Abstract
We review our recent work on adenosine receptors, a family of GPCRs; focusing our attention on A3 adenosine receptor, we have demonstrated that the reciprocal integration of different theoretical and experimental disciplines can be very useful for the successful protein-based design of new, potent and selective receptor ligands.
G Protein-coupled receptors: new opportunities for drug design
G Protein-coupled receptors (GPCRs) are considered to be one of the most significant groups of drug targets.1–7 This is because GPCRs are implicated in a very wide range of body functions and processes, including those of the nervous, cardiovascular, endocrine, and immune systems, and modulation of GPCRs has implications for treatment of major diseases such as hypertension, cardiac dysfunction, depression, eating disorders (obesity), certain types of cancer, pain, schizophrenia, and viral infection. Thus, while GPCRs are only a small subset of the human genome (2–3%); nevertheless, they constitute about 50% of the drug targets that are of interest to the pharmaceutical industry.1–7
Based on nucleotide and amino acid sequence similarity, the superfamily of GPCRs can be subdivided into six families of receptors whose protein sequences share significant similarity.1–7 The main family (family A) is of the rhodopsin/adrenergic receptors, which consists of the majority of G-protein-coupled receptors identified to date. This family is the best studied from both structural and functional points of view. Receptors belonging to this family are activated by a variety of stimuli including photons, odorants, hormones, and neurotransmitters with molecular structures ranging from small biogenic amines (e.g., catecholamines and histamine) to peptides (e.g., gonadotropin-releasing hormone (GnRH), thyrotropin-releasing hormone (TRH)), and complex glycoproteins, such as luteinizing hormone (LH), follicle-stimulating hormone (FSH), and thyroid-stimulating hormone (TSH).1–7 The other subfamilies are the secretin/vasointestinal peptide (VIP) family (family B), which binds several neuropeptides and peptide hormones, the metabotropic glutamate receptor family (family C), which comprises at least six closely related subtypes of receptors that bind glutamate, the major excitatory neurotransmitter in the central nervous system.1–7 Three additional GPCR families are the fungal pheromone P and a-factor (STE2/MAM2) family (family D), the fungal pheromone A and M-factor (STE3/mAp3) receptors (family E), and the cyclic adenosine monophosphate (cAMP) receptors of Dictyostelium (family F).1–7
A number of new putative GPCR families have been discovered with varying degrees of similarity to the established families, including frizzled, smoothened, basal vomeronasal receptors, and bride of sevenless (BOSS) of Drosophila and mammals, latrophilin, several plant GPCRs, another yeast GPCR, GPR1, as well as other mammalian sequences, p40 and pm1.1–7
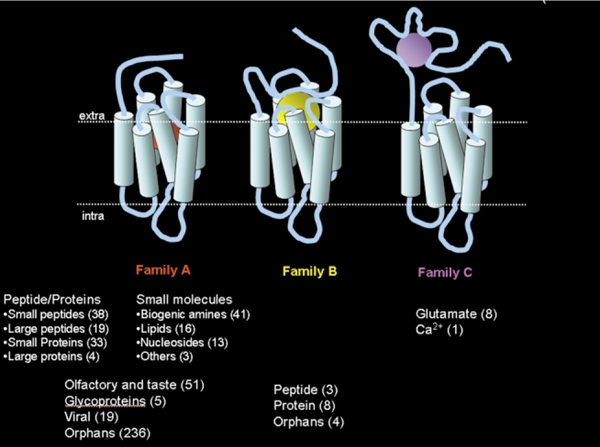
Fig. 1 depicts the distribution of the 508 human GPCRs identified so far, grouped according to the type of ligand. Approximately half of these GPCRs are still orphans, indicating that their function is yet unknown. It is expected that by the completion of the analysis of the human genome project even more GPCRs, that are potential new drug targets, will be discovered.
Fig. 1.
The “world of human GPCRs”. A distribution of the 508 human GPCRs that have been discovered so far, grouped according to the type of natural ligand that binds to them. Orphan receptors are receptors of yet unknown function.
From a biophysical perspective, members of the GPCR family are characterized by seven regions, each 20–25 amino acid sequences in length, that represent the transmembrane (TM) hydrophobic regions of the proteins. The seven TM domains are thought to form a barrel shape, oriented roughly perpendicular to the plane of the membrane in a counter clockwise manner, as viewed from the extracellular surface. Each receptor is believed to have an extracellular N-terminal region that varies in length from less than 10 amino acids (e.g., adenosine receptors) to several hundred amino acids (e.g., metabotropic glutamate receptors) and an intracellular C-terminal region. The majority of intra- and extra-cellular loops are thought to be 10–40 amino acids long, although the third intracellular loop and the C-terminal sequence may have more than 150 residues. The overall size of GPCRs varies significantly from less than 300 amino acids, in the case of adrenocorticotrophin hormone receptor, to more than 1100 amino acids for the metabotropic glutamate receptors.1–7
Most of the primary sequence homology among the different groups of GPCRs is contained within the TM domains. The most conserved residues among the GPCR superfamily are located within the TM domains and apparently represent essential structural determinants of receptor structure and function.
The evolution of the field of GPCR modeling has depended on the availability of suitable molecular templates. Due to technical difficulties, which complicate experimental X-ray crystallography and NMR structure determination of GPCRs; the 3D structure of most GPCRs is still unknown. The only known GPCR structure, a 2.8 Å resolution structure of rhodopsin, was published only recently by Palczewski et al.8
This structure sheds light on the mechanism of receptor activation and on specific receptor ligand interactions. Until the publication of this work the only structural information that existed about any GPCR was the low-resolution structure of rhodopsin that was solved by cryoelectron microscopy.9 Another structural template used in previous GPCR modeling was the 2.5 Å resolution X-ray structure of bacteriorhodopsin, obtained from microcrystals grown in the lipid cubic phase, that was determined a few years ago, even though bacteriorhodopsin is not a GPCR.10 The projection maps of bacteriorhodopsin and rhodopsin clearly showed the presence of seven TMs and confirmed the basic seven-helix bundle structure. However, the spatial organization of the TMs in rhodopsin is different from that in bacteriorhodopsin. To date, rhodopsin is still the only GPCR of known 3D structure.
Structure-based drug discovery is an alternative approach to conventional drug discovery, which relies on the fact that interactions of molecules within the human body take place in three dimensions. Drug molecules compete with natural ligands by inserting themselves into the functional site of the target protein and inducing (agonists) or inhibiting (antagonists) its activity. The affinity of these drugs to their respective target proteins is due to structural and chemical complementarity, and can be explored by computational methods. This allows for using relatively inexpensive and efficient computational screening technology instead of the costly and low yield experimental high-throughput screening (HTS) techniques. Furthermore, structure-based drug discovery can identify possible binding modes for ligands within the receptor cavity, typically through the identification of pharmacophoric centers complementary in character to the centers found on the surface of the receptor.
Here we review our recent work on adenosine receptors, a family of GPCRs. Focusing our attention on the A3 adenosine receptor, we have demonstrated that the reciprocal integration of different theoretical and experimental disciplines can be very useful for the successful protein-based design of new, potent and selective receptor ligands.
Adenosine receptors: a fascinating key study
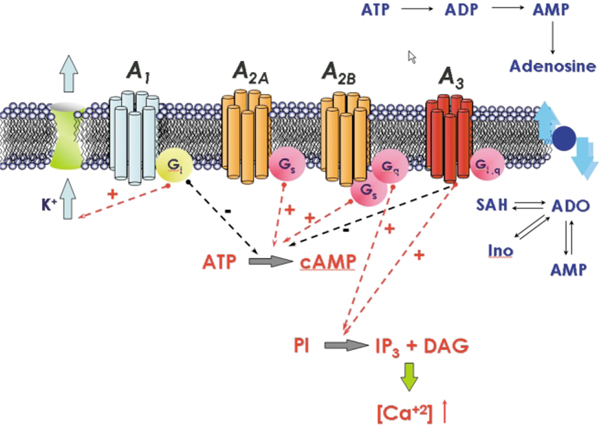
Ever since the discovery of the hypotensive and bradycardiac effects of adenosine in the circulation, adenosine receptors continue to represent promising drug targets.11,12 Firstly, this is due to the fact that the receptors are expressed in a large variety of cells; in particular, the actions of adenosine (or, respectively, of the antagonistic methylxanthines) in the central nervous system, in the circulation, on immune cells and on other tissues can be beneficial in certain disorders.11,12 Secondly, there exists a large number of ligands, which have been generated by introducing several modifications in the structure of the lead compounds (adenosine and methylxanthine), some of them highly specific.11,12 Four adenosine receptor subtypes (A1, A2A, A2B and A3) have been identified by molecular cloning. They are encoded by distinct genes. Originally, the receptors were classified based on their belonging to the family of G protein-coupled receptors, which transfer signals by activating heterotrimeric G proteins.11,12 As shown in Fig. 2, adenosine receptors can also be differentiated according to their preferred mechanism of signal transduction: A1 and A3 receptors interact with pertussis toxin-sensitive G proteins of the Gi and Go family; the canonical signalling mechanism of the A2A and of the A2B receptors is stimulation of adenylyl cyclase via Gs. A2B receptors can, in addition, also activate phospholipase C and this is thought to be mediated by activation of Gq.11,12
Fig. 2.
Signal transduction pathways associate with the activation of the human adenosine receptors.
The A3 adenosine receptor, which is the most recently identified adenosine receptor is implicated in a variety of important physiological processes.11–14 Activation of A3 adenosine receptors increases the release of inflammatory mediators, such as histamine, from rodent mast cells and inhibits the production of tumor necrosis factor-α. The activation of the A3 adenosine receptors is also suggested to be involved in immunosuppression and in the response to ischemia of the brain and heart.13,14 It is becoming increasingly apparent that agonists or antagonists of A3 adenosine receptors have potential as therapeutic agents for the treatment of ischemic and inflammatory diseases.13,14
As we will summarize in thisIt is becoming increasingly apparent article, the development of agonists and antagonists for the A3 receptors has so far been directed by traditional medicinal chemistry. The availability of genetic information promises to facilitate understanding of the drug-receptor interaction leading to the rational design of a potentially therapeutically important class of drugs. Moreover, molecular modeling may further rationalize observed interactions between the receptor and a ligand.
Topology of the TM domain of the A3 receptor: the homology modeling approach
Due to the lack of experimental three dimensional structures of the majority of GPCRs, structural insights must be inferred with the aid of three dimensional computer models.15–17 As discussed above, the structures of only two heptahelical membrane proteins have been determined at high resolution to date, rhodopsin and bacteriorhodopsin.
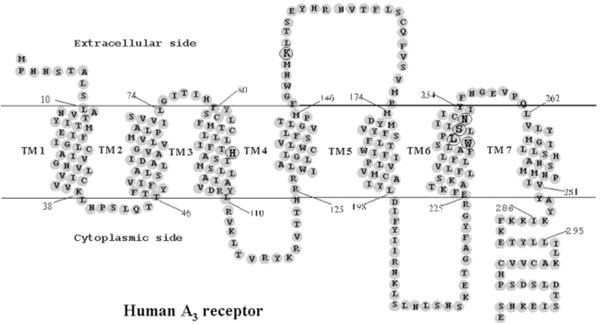
Therefore, rhodopsin (and before it bacteriorhodopsin) is widely used as a template for modeling the backbone structures of many GPCRs using homology-modeling techniques.15–17 Homology modeling describes an extended collection of techniques with the goal of modeling the 3D structure of a protein with an unknown structure, based on the known structures of related proteins. The accuracy of the prediction relies heavily on the number of structures that serve as a template and on their homology to the protein of interest (typically it requires more than 35% sequence identity).15–17 This method has proven very successful in modeling certain types of globular proteins. However, applying homology modeling to GPCRs is hampered by the low sequence identity between most GPCRs and rhodopsin (or bacteriorhodopsin). Furthermore, the great diversity of ligands that bind to GPCRs and the known diversity in binding sites (discussed above) suggest that ligands may interact with the receptor in different and diverse ways. Since at present the main purpose of GPCR modeling is to describe the ligand binding sites, the homology modeling approaches are clearly focused on their ability to predict novel binding pocket structures for the vast space of GPCR ligands. In this article, we focus our attention on the molecular architecture of GPCRs, with a special emphasis on our work on the human A3 receptor. A schematic representation (snake model) of the human A3 topology is shown in Fig. 3.
Fig. 3.
Heptahelical diagram of the human A3 adenosine receptor. The putative transmembrane domains were modified according to the high resolution rhodopsin model.8 Amino acids mutated in the present study are circled. Residues 286–295 correspond to an extra helical domain in rhodopsin, which is discontinuous from TM7.
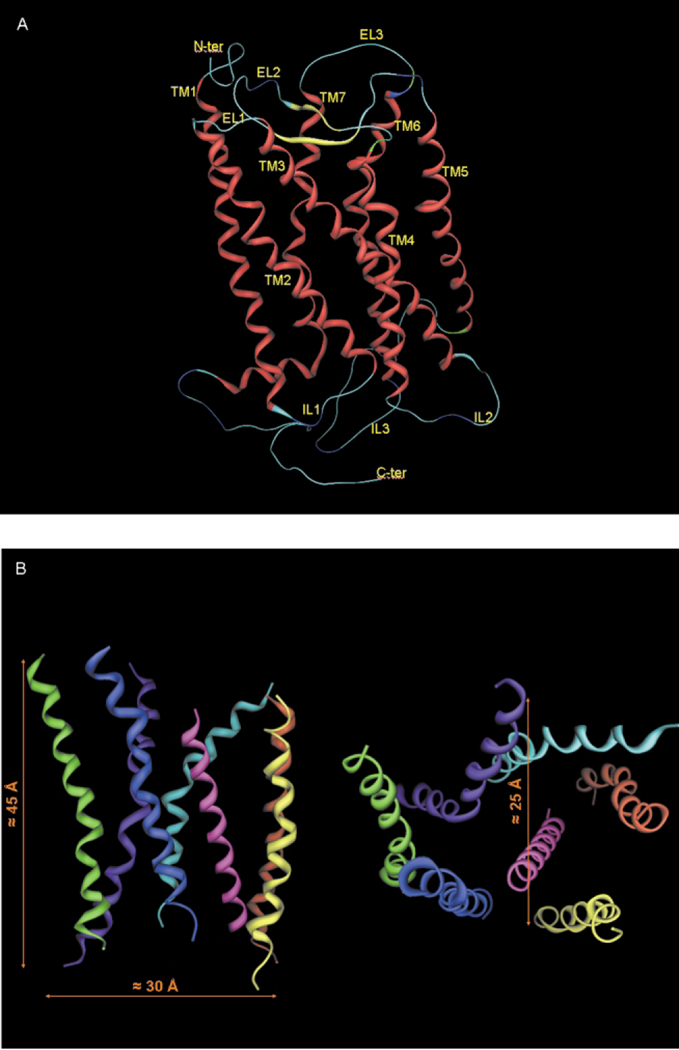
The first model of the TM segments of A3 receptor cloned from rat was proposed by van Galen et al.18 The model was based on the primary sequence and structural homology with bacteriorhodopsin, in analogy to the method proposed by Hibert et al.19 A putative ligand binding region in biogenic amine receptors and other GPCRs was originally proposed and explored by docking several antagonists into the TM domain model. In particular, this model suggested that TM3, TM6 and TM7 might be involved in ligand binding. However, bacteriorhodopsin is a proton pump in the outer membrane of Halobacterium halobium, and lacks any functional or sequence homology with GPCRs. In the interim, new inferences about the structure of these receptors have been based on cryomicroscopic studies of rhodopsin that indicated the existence of seven transmembrane segments and gave an indication of the relative disposition of these TMs.9 Using the low resolution structure of rhodopsin, we have extended the original model of van Galen et al. improving the description of ligand/receptor interactions by introduction of a simple means to simulate the reorganization of the native receptor structure induced by ligand coordination (cross-docking methodology).20,21 Our results illustrate that cross-docking can be used to predict local structural changes induced by ligand in the receptor binding site. The presence of both agonist and antagonist within the receptor could produce a simultaneous adjustment in the orientation of TM3, TM5, TM6, and TM7.21 Rotations and translations of the TM domains are crucial factors in the ligand recognition process in different GPCRs, as described by Gouldson et al.22 Consequently, our approach to docking is designed to mimic the natural domain movement within the receptors. Like other G protein-coupled receptor models, the length of the membrane-spanning region is about 40 Å (Fig. 4). The interhelical distance between pairs of adjacent helical axes is roughly 10Å, consistent with a common interhelical contact distance. The interhelical angles, measured between the principal axes of adjacent helices, range between − 150 to 170° for antiparallel, and 10 to 25° for the parallel helices. This is typical of a 3–4 type helix-helix contact associated with optimal interactions between nearly parallel aligned helices. Each helix maintained almost the same position and tilt angle after cross-docking.
Fig. 4.
A) Stereoview of the complete topology of human A3 receptor obtained by a homology modeling approach. B) Stereoview of the human A3 receptor transmembrane helical bundle model viewed along the helical axes from the extracellular end (right) and perpendicular to the helical axes (left).
Very recently, we have built an improved model of the transmembrane region of the human A3 receptor, using the high resolution crystal structure of bovine rhodopsin as a template, which can be considered a further refinement in building the hypothetical binding site of the A3 receptor antagonists already proposed.23–25 In fact, the ligand-binding sites on the A1, A2A, and A2B receptors have been experimentally characterized previously using site-directed mutagenesis.26–30 However, the molecular basis for ligand recognition in the A3 adenosine receptor remains largely unknown. Only very recently, Jacobson and collaborators created a “neoceptor” and several constitutively active mutant human A3 adenosine receptors by site-directed mutagenesis,31 which provided new insight into the molecular recognition in the A3 receptor. In order to provide additional insights into ligand-A3 adenosine receptor interactions, site-directed mutagenesis was used to study the role of a number of residues in the transmembrane (TM) domains and the second extracellular loop (EL2).32 We mutated several residues of the human A3 adenosine receptor within transmembrane domains 3 and 6 and the second extracellular loop, which have been predicted by previous molecular modeling to be involved in the ligand recognition, including His95, Trp243, Leu244, Ser247, Asn250, and Lys152. The N250A mutant receptor lost the ability to bind both radiolabeled agonist and antagonist.32 The H95A mutation significantly reduced the affinity of both agonists and antagonists. In contrast, the K152A (EL2), W243A (6.48), and W243F (6.48) mutations did not significantly affect the agonist binding but decreased antagonist affinity by 3–38-fold, suggesting that these residues were critical for the high affinity of A3 adenosine receptor antagonists.32 The A3 agonist 2-chloro-N6-(3-iodobenzyl)-5′-N-methylcarbamoyladenosine stimulated phosphoinositide turnover in the wild-type but failed to evoke a response in cells expressing W243A and W243F mutant receptors, in which agonist binding was less sensitive to guanosine 5′-γ-thiotriphosphate than in the wild-type. Thus, although not important for agonist binding, Trp243 was critical for receptor activation. The results were successfully interpreted using a rhodopsin-based model of ligand–A3 receptor interactions.33
From receptor topology to the synthesis of new potent and selective inhibitors
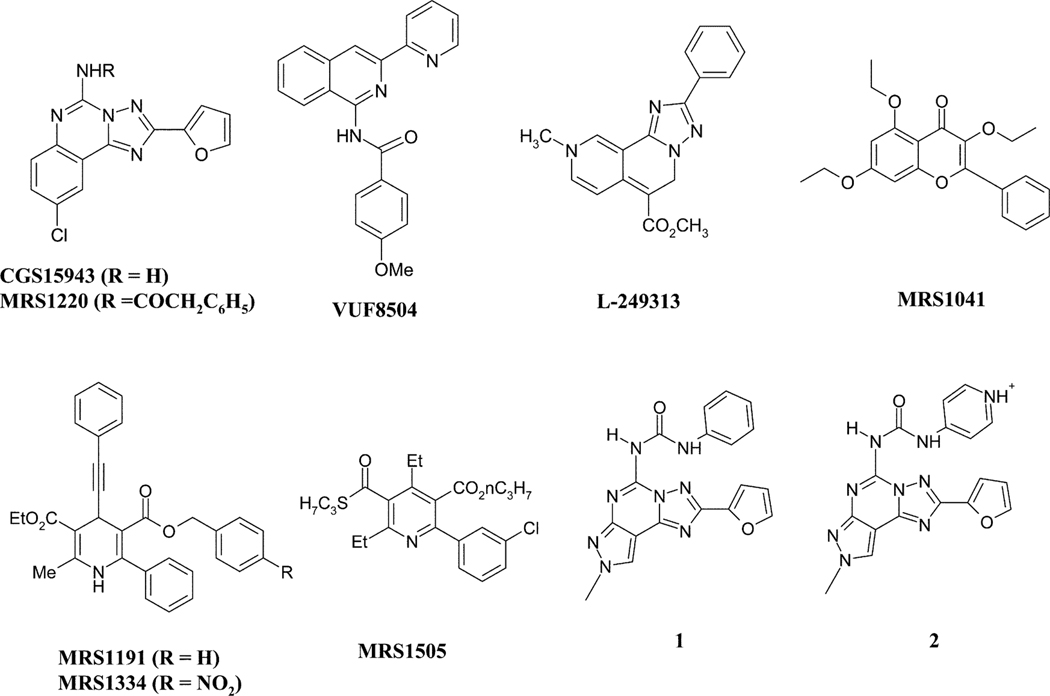
As already mentioned, the increasing knowledge of GPCRs (biological target space) and their ligands (chemical ligand space) enables novel drug design strategies to accelerate the finding and optimization of GPCR leads. The crystal structure of rhodopsin provides the first three dimensional GPCR information, which now supports homology modeling studies and structure-based drug design approaches within the GPCR target family. On the other hand, the classical ligand-based design approaches (for example, virtual screening, pharmacophore modeling, quantitative structure-activity relationships (QSAR)) are still powerful methods for lead finding and optimization. In addition, the cross-target analysis of GPCR ligands has revealed more and more common structural motifs and three dimensional pharmacophores. Such GPCR privileged structural motifs have been successfully used by many pharmaceutical companies to design and synthesize combinatorial libraries, which are subsequently tested against novel GPCR targets for lead finding. In the past few years, we have used both ligand-based and protein-based drug design approaches to discover new potent and selective antagonists of the A3 receptor. Fig. 5 summarizes the most representative members of the several classes of heterocyclic compounds identified as A3 adenosine antagonists.
Fig. 5.
Chemical structures of the most representative members of the several classes of heterocyclic compounds identified as A3 adenosine antagonists.
Attempts to identify leads for selective xanthine-based antagonists of the A3 receptor have not yet been productive. At human, dog, and sheep A3 receptors, certain xanthines are of intermediate affinity (Ki < 1 μM), however none have been found to be highly selective.
Promising leads for A3 antagonists have appeared among non-xanthine heterocyclic compounds. For example, the triazoloquinazoline CGS 15943 (9-chloro-2-(2-furanyl)[1,2,4]triazolo[1,5-c]quinazolin-5-amine), bound non-selectively to human adenosine receptors (almost equipotent at all four subtypes, with Ki values of ~ 1–10 nM), and thus has served as a lead for antagonists of the less well characterized subtypes. A N5-phenylacetyl derivative, MRS 1220, was a moderately A3 selective antagonist of very high affinity (Ki = 0.65 nM) at the human A3 receptor,34 but was of limited use in other species due to reduced A3 receptor affinity in rat, etc.
Since xanthines and other known adenosine antagonists provide only limited leads for A3 selective antagonists, an alternate strategy to identify selective antagonists is to exploit molecular diversity through screening chemical libraries, ligand-based and protein-based drug design approaches.
Novel structures have been identified through library screening in both industrial and academic laboratories.13 Among the first high affinity, A3 selective antagonists to be defined screening of chemically diverse compound collections were a triazolonaphthyridine (L-249313), a thiazolopyrimidine (L-268605), and pyridylisoquinolines (e.g. VUF 8504).35 Chemical structures of the most representative members of the several classes of heterocyclic compounds identified as A3 adenosine antagonists are shown in Fig. 5.
We have assembled a small, biased library of pure substances, containing many known pharmaceutical agents and structures that resemble purines in one or more features. The primary screen at the human A3 adenosine receptor consisted of single point binding assays at a fixed concentration (10 μM) using the radioligand 125I-AB-MECA. Once we discovered structural principles of recognition by the A3 adenosine receptor in these novel antagonist classes, we subjected the leads to structural optimization through iterative chemical synthesis and pharmacological testing.
Naturally-occurring phenolic derivatives, e.g. flavones and flavonols, provided a structural lead for A3 adenosine receptor antagonists. The affinity of common phytochemicals at adenosine receptors suggested that a wide range of natural substances in the human diet may potentially antagonize the effects of endogenous adenosine, including those mediated via the A3 subtype. The flavonoid class was chemically optimized in the form of MRS 1067 (3,6-dichloro-2′-(isopropoxy)-4′-methyl-flavone), which was 200-fold selective for human A3- vs. A1-adenosine receptors.36 Classical SAR studies performed on this natural class of compounds indicated that: i) the hydroxyl groups of flavonoid are not essential for A3 affinity and selectivity; ii) the etherification of hydroxyl functions with alkyl groups increased both potency and selectivity; iii) the introduction of chlorine at 3 and 6 positions affords the most potent and selective compounds of this class at hA3 adenosine receptor subtype;36 iv) the reduction of double bond at 2,3 position does not modify significantly the biological profile.36
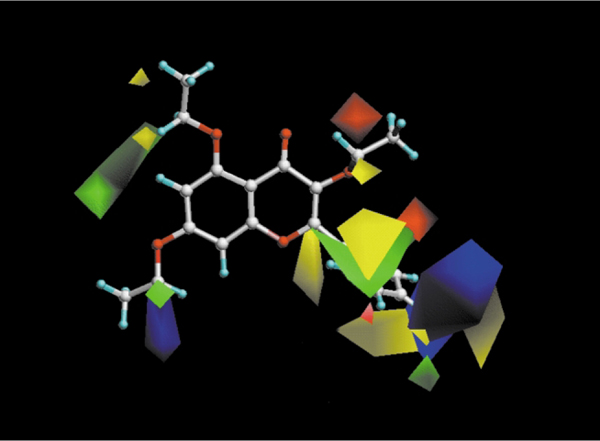
A quantitative explanation of the effect of these substitutions on the affinity at the human A3 adenosine receptor subtypes has been given through the help of a computational approach, in particular using CoMFA (Comparative Molecular Field Analysis).36 The proposed model clearly indicates that bulky groups at the C2 position of the chromone ring are not tolerated, while bulky substituents at the C6 and ortho-position of the phenyl ring and at the C2 position, are not only well tolerated but also increase A3 affinity (Fig. 6). This CoMFA model, also permitted discrimination of the steric and electronic contributions necessary for the receptor binding site of multiple subtypes of adenosine receptors.36
Fig. 6.
CoMFA steric and electrostatic contour plots from the analysis based on the A3 receptor 3D-QSAR without cross-validation. Compound MRS1041 shown inside the field. Favoring activity: green, bulky group (contribution level 80%); yellow, less bulky; blue, positive charge (contribution level 70%); red, negative charge.
Optimization of leads through a ‘multidimensional’ template approach — the pyridine family
The family of substituted pyridines and 1,4-dihydropyridines (DHPs) has given rise to a large number of novel adenosine receptor antagonists. DHPs are privileged structures in medicinal chemistry, i.e. they display low affinity binding at a variety of receptor and ion channel sites.37 Varying substituents on the DHP template can have dramatic effects on their interactions with these receptors. The corresponding, oxidized pyridines have provided a separate library of A3 adenosine receptor antagonists, which have SAR in some ways similar to, but overall distinct from the SAR for DHPs.
Dihydropyridines.
DHPs, known as potent blockers of L-type calcium channels and used widely in treating coronary heart disease, were found to bind appreciably to human adenosine A3 receptors. Common DHP drugs typically bound either non-selectively (for example, nifedipine, with a Ki value at human A3 receptor of 8.3 μM) or in some cases with selectivity for the A3 vs. other adenosine receptor subtypes (for example, S-niguldipme, with a Ki value at human A3 receptor of 2.8 μM).
We have used the 1,4-DHP nucleus as a template for probing structure-activity relationships (SAR) at the subtypes of adenosine receptors. We observed that at adenosine receptors, the affinity of many DHPs, even commercial L-type calcium channel blockers, is in the micromolar range at the human A3 receptor, and we optimized this lead in the design of more selective antagonists. The essential modifications leading to A3 selectivity and preventing binding to L-type calcium channels and other sites were: a phenyl ring at the 6-position and either a styryl or phenylethynyl group at the 4-position. Thus, DHP derivatives, such as MRS 1191 (3-ethyl-5-benzyl-2-methyl-6-phenyl-4-phenylethynyl-1,4-(±)-dihydropyridine-3,5-dicarboxylate), appear to be useful as A3 adenosine receptor antagonists across species, although there is still a need for high affinity antagonists of rat A3 adenosine receptors.
The structure-activity relationships of analogues of MRS 1191, containing both subtle and drastic structural changes at various positions of the DHP ring (its 3- and 5-acyl substituents, the 4- and 6-aryl/alkyl substituents, and the 2-methyl group), were systematically investigated. Substitutions of a 5-benzyl ester group provided the greatest versatility for achieving A3 receptor selectivity of > 30000-fold and nanomolar potency. Affinity and selectivity for the human A3 adenosine receptor within this series was optimal in MRS 1334 (3-ethyl 5-(4-nitrobenzyl) 2-methyl-6-phenyl-4-phenylethynyl-1,4-(±)-dihydropyridine-3,5-dicarboxylate) which had a Ki value of 2.7 nM at the human A3 receptor.
While the stereospecificity of the binding of chiral ligands is a general property of receptors, the enantiomeric ratios of affinities may vary widely. Since racemic 6-phenyl-4-phenylethynyl-1,4-dihydropyridine derivatives had displayed exceptionally high A3 adenosine receptor selectivity, compounds in this series were chemically resolved in order to study the biological properties of pure stereoisomers. Such a method for resolving the enantiomers at the C4 position consisted of either the selective crystallization or HPLC purification of diastereomeric ester derivatives. The chiral ester moieties, derivatives of 2,2-dimethyl-1,3-dioxolane-4-methanol and 2,3-O-isopropylidene-D-threitol, could have been selectively replaced with an ethyl moiety in two steps, i.e. deprotection of a diol followed by transesterification in ethanol. Binding studies with the pure enantiomers of MRS 1191 had indicated a 35-fold stereoselectivity for the (4-S-isomer at the human A3 adenosine receptor.38
Pyridines.
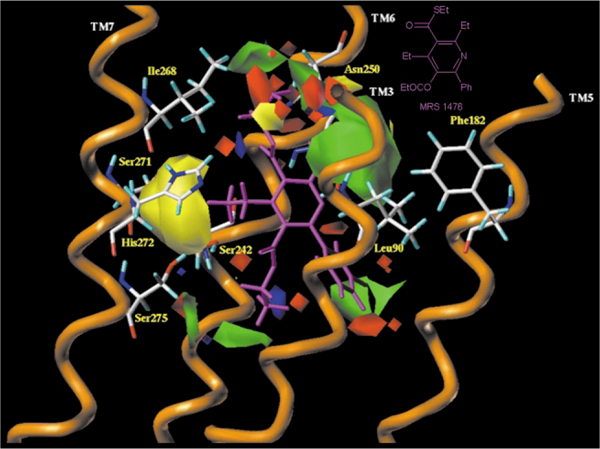
We explored structure-activity relationships (SAR) for the oxidized forms of the 1,4-DHps, i.e. 3,5-diacylpyridines, as highly selective antagonists for the human A3 adenosine receptor.39 Moderate selectivity for the rat A3 adenosine receptor was also present. The rationale of this pharmacologic difference between rat and human is still under investigation. As for the DHPs, a 6-aryl substituent present in the oxidized pyridines favored A3 adenosine receptor selectivity. However, in contrast to DHPs, only small alkyl groups such as propyl were tolerated at the 4-position and 5-ester position. MRS 1505 (2,4-diethyl-3,5-dipropyl-6-(3-chlorophenyl)pyridine-3-thiocarboxylate-5-carboxylate) shown in Fig. 5 is an example of a potent and selective antagonist at the human A3 receptor. Furthermore, a 3-thioester enhanced A3AR selectivity. Fluoro-, hydroxy- and other polar groups were introduced in this series, which otherwise had a highly hydrophobic character, and affinity as high as 4 nM was achieved. Sterically constrained (bicyclic) analogues were designed and prepared for purposes of conformational analysis, demonstrating a preference for the 3-carbonyl group in the “down” orientation. New pyridine derivatives with higher affinity and selectivity for the A3 adenosine receptor and potentially improved pharmacodynamic properties were obtained. Through in tandem CoMFA and docking studies on a model of human A3 adenosine receptor derived from rhodopsin, has been observed that a different steric control around the 4-position between 1,4-dihydropyridine and pyridine derivatives is present. As shown in Fig. 7 where MRS1476 (2,3,4,5-tetraethyl-6-phenyl-pyridine-3-thiocarboxylate-5-carboxylate) has been docked into human A3 adenosine receptor, bulky 4-position substituents, which are affinity-enhancing in the 1,4-dihydropyridines, are not well tolerated in the pyridine series.12 This has been explained with the change of the C4-hybridization from sp3 to sp2, which change the C5-C4-R4 angle from 68.1 to 0.2°. On this basis, it is evident that in order to retain affinity and selectivity at hA3 adenosine receptors, small groups at the 4-position are essential.39
Fig. 7.
Side view of the human A3–MRS1476 complex model. The side chains of the important residues in proximity ( < 5 Å) to the docked pyridine molecule are highlighted and labeled: Leu90 (TM3), Phe182 (TM5), Ser242 (TM6), Ser247 (TM6), Asn250 (TM6), Ser271 (TM7), His272 (TM7) and Ser275 (TM7). The steric and the electrostatic contour plots, obtained from the CoMFA analysis, are included into the ligand binding cavity.
A new class of potent and selective human A3 antagonist: pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidine derivatives
A fruitful synergism between synthetic and theoretical chemists has been reported for a large series of 5-[[(substituted-phenyl)amino]carbonyl]amino-8-alkyl-2-(2-furyl)-pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidine which proved to be potent and selective human A3 adenosine receptor antagonists.23–25 In particular the optimization of both N8 and N5 substitution have been performed with the fundamental help of docking studies performed on a new model of the transmembrane region of the human A3 receptor, using the crystal structure of rhodopsin as a template, which can be considered a further refinement in building the hypothetical binding site of the A3 receptor antagonists previously proposed.23–25
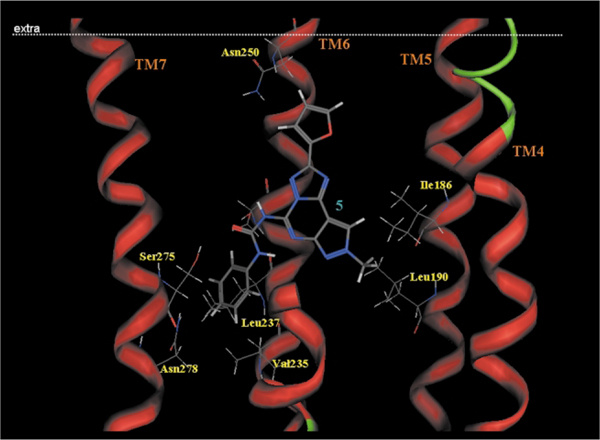
As shown in Fig. 8, we identified the hypothetical binding site of the pyrazolo-triazolo-pyrimidine nucleus surrounded by TMs 3, 5, 6, and 7 with the furan ring pointing towards the extracellular environment.23–25
Fig. 8.
Side view of the human A3–5 complex model. The side chains of the important residues in proximity to the docked 5 molecule are highlighted and labeled.
Around the N8 pyrazole position, two hydrophobic pockets are likely present, where an alkyl/aryl side chain would bind (Leu90, TM3 and Phe182, tM5). This small hydrophobic pocket clearly indicates that only small alkyl chains (e.g. methyl is better) at the N8 pyrazole nitrogen fit very well with the shape of the receptor pocket.
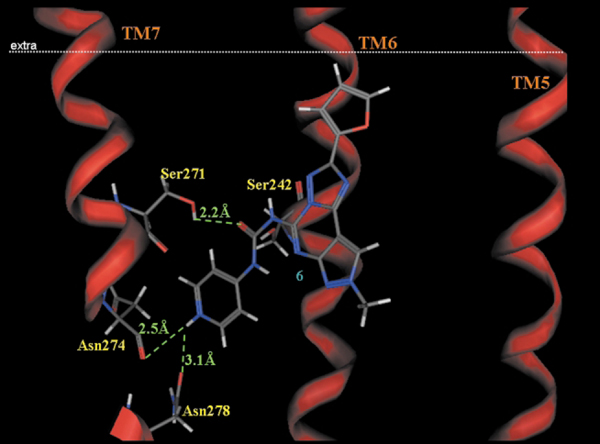
A similar approach has been utilized for the optimization of the substituent at the N5 position. In particular, the carbamoyl moiety in the 5-position of the pyrazolo[4,3-e]1,2,4-triazolo[1,5-c]pyrimidine structure surrounded by four polar aminoacids: Ser242 (TM6), Ser271 (TM7), His274 and Ser275 (TM7) affording strong interactions, orienting the carbamoyl phenyl ring in the middle of TM6 and TM7. Analysis of the phenyl substituent at the N5 position was very interesting. In fact, the receptor region around the para and meta positions (TM6 and TM7 regions) of the phenyl ring is mostly hydrophobic (Val235[TM6], Leu236[TM6], and Asn278[TM7]) and reveals that very limited empty space is present suggesting that steric control seems to be taking place around this position of the phenyl ring. In contrast, substituents at the ortho position seem to occupy an empty region of the binding cavity.23–25 These experimental observations not only explain the great potency and selectivity of compound 1 (Fig. 5), but induce the authors to avoid any kind of substitutions on the phenyl ring to increase water solubility. With the aim of achieving a water soluble compound with retention of affinity and selectivity Spalluto and coworkers25 conjectured the bioisosteric replacement of a phenyl ring with a pyridine nucleus. The obtained compounds 2 (Fig. 5), not only resulted to be highly water soluble, but showed an increased potency of about 16 fold. In a similar manner, for better understanding this increasing of potency has been examined through the use of docking studies. As shown in Fig. 9, the increase of the affinity of compound 2 seems to be attributed to additional strong electrostatic interactions between the positively charged pyridinium moiety of 2 and the carbonyl oxygen atoms of Asn274 (N+H···OC distance = 2.5 Å) and Asn278 (N+H···OC distance = 3.1 Å), both located on TM7. Thus, in view of the potency, selectivity, and water solubility, derivative 2 could be an ideal candidate for pharmacological and clinical investigation of the hA3 adenosine receptor subtype.
Fig. 9.
Side view of the human A3–6 complex model. The side chains of the important residues in proximity to the docked 6 molecule are highlighted and labeled.
Conclusion
In conclusion, in this review we have attempted to describe how different molecular modeling approaches can help the investigation of both receptor architecture and ligand/receptor molecular recognition. However, fundamental understanding of the molecular details of ligand/GPCR interactions remains very rudimentary. How agonist binding transforms a resting GPCR into its active form and the microscopic basis of binding site blockade by an antagonist are generally still unclear. In the absence of high-resolution structural knowledge of the different conformational states of different GPCRs, such questions can only be addressed by building models, which are tested through pharmacological and biochemical studies. Structural models can be used to describe the interactions between a ligand and its receptor. However, even if X-ray crystal structures were available, problems would still arise in the modeling due to the dynamic nature of the ligand recognition process. Indeed, molecular modeling is also an interesting tool to the rational design of new, potent and selective ligands. The increasing knowledge of GPCRs (biological target space) and their ligands (chemical ligand space) enables novel drug design strategies to accelerate the finding and optimization of GPCR leads.
Biography
Stefano Moro is an Assistant Professor of medicinal chemistry at Padova University, Italy. He received a B.Sc. degree in medicinal chemistry and a Ph.D. degree in physical organic chemistry from the University of Padova, Italy, and was a Postdoctoral Fellow at the National Institutes of Health in Bethesda, MD. He was Visiting Professor at ETH of Zurich, Switzerland. He is the chief of the Molecular Modeling Section at Padova University. His main research interest concerns the application of computational approaches to medicinal chemistry. He is the recipient of the National Award in Medicinal Chemistry for Excellent Research (2002).
Francesca Deflorian is a Ph.D. student in the Department of Pharmaceutical Sciences, Padova University, Italy. She received a B.Sc. degree and a M.Sc. degree in Medicinal Chemistry from Padova University, Italy. She is a computational chemist and she has been working on GPCR protein design and on rational design of new adenosine receptor antagonists.
Giampiero Spalluto received his degree in Chemistry and Pharmaceutical Technology in 1987from the University of Ferrara. He obtained his Ph.D. in Organic Chemistry from the University of Parma in 1992. Between 1995 and 1998 he was Assistant Professor of Medicinal Chemistry at the University of Ferrara. From November 1998 to the present he has held the position of Associate Professor of Medicinal Chemistry at the University of Trieste. His scientific interests have focused on the medicinal chemistry of ligands for adenosine receptor subtypes.
Giorgia Pastorin received her degree in Chemistry and Pharmaceutical Technology in 2000 from the University of Trieste. She is now carrying out her Ph.D. work on the design and synthesis of new ligands for adenosine receptor subtypes.
Barbara Cacciari received her degree in Chemistry and Pharmaceutical Technology in 1990 from the University of Bologna. She obtained her Ph.D. in Medicinal Chemistry from the University of Ferrara in 1994. Between 1992 and 1993 she worked at the Scripps Research Institute in La Jolla, CA, USA, with Professor Dale L. Boger. From 1995 to the present she has held a position of Assistant Professor in Medicinal Chemistry at the University of Ferrara. Her scientific interests focus on adenosine receptor ligands.
Soo Kyung Kim is a Visiting Fellow in the Molecular Recognition Section, National Institute of Diabetes, Digestive, and Kidney Diseases, at the National Institutes of Health in Bethesda, MD. She is a computational chemist and has been modeling adenosine receptors in conjunction with site directed mutagenesis experiments.
Kenneth A. Jacobson is Chief of the Molecular Recognition Section and Director of the Chemical Biology Laboratory, National Institute of Diabetes, Digestive, and Kidney Diseases, at the National Institutes of Health in Bethesda, MD. He is a medicinal chemist with interests in the structure and pharmacology of G protein-coupled receptors, particularly receptors for the purine nucleosides and nucleotides. His interdisciplinary approach involves both synthesis of small molecule ligands and characterization of their protein targets (receptors).
Notes and references
- 1.Wilson S, Bergsma DJ, Chambers JK, Muir AI, Fantom KG, Ellis C, Murdock PR, Herrity NC and Stadel JM, Br. J. Pharmacol, 1999, 125, 1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilson S. and Bergsma D, Drug Des. Discov, 2000, 17, 105–114. [PubMed] [Google Scholar]
- 3.Civelli O, Nothacker HP, Saito Y, Wang Z, Lin SH and Reinscheid RK, Trends Neurosci. , 2001, 24, 230–237. [DOI] [PubMed] [Google Scholar]
- 4.Marchese A, George SR, Kolakowski LF Jr, Lynch KR and O’Dowd BF, Trends Pharmacol. Sci, 1999, 20, 370–375. [DOI] [PubMed] [Google Scholar]
- 5.Lee DK, George SR, Evans JF, Lynch KR and O’Dowd BF, Curr. Opin. Pharmacol, 2001, 1, 31–39. [DOI] [PubMed] [Google Scholar]
- 6.Howard AD, McAllister G, Feighner SD, Liu Q, Nargund RP, Van der Ploeg LH and Patchett AA, Trends Pharmacol. Sci, 2001, 22, 132–140. [DOI] [PubMed] [Google Scholar]
- 7.Klabunde T. and Hessler G, Chembiochem. , 2002, 10, 928–944. [DOI] [PubMed] [Google Scholar]
- 8.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M. and Miyano M, Science, 2000, 289, 739–742. [DOI] [PubMed] [Google Scholar]
- 9.Unger VM, Hargrave PA, Baldwin JM and Schertler GF, Nature, 1997, 389, 203–207. [DOI] [PubMed] [Google Scholar]
- 10.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E. and Downing KH, J. Mol. Biol, 1990, 213, 899. [DOI] [PubMed] [Google Scholar]
- 11.Klotz KN, Naunyn-Schmiedeberg’s Arch. Pharmacol, 2000, 362, 382–391. [DOI] [PubMed] [Google Scholar]
- 12.Klinger M, Freissmuth M. and Nanoff C, Cell. Signalling, 2002, 14, 99–108. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson KA, Trends Pharmacol. Sci, 1998, 19, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller CE, Mini Rev. Med. Chem, 2001, 1, 417–427. [DOI] [PubMed] [Google Scholar]
- 15.Shacham S, Topf M, Avisar N, Glaser F, Marantz Y, Bar-Haim S, Noiman S, Naor Z. and Becker OM, Med. Res. Rev, 2001, 21, 472–483. [DOI] [PubMed] [Google Scholar]
- 16.Okada T. and Palczewski K, Curr. Opin. Struct. Biol, 2001, 11, 420–426. [DOI] [PubMed] [Google Scholar]
- 17.Gershengorn MC and Osman R, Endocrinology, 2001, 142, 2–10. [DOI] [PubMed] [Google Scholar]
- 18.van Galen PJ, van Bergen AH, Gallo-Rodriguez C, Melman N, Olah ME, IJzerman AP, Stiles GL and Jacobson KA, Mol. Pharmacol, 1994, 45, 1101–1111. [PMC free article] [PubMed] [Google Scholar]
- 19.Hibert MF, Trumpp-Kallmeyer S, Bruinvels A. and Hoflack J, Mol. Pharmacol, 1991, 40, 8. [PubMed] [Google Scholar]
- 20.Moro S, Guo D, Camaioni E, Boyer JL, Harden TK and Jacobson KA, J. Med. Chem, 1998, 41, 1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moro S, Li AH and Jacobson KA, J. Chem. Inf. Comput. Sci, 1998, 38, 1239–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouldson PR, Snell CR and Reynolds CA, J. Med. Chem, 1997, 40, 3871. [DOI] [PubMed] [Google Scholar]
- 23.Baraldi PG, Cacciari B, Moro S, Romagnoli R, Ji XD, Jacobson KA, Gessi S, Borea PA and Spalluto G, J. Med. Chem, 2001, 44, 2735–2742. [DOI] [PubMed] [Google Scholar]
- 24.Baraldi PG, Cacciari B, Moro S, Spalluto G, Pastorin G, Da Ros T, Klotz KN, Varani K, Gessi S. and Borea PA, J. Med. Chem, 2002, 45, 770–780. [DOI] [PubMed] [Google Scholar]
- 25.Maconi A, Pastorin G, Da Ros T, Spalluto G, Gao ZG, Jacobson KA, Baraldi PG, Cacciari B, Varani K, Moro S. and Borea PA, J. Med. Chem, 2002, 45, 3579–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barbhaiya H, McClain R, IJzerman AP and Rivkees SA, Mol. Pharmacol, 1996, 50, 1635–1642. [PubMed] [Google Scholar]
- 27.Tucker AL, Robeva AS, Taylor HE, Holeton D, Bockner M, Lynch KR and Linden J, J. Biol. Chem, 1994, 269, 27900–27906. [PubMed] [Google Scholar]
- 28.Kim J, Wess J, van Rhee AM, Schoneberg T. and Jacobson KA, J. Biol. Chem, 1995, 270, 13987–13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao ZG, Jiang Q, Jacobson KA and IJzerman AP, Biochem. Pharmacol, 2000, 60, 661–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beukers MW, den Dulk H, van Tilburg EW, Brouwer J. and IJzerman AP, Mol. Pharmacol, 2000, 58, 1349–1356. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson KA, Gao ZG, Chen A, Barak D, Kim SA, Lee K, Link A, van Rompaey P, van Calenbergh S. and Liang BT, J. Med. Chem, 2001, 44, 4125–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao ZG, Chen A, Barak D, Kim SK, Muller CE and Jacobson KA, J. Biol. Chem, 2002, 277, 19056–19063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao ZG, Kim SK, Biadatti T, Chen W, Lee K, Barak D, Kim SG, Johnson CR and Jacobson KA, J. Med. Chem, 2002, 45, 4471–4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim YC, de Zwart M, Chang L, Moro S, von JK Frijtag Drabbe Kunzel, N. Melman, A. P. IJzerman and K. A. Jacobson, J. Med. Chem, 1998, 41, 2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Muijlwijk-Koezen JE, Timmerman H, Link R, Van der Goot H. and IJzerman AP, J. Med. Chem, 1998, 41, 3994–4000. [DOI] [PubMed] [Google Scholar]
- 36.Jacobson KA, Moro S, Manthey JA, West PL and Ji XD, Adv. Exp. Med. Biol, 2002, 505, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Triggle DJ, in Comprehensive Medicinal Chemistry, 1985, vol. 3, Emmett JC, ed., Pergamon Press, London, pp. 1047–1099. [Google Scholar]
- 38.Jiang JL, Li A-H, Jang SY, Chang L, Melman N, Moro S, Ji X-D, Lobkovsky EB, Clardy JC and Jacobson KA, J. Med. Chem, 1999, 42, 3055–3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li A-H, Moro S, Forsyth N, Melman N, Ji XD and Jacobson KA, J. Med. Chem, 1999, 42, 706–721. [DOI] [PMC free article] [PubMed] [Google Scholar]