Significance
Emerging zoonotic viruses are at the forefront due to the ongoing COVID-19 pandemic. Bunyaviruses are a large group of diverse, arthropod-borne viruses that present concern due to reassortment and evolutionary capacity of their segmented RNA genomes. Here, we demonstrate that the conserved host cell surface receptor low-density lipoprotein receptor-related protein (Lrp1) facilitates efficient cellular infection by the South American bunyavirus Oropouche virus (OROV). Therefore, Lrp1 is a host factor for multiple bunyaviruses, including OROV and Rift Valley fever virus (RVFV), and it plays a broader role in bunyavirus infection than has been previously known. This is the first study to identify a pan-bunyaviral host factor with significant implications for therapeutic targets.
Keywords: Oropouche virus, bunyavirus, Lrp1, lipoprotein, entry factor
Abstract
Oropouche orthobunyavirus (OROV; Peribunyaviridae) is a mosquito-transmitted virus that causes widespread human febrile illness in South America, with occasional progression to neurologic effects. Host factors mediating the cellular entry of OROV are undefined. Here, we show that OROV uses the host protein low-density lipoprotein–related protein 1 (Lrp1) for efficient cellular infection. Cells from evolutionarily distinct species lacking Lrp1 were less permissive to OROV infection than cells with Lrp1. Treatment of cells with either the high-affinity Lrp1 ligand receptor-associated protein (RAP) or recombinant ectodomain truncations of Lrp1 significantly reduced OROV infection. In addition, chimeric vesicular stomatitis virus (VSV) expressing OROV glycoproteins (VSV-OROV) bound to the Lrp1 ectodomain in vitro. Furthermore, we demonstrate the biological relevance of the OROV-Lrp1 interaction in a proof-of-concept mouse study in which treatment of mice with RAP at the time of infection reduced tissue viral load and promoted survival from an otherwise lethal infection. These results with OROV, along with the recent finding of Lrp1 as an entry factor for Rift Valley fever virus, highlight the broader significance of Lrp1 in cellular infection by diverse bunyaviruses. Shared strategies for entry, such as the critical function of Lrp1 defined here, provide a foundation for the development of pan-bunyaviral therapeutics.
Bunyaviruses are a large group of related viruses with single-stranded, segmented, negative, or ambisense RNA genomes (1, 2). Within the order Bunyavirales, the Peribunyaviridae family contains viruses that infect humans and animals with confirmed or potential zoonotic transmission (2, 3). Oropouche virus (OROV; Orthobunyavirus genus; Simbu serogroup) is found primarily in the South American regions of Brazil, Trinidad, Peru, Panama, and Tobago (4). OROV has caused more than 30 epidemics, resulting in excess of 500,000 total cases of human febrile illness, making it the second most common arboviral disease in Brazil, behind Dengue fever (4–6). The true case number is likely higher as clinical testing for OROV is lacking and patients are often misdiagnosed as having Chikungunya or Dengue fevers (4). The arthropod vectors for OROV include Culicoides midges and Culex mosquitoes. In humans, OROV causes a febrile illness that manifests as fever, intense headache, myalgia, joint pain, retro-orbital pain, and photophobia, which can further develop into encephalitis or meningitis (5–7). Systemic infection manifests as rash, nausea, vomiting, and diarrhea. Viremia and leukopenia are common features (6), and virus can be detected in the cerebrospinal fluid (8, 9). In mice, the virus replicates in the liver and spleen after either subcutaneous or intracerebral infection (10).
Due to the broad cellular tropism and ability to infect a variety of species, bunyaviruses are thought to use multiple receptors or attachment factors for entry and/or a protein that is widely expressed across different tissues and conserved across species. Recently, using a CRISPR-Cas9 screen, the conserved host protein low-density lipoprotein receptor (LDLR)-related protein-1 (Lrp1) was reported to mediate cellular infection with Rift Valley fever virus (RVFV), a phlebovirus within the Bunyavirales order (11). Lrp1 (also known as alpha-2-macroglobulin receptor or CD91) is a highly conserved multifunctional member of the LDLR family of transmembrane surface proteins. Lrp1 is important for ligand endocytosis, cell signaling, lipoprotein metabolism, blood–brain barrier maintenance, and angiogenesis (12–15). Homozygous deletion of Lrp1 is embryonically lethal in mice (16), further supporting the critical nature of Lrp1 in homeostatic functions.
The M segment of Bunyavirales encodes the surface glycoproteins Gn and Gc, which form heterodimers and multimerize on the surface of the virion. Few studies have been conducted on the binding and entry mechanisms facilitated by OROV Gn/Gc. Given the conserved nature of Lrp1 across taxonomically distinct species and its expression in different tissues, we investigated whether OROV, a bunyavirus distantly related to RVFV, also requires Lrp1 for efficient infection of host cells. Despite having a similar genome organization among members of the Bunyavirales, many of the virally encoded sequences show little sequence homology. Therefore, studies to define similar host protein usage by these two distantly related viruses would have significant implications for pan-bunyavirus therapeutic and diagnostic development.
Here, we used Lrp1 knockout (KO) cell lines to show that OROV infection is decreased compared to parental cells expressing Lrp1. Pretreatment of cells with varying concentrations of the high-affinity Lrp1-binding protein receptor-associated protein (RAP) significantly reduced OROV infection. Zika virus (ZIKV), an arbovirus outside the Bunyavirales order, was unaffected by the loss of Lrp1 or by treatment with Lrp1-binding RAP. Chimeric virions expressing OROV glycoproteins bound to the Lrp1 ectodomain. Finally, the role of Lrp1 in OROV infection was validated in vivo, whereby RAP treatment was able to reduce viral tissue titers and rescue mice from lethal intracerebral infection with OROV. Based on our findings, Lrp1 is a host factor for multiple bunyaviruses, presenting a potential therapeutic approach to address this important group of emerging arboviruses. This work also paves the way for future studies to understand the mechanism of OROV binding to Lrp1.
Results
OROV Infection Is Reduced in Lrp1 KO Cell Lines.
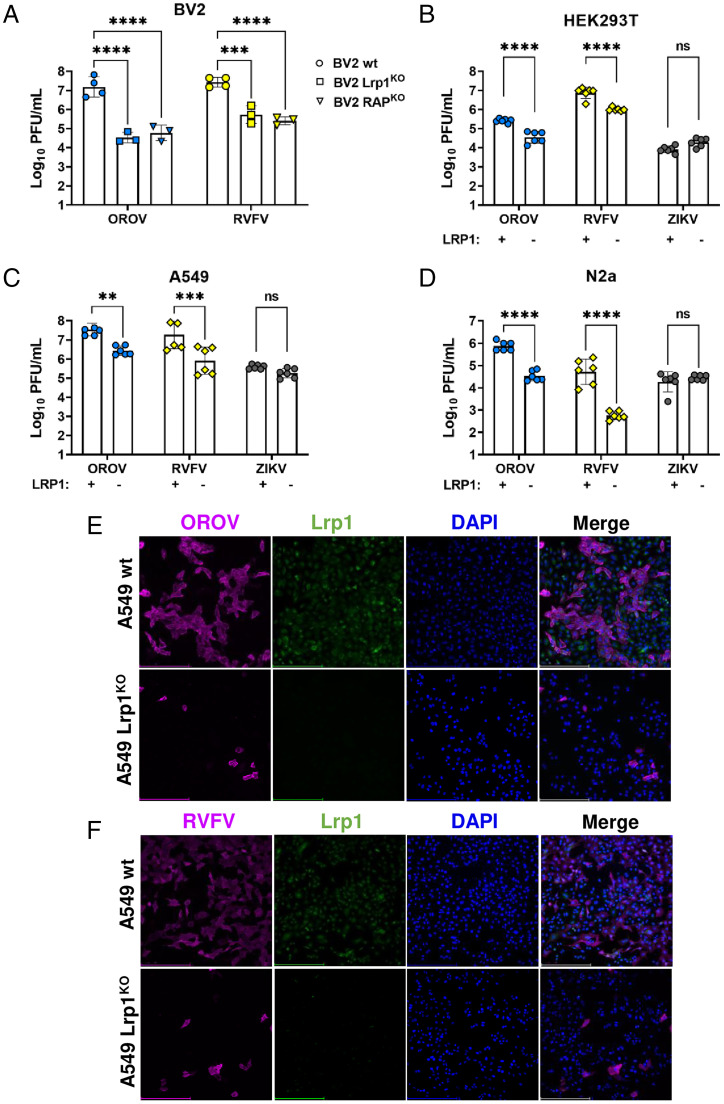
OROV strain BeAn19991 (17) was grown in mouse microglial BV2 cells at a multiplicity of infection (MOI) of 0.1 and 0.01 along with RVFV strain ZH501 and ZIKV strain PRVABC59 for comparison (SI Appendix, Fig. S1A). While ZIKV did not replicate well in BV2 cells, OROV and RVFV reached 106 PFU/mL by 24 h postinfection (hpi) at MOI 0.1, and these parameters were used for the remaining cellular infection studies. Clonal BV2 KO cell lines that are deleted for either Lrp1 or the Lrp1 chaperone protein RAP express significantly reduced levels of Lrp1, and this was visualized and quantified using immunofluorescence and western blot (SI Appendix, Fig. S1B–D) (11). Infection of both Lrp1 and RAP clonal KO cell lines with OROV resulted in significantly less infectious virus produced by 24 hpi when compared to the infection of BV2 wild-type (WT) cells (Fig. 1A), with reductions of 2 to 3 log in titer for both OROV and RVFV as a comparator. Thus, OROV requires Lrp1 or related proteins for efficient cellular infection and production of infectious virus from BV2 cells.
Fig. 1.
OROV and RVFV show reduced infection in multiple cell lines that are KO for Lrp1. (A) Infection of Lrp1 KO and RAP KO cell lines described in Ganaie et al. (11), with RVFV and OROV at MOI 0.1. Infection of WT and Lrp1 KO versions of (B) HEK293T, (C) A549, and (D) N2a cells with OROV, RVFV, and ZIKV at MOI 0.1. OROV and RVFV samples were harvested at 24 hpi, and infectious virus was measured by viral plaque assay. ZIKV samples were harvested at 48 hpi and viral RNA (vRNA) was evaluated by qRT-PCR. Fluorescent microscopy (20×) of (E) OROV and (F) RVFV infection of A549 WT and Lrp1 KO cells at MOI 0.1 at 24 hpi. Scale bars, 250 μm. Statistical significance was determined using an unpaired t test on log-transformed data. Experiments were repeated three times. **P < 0.01; ***P < 0.001; ****P < 0.0001.
Additional clonal Lrp1 KO cell lines were established in human HEK293T, A549, and murine N2a cell lines, with the loss of Lrp1 verified by western blot (SI Appendix, Fig. S1D). As with BV2 cells, Lrp1 KO resulted in significantly reduced OROV infection across all cell lines. While similar reductions were seen with RVFV in Lrp1 KO cells, no significant difference in virus infection or production was observed in cells infected with Zika virus (ZIKV), a flavivirus used as a control (Fig. 1 B–D). The titers for both OROV and RVFV infection of KO cell lines were 10- to 100-fold lower than WT cell lines. By immunofluorescence, Lrp1 was detectable in WT parental A549 cells but was absent from Lrp1 KO lines (Fig. 1 E and F). The number of OROV-infected cells at 24 hpi was reduced at least 10-fold in the KO line (Fig. 1E), which corresponds to the observed reduction in titers.
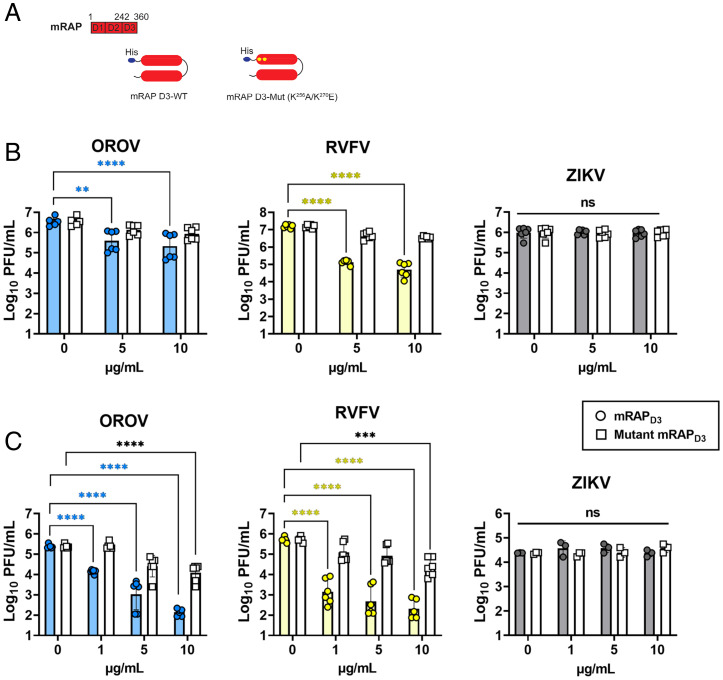
The Lrp1-Binding Chaperone Protein RAP Inhibits OROV Infection of Cell Lines from Taxonomically Distinct Species.
RAP (or Lrpap1) is a high-affinity Lrp1 ligand and critical chaperone of Lrp1 and other LDLR family members (18). Domain 3 of RAP (RAPD3) (Fig. 2A) specifically binds to two extracellular cluster domains of Lrp1 (CLII and CLIV) and competes for attachment with other compatible ligands while chaperoning the protein through the endoplasmic reticulum (ER) to the cell surface (18). Here, we tested the ability of exogenous mouse RAPD3 (mRAPD3) to block OROV infection by adding it before infection, at the time of infection, or following infection (SI Appendix, Fig. S2A). After determining that pretreatment most effectively blocked infection, mRAPD3 was added to murine BV2 microglial cells, monkey Vero E6 kidney cells, and human SH-SY5Y neuroblastoma cells at various concentrations 1 h before infection with MOI 0.1 of OROV, RVFV, or ZIKV as comparators (Fig. 2 B and C and SI Appendix, Fig. S2B). At 24 hpi, samples were tested for infectious virus by plaque assay. As a control, mutant mRAPD3 in which two lysines were changed (Fig. 2A) was tested in parallel, as these mutations have been shown previously to reduce the binding of RAP to CLII and CLIV of Lrp1 (11, 19, 20). In all of the cell lines, treatment with mRAPD3 significantly reduced OROV infection compared to untreated cells at all concentrations, and the reduction in infection was comparable to RVFV (Fig. 2 B and C and SI Appendix, Fig. S2B). The mutant mRAPD3 was less effective at inhibiting both viruses at lower concentrations, with significant inhibition of both OROV and RVFV at the highest concentrations tested (10 μg/mL). ZIKV does not efficiently infect BV2 cells, but it does infect SH-SY5Y and Vero cells (SI Appendix, Fig. S1A). Neither mRAPD3 nor mutant mRAPD3 inhibited the ZIKV infection of SH-SY5Y or Vero cells (Fig. 2 B and C).
Fig. 2.
The Lrp1-binding chaperone RAP can inhibit OROV infection of Vero E6 cells and undifferentiated SH-SY5Y cells. (A) RAP is a 39-kDa ER-resident protein consisting of 3 domains (D1-3) that chaperones LDLR family proteins, including LRP1. Recombinant mRAPD3 and mutant mRAPD3 (K256A and K270E) were expressed and purified from BL21 (DE3) cells using an N-terminal His-tag. mRAPD3 or mutant mRAPD3 was added to (B) Vero E6 nonhuman primate cells or (C) SH-SY5Y human neuroblastoma cells 1 h before infection with MOI 0.1 of RVFV, OROV, or ZIKV. Samples were harvested at 24 hpi (for RVFV and OROV) or 48 hpi (for ZIKV), and infectious virus was measured by plaque assay or qRT-PCR. Statistical significance was determined using two-way ANOVA on log-transformed data. Experiments were repeated three times. **P < 0.01; ****P < 0.0001.
OROV Interaction with Lrp1 Is Dependent on Viral Glycoproteins.
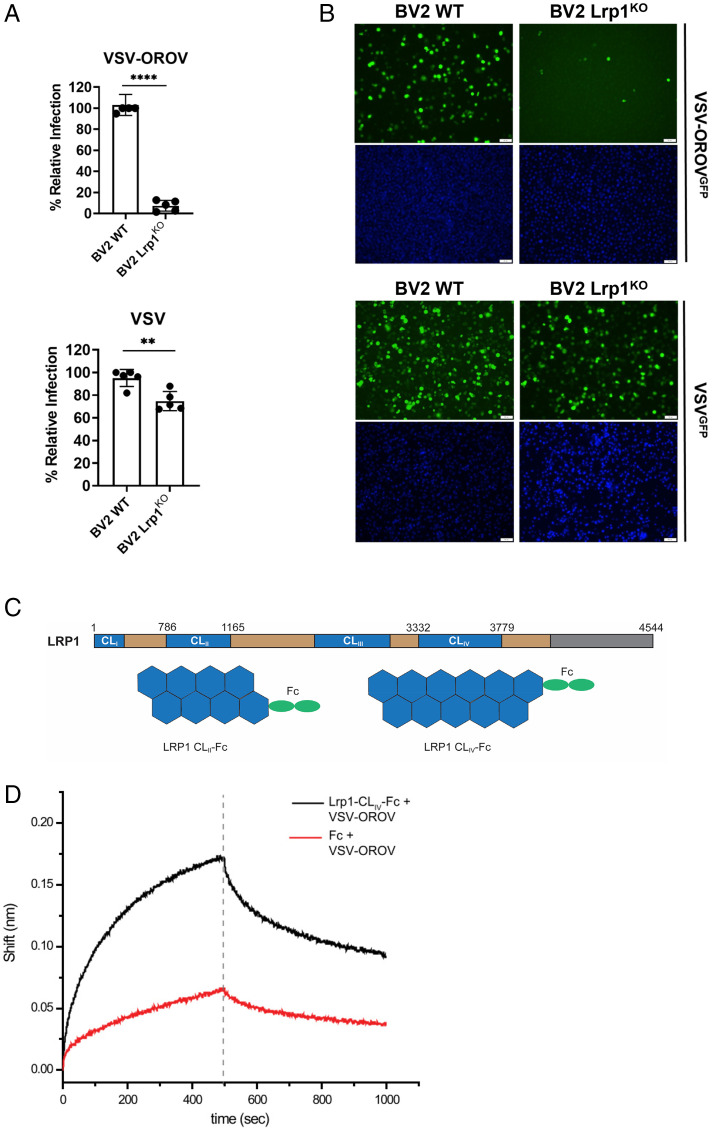
To determine whether the restriction in OROV infection in Lrp1-deficient cells is at the level of the surface glycoproteins, we used a chimeric vesicular stomatitis virus (VSV) expressing green fluorescent protein (GFP) and the OROV glycoproteins Gn and Gc (VSV-OROV). Purified VSV-OROV or VSV control virions were used to infect BV2 WT cells or BV2 Lrp1 KO cells. Samples were collected at 6 to 8 hpi and analyzed for GFP expression by flow cytometry (Fig. 3A and SI Appendix, Fig. S3) or imaging by fluorescent microscopy (Fig. 3B). VSV-OROV infection was significantly reduced in BV2 Lrp1 KO cells. VSV infection was also significantly reduced but to a lesser degree, likely due to its utilization of other LDLR family members for viral entry (21). The reduction in VSV-OROV infection was confirmed by immunofluorescent microscopy (Fig. 3B).
Fig. 3.
Lrp1 KO reduces VSV-OROV infection in BV2 cells and VSV-OROV binds to Lrp1 CLIV. BV2 WT and BV2 Lrp1 KO cells were infected with MOI 1 of VSV or MOI of 5 of VSV-OROV. Samples were collected at 6 and 8 hpi to be processed by (A) flow cytometry or (B) imaging by fluorescent microscopy (20×). Scale bars, 50 μm. (C) LRP1 consists of a 515-kDa extracellular alpha chain (blue/tan) and an 85-kDa intracellular beta chain (not shown) connected by a transmembrane domain (gray). The alpha chain is further divided into four complement-type repeat clusters (CLI-IV; blue), and epidermal growth factor (EGF)-like and YWTD domains (tan). Recombinant Fc-fused LRP1 CLII and CLIV were expressed and purified from Expi293 cells for the experiments presented here. (D) AHC sensors coated with either Fc or Lrp1-CLIV-Fc and incubated with VSV-OROV particles. Sensograms show the over-time association and dissociation of virus particles to coated sensors. Significance was determined using an unpaired t test. Experiments were repeated two times. **P < 0.01; ****P < 0.0001.
Furthermore, because mRAPD3 is known to bind to Lrp1 CLIV and is able to block OROV infection (Fig. 2), we used biolayer interferometry to determine whether chimeric VSV-OROV binds to Lrp1 CLIV. To do this, we used a recombinant Fc-fusion of the Lrp1 CLIV domain (Fig. 3C), which was previously shown to block RVFV infection (11). We found that VSV-OROV virions bound to immobilized Fc-Lrp1 CLIV but not to Fc control (Fig. 3D).
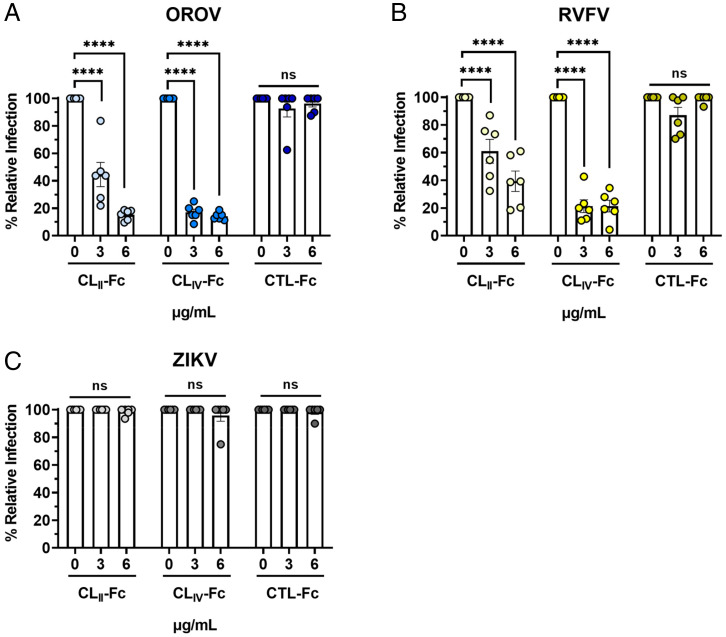
Lrp1 Cluster Domains CLII and CLIV Inhibit OROV Infection.
Many ligands of Lrp1 bind to the CLII and CLIV extracellular domains, including mRAPD3. Given the results showing VSV-OROV binding to CLIV (Fig. 3D), we treated Vero E6 cells with soluble Fc-fused CLII and CLIV proteins (Fig. 3C) (11) and compared the relative infection to untreated cells and Fc-control treated cells. We observed that Fc-fused CLII and CLIV treatment significantly reduced OROV infection compared to the Fc-control treated cells (Fig. 4A). These results are comparable to those of treated cells infected with RVFV at the same MOI (Fig. 4B). In addition, ZIKV infection of Vero E6 cells was unaffected by treatment with any Fc-bound Lrp1 proteins (Fig. 4C).
Fig. 4.
Soluble Fc-bound Lrp1 CLII and CLIV inhibit cellular infection by OROV. (A) Soluble Fc-bound CLII, CLIV, or Fc control proteins were added to Vero E6 cells 1 h before infection with (A) OROV, (B) RVFV, or (C) ZIKV at MOI 0.1. Samples were harvested at 24 hpi (OROV and RVFV) or 48 hpi (ZIKV), and virus was measured by plaque assay and qRT-PCR. Data are expressed as a percentage of untreated control titers. Statistical significance was determined using two-way ANOVA. Experiments were repeated two times. ****P < 0.0001.
OROV Infection Is Inhibited by the Glycoprotein Gn from RVFV.
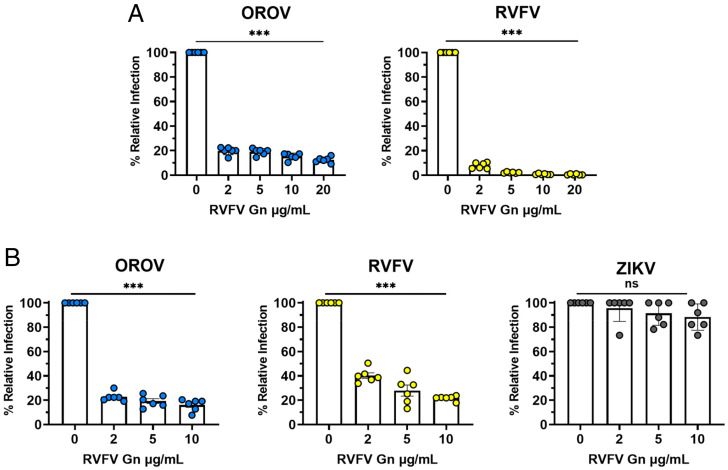
We previously determined that the RVFV Gn protein binds to CLII and CLIV of Lrp1, and that mRAPD3 can compete with RVFV Gn for Lrp1 binding, indicating overlapping binding sites (11). Furthermore, the addition of soluble RVFV Gn was able to block RVFV infection (11). Since mRAPD3 was also able to block OROV infection and VSV-OROV bound to CLIV, we next determined whether addition of RVFV Gn can similarly block OROV infection. We treated BV2 cells with RVFV Gn 1 h before OROV infection and kept it in the media for the duration of the experiment. At 24 hpi, we evaluated OROV titers by plaque assay. We compared RVFV Gn blocking of RVFV infection as a control. In BV2 cells, we found that pretreatment of cells with RVFV Gn at concentrations of 2, 5, 10, and 20 μg/mL significantly reduced infectious titers of both OROV and RVFV (Fig. 5A). This experiment was repeated in Vero E6 cells with the addition of ZIKV as a control. RVFV Gn significantly reduced OROV and RVFV infection in Vero E6 cells and showed no significant effect on ZIKV infection (Fig. 5B).
Fig. 5.
RVFV Gn inhibits cellular infection by OROV. (A) RVFV Gn was added to BV2 mouse microglia cells 1 h before infection with MOI 0.1 of OROV or RVFV ZH501. Samples were collected at 24 hpi and processed by viral plaque assay. (B) RVFV Gn was added to Vero E6 nonhuman primate cells 1 h before infection with MOI 0.1 of OROV, RVFV, or ZIKV. Samples were harvested at 24 hpi (for RVFV and OROV) or 48 hpi (for ZIKV) and infectious virus was measured by plaque assay and qRT-PCR. Data are expressed as a percentage of untreated control titers. Statistical significance was determined using one-way ANOVA. Experiments were repeated two times. ***P < 0.001.
mRAPD3 Treatment Rescues Mice from Lethal OROV Infection.
Because mRAPD3 reduced OROV infection in vitro, we used a proof-of-concept experiment to determine the in vivo relevance of this interaction. OROV does not cause lethal disease in adult mice when administered subcutaneously. However, the median lethal dose (LD50) of OROV administered by intracerebral (IC) injection in young adult mice is <5 PFU (SI Appendix, Fig. S4A). Mice succumbed to infection with an average time to death of 4.5 days postinfection (dpi) and viral titers in the brain at the time of death were >107 PFU/g tissue (SI Appendix, Fig. S4B).
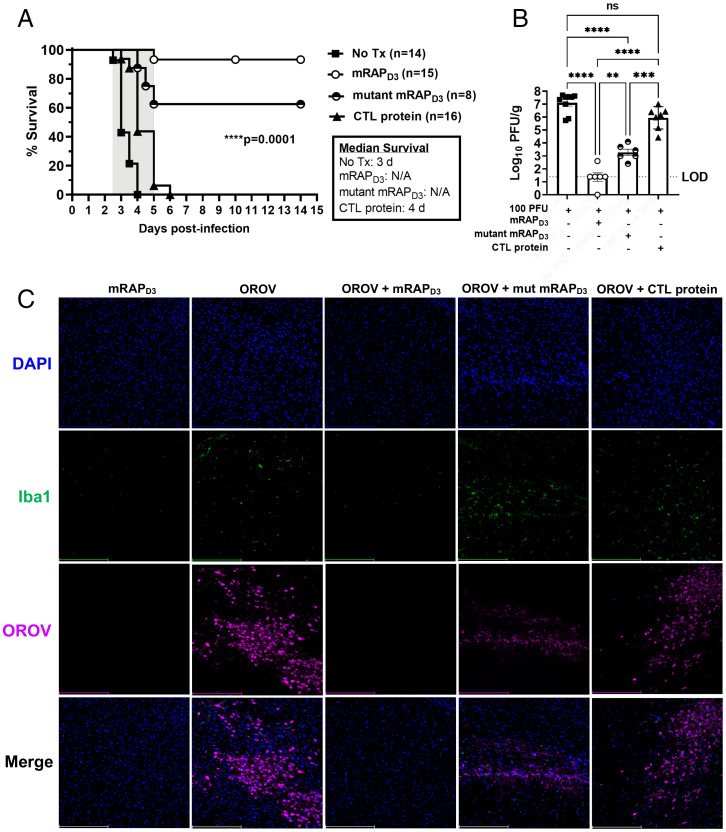
Based on the LD50, a dose of 100 PFU (at least 20× OROV IC LD50) was chosen for the mRAPD3 treatment experiments. OROV was administered IC in conjunction with 215 μg mRAPD3, mutant mRAPD3, or a similar-sized, unrelated control protein (Ebola VP30) to C57BL/6J mice in a proof-of-concept experiment (Fig. 6A). All untreated and control protein-treated mice succumbed by 4 to 6 dpi. Of the mice that received mRAPD3 treatment, 90% survived, while 60% of mice that received mutant mRAPD3 also survived.
Fig. 6.
mRAPD3 protects mice from lethal OROV IC infection and significantly reduces infectious virus in the brain at 3 dpi. (A) Mice were infected with 100 PFU of OROV IC alone or in combination with either mRAPD3, mutant mRAPD3, or the control protein VP30. They were monitored for 15 d to determine percentage of survival in each group. (B) A subset of mice from each group was euthanized at 3 dpi to collect brain tissue, which was processed by viral plaque assay. (C) Immunofluorescent microscopy of brain tissues (cerebral cortex) from mice euthanized at 3 dpi (20×). Scale bars, 250 μm. Statistical significance was determined using a Mantel-Cox test for survival and two-way ANOVA for log-transformed data. Experiments were repeated four times. **P < 0.01; ***P < 0.001; ****P < 0.0001. No tx, No treatment; mut, mutant.
A cohort of mice underwent planned euthanasia at 3 dpi to directly compare tissue titers across treatment groups. The untreated and control protein-treated mice had high viral titers in their brains (106 to 108 PFU/g tissue), while the mRAPD3-treated mice had levels of infectious virus that were near or at the limit of detection (Fig. 6B). Fitting with the survival data and in vitro data (Fig. 2C), mutant mRAPD3-treated mice had intermediate amounts of virus in the brain compared to mRAPD3 and control animals (Fig. 6B). This can be visualized by immunofluorescence microscopy using an anti-OROV N polyclonal antibody on cryosections of the cerebral cortex from 3 dpi brain tissues. The mRAPD3-treated mice have little to no staining for OROV N protein in the brain at 3 dpi (Fig. 6C and SI Appendix, Fig. S5A), whereas the untreated and control protein-treated mice had abundant, diffuse OROV staining in the brain (Fig. 6C). The mutant mRAPD3-treated mice had reduced, focal regions of OROV staining in the brain at 3 dpi, which was substantially more than mRAPD3-treated mice. Thus, the reduced inhibition of OROV infection displayed by the mutant mRAPD3 protein (Fig. 2) corresponds to reduced binding affinity to CLII and CLIV, intermediate tissue viral loads, and concomitant intermediate levels of survival in IC-infected mice.
We also stained the brain tissues with the microglial marker Iba-1 to examine immune activation within the tissue. Brains from mice receiving mRAPD3 treatment alone or mRAPD3 + OROV expressed low levels of Iba-1+ cells, indicative of a normal resting state in the brain. However, OROV infected and untreated, mutant mRAPD3-treated, or control protein-treated brain sections had more activated microglia (Iba1+ cells), indicating higher levels of inflammatory activation and recruitment (Fig. 6C).
Finally, serum samples from the surviving mRAPD3 and mutant mRAPD3-treated mice were tested for neutralizing activity against OROV using a plaque-reduction neutralization assay (PRNT50). Titers in the surviving mice were ≥1:40, while the uninfected control animals had no detectable neutralizing titer, thus confirming that these exposed yet surviving animals were infected with OROV (SI Appendix, Fig. S4C).
Discussion
Our data show that a highly conserved protein Lrp1 is a host factor that supports efficient cellular infection by Oropouche orthobunyavirus. We demonstrate that OROV infection of Lrp1-deficient cells was significantly decreased. We also show that high-affinity Lrp1-binding RAP significantly reduced OROV infection in vitro and in vivo. Direct association between OROV and Lrp1 was established with binding assays using VSV-OROV. The relevance of our finding is also supported by the in vivo studies, in which intracerebral infection of mice with OROV in the presence of mRAPD3 rescued the animals from an otherwise lethal infection and significantly reduced viral titers in the brain. Taken together, these data support a role for Lrp1 in OROV infection.
Lrp1 mediates cellular infection by the phlebovirus RVFV (11, 22). Lrp1 is needed for efficient in vitro cellular infection and to promote lethal RVFV infection in vivo in a mouse model. While important, the previous study (11) was focused on RVFV and did not implicate Lrp1 as having a broader impact. Here, we reveal that the orthobunyavirus OROV, while classified in a different family than RVFV, also uses Lrp1 to efficiently infect cells and cause disease in vivo, thus implicating Lrp1 as a much broader host factor for bunyaviral infection. Both viruses bind similar and potentially overlapping regions within Lrp1 extracellular domains CLII and CLIV. This finding implies some structural similarities between the two Gn proteins despite sequence diversity.
Consistent with our results, recent studies have examined the role of the LDLR family and related proteins in the context of viral infections. These efforts led to several reports, including a role for LDLR in binding and entry of dengue virus (23), hepatitis C virus (24), VSV (25), and rhinovirus (26). The LDLR-related protein LDLRAD3 was recently reported to facilitate the entry of Venezuelan equine encephalitis virus (27). Finally, VLDLR and ApoER2 were recently identified as entry receptors for multiple alphaviruses, including Semliki Forest virus, eastern equine encephalitis virus, and Sindbis virus, despite differences in E2/E1 amino acid sequence homology (28). These findings of related LDLRs, but not Lrp1, further support a wide-ranging role for this family of receptors in viral infections. However, our studies define a role for Lrp1 in mediating efficient infection by both OROV and RVFV. While consistent with this broader role of LDLRs in viral infections, this study highlights distinct drivers of specificity in the viral entry of bunyaviruses that are related to Lrp1.
The strain of OROV used here, BeAn 19991, was isolated from a sloth in Brazil in 1960 and falls within Lineage I (29). This is also the strain that served as the basis for the OROV reverse genetics system (17). While further studies testing OROV strains from Lineage II and III strains would be informative, the conserved nature of the interactions and our studies using heterologous proteins such as RVFV Gn and RAPD3 to inhibit OROV infection suggest that Lrp1 may support infection by multiple OROV lineages.
Lrp1 has a large ectodomain and binds many different ligands. RAP is one such ligand, and the mutant mRAPD3 reduces affinity for Lrp1, but it does not abolish binding (19). Here, we observed an intermediate reduction in both OROV and RVFV infection in the presence of the mutant mRAPD3 protein at the highest concentrations tested (10 μg/mL), whereas the mRAPD3 inhibits both viruses at 1 μg/mL. The intermediate in vitro phenotype of the mutant mRAPD3 correlated well with the in vivo OROV findings, in which we see intermediate levels of virus in the brain as well as 60% survival of the mice (compared to 93% survival with mRAPD3). In contrast to the in vitro data presented here, mutant mRAPD3 is unable to out compete RVFV in vivo (11). This observation may be attributable to either differences in the affinity of each of the Gn proteins for Lrp1 or, alternatively, to differences in inherent pathogenicity between OROV and RVFV, as RVFV is much more pathogenic. Studies to address these key observations and differences are ongoing.
The work presented here underscores a previously unappreciated role played by Lrp1 in cellular infection by diverse bunyaviruses. Our results with OROV and RVFV suggest that it is likely that Lrp1 is used by other members of the Bunyavirales order. Importantly, given the need to identify broadly acting inhibitors of emerging viruses, these results highlight the feasibility of Lrp1 as a pan-bunyaviral target. The dependence on Lrp1 by OROV (Peribunyaviridae) and RVFV (Phenuiviridae) suggest common structural elements that transcend sequence homology. Future studies to characterize the binding and internalization mechanisms of both OROV and RVFV will also identify potentially conserved binding epitopes as targets for therapeutic strategies.
Materials and Methods
Biosafety.
Work with OROV and ZIKV was completed in a Biosafety Level 2 (BSL-2) laboratory following all university biosafety guidelines. Work with RVFV was completed at BSL-3 in the Regional Biocontainment Laboratory at the University of Pittsburgh. The authors have approval from the Federal Select Agent Program to work with RVFV in the described facilities.
Animal Study Oversight.
The University of Pittsburgh is fully accredited by the Assessment and Accreditation of Laboratory Animal Care (AAALAC). The Guide for the Care and Use of Laboratory Animals published by the NIH was adhered to throughout the duration of the animal work. The Institutional Animal Care and Use Committee (IACUC) at the University of Pittsburgh oversaw and approved this work under the protocol number 19114577.
Cells.
The LPR1KO R4 and RAPKO A7 cells were generated, as previously described (11). All BV2 and Vero cells (American Type Culture Collection [ATCC], CRL-1586) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (ATCC, 30-2002) supplemented with 1% penicillin/streptomycin (Pen/Strep), 1% l-glutamine (l-Glut), and either 2% (D2), 10% (D10), or 12% (D12) fetal bovine serum (FBS). SH-SY5Y cells were obtained from ATCC (CRL-2266) and cultured in D10/F12 media (ATCC, 30-2006) supplemented with 1% Pen/Strep and 1% l-Glut. HEK293T, A549, and N2a clonal KO cells were generated by CRISPR-Cas9 using ribonucleoprotein complexes of Cas9 and Lrp1-specific guide RNAs, as previously described (11). The resulting cells were subcloned and subjected to next-generation sequencing analysis and short tandem repeat profiling to confirm the deletion and homogeneity of the clones. HEK293T Lrp1 KO cells were maintained in D10 media, A549 Lrp1 KO cells were maintained in D12/F12 media, and N2a Lrp1 KO cells were maintained in Eagle’s minimum essential medium (ATCC, 30-2003) with 10% FBS.
Viruses.
The BeAn19991 strain of OROV was rescued through reverse genetics and was generously provided by Paul Duprex and Natasha Tilston-Lunel (Pitt Center for Vaccine Research) (17). RVFV ZH501 was rescued through reverse genetics and provided by Stuart Nichol (Centers for Disease Control and Prevention [CDC]). The PRVABC59 (Human/2015/Puerto Rico) strain of ZIKV was obtained from BEI Resources (NR-50240) from the Arbovirus Reference Collection (CDC, Fort Collins, CO, USA). VSV-OROV virus was generated as described previously (30). All of the viruses were propagated in Vero E6 cells with standard culture conditions using D2 media supplemented with 1% Pen/Strep and 1% l-Glut. A standard viral plaque assay (VPA) was used to determine the titer of the stocks. The agar overlay for the VPA was comprised of 1× minimal essential medium, 2% FBS, 1% Pen/Strep, 1% HEPES buffer, and 0.8% SeaKem agarose; the assay incubated at 37°C for 4 d (OROV) and 3 d (RVFV), followed by visualization of plaques with 0.1% crystal violet.
Antibodies.
The following antibodies were used: rabbit anti-Lrp1 (Abcam ab92544), mouse anti-Lrp1 (Santa Cruz, sc-57353), rabbit anti-OROV N (Custom Genescript), and mouse anti-RVFV N (BEI Resources, NR-43188) for fluorescence immunostaining, and rabbit anti-Lrp1 (Cell Signaling Technology, 64099S) and rabbit anti-GAPDH (Thermo Fisher, PA1-987) for western blots.
Neutralization Assays with mRAPD3, RVFV Glycoprotein Gn, and Lrp1 clusters.
Cells were seeded in a 24-well plate at a density of 2.4E−5 cells/mL in D10 the day before infection. The following day, the media was removed and the virus was diluted in D2 (MOI 0.1) and added to the cell monolayer in a 200-μL volume. Following a 1-h incubation at 37 °C, the inoculum was removed. The monolayer was washed with 1× Dulbecco’s phosphate-buffered saline (dPBS), and fresh D2 media was added to each well. At the designated timepoint(s), supernatants were collected and infectious virus titers were determined by VPA. In the treatment assays, mRAPD3, mutant mRAPD3, RVFV Gn, Fc-CLII, IV, or Fc-control (trastuzumab) were diluted in D2 and added to the cell monolayer, followed by a 1-h incubation at 37 °C. Following the incubation, virus diluted in D2 (MOI 1, 0.1) was added to the media and incubated for 1 h at 37 °C. After the absorption period, the inoculum was removed, the cell monolayer was washed once with 1× dPBS, and D2 media containing the designated proteins was added in a 500-μL volume. Supernatants were collected at 24 hpi (OROV and RVFV) or 48 hpi (ZIKV) and viral titers were determined through VPA. Vial titers for RVFV and ZIKV were also analyzed through qRT-PCR as previously described (31–33).
Immunofluorescence.
Cover glass (FisherBrand, 12-546-P) was sterilized in 70% EtOH and coated with BME Cultrex (R&D Systems, 3432-010-01) before seeding. Cells were seeded the day before staining at a density of 1E−5 cells/mL in D10 media. Upon harvest, virus-infected cells were fixed in 4% paraformaldehyde for 20 min, followed by permeabilization with 0.1% Triton X-100 detergent + 1xPBS for 15 min at room temperature (RT). Cells were blocked using 5% normal goat serum (Sigma, G9023) for 1 h at RT, followed by incubation with the primary antibodies rabbit anti-LRP1 at 1:200, mouse anti-Lrp1 at 1:50, mouse anti-RVFV N at 1:200, or rabbit anti-OROV N at 1:200 for 1 h at RT. The secondary antibodies goat anti-mouse Cy3 (JacksonImmuno, 115-165-003), goat anti-rabbit 488 (JacksonImmuno, 111-545-003), goat anti-mouse 488 (JacksonImmuno, 115-545-003), or goat anti-rabbit Cy3 (JacksonImmuno, 111-165-144) were added (1:500 dilution) for 1 h at RT. The cells were counterstained with Hoescht and mounted using Gelvatol.
Brain tissue collected from mice at 3 dpi was fixed in 4% paraformaldehyde for 24 h, washed in 1xPBS, and submerged in the following sucrose concentrations for 24 h each: 10%, 20%, 40%. Brains were embedded in OCT (Fisher 23-730-571) and cerebral cortical regions were sliced at 6 μm on a cryostat. Brain sections were permeabilized in 0.01% Triton X-100 detergent + 1xPBS for 15 min at RT. Tissue sections were blocked with 5% normal donkey serum (Sigma, D9663) for 1 h at RT, washed with 1xPBS + 0.5% bovine serum albumin (BSA) (PBB) and probed with rabbit anti-OROV N and/or goat anti-IBA-1 (Novus NB-100-1028) at 1:200 for 1 h at RT. Following washes with 1xPBS + 0.5% BSA (PBB), the tissue sections were probed with donkey anti-rabbit Cy3 (Jackson Immuno, 711-165-152) or donkey anti-goat 488 (Jackson Immuno, 705-545-003) secondary antibodies. Slides were washed with 1xPBS and stained with Hoescht for 30 s at RT before being mounted with Gelvatol. Fluorescent slides were imaged using a Leica DMI8 inverted fluorescent microscope and denoised using the Leica Application Suite X software provided by the Center for Vaccine Research. Images were processed and quantified using ImageJ.
Animal studies.
Female, 3- to 4-wk-old C57BL/6J were obtained from Jackson Laboratories. Previous studies have shown no sex-based differences in susceptibility to RVFV infection or disease outcome (34). Mice were anesthetized with isoflurane before infection. Virus and 215 μg of respective protein treatments were diluted in D2 to a final volume of 15 μL and injected IC using a Hamilton syringe with a 27½-G needle. Following infection, mice were monitored daily for clinical signs of disease. Mice were euthanized when they met IACUC euthanasia criteria. Blood was collected for serum analysis from the surviving mice. At the 3 dpi planned euthanasia, mice were anesthetized and blood was collected via cardiac puncture. The mice were then euthanized and necropsied to harvest brain tissue, which was homogenized and titered using a VPA.
Protein Expression and Purification.
Recombinant protein expression and purification was performed as described previously (11). Briefly, expression plasmids containing mRAPD3 or mutant mRAPD3 were transformed in BL21(DE3) Escherichia coli cells (Novagen), cultured in Luria Broth media at 37 °C to an optical density 600 of 0.6 and induced with 0.5 mM isopropyl-b-d-thiogalactoside for 12 h at 18 °C. Cells were harvested, resuspended in lysis buffer containing 20 mM Tris⋅HCl (pH 8.0), 500 mM NaCl, and 5 mM 2-mercaptoethanol, and lysed using an EmulsiFlex-C5 homogenizer (Avestin). The resulting pellet was resuspended in buffer containing 2 M urea, 20 mM Tris⋅HCl (pH 8.0), 500 mM NaCl, and 2% Triton X-100 before centrifugation at 47,000 × g at 4 °C for 10 min. Inclusion bodies were isolated after repeated rounds of resuspension in urea and centrifugation. The final pellet was resuspended in 20 mM Tris⋅HCl (pH 8.0), 500 mM NaCl, 5 mM imidazole, 8 M urea, and 1 mM 2-mercaptoethanol and refolded on a NiFF (GE Healthcare) column using a reverse linear urea gradient with imidazole. Endochrome-K kit (Charles River Laboratories) was used, following the manufacturer’s instructions to determine endotoxin levels for purified mRAPD3 proteins and the control protein.
Expression plasmids encoding CLII or CLIV with an Fc- and His6-tag were transfected in Expi293F expression cells (Gibco-Thermo Fisher) using the ExpiFectamine293 transfection kit. Cells were cultured in Expi293 Expression Media at 37 °C and 8% CO2 for 5 d. Cells were separated from the supernatant by centrifugation and supernatant containing the secreted proteins was loaded onto a NiFF (GE Healthcare) followed by ion exchange and size exclusion column as the final step.
Virus-Binding Assays Using Biolayer Interferometry.
VSV-OROV was grown in BSR cells in DMEM media containing 2% FBS, as described previously (30). The supernatants containing the virus from two 150-mm dishes were collected, filtered, and concentrated by high-speed ultracentrifugation. The virus pellet was resuspended in 1 mL of BLI binding buffer (PBS, containing 10 mg/mL BSA and 0.1% Tween 20). For binding assays, anti-human Fc capture (Sartorius 18-5060) AHC sensors were coated with either Fc or CLIV-fused Fc and incubated with 20 µL concentrated VSV-OROV and monitored over time for association and dissociation.
Supplementary Material
Acknowledgments
We thank members of our research groups for helpful discussions and support. We thank W. Paul Duprex and Natasha Tilston-Lunel (University of Pittsburgh) for providing the OROV virus used in these studies. The authors also thank Stacey Barrick for coordinating housing for the animal studies. The following reagents were obtained through BEI Resources, National Institute of Allergy and Infectious Diseases, NIH: Zika virus, PRVABC59, NR-50240; RVFV anti-N mAb, Clone 1D8, NR-43188. Research was supported by NIH grants (R01AI161765 and R21AI163603, to G.K.A. and A.L.H.; R01NS101100, to A.L.H.; P01AI120943, to G.K.A.; R01AI140758, to D.W.L.; and R01AI130152, to T.E.). D.A.P. was supported by R01AI40758-S1.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This manuscript is on the following preprint server: bioRxiv 2022.02.26.482111; doi: https://doi.org/10.1101/2022.02.26.482111 under a CC-BY-NC-ND 4.0 International license.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2204706119/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Abudurexiti A., et al. , Taxonomy of the order Bunyavirales: Update 2019. Arch. Virol. 164, 1949–1965 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barr J. N., Weber F., Schmaljohn C. S., “Bunyavirales: The viruses and their replication” in Fields Virology Volume 1: Emerging Viruses, Knipe D. M., Howley P., Whelan S. P., Eds. (Wolters Kluwer, Philadelphia, 2020), chap. 16. pp 707–749. [Google Scholar]
- 3.McMullan L. K., et al. , A new phlebovirus associated with severe febrile illness in Missouri. N. Engl. J. Med. 367, 834–841 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Sakkas H., Bozidis P., Franks A., Papadopoulou C., Oropouche fever: A review. Viruses 10, 175 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mourãão M. P., et al. , Oropouche fever outbreak, Manaus, Brazil, 2007-2008. Emerg. Infect. Dis. 15, 2063–2064 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinheiro F. P., et al. , Oropouche virus. I. A review of clinical, epidemiological, and ecological findings. Am. J. Trop. Med. Hyg. 30, 149–160 (1981). [PubMed] [Google Scholar]
- 7.Pinheiro F. P., et al. , Meningitis associated with Oropouche virus infections. Rev. Inst. Med. Trop. São Paulo 24, 246–251 (1982). [PubMed] [Google Scholar]
- 8.Bastos Mde. S., et al. , Identification of Oropouche Orthobunyavirus in the cerebrospinal fluid of three patients in the Amazonas, Brazil. Am. J. Trop. Med. Hyg. 86, 732–735 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang J. O., et al. , Neurological disease caused by Oropouche virus in northern Brazil: should it be included in the scope of clinical neurological diseases? J. Neurovirol. 27, 626–630 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Proenca-Modena J. L., et al. , Interferon-regulatory factor 5-dependent signaling restricts Orthobunyavirus dissemination to the central nervous system. J. Virol. 90, 189–205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganaie S. S., et al. , Lrp1 is a host entry factor for Rift Valley fever virus. Cell 184, 5163–5178.e24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonias S. L., Campana W. M., LDL receptor-related protein-1: a regulator of inflammation in atherosclerosis, cancer, and injury to the nervous system. Am. J. Pathol. 184, 18–27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heissig B., Salama Y., Takahashi S., Osada T., Hattori K., The multifaceted role of plasminogen in inflammation. Cell. Signal. 75, 109761 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potere N., Del Buono M. G., Mauro A. G., Abbate A., Toldo S., Low Density Lipoprotein Receptor-Related Protein-1 in Cardiac Inflammation and Infarct Healing. Front. Cardiovasc. Med. 6, 51 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bres E. E., Faissner A., Low density receptor-related protein 1 interactions with the extracellular matrix: More than meets the eye. Front. Cell Dev. Biol. 7, 31 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herz J., Clouthier D. E., Hammer R. E., LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell 71, 411–421 (1992). [DOI] [PubMed] [Google Scholar]
- 17.Tilston-Lunel N. L., Acrani G. O., Randall R. E., Elliott R. M., Generation of recombinant Oropouche viruses lacking the nonstructural protein NSm or NSs. J. Virol. 90, 2616–2627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bu G., The roles of receptor-associated protein (RAP) as a molecular chaperone for members of the LDL receptor family. Int. Rev. Cytol. 209, 79–116 (2001). [DOI] [PubMed] [Google Scholar]
- 19.Migliorini M. M., Behre E. H., Brew S., Ingham K. C., Strickland D. K., Allosteric modulation of ligand binding to low density lipoprotein receptor-related protein by the receptor-associated protein requires critical lysine residues within its carboxyl-terminal domain. J. Biol. Chem. 278, 17986–17992 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Rauch J. N., et al. , LRP1 is a master regulator of tau uptake and spread. Nature 580, 381–385 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finkelshtein D., Werman A., Novick D., Barak S., Rubinstein M., LDL receptor and its family members serve as the cellular receptors for vesicular stomatitis virus. Proc. Natl. Acad. Sci. U.S.A. 110, 7306–7311 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devignot S., et al. , Low density lipoprotein receptor-related protein 1 (LRP1) is a host factor for RNA viruses including SARS-CoV-2. bioRxiv [Preprint] (2022). 10.1101/2022.02.17.480904, Accessed 18 February 2022. [DOI]
- 23.Tree M. O., et al. , Dengue virus reduces expression of low-density lipoprotein receptor-related protein 1 to facilitate replication in Aedes aegypti. Sci. Rep. 9, 6352 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molina S., et al. , The low-density lipoprotein receptor plays a role in the infection of primary human hepatocytes by hepatitis C virus. J. Hepatol. 46, 411–419 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Nikolic J., et al. , Structural basis for the recognition of LDL-receptor family members by VSV glycoprotein. Nat. Commun. 9, 1029 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofer F., et al. , Members of the low density lipoprotein receptor family mediate cell entry of a minor-group common cold virus. Proc. Natl. Acad. Sci. U.S.A. 91, 1839–1842 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma H., et al. , LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 588, 308–314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark L. E., et al. , VLDLR and ApoER2 are receptors for multiple alphaviruses. Nature 602, 475–480 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeed M. F., et al. , Nucleotide sequences and phylogeny of the nucleocapsid gene of Oropouche virus. J. Gen. Virol. 81, 743–748 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Stubbs S. H., et al. , Vesicular stomatitis virus chimeras expressing the Oropouche virus glycoproteins elicit protective immune responses in mice. MBio 12, e0046321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waggoner J. J., Pinsky B. A., Zika virus: Diagnostics for an emerging pandemic threat. J. Clin. Microbiol. 54, 860–867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carossino M., et al. , Evaluation of a field-deployable reverse transcription-insulated isothermal PCR for rapid and sensitive on-site detection of Zika virus. BMC Infect. Dis. 17, 778 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMillen C. M., et al. , Rift Valley fever virus induces fetal demise in Sprague-Dawley rats through direct placental infection. Sci. Adv. 4, eaau9812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cartwright H. N., Barbeau D. J., McElroy A. K., Rift valley fever virus is lethal in different inbred mouse strains independent of sex. Front. Microbiol. 11, 1962 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.