Significance
The early B cell factor (EBF) family of transcription factors has important regulatory roles in different cell lineages. We show here that EBF4, a little-studied family member expressed in human but not mouse immune cells, has a modulatory role in Fas-induced apoptosis and contributes to the expression of genes encoding critical factors for the development of CD8+ T and natural killer lymphocytes in the human immune system. We also found that expression of the EBF4 gene itself is turned off after T cell receptor stimulation and remains silenced for an extended time, presumably to control the intensity of cytolytic responses. Thus, unlike EBF1, which controls B lymphocyte differentiation, EBF4 has a unique role in cytotoxic lymphocyte function.
Keywords: apoptosis, cytotoxic lymphocyte, EBF
Abstract
Apoptosis is a genetically regulated program of cell death that plays a key role in immune disease processes. We identified EBF4, a little-studied member of the early B cell factor (EBF) family of transcription factors, in a whole-genome CRISPR screen for regulators of Fas/APO-1/CD95-mediated T cell death. Loss of EBF4 increases the half-life of the c-FLIP protein, and its presence in the Fas signaling complex impairs caspase-8 cleavage and apoptosis. Transcriptome analysis revealed that EBF4 regulates molecules such as TBX21, EOMES, granzyme, and perforin that are important for human natural killer (NK) and CD8+ T cell functions. Proximity-dependent biotin identification (Bio-ID) mass spectrometry analyses showed EBF4 binding to STAT3, STAT5, and MAP kinase 3 and a strong pathway relationship to interleukin-2 regulated genes, which are known to govern cytotoxicity pathways. Chromatin immunoprecipitation and DNA sequencing analysis defined a canonical EBF4 binding motif, 5′-CCCNNGG/AG-3′, closely related to the EBF1 binding site; using a luciferase-based reporter, we found a dose-dependent transcriptional response of this motif to EBF4. We also conducted assay for transposase-accessible chromatin sequencing in EBF4-overexpressing cells and found increased chromatin accessibility upstream of granzyme and perforin and in topologically associated domains in human lymphocytes. Finally, we discovered that the EBF4 has basal expression in human but not mouse NK cells and CD8+ T cells and vanishes following activating stimulation. Together, our data reveal key features of a previously unknown transcriptional regulator of human cytotoxic immune function.
Apoptosis is a genetically regulated program of cell death that plays a key role in cell and organ homeostasis and the prevention of cancer (1–3). Abnormalities of apoptosis affect many biological processes, including aging, organismal degeneration, and disease (4). In particular, the autoimmune lymphoproliferative syndrome (ALPS), contributing to expansion of the secondary lymphoid tissue and autoimmune cytopenias, is caused by germline mutations in FAS, FASL, CASP10, CASP8, and FADD, which are involved in the Fas (CD95)/tumor necrosis factor apoptosis pathway (5–7). The proteins encoded by these genes associate together once the Fas ligand (FASL) engages the Fas receptor to form the death-inducing signaling complex (DISC) (Fig. 1D) (8). The result of DISC formation is the proteolytic process converting caspase-8 and caspase-10 zymogens into highly active and soluble aspartate proteases. These proteases cleave a limited number of specific substrates, leading to apoptotic cell death. The disruption of DISC formation or the inhibition of these proteases prevents apoptosis. However, identification of the molecular checkpoints of DISC and Fas-mediated death is still incomplete. In addition to FL-killing, cytotoxic CD8+ T cells and natural killer (NK) cells secrete granules containing granzyme proteases and perforin (9). Perforin allows granzymes to enter virus-infected or malignant target cells and trigger apoptosis and cell death (10, 11). The transcription factors controlling the development of immune cytolytic cells are not completely understood.
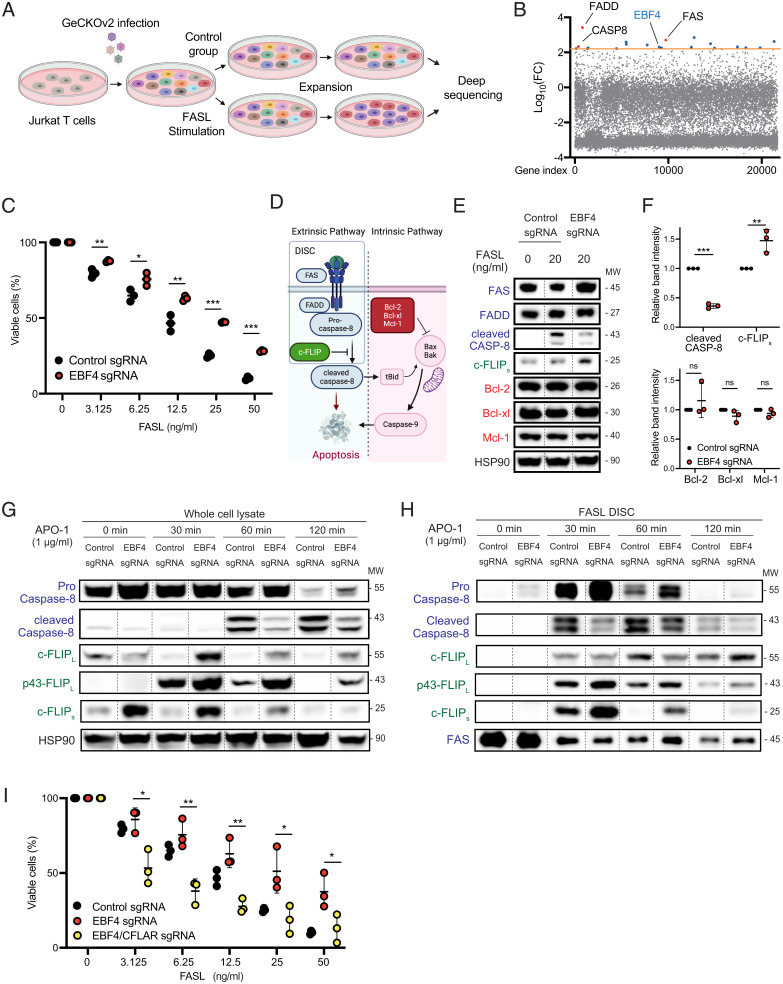
Fig. 1.
The whole-genome CRISPR screen shows that EBF4 KO attenuates Fas-induced apoptosis. (A) Scheme of whole-genome CRISPR screen to identify genes whose deletion causes resistance to Fas-induced apoptosis. (B) Graph of log10- FC difference vs. gene index from DNA samples after Fas stimulation compared to unstimulated cells. The orange line denotes the significance cutoff of our CRISPR screen. (C) The percentage of viable WT and EBF4 KO Jurkat T cells after different concentrations of FASL stimulation. (D) Scheme of Fas-mediated apoptosis. (E) Immunoblot of key Fas canonical regulators with or without stimulation with FasL for 4 h. Lanes have been rearranged along dashed lines but sourced from the same gel, membrane, and exposure time. (F) Intensity of immunoblot bands of cleaved caspase-8, c-FLIPs, Bcl-2, Bcl-xl, and Mcl-1 following FasL stimulation relative to HSP90 loading control. (G) Immunoblot of Fas canonical regulators following stimulation with 1 μg/mL APO-1 to APO-3 for 0 to 120 min. Lanes have been rearranged along dashed lines but sourced from the same gel, membrane, and exposure time. (H) CD95-immunoprecipitation was performed, and immunoprecipitated members of the DISC were analyzed with the same condition in G. Lanes have been rearranged along dashed lines but sourced from the same gel, membrane, and exposure time. (I) The percentage of viable EBF4 KO and EBF4 CFLAR double-KO cells compared to WT following FASL from experiments represented in C. All experiments are representative of at least three independent experiments with similar findings. Two-way comparisons were calculated using a two-tailed, unpaired Student’s t test. MW, Molecular weight; *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
One checkpoint protein for Fas apoptosis is cellular FLICE inhibitory protein (c-FLIP), encoded by the CFLAR gene, for which human genetic deficiencies have not yet been reported (12). c-FLIP is a catalytically inactive evolutionary derivative of the progenitor molecule of caspase-8 and caspase-10 in humans. Genes for all three molecules are found tightly linked on chromosome 2. c-FLIP shares the caspase subunit structure and can be cleaved by caspases despite having amino acid substitutions that preclude its own enzymatic function. The structure of c-FLIP also preserves the death effector domain (DED), which permits it to associate with DISC DED-containing proteins and serve as a dominant inhibitor of caspase activation and Fas apoptosis (13). Due to its apoptosis-inhibitory function, c-FLIP contributes to cancer progression and metastases, and therefore it is important to understand its molecular regulation (14).
The early B cell factor (EBF; also called Olf) family consists of EBF 1, 2, 3, and 4 in both mice and humans (15–19). EBF family proteins contain a DNA binding domain comprising a zinc finger motif as well as a helix–loop–helix (HLH) domain, which enables them to interact with specific short DNA sequences, generally palindromic variants of the motif 5′-GGGNNCCC-3′ (20–22). EBF1 is the best studied and is known as a central transcription factor in B cell differentiation (23, 24). In addition, EBF2 and EBF3 proteins have roles in neuronal cell differentiation (7). However, as the last member of this family, EBF4 is not well investigated and its immune function remains unknown (18).
In this paper, we show that EBF4 plays a small regulatory role in FAS-mediated apoptosis using a whole-genome CRISPR screening system. This was due to the ability of EBF4 to control the degradation of the anti-apoptotic molecule c-FLIPs. Further investigation, however, revealed that it is selectively expressed in NK and CD8+ T cells and regulates cytotoxic and other key molecules. These findings shed light on the role of EBF4 in immunity.
Results
Whole-genome CRISPR screen reveals that EBF4 mediates Fas-induced apoptosis.
Genetic abnormalities in the canonical Fas apoptosis pathway cause ALPS (7). Because there is incomplete penetrance of Fas mutations, we sought to identify additional genes influencing this pathway through a whole-genome CRISPR screen. We therefore transduced the human CRISPR Genome-Scale CRISPR Knock-Out (GeCKO) v2 guide RNA library into Jurkat T cells (Fig. 1A). We found that deletion in the FAS, FADD, and CASP8 genes encoding the known Fas signaling DISC proteins attenuates FASL-induced death. Enriched gene barcodes were plotted as the log10-fold change (FC) difference of gene-associated bar codes vs. the gene index, using a screening cutoff value for log10(FC) of 2.2 (Fig. 1B and SI Appendix, Table 1). One highly ranking gene that we identified was EBF4, a little-studied member of the EBF transcription factor family (18, 25). Because of the important role of EBF1 in the immune system, specifically B cell development, we believed that EBF4 could also play a significant role in immune cell function and development. By performing a selected gene knockout (KO) in Jurkat T cells (EBF4 KO), we confirmed that the loss of EBF4 partially attenuated Fas apoptosis (Fig. 1C).
Fas initiates the extrinsic apoptosis pathway when FasL triggers the recruitment of FADD, procaspase-8, and c-FLIP into the DISC, leading to the processing and release of caspase-8 or caspase-10 as a highly active, soluble aspartate protease (Fig. 1D) (8). This can elicit caspase-9 cleavage through the intrinsic pathway regulated by B-Cell CLL/Lymphoma 2 (Bcl-2) family members (Fig. 1D). To elucidate the mechanism of apoptosis resistance in EBF4 KO cells, we investigated key proteins of the Fas pathway (Fig. 1 E and F). After FasL stimulation, the control cells showed the expected increase in cleaved caspase-8, but this was reduced in EBF4 KO cells (Fig. 1 E and F). Conversely, we found that the “short” low molecular weight variant of c-FLIP (c-FLIPs) was reproducibly increased (Fig. 1 E and F). The levels of Bcl-2, Bcl-xl, and Mcl-1, as well as the HSP90 control, remained unchanged (Fig. 1E). We obtained the same reduction in cleaved caspase-8 accompanied by an increase in c-FLIP variants over a time course of stimulation using an FAS agonistic antibody (Fig. 1G and SI Appendix, Fig. 1A). Thus, we hypothesized that increased c-FLIP inhibits Fas apoptosis in EBF4 KO cells.
We next analyzed the DISC assay using APO-1, an agonist monoclonal antibody that triggers the assembly and precipitation of the DISC (Fig. 1H and SI Appendix, Fig. 1B) (26). From 0 to 120 min, APO-1 induced cleaved caspase-8 in the DISC while procaspase-8 expression accordingly dropped. In EBF4 KO cells, cleaved p43/p41 caspase-8 was decreased in the DISC and uncleaved caspase-8 was correspondingly higher. Notably, the DISC also showed increases in c-FLIPs in EBF4 KO cells at all time points (Fig. 1H). Finally, knocking out the CFLAR gene encoding c-FLIP in EBF4 KO cells rescued full FASL death induction (Fig. 1I and SI Appendix, Fig. 1C). Thus, the EBF4 KO inhibits apoptosis by inducing c-FLIP, altering the stoichiometry of its binding at the DISC and reducing the production of cleaved caspase-8.
EBF4 is found primarily in cytotoxic immune cells.
We further investigated the basis for elevated c-FLIP expression. Unexpectedly, we found that CFLAR messenger RNA (mRNA) was modestly decreased in EBF4 KO cells (Fig. 2A). Since it is well established that c-FLIP expression is regulated posttranscriptionally, especially by degradation, we investigated protein turnover (27, 28). c-FLIP has a naturally high turnover rate, so presumably it has a high translation rate to maintain steady-state levels, and any differences in the degradation rate will be magnified (29). We therefore measured c-FLIP turnover using a cycloheximide (CHX) chase assay. After 2 h CHX treatment, the EBF4 KO cells had an average of 30% of their original c-FLIPs levels left, while the wild-type (WT) cells retained an average of 15% (Fig. 2 B and C). Thus, the loss of EBF4 caused the c-FLIPs protein to be degraded at a slower rate (Fig. 2 B and C). The ubiquitination of c-FLIP reduces its stability by leading to proteasomal degradation (30). We therefore examined ubiquitin–protein that could contribute to c-FLIP protein degradation. However, we found small or no differences in these factors between WT and EBF4 KO Jurkat T cells (SI Appendix, Fig. 2B).
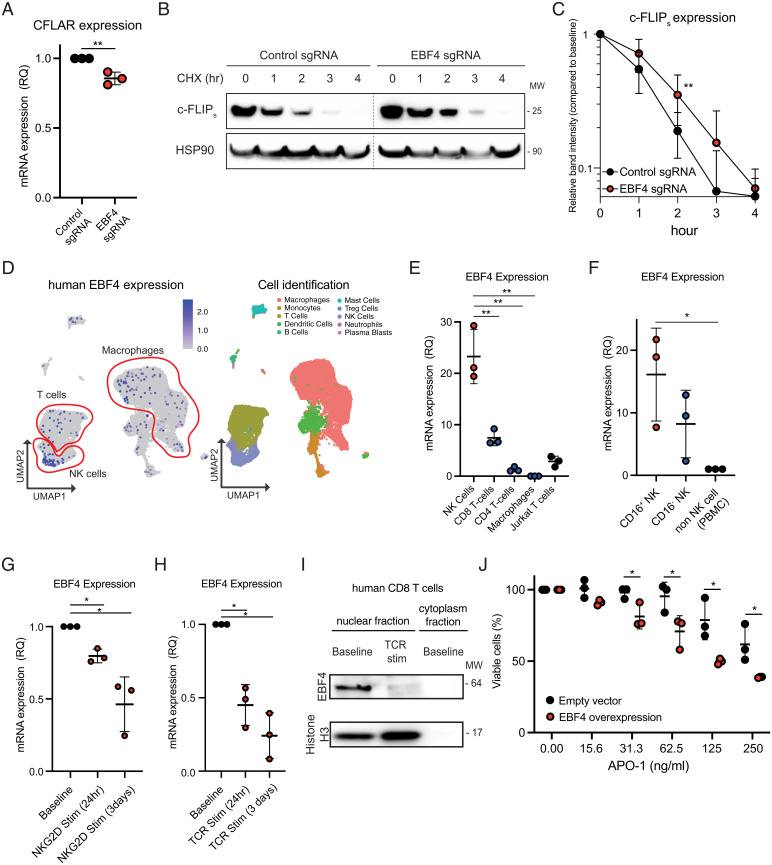
Fig. 2.
EBF4 has low selective expression in primary cells but is most highly expressed in cytotoxic cells. (A) mRNA levels of CFLAR. (B) Immunoblot of c-FLIPs and HSP90 loading control in Jurkat T cells transfected with Cas9 loaded with either a control EBF4-targeting sgRNA and then treated with 10 μg/mL CHX for 0 to 4 h. Lanes have been rearranged along dashed lines but sourced from the same gel, membrane, and exposure time. (C) Calculated percentage of c-FLIP protein remaining compared to baseline expression following CHX based upon relative intensity of immunoblot bands in B. (D) Human single-cell RNA-seq showing EBF4 expression in different immune cell subsets. (E) EBF4 expression quantified by RT-qPCR in primary NK, CD8+, and CD4+ T cells compared to Jurkat T cells. Each immune cell subset had three independent donors. (F) RT-qPCR results of EBF4 mRNA levels in purified CD16+ NK cells, CD16− NK cells, and NK cell–deficient PBMCs. (G) RT-qPCR results of EBF4 mRNA levels in NK cells before and after 24 and 72 h NKG2D stimulation (Stim). (H) RT-qPCR results of EBF4 mRNA levels in CD8+ T cells before and after 24 and 72 h CD3/CD28 TCR stimulation. (I) Immunoblot of EBF4 in CD8+ T cells following 24 h CD3/CD28 TCR stimulation. (J) The percentage of viable WT and EBF4 OE primary CD8+ T cells after different concentrations of soluble APO-1 to APO-3 stimulation. All experiments are representative of at least three independent experiments with similar findings. Two-way comparisons were calculated using a two-tailed, unpaired Student’s t test. RQ, relative quantification; PBMC, peripheral blood mononuclear cells; UMAP, uniform manifold approximation and projection. *P ≤ 0.05 and **P ≤ 0.01.
EBF4 (Olf-1/EBF-Like 4) was originally identified as an olfactory neuron–enriched transcription factor and has strong homology to the three other HLH genes, including the B cell regulator EBF1 (25). We examined the human expression quantitative trait loci (eQTL) database and found that EBF1 is highly and solely expressed in B cells, EBF2 and EBF3 are absent from immune cells altogether, and EBF4 is most highly expressed in NK and cytotoxic CD8+ T lymphocytes (SI Appendix, Fig. 2C). Human single-cell RNA sequencing (RNA-seq) also showed preferential expression in NK cells, CD8+ T cells, and a small fraction of monocyte/macrophages, mast cells, and B cells (Fig. 2D). We experimentally confirmed these data by RT-qPCR on purified primary cells from healthy human donors by showing that EBF4 is most highly expressed in cytotoxic NK cells, especially CD16+ NK cells, followed by CD8+ T cells, and much less in CD4 or Jurkat T cells (Fig. 2 E and F). Interestingly, stimulating purified NK cells with natural killer group 2D (NKG2D) and natural killer cell receptor 2B4 (2B4) caused EBF4 mRNA levels to decrease (Fig. 2G). Similarly, anti-CD3/CD28 stimulation of purified CD8+ T cells caused a rapid drop and near-disappearance of both the EBF4 mRNA and protein levels (Fig. 2 H and I). Since the loss of EBF4 causes resistance, we tested the effect of EBF4 overexpression in CD8+ T cells and found that it increased sensitivity to Fas apoptosis (Fig. 2J). Thus, EBF4 is necessary and sufficient to achieve enhanced Fas-mediated T cell death under these conditions.
EBF4 regulates cytotoxic molecules via interleukin-2 signaling.
To further elucidate the role of EBF4 in immune cells, we performed transcriptome analysis that compared overexpressed (OE) EBF4 to EBF4 KO Jurkat T cells prior to any stimulation. A volcano plot showed that transcripts from genes encoding cytotoxic proteins such as granzyme A (GZMA), granzyme K (GZMK), and perforin (PRF1) as well as the differentiation factors for NK cells and the CD8+ T cells eomesodermin (EOMES) and T-bet (TBX21) were up-regulated in the OE EBF4 Jurkat T cells (Fig. 3A, Left) and down-regulated in the EBF4 KO (Fig. 3A, Right) Jurkat T cells, indicating that EBF4 positively regulates the expression of these genes. These data are consistent with higher expression of the EBF4 gene itself in NK and CD8+ T cells.
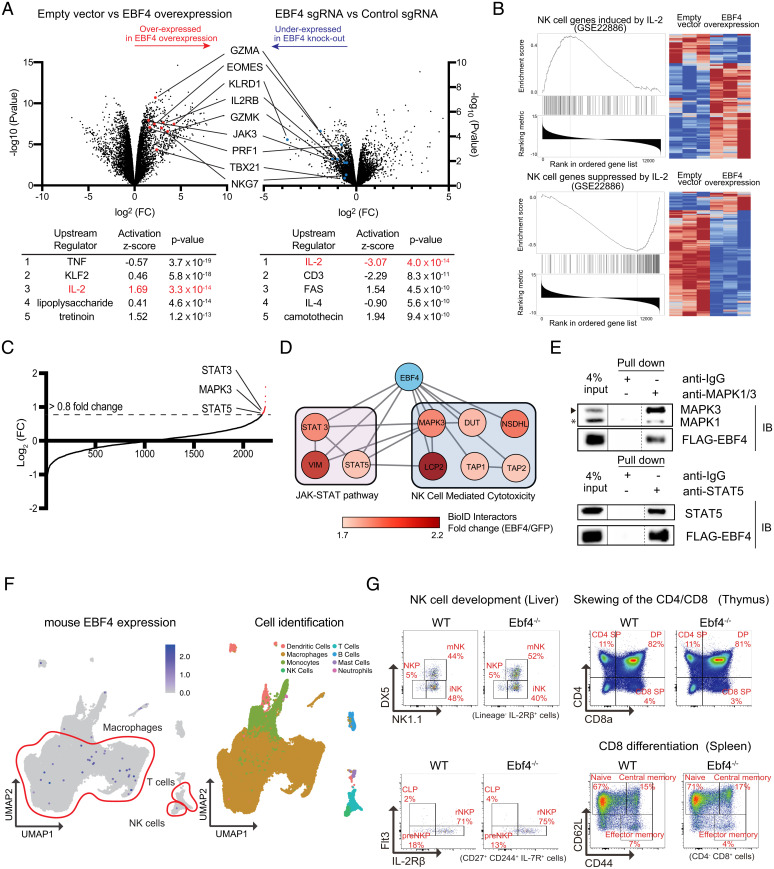
Fig. 3.
EBF4 regulates cytotoxic molecules via IL-2 signaling. (A) Volcano plots of mRNA sequence data from (Left) empty vector vs. EBF4 OE cells or (Right) control sgRNA vs. EBF4 KO cells. Table shows the results of Ingenuity upstream regulator analysis. TNF = tumor necrosis factor. (B) GSEA of mRNA-seq data from empty vector vs. EBF4 OE cells for NK cell genes (Top) induced by IL-2 or (Bottom) suppressed by IL-2, where blue indicates low counts and red indicates high counts. (C) Protein–protein interaction detected by Bio-ID mass spectrometry. Significance of interacting proteins was a log2-FC cutoff of 0.8 or higher (dashed line). (D) Binding proteins are analyzed using STRING and visualized by CytoScape software (The darker the color, the stronger the interaction). (E) Anti-FLAG immunoprecipitation of MAPK3/ERK1 and STAT5 by FLAG-tagged EBF4 with a rabbit IgG control. IB = immunoblot. Lanes have been rearranged along dashed lines but sourced from the same gel and membrane. A solid line between lanes denotes different exposure times. (F) Mouse single-cell RNA-seq showing EBF4 expression in different immune cell subsets. (G) Flow cytometry dot plots using the indicated surface markers for NK, CD8+, and CD4+ precursor and mature cell subsets in thymus, spleen, and liver of WT and EBF4 KO mice. Each group had n = 5 mice. STRING, search tool for the retrieval of interacting genes/proteins; DX5, Cd49b; NK1.1, killer cell lectin-like receptor subfamily B member 1C; Flt3, fms related receptor tyrosine kinase 3; IL-2Rbeta, IL-2 receptor beta; mNK, mature NK; NKP, NK progenitor; iNK, immature NK; SP, single positive; DP, double positive.
In addition, ingenuity upstream regulator analysis predicted that interleukin-2 (IL-2) was a positive upstream regulator in EBF4 OE cells and a negative upstream regulator in EBF4 KO cells (Fig. 3A, Bottom). Gene set enrichment analysis (GSEA) further revealed that EBF4 OE induces the same gene sets that IL-2 induces in NK cells and decreases those that IL-2 reduces in NK cells (Fig. 3B). Furthermore, we performed Proximity-dependent biotin identification (Bio-ID) mass spectrometry analysis to investigate proteins interacting with EBF4 (Fig. 3C and SI Appendix, Table 2). Filtering out potential contaminants and evaluating the remaining candidates using the National Institute of Allergy and Infectious Disease Selection by Iterative Pathway Group and Network Analysis Looping database to classify Kyoto Encyclopedia of Genes and Genomes biological pathways (Fig. 3D), we found that EBF4 is potentially involved in the JAK-STAT pathway and NK cell cytotoxicity. Based on our RNA-seq data implicating IL-2 signaling, we immunoprecipitated key members of this pathway, MAPK3 and STAT5, and found that EBF4 was specifically coprecipitated with these factors (Fig. 3E). These data suggest that EBF4 has an important role in controlling cytotoxic molecules in conjunction with the common gamma chain cytokine pathway.
To investigate the function of EBF4 in the CD8+ and NK cells, we next examined mice. The EBF4 gene is highly conserved in mice; however, the expression was not detected in mouse immune cell public databases (Fig. 3F and SI Appendix, Fig. 3A). Although EBF4 was strongly detected in purified WT mouse olfactory epithelium, no EBF4 expression was detected by RT-qPCR in NK cells and CD8+ T cells (SI Appendix, Fig. 3B). We next established an EBF4 KO mouse line to determine whether the developmental stages of NK and CD 8+ T cells depend on EBF4. However, we detected no differences in the NK, CD8+, and CD4+ precursor and mature cell subsets in the thymus, spleen, or liver between WT and EBF4 KO mice (Fig. 3G and SI Appendix, Fig. 3C). From these data, we conclude that the function of EBF4 in immune cells is different between humans and mice.
EBF4 binds the promoters of human cytotoxicity mediators.
Next, we performed chromatin immunoprecipitation and DNA sequencing (ChIP-seq) to analyze EBF4 DNA binding regions. Our ChIP-seq analysis of Jurkat T cells expressing FLAG-tagged EBF4 captured DNA sequences that defined an EBF4 binding motif, 5′-CCCNNGG/AG-3′, that was closely related to the binding site of EBF1 and other EBF family proteins (Fig. 4A). Furthermore, more than half of the sites were located in either intronic or intergenic regions and ∼18% were near promoters/transcription start sites (Fig. 4B). This finding could suggest that EBF4 mainly regulates enhancers/intergenic control regions. Next, we tested the transcriptional function of this motif by transfecting 293T cells with a minimal promoter–luciferase reporter construct containing five upstream tandem repeats of the EBF4 binding sequence, 5′-CCCAGGGG-3′. We then cotransfected increasing amounts of an EBF4 expression construct and measured a dose-dependent increase in the luciferase response (Fig. 4C). These findings support the conclusion that this site could respond to EBF4 and perhaps other members of the EBF/HLH transcription factor families.
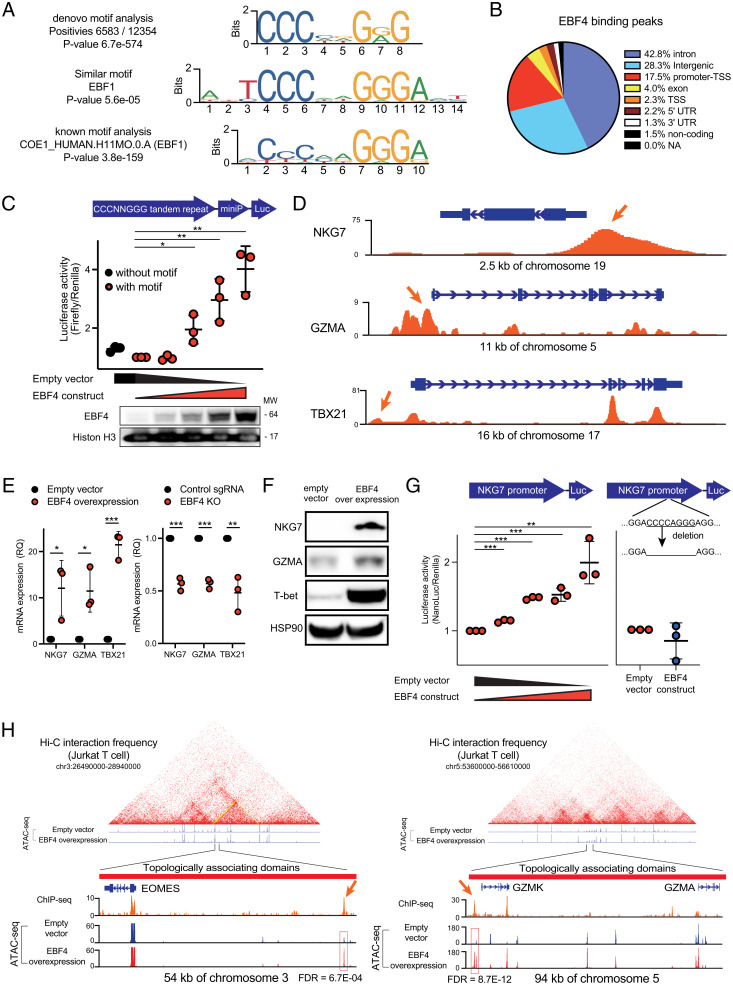
Fig. 4.
ChIP-seq and ATAC-seq analysis reveal which regions of DNA bind EBF4. (A) The EBF4 binding motif revealed by ChIP-seq. Top: most significant De novo motif identified in peaks; Middle: known transcription factor binding motif most like the De novo motif; Bottom: known transcription factor binding motif most significantly enriched in peaks. (B) Genomic features associated with EBF4 binding sites. (C) Luciferase assay of CCCAGGGG 5 tandem repeats transfected in 293 T cells. Bottom: immunoblot of EBF4 in the nucleus. (D) ChIP-seq tracks from FLAG-tagged EBF4 Jurkat T cells for the NKG7, GZMA, and TBX21 loci normalized by input. (E) mRNA expression of NKG7, GZMA, and TBX21 quantified by RT-qPCR in Jurkat cells for EBF4 OE vs. control (Left) and control vs. EBF4 KO (Right). (F) Immunoblot of protein levels of NKG7, TBX21, and GZMA. Data from NKG7 and HSP90 are sourced from a different blot than GZMA and T-bet. (G) Luciferase assay of NKG7 promoter region without deletion of CCCAGGGG (Left) or with deletion (Right) transfected in 293 T cells. (H) ATAC-seq tracks from WT and EBF4 OE Jurkat T cells for the EOMES and GZMK/GZMA loci and their topologically associating domains. ChIP-seq tracks are also shown from experiments represented in (D). FDR = false discovery rate. All experiments are representative of at least three independent experiments with similar findings. Two-way comparisons were calculated using a two-tailed, unpaired Student’s t test. MW, Molecular weight; miniP, Minimal promoter; RQ, relative quantification; T-bet, T box expressed in T cells. *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.001.
We next compared the binding of EBF4 to promoters to the transcriptomic analysis with the hypothesis that genes with altered expression in the EBF4 OE and KO conditions could be direct targets of EBF4. A total 279 genes were up-regulated in the OE cells and underexpressed in the KO cells. Among them, EBF4 binds only to the promoter region in 27 genes and to both promoter and intergenic or intron regions in 21 other genes (SI Appendix, Table 3). Three immunoregulatory genes for cytotoxic CD8+ or NK lymphocytes, NKG7, GZMA, and TBX21, illustrate our findings (Fig. 4D). EBF4 OE or KO in Jurkat T cells caused changes in mRNA corresponding to a positive regulatory role for EBF4 (Fig. 4E). In addition, EBF4 overexpression in Jurkat T cells increased NKG7, GZMA, and T-bet protein expression (Fig. 4F). We therefore performed luciferase reporter assays with the NKG7 promoter region containing the EBF4 binding site. We found a dose-dependent inducible effect of EBF4 on NKG7 activity that was completely abrogated by an 8–base pair deletion of the EBBF4 binding site (Fig. 4G). Our data show that EBF4 can regulate the promoter region of NKG7, a gene important to NK and CD8+ T cell function, using the sequence CCCNNGGG.
We also found that 122 genes (including EBF4 itself) with intergenic or intron EBF4 binding sites were transcriptionally regulated by EBF4 (SI Appendix, Table 4). Among them, we noticed several important molecules for NK and CD8+ T cell function such as GZMK, PRF1, and EOMES. Therefore, we conducted assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-seq) in EBF4 OE cells and found increased chromatin accessibility upstream of these molecules and in topologically associated domains (Fig. 4H). EBF4 binding on these sites was also confirmed by ChIP-seq (Fig. 4H and SI Appendix, Fig. 4A). Of note, the GZMK and GZMA genes are in close proximity in the genome, and they may be coregulated by EBF4 via the same open chromatin site (Fig. 4H). Collectively, we concluded that EBF4 regulates cytotoxic molecules by binding to their promoter or enhancer regions. Moreover, we found that EBF4 binds to the promoter region for three transcriptional factors, binds to the intergenic or intron region for 15 transcriptional factors, and binds to both the promoter region and intergenic or intron regions for one transcriptional factor among 279 genes, suggesting that EBF4 may be part of a hierarchical gene regulatory network and may have indirect effects on chromatin and gene accessibility, as previously discovered for the related factor EBF1 (31, 32).
Discussion
In this study, we uncovered an immunoregulatory role for EBF4, a member of the EBF family, that has regulatory effects in human CD8+ and NK cytotoxic lymphocytes. EBF4 is a previously unexplored transcription factor. We were surprised by the fact that since its discovery in olfactory epithelium in the early 1990s, there were no in-depth studies of this protein even though it is a close homolog of EBF1, which has a critical role in B cell differentiation. The EBF family of HLH transcription factors consists of EBF1 through EBF4, which have been shown to have important roles in various cell lineages. Among them, EBF1 has been the best studied in its crucial role as a B cell lineage developmental factor (23, 33, 34). EBF2 is an essential mediator of brown adipocytes (35), whereas EBF3 is a regulator of inhibitory GABAergic neuronal development (36). In humans, EBF3 loss causes a genetic neurodevelopmental syndrome (37). By comparison, there is only a single report on EBF4, showing its cloning from olfactory cells (18). We now recognize that a major obstacle has been that the gene and protein are expressed little, if at all, in immune or other tissues in C57BL/6 experimental mice. Using human samples, we were able to identify functions in Fas-induced apoptosis and in cytotoxic lymphocytes. Thus, EBF4 is like EBF1 in that it has a role in the immune system but predominates in T and NK lymphocytes rather than B lymphocytes.
EBF4 first came to our attention in a whole-genome CRISPR screen showing that FAS-mediated apoptosis was impaired by an EBF4 KO in Jurkat T cells. Biochemical analyses revealed that the loss of EBF4 causes an increase in the endogenous protein levels of c-FLIP due to a reduced turnover of the protein. c-FLIP competitively binds to the DISC formed after FasL binding to the Fas receptor, and we found that its increased level in EBF4 KO Jurkat T cells impaired caspase-8 cleavage, thereby reducing apoptosis (38, 39). Transcriptome analysis comparing EBF4 KO cells to WT cells revealed that three different E3 ligases showed a more than 25% decrease in the EBF4 KO cells, such as RNF212, ZNF521, and RNF125, among 390 human E3 ubiquitin ligases (SI Appendix, Table 5). We subsequently knocked out these molecules to check the c-FLIP expression; however, these E3 ligases did not affect the rate of cFLIPs protein degradation. Further investigation will be required to reveal the underlying mechanism of how EBF4 regulates c-FLIP degradation. FAS and its signals are important for apoptosis induction in cancer cells. The Cancer Genome Atlas database revealed that lower EBF4 expression in glioma/glioblastomas correlated with higher mortality (SI Appendix, Fig. 4B). Conversely, higher CFLAR (c-FLIP) expression correlated with higher mortality (SI Appendix, Fig. 4C). We have shown that a loss of EBF4 results in a loss of sensitivity to stimulation that induces apoptosis via the induction of c-FLIP expression. We conjecture that this anti-apoptotic mechanism may manifest during the proliferation and survival of cancer cells. When there is a loss of EBF4, the cancer cells that have an increased level of c-FLIP may not respond normally to apoptotic signals and have better survival. Nevertheless, despite the clear changes in c-FLIP, the effects of EBF4 on cleaved caspase-8 levels and apoptosis are mild, and EBF4 KO cells can be killed by increased FAS stimulation. Thus, we believe that this mechanism is a modulatory influence but not a primary regulator of Fas homeostasis.
We gained insight into EBF4 function by first conducting transcriptome analysis. This brought to our attention that human CD8+ and NK lymphocytes had high expression. Our mRNA sequencing data revealed that EBF4 regulates molecules such as TBX21 and EOMES, which are important for the development of NK and CD8+ T cells, as well as granzyme and perforin, which are important for immune cytotoxicity. In addition, our data and a previous report showed that the CD56dim CD16+ human NK cell subset had a higher expression of EBF4 (40). Further studies will be needed to reveal whether a specific cell subset among CD8 and NK cells highly expresses EBF4. Interestingly, we found that EBF4 mRNA expression was decreased by T cell receptor (TCR) activation in CD8+ T cells or following NKG2D stimulation of NK cells. This finding was particularly striking in how swiftly EBF4 expression was down-regulated. Although we do not really understand the regulatory significance, previous papers showed that the release of FasL from NK cells also leads to NK cell death (41). This leads us to conclude that activation-induced suppression of EBF4 could contribute to regulating cell death mechanisms during immune responses. However, most functional assays were performed with Jurkat T cells, and thus further experimentation is necessary to address the expression of EBF4 in human NK and CD8+ T cells or other rare immune subsets that could yield fresh insight into its immune function. The uniqueness of this observation invites further investigation especially because we found that the levels of EBF4 are not restored for an extended time. Hence, the response resembles that of a developmental or differentiation switch—once turned, it locks in the cells to a particular phenotype by the presence or absence of specific transcription factors.
Materials and Methods
Jurkat T cells were transduced with a CRISPR KO pooled library and selected for response to FASL, and the small-guide RNA (sgRNA) sequences were deep-sequenced. EBF4 OE Jurkat T cells were generated through lentiviral transduction, while EBF4 KO cells were generated with a CRISPR KO system either through lentiviral transduction or electroporation. EBF4-deficient mice were generated using the CRISPR-Cas9 genome–editing method. Animal procedures were performed under protocols approved by the National Institute of Allergy and Infectious Disease Animal Care and Use Committee. Under various conditions, cells were subjected to immunoprecipitation, immunoblotting, FAS killing assays, qPCR, mass spectrometry, transcriptome analysis, DNA sequencing, ChIP-seq analysis, ATAC-seq analysis, and luciferase reporter assays. A more detailed explanation of methods can be found in the SI Appendix.
Supplementary Material
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH. SK was supported by the Japan Research Foundation for Clinical Pharmacology. We thank Brittany Dulek and Paul Schaughency for processing the ATAC-seq data.
Footnotes
Reviewers: R.G., Max Planck Institute of Immunobiology and Epigenetics; and C.M., University of California, San Diego
Competing interest statement: T.O. received research funding from Gilead, Merck KGaA, and the German Research Foundation (SFB1530), and is a consultant for Roche and Merck KGaA, neither of which has anything to do with this manuscript.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2208522119/-/DCSupplemental.
Data Availability
Raw data for RNA-seq, ChIP-seq, and ATAC-seq have been deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE206222) (42).
All study data are included in the article and/or SI Appendix.
References
- 1.Metzstein M. M., Stanfield G. M., Horvitz H. R., Genetics of programmed cell death in C. elegans: Past, present and future. Trends Genet. 14, 410–416 (1998). [DOI] [PubMed] [Google Scholar]
- 2.Nicholson D. W., From bench to clinic with apoptosis-based therapeutic agents. Nature 407, 810–816 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Opferman J. T., Korsmeyer S. J., Apoptosis in the development and maintenance of the immune system. Nat. Immunol. 4, 410–415 (2003). [DOI] [PubMed] [Google Scholar]
- 4.Fuchs Y., Steller H., Programmed cell death in animal development and disease. Cell 147, 742–758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fisher G. H., et al. , Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell 81, 935–946 (1995). [DOI] [PubMed] [Google Scholar]
- 6.Sneller M. C., et al. , Clinical, immunologic, and genetic features of an autoimmune lymphoproliferative syndrome associated with abnormal lymphocyte apoptosis. Blood 89, 1341–1348 (1997). [PubMed] [Google Scholar]
- 7.Price S., et al. , Natural history of autoimmune lymphoproliferative syndrome associated with FAS gene mutations. Blood 123, 1989–1999 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lavrik I. N., Krammer P. H., Regulation of CD95/Fas signaling at the DISC. Cell Death Differ. 19, 36–41 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voskoboinik I., Whisstock J. C., Trapani J. A., Perforin and granzymes: Function, dysfunction and human pathology. Nat. Rev. Immunol. 15, 388–400 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Trapani J. A., Smyth M. J., Functional significance of the perforin/granzyme cell death pathway. Nat. Rev. Immunol. 2, 735–747 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Smyth M. J., Trapani J. A., Granzymes: Exogenous proteinases that induce target cell apoptosis. Immunol. Today 16, 202–206 (1995). [DOI] [PubMed] [Google Scholar]
- 12.Algeciras-Schimnich A., Griffith T. S., Lynch D. H., Paya C. V., Cell cycle-dependent regulation of FLIP levels and susceptibility to Fas-mediated apoptosis. J. Immunol. 162, 5205–5211 (1999). [PubMed] [Google Scholar]
- 13.Kataoka T., The caspase-8 modulator c-FLIP. Crit. Rev. Immunol. 25, 31–58 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Shirley S., Micheau O., Targeting c-FLIP in cancer. Cancer Lett. 332, 141–150 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Wang M. M., Tsai R. Y., Schrader K. A., Reed R. R., Genes encoding components of the olfactory signal transduction cascade contain a DNA binding site that may direct neuronal expression. Mol. Cell. Biol. 13, 5805–5813 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang S. S., Tsai R. Y., Reed R. R., The characterization of the Olf-1/EBF-like HLH transcription factor family: Implications in olfactory gene regulation and neuronal development. J. Neurosci. 17, 4149–4158 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagman J., Belanger C., Travis A., Turck C. W., Grosschedl R., Cloning and functional characterization of early B-cell factor, a regulator of lymphocyte-specific gene expression. Genes Dev. 7, 760–773 (1993). [DOI] [PubMed] [Google Scholar]
- 18.Wang S. S., Betz A. G., Reed R. R., Cloning of a novel Olf-1/EBF-like gene, O/E-4, by degenerate oligo-based direct selection. Mol. Cell. Neurosci. 20, 404–414 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Wang M. M., Reed R. R., Molecular cloning of the olfactory neuronal transcription factor Olf-1 by genetic selection in yeast. Nature 364, 121–126 (1993). [DOI] [PubMed] [Google Scholar]
- 20.Hagman J., Gutch M. J., Lin H., Grosschedl R., EBF contains a novel zinc coordination motif and multiple dimerization and transcriptional activation domains. EMBO J. 14, 2907–2916 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murre C., Helix-loop-helix proteins and the advent of cellular diversity: 30 years of discovery. Genes Dev. 33, 6–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murre C., Helix-loop-helix proteins and lymphocyte development. Nat. Immunol. 6, 1079–1086 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Boller S., Li R., Grosschedl R., Defining B cell chromatin: Lessons from EBF1. Trends Genet. 34, 257–269 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Györy I., et al. , Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev. 26, 668–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao D., Emerging roles of the EBF family of transcription factors in tumor suppression. Mol. Cancer Res. 7, 1893–1901 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trauth B. C., et al. , Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science 245, 301–305 (1989). [DOI] [PubMed] [Google Scholar]
- 27.Safa A. R., Day T. W., Wu C. H., Cellular FLICE-like inhibitory protein (C-FLIP): A novel target for cancer therapy. Curr. Cancer Drug Targets 8, 37–46 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Micheau O., Cellular FLICE-inhibitory protein: An attractive therapeutic target? Expert Opin. Ther. Targets 7, 559–573 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ivanisenko N. V., et al. , Regulation of extrinsic apoptotic signaling by c-FLIP: Towards targeting cancer networks. Trends Cancer 8, 190–209 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Oztürk S., Schleich K., Lavrik I. N., Cellular FLICE-like inhibitory proteins (c-FLIPs): Fine-tuners of life and death decisions. Exp. Cell Res. 318, 1324–1331 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Treiber T., et al. , Early B cell factor 1 regulates B cell gene networks by activation, repression, and transcription- independent poising of chromatin. Immunity 32, 714–725 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Romanow W. J., et al. , E2A and EBF act in synergy with the V(D)J recombinase to generate a diverse immunoglobulin repertoire in nonlymphoid cells. Mol. Cell 5, 343–353 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Lin H., Grosschedl R., Failure of B-cell differentiation in mice lacking the transcription factor EBF. Nature 376, 263–267 (1995). [DOI] [PubMed] [Google Scholar]
- 34.Kee B. L., Murre C., Transcription factor regulation of B lineage commitment. Curr. Opin. Immunol. 13, 180–185 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Wang W., et al. , Ebf2 is a selective marker of brown and beige adipogenic precursor cells. Proc. Natl. Acad. Sci. U.S.A. 111, 14466–14471 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S. S., Lewcock J. W., Feinstein P., Mombaerts P., Reed R. R., Genetic disruptions of O/E2 and O/E3 genes reveal involvement in olfactory receptor neuron projection. Development 131, 1377–1388 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Chao H. T., et al. ; Undiagnosed Diseases Network, A syndromic neurodevelopmental disorder caused by de novo variants in EBF3. Am. J. Hum. Genet. 100, 128–137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Budd R. C., Yeh W. C., Tschopp J., cFLIP regulation of lymphocyte activation and development. Nat. Rev. Immunol. 6, 196–204 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Krueger A., Schmitz I., Baumann S., Krammer P. H., Kirchhoff S., Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J. Biol. Chem. 276, 20633–20640 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Dogra P., et al. , Tissue determinants of human NK cell development, function, and residence. Cell 180, 749–763.e13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spaggiari G. M., et al. , Soluble HLA class I induces NK cell apoptosis upon the engagement of killer-activating HLA class I receptors through FasL-Fas interaction. Blood 100, 4098–4107 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Kubo S., et al. , Early B cell factor 4 modulates FAS-mediated apoptosis and promotes cytotoxic function in human immune cells. GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE206222. Deposited 15 June 2022. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data for RNA-seq, ChIP-seq, and ATAC-seq have been deposited in GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE206222) (42).
All study data are included in the article and/or SI Appendix.