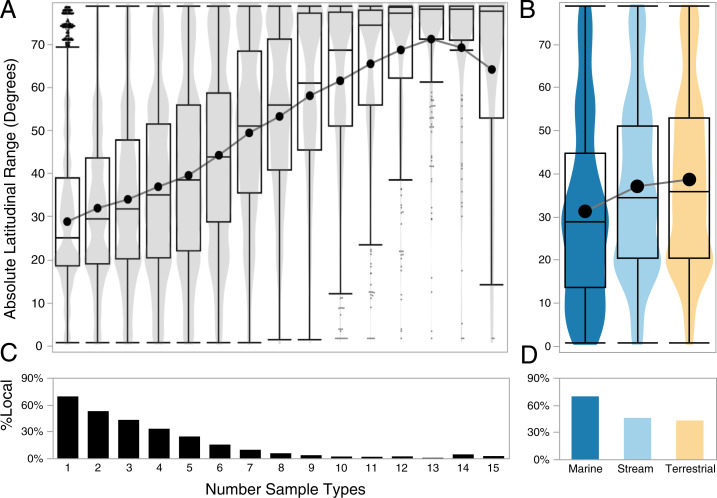
We found strong support for our fourth prediction, that a microbe’s local niche breadth can predict its global distribution. Although modeling the degree of microbial overlap among guilds or habitats is useful for identifying microbiome sources, it does little to predict the composition of microbes they contain. Because of the high host, habitat, and environmental diversity of our model ecosystem, our study affords a unique opportunity to measure niche breadth (defined here as either the number of sample types occupied by a microbe [Fig. 4A] or a microbe’s elevational range within the watershed; SI Appendix, Fig. S3). Using the Earth Microbiome Project database as a reference, we found that local occurrence is a useful predictor of global microbial distributions. The global distribution of a microbe (measured as latitudinal range) that occurs among all sample types in our study system, was, on average, more than twice that of a microbe occurring in a single sample type (Fig. 4A). This relationship indicates an important opportunity, and constraint, for microbiome engineering since the most generalist microbes within a given location are also the most transportable around the world. The relationship also illustrates the interplay between host/environmental specialization, patch dynamics, and microbial distributions. For example, globally distributed microbes (in our dataset, dominated by members of the Gammaproteobacteria and Alphaproteobacteria) (SI Appendix, Figs. S2 and S11) might use a wide variety of hosts and substrates as stepping-stones to enable stepwise dispersal over large distances and across disjunct habitats.
Fig. 4.
Local sample breadth predicts global distributions of microbes. (A) Violin contour densities of latitudinal range in the EMP dataset, binned by the number of EMPO 3 categories in which ASVs occur in Hawaiʻi. Data depicts all ASVs found in both the Hawaiʻi and EMP datasets (n = 116,507). Quantile box plots are overlaid as in Fig. 2) The line tracks the mean. (B) Histograms indicate the proportion of ASVs from the combined dataset (n = 1,911,880) unique to the Hawaiʻi dataset (local) as a function of EMPO 3 category breadth. (C) Latitudinal ranges of ASVs differed significantly by habitat (ANOVA F = 1,279, P > 0.0001, Tukey post hoc all pairs P < 0.0001). (D) Marine samples contained the highest percentage of local ASVs.
We also found significant differences between the mean latitudinal range of microbial ASVs in different habitats in the Waimea watershed. The average latitudinal range of a microbe was comparable between terrestrial and stream habitats; however, the average range size of microbes in marine systems was significantly smaller. While this might indicate a fundamental difference between dispersal ability or niche breadth of microbes among these habitats, other factors might account for this difference. First, the geographic ranges of marine hosts might be more limited than those sampled on land or in streams. Second, the fact that terrestrial and stream microbiomes were sampled along a strong elevation gradient might have contributed to their increased range size overall. The positive relationship between a site’s elevation at Waimea and the global range size of microbes it contains (SI Appendix, Fig. S3B) is congruent with an extension of Rapoport’s rule (39) that suggests because residents of higher elevations are subject to a greater seasonal and diurnal breadth of climatic conditions than those at sea level, they should occur over greater latitudinal extents. In contrast, marine microbes were sampled at a single, tropical location with comparatively little diurnal or seasonal variance. Studies show that pelagic marine bacteria from the tropics have the smallest ranges (3, 40), and we might expect that samples collected from higher latitudes would have larger range sizes.
A Watershed as a Microbial Mesocosm.
Our study highlights the interconnectedness of hosts and environments that is overlooked when microbiomes of ecosystem components are examined in isolation. Our landscape-scale study demonstrates that across major habitat divides and steep abiotic gradients, environmental microbiomes are taxonomically rich relative to those associated with hosts, and are potentially a significant source pool for microbiomes throughout foodwebs, particularly in aquatic habitats. We anticipate that these linkages are important in similarly connected landscapes at much broader geographic scales. Our results provide a useful and important framework to understand microbiome dynamics from individual hosts to entire ecosystems as structured within their respective foodwebs. We provide evidence for the contribution of the environment to microbiome compositional stability and stepwise dispersal, which might hold the key to harnessing microbiomes for beneficial engineering and restoration efforts.
Materials and Methods
Sample Collection and Library Preparation.
Plots were selected along the gradient for their accessibility, spacing, and adjacency of suitable stream and terrestrial habitat. Plots were delineated as 20-m-diameter circles in terrestrial and marine habitats, and 100 m stretches along the perennial Kamananui stream. Biological samples were collected using a stratified random design balancing sample types and approximating trophic web abundance distributions (SI Appendix, Methods and https://www.protocols.io/view/waimea-field-sampling-cadbsa2n). DNA was extracted using sample-specific protocols, and negative controls were included at random positions in every PCR and extraction plate (19 PCR negatives, 21 extraction negatives, 2 sterile filter negatives). Samples were sequenced across three lanes of a HiSEq. 2500 at GENEWIZ (South Plainfield, NJ, USA) using 2 × 250 bp reads (SI Appendix, Methods).
Data Processing.
Sequences were demultiplexed and processed using the MetaFlow|mics analysis pipeline (41), which uses DADA2 (42) to filter low-quality reads, denoise the data, and merge forward and reverse reads. ASVs generated by DADA2 were subsequently processed using mothur (43) along with the Silva database v138 to filter and annotate sequences. We removed potential chimeras with VSEARCH (44). Finally, we used the LULU algorithm (45) with default settings to collapse putative within-genome ribotype variants into a single ASV. Samples with <10,000 reads were discarded. The complete dataset contained 1,562 samples consisting of 355,693 ASVs and a mean sequencing depth of 96,906 +/− 89,508 (SD). Sequencing depth comprises >99% of hypothesized ASV diversity within samples across all sample types (SI Appendix, Fig. S1). This method does not distinguish between DNA in viable and nonviable cells, and occurrence counts reflect both, potentially inflating the frequency of occurrence of some ASVs (46). Mean ASV richness in negative controls (44.3) was two orders of magnitude lower than in biological samples (1,040.8) and represented 0.4% of the overall diversity detected in the study (1,391 ASVs out of 355,693). A reduced version of the dataset was generated for analyses that relied on distance matrices of entire microbial communities (ordinations, clustered heatmaps, and PERMANOVAs) to reduce computational time.
Statistical Analyses.
To assess adequacy of sequencing depth we calculated rarefaction curves. Extrapolations indicated that sequencing likely captured >99% of estimated microbial diversity, and for this reason, no normalization or downsampling steps were undertaken on the complete dataset to account for differential sequencing effort, and observed richness was deemed appropriate.
To rank sample type categories by their contributions to cumulative maximized ecosystem richness, each sample was downsampled to 6,000 sequences and ten samples were randomly selected from each sample type. Sample types were then ordered by their contribution to cumulative richness. This process was repeated over 1,000 bootstraps to achieve a hierarchy and distribution of microbial richness.
Non metric multidimensional scaling ordinations were performed in Vegan (47) on Bray-Curtis distances of the reduced dataset using relative abundance-transformed data. PERMANOVA analyses were calculated using type II sum of squares on the reduced dataset using the R package RVAideMemoire (48). To evaluate the effects of the environmental gradient, a second PERMANOVA was used on the same distance matrix in which marine samples were excluded.
To evaluate how compositional overlap of stream and terrestrial communities varied across the transect, we computed Bray-Curtis dissimilarity between all pairs of stream and terrestrial samples within a site (i.e., each “Beach” stream microbiome compared to each “Beach” terrestrial microbiome). These values were analyzed using GAM beta regression, parameterized for 1-inflated data, using both geographic position and trophic guild as predictors (SI Appendix, Methods).
To evaluate patterns of nestedness, we created a series of matrices in which samples were summed by sample types and separated by site location and habitat. These contingency tables were used to create bipartite networks in order to calculate network indices WNODF (an implementation of nestedness metric based on overlap and decreasing fill that is weighted by sequence abundance) and H2. The WNODF (49) indicates the average proportion of a lower richness subset that is contained in a higher richness subset, weighted by abundance, when all pairwise combinations of subsets in a network are considered. The analysis can be partitioned into “columns” (a measure of compositional overlap of samples, rows (a measure of overlap of incidence of ASVs), or a combination of the two. Because we were most interested in compositional redundancies among microbiomes and sample types, we chose to restrict the analysis to columns. Values for WNODFc range from 0 (no nestedness) to 100 (perfect nestedness). To evaluate specialization of the same networks, we calculated the H2’ index (50), which is a network-wide measure of interaction specialization among hosts and symbionts. The index calculates the extent to which species interaction deviates from random association with potential network members. The index ranges from 0 (no specialization) to 1 (perfect specialization). WNODFc and H2 values were compared among habitats using a one-way ANOVA.
To evaluate how EMPO 3 occupancy in Waimea predicts global distributions, we calculated the absolute latitudinal ranges of ASVs present in both the Earth Microbiome Project and Waimea datasets. These were calculated by subtracting the absolute minimum latitude from the absolute maximum latitude as recorded in the Qiita metadata. To generate a composite dataset, including samples from both the EMP and this study, raw FASTQ data were processed using the Qiita (51) portal following the methods of Thompson (26). The combined dataset contained 28,841 samples consisting of 1,911,880 ASVs and a mean sequencing depth of 48,603 +/− 50,696 (SD). Of those ASVs, 16% (309,467) were present in at least one Waimea sample. A total of 136,432 of the ASVs in the composite dataset were present in both the Waimea and at least one EMP sample, representing 44% of the Waimea ASVs. Mean differences between range size of microbes residing among habitats were evaluated with a one-way ANOVA, and pairwise differences were calculated using a Tukey test.
Supplementary Material
Acknowledgments
We thank Eoin Brodie, Pieter Dorrestein, Jannet Janssen, Rob Knight, Jennifer Martiny, Monique Chyba, and Edward Ruby for their input; Cedric Aridakessian for assistance with data processing; and Tanja Lantz Hirvonen, Reece Kilbey, Brennan Hee, Kahiwahiwa Davis, Joma Santos, Leina Uemura, Nicole Yoneishi, Anastasia Morse, Shayle Matsuda, Campbell Gunnel, David Pence, Chris Wall, and Jeff Kuwabara for assistance in the laboratory and field. We thank Richard Pezzulo, Chad Durkin, Josie Hoh, and Laurent Pool of Hi’ipaka LLC and Waimea Botanical Garden for their assistance. This content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204146119/-/DCSupplemental.
Data, Materials, and Software Availability
Code for reproducing sequence processing, data analysis, and figure generation is provided at Github (https://github.com/soswift/microbial_mapping) (52) and is archived at Figshare (https://doi.org/10.6084/m9.figshare.14497992) along with sample by ASV matrices, FASTA sequences, and sampling data (53). Sequence files and sample metadata that support the findings of this study are available from SRA BioProject with project No. PRJNA701450 (54) and from Qiita with study ID 13115.
References
- 1.Shade A., Caporaso J. G., Handelsman J., Knight R., Fierer N., A meta-analysis of changes in bacterial and archaeal communities with time. ISME J. 7, 1493–1506 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fierer N., Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Amend A. S., et al. , Macroecological patterns of marine bacteria on a global scale. J. Biogeogr. 40, 800–811 (2013). [Google Scholar]
- 4.Tedersoo L., Bahram M., Zobel M., How mycorrhizal associations drive plant population and community biology. Science 367, eaba1223 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Song S. J., et al. , Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. MBio 11, e02901-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks A. W., Kohl K. D., Brucker R. M., van Opstal E. J., Bordenstein S. R., Phylosymbiosis: Relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 14, e2000225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locey K. J., Lennon J. T., Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. U.S.A. 113, 5970–5975 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kembel S. W., et al. , Relationships between phyllosphere bacterial communities and plant functional traits in a neotropical forest. Proc. Natl. Acad. Sci. U.S.A. 111, 13715–13720 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guittar J., Shade A., Litchman E., Trait-based community assembly and succession of the infant gut microbiome. Nat. Commun. 10, 512 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elton C. S., Animal Ecology, by Charles Elton; With an Introduction by Julian S. Huxley (Macmillan Co., New York, 1927). [Google Scholar]
- 11.Lindeman R. L., The trophic-dynamic aspect of ecology. Ecology 23, 399–417 (1942). [Google Scholar]
- 12.Odum E. P., The strategy of ecosystem development. Science 164, 262–270 (1969). [DOI] [PubMed] [Google Scholar]
- 13.Holmes R. T., Likens G. E., Hubbard Brook (Yale University Press, 2020). 10.12987/9780300220780. [DOI] [Google Scholar]
- 14.Likens G. E., Driscoll C. T., Buso D. C., Long-term effects of acid rain: Response and recovery of a forest ecosystem. Science 272, 244–246 (1996). [Google Scholar]
- 15.Groffman P. M., et al. , Colder soils in a warmer world: A snow manipulation study in a northern hardwood forest ecosystem. Biogeochemistry 56, 135–150 (2001). [Google Scholar]
- 16.Wiens J. A., Stenseth N. C., Van Horne B., Ims R. A., Ecological mechanisms and landscape ecology. Oikos 66, 369 (1993). [Google Scholar]
- 17.Fahrig L., Effects of habitat fragmentation on biodiversity. Annu. Rev. Ecol. Evol. Syst. 34, 487–515 (2003). [Google Scholar]
- 18.Mony C., Vandenkoornhuyse P., Bohannan B. J. M., Peay K., Leibold M. A., A landscape of opportunities for microbial ecology research. Front. Microbiol. 11, 561427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller E. T., Bohannan B. J. M., Life between patches: Incorporating microbiome biology alters the predictions of metacommunity models. Front. Ecol. Evol. 7, 276 (2019). [Google Scholar]
- 20.Bittleston L. S., Gralka M., Leventhal G. E., Mizrahi I., Cordero O. X., Context-dependent dynamics lead to the assembly of functionally distinct microbial communities. Nat. Commun. 11, 1440 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Green J. L., Bohannan B. J. M., Whitaker R. J., Microbial biogeography: From taxonomy to traits. Science 320, 1039–1043 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Tipton L., et al. , Fungal aerobiota are not affected by time nor environment over a 13-y time series at the Mauna Loa Observatory. Proc. Natl. Acad. Sci. U.S.A. 116, 25728–25733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martiny J. B. H., et al. , Microbial biogeography: Putting microorganisms on the map. Nat. Rev. Microbiol. 4, 102–112 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Brown J. H., On the relationship between abundance and distribution of species. Am. Nat. 124, 255–279 (1984). [Google Scholar]
- 25.Slatyer R. A., Hirst M., Sexton J. P., Niche breadth predicts geographical range size: A general ecological pattern. Ecol. Lett. 16, 1104–1114 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Thompson L. R., et al. ; Earth Microbiome Project Consortium, A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pimm S. L., Food Webs (University of Chicago Press, Chicago, IL, 2002). [Google Scholar]
- 28.Bernard J., et al. , Plant part and a steep environmental gradient predict plant microbial composition in a tropical watershed. ISME J. 15, 999–1009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadro S., Nelson C. E., Melack J. M., The influence of landscape position and catchment characteristics on aquatic biogeochemistry in high-elevation lake-chains. Ecosystems (N.Y.) 15, 363–386 (2012). [Google Scholar]
- 30.Kling G. W., Kipphut G. W., Miller M. M., O’Brien W. J., Integration of lakes and streams in a landscape perspective: The importance of material processing on spatial patterns and temporal coherence. Freshw. Biol. 43, 477–497 (2000). [Google Scholar]
- 31.Nelson C. E., Sadro S., Melack J. M., Contrasting the influences of stream inputs and landscape position on bacterioplankton community structure and dissolved organic matter composition in high-elevation lake chains. Limnol. Oceanogr. 54, 1292–1305 (2009). [Google Scholar]
- 32.Crump R. C., Adams H. E., Hobbie J. E., Kling G. W., Biogeography of bacterioplankton in lakes and streams of an Arctic tundra catchment. Ecology 88, 1365–1378 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Stadler M., Del Giorgio P. A., Terrestrial connectivity, upstream aquatic history and seasonality shape bacterial community assembly within a large boreal aquatic network. ISME J. 16, 937–947 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinstein S. B., et al. , Microbiome stability and structure is governed by host phylogeny over diet and geography in woodrats (Neotoma spp.). Proc. Natl. Acad. Sci. U.S.A. 118, e2108787118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amend A. S., et al. , Phytobiomes are compositionally nested from the ground up. PeerJ 7, e6609 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christian N., Herre E. A., Mejia L. C., Clay K., Exposure to the leaf litter microbiome of healthy adults protects seedlings from pathogen damage. Proc. Biol. Sci. 284, 20170641 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shade A., Jacques M.-A., Barret M., Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 37, 15–22 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Sprent J. I., James E. K., Legume evolution: Where do nodules and mycorrhizas fit in? Plant Physiol. 144, 575–581 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stevens G. C., The elevational gradient in altitudinal range: An extension of Rapoport’s latitudinal rule to altitude. Am. Nat. 140, 893–911 (1992). [DOI] [PubMed] [Google Scholar]
- 40.Sul W. J., Oliver T. A., Ducklow H. W., Amaral-Zettler L. A., Sogin M. L., Marine bacteria exhibit a bipolar distribution. Proc. Natl. Acad. Sci. U.S.A. 110, 2342–2347 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arisdakessian C., Cleveland S. B., Belcaid M., Practice and Experience in Advanced Research Computing (ACM, New York, NY, 2020). 10.1145/3311790.3396664. [DOI] [Google Scholar]
- 42.Callahan B. J., et al. , DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schloss P. D., et al. , Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rognes T., Flouri T., Nichols B., Quince C., Mahé F., VSEARCH: A versatile open source tool for metagenomics. PeerJ 4, e2584 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frøslev T. G., et al. , Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun. 8, 1188 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lennon J. T., Muscarella M. E., Placella S. A., Lehmkuhl B. K., How, when, and where relic DNA affects microbial diversity. MBio 9, e00637-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixon P., VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930 (2003). [Google Scholar]
- 48.Hervé M., RVAideMemoire: Testing and plotting procedures for biostatistics. R Package Version 0. 9–69. https://CRAN.R-project.org/package=RVAideMemoire (2018). Accessed 15 January 2021.
- 49.Almeida-Neto M., Ulrich W., A straightforward computational approach for measuring nestedness using quantitative matrices. Environ. Model. Softw. 26, 173–178 (2011). [Google Scholar]
- 50.Blüthgen N., Menzel F., Blüthgen N., Measuring specialization in species interaction networks. BMC Ecol. 6, 9 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonzalez A., et al. , Qiita: Rapid, web-enabled microbiome meta-analysis. Nat. Methods 15, 796–798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Swift S. O. I., Microbial_mapping. GitHub. https://github.com/soswift/microbial_mapping. Deposited 22 April 2021.
- 53.Amend A. S., et al., Waimea Main Dataset. Figshare. https://figshare.com/articles/dataset/Waimea_Main_Dataset/14497992. Accessed 27 April 2021.
- 54.Amend A. S., Swift S. I. O, A whole-watershed microbiome reveals a cross-biome role in symbiont sources and sinks. NCBI:BioProject. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA701450. Accessed 11 February 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Code for reproducing sequence processing, data analysis, and figure generation is provided at Github (https://github.com/soswift/microbial_mapping) (52) and is archived at Figshare (https://doi.org/10.6084/m9.figshare.14497992) along with sample by ASV matrices, FASTA sequences, and sampling data (53). Sequence files and sample metadata that support the findings of this study are available from SRA BioProject with project No. PRJNA701450 (54) and from Qiita with study ID 13115.