Abstract
The recent revolution in cryo-electron microscopy (cryo-EM) has made it possible to determine macromolecular structures directly from cell extracts. However, identifying the correct protein from the cryo-EM map is still challenging and often needs additional sequence information from other techniques, such as tandem mass spectrometry and/or bioinformatics. Here, we present DeepTracer-ID, a server-based approach to identify the candidate protein in a user-provided organism de novo from a cryo-EM map, without the need for additional information. Our method first uses DeepTracer to generate a protein backbone model that best represents the cryo-EM map, and this model is then searched against the library of AlphaFold2 predictions for all proteins in the given organism. This method is highly accurate and robust for high-resolution cryo-EM maps: in all 13 experimental maps tested blindly, DeepTracer-ID identified the correct proteins as the top candidates. Eight of the maps were of known structures, while the other five unpublished maps were validated by prior protein annotation and careful inspection of the model refined into the map. The program also showed promising results for both homomeric and heteromeric protein complexes. This platform is possible because of the recent breakthroughs in large-scale three-dimensional protein structure prediction.
Significance
While it has now become routine for cryo-EM maps of proteins to reach a near-atomic resolution, potentially allowing for reliable atomic models to be built, there is a growing number of instances where the protein identity may not be known. Without knowing the protein sequence, it is almost impossible to build an atomic model. DeepTracer-ID is a server-based approach to surmount this problem by identifying the proteins in a given organism that are found in the cryo-EM map. A free web service for global academic access is provided.
Introduction
Over the past 9 years, there has been a resolution revolution in cryo-electron microscopy (cryo-EM), with exponential growth in the number of determined atomic structures per year (1, 2, 3). Unlike X-ray crystallography or nuclear magnetic resonance, which often require a large amount of sample, a high concentration, and a high degree of sample purity, using cryo-EM it is possible to capture structures of macromolecular complexes under conditions closer to the in situ environments with fewer restraints on volume, sample purity, and concentration (4, 5, 6). With recent software developments (7,8), cryo-EM can routinely sort out different conformational states of single macromolecules (9) and even unrelated complexes within the same data set (10,11). When a cryo-EM map of unknown components reaches ≈4.2 Å resolution or better where β-sheets are well resolved, one can typically trace the Cα backbone directly from the map. The remaining challenge is identifying the correct protein sequence from the organism’s proteome.
A typical workflow that has been used to address this problem is first to detect which proteins are present in the sample by tandem mass spectrometry (MS/MS), then threading the sequences of detected proteins into the cryo-EM map to identify the best match (12, 13, 14). However, this approach first relies on the target protein being detected by MS/MS, which is sometimes not the case due to, for example, the lack of digestion sites (15) or a high degree of post-translational modifications (16). Thousands of proteins can be detected if the target of interest has been imaged directly from cell extracts. Under such circumstances, possible sequence targets have to be selected based on the structural features of the map. Each sequence must then be built into the map to identify the best match by trial and error.
A second approach is using model-building tools, such as DeepTracer (17), to perform protein sequence prediction based on an input cryo-EM map. The predicted sequence is then used for BLAST analysis to identify potential hits in the selected organism (18). This approach is greatly limited by the accuracy of the backbone trace and the quality of side-chain densities of the input cryo-EM map. In addition, three-dimensional (3D) structural information is not used in this type of search, and the BLAST results may be very difficult to interpret.
Another approach to identifying unknown proteins is to trace the backbone from the cryo-EM map and then find the best matches against the experimentally determined structures in the Protein Data Bank (PDB). One example of this approach is FindMySequence (19). However, this method is likely to be limited to detecting proteins of known structure or proteins that are structurally similar to the known structures. Fortunately, with recent breakthroughs in protein 3D structure prediction by AlphaFold2, it is possible to obtain accurate protein structure predictions even when no homologous structure is available (20).
Here, we describe a de novo protein identification approach, DeepTracer-ID, for cryo-EM maps with better than 4.2 Å resolution. Our program first automatically generates a backbone tracing of the cryo-EM map using DeepTracer. The AlphaFold2 predicted models are then aligned to the DeepTracer backbone using three different algorithms, PyMOL-align (21), PyMOL-cealign (22) and FATCAT (23). The version of PyMOL being used is open-source 2.6.0a0. On a benchmark set of 11 experimental segmented cryo-EM maps, including three previously unsolved structures, we show that our program, DeepTracer-ID, can identify the correct protein and their closely related homologs in a pool of AlphaFold2 predicted models. Furthermore, we show that DeepTracer-ID can detect the correct protein from maps of homomeric protein complexes and also identify the largest components within maps of massive heteromeric complexes.
Materials and methods
Pipeline inputs of DeepTracer-ID
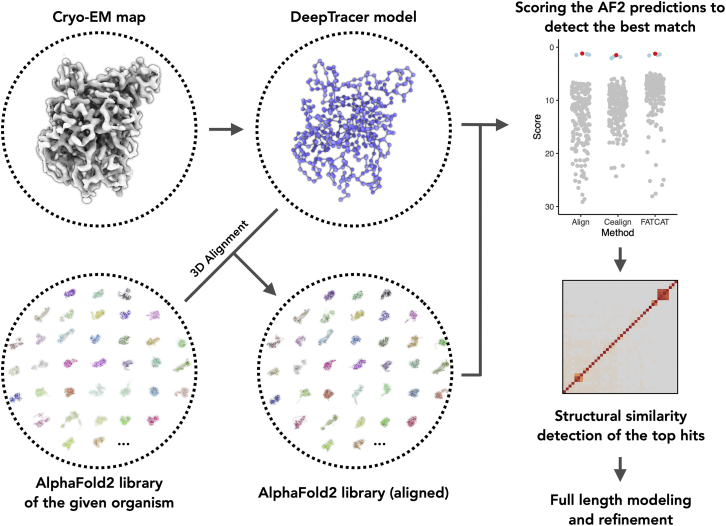
Two inputs are needed to use the pipeline: 1) a cryo-EM map, segmented to correspond to a single protein subunit, which is preferred but not necessary; and 2) a pre-calculated AlphaFold2 protein library to search against (Fig. 1). The input cryo-EM map is used to generate a 3D model trace by DeepTracer (17). In a rare case when the cryo-EM map does not contain a single α-helix, there is an ambiguity in the absolute hand of the cryo-EM map (24). In this case, the users are encouraged to mirror the input map and run DeepTracer-ID twice (for the original and mirrored map) for the best results. The proteomes from a number of model organisms and pathogens important to global health, such as Homo sapiens, Escherichia coli, Arabidopsis thaliana, Caenorhabditis elegans, Campylobacter jejuni, and Streptococcus pneumoniae, have already been pre-calculated (25), and for those organisms an AlphaFold2 protein library is saved on the DeepTracer-ID server and is thus not required from the user side. For other organisms, the user can either generate their AlphaFold2 library locally or use an online service such as ColabFold (26).
Figure 1.
DeepTracer-ID: De novo protein identification from cryo-EM density maps. Starting with a cryo-EM map and a given organism, DeepTracer-ID identifies the protein in three steps: 1) the initial model is generated from the cryo-EM map using deep learning based DeepTracer; 2) the AlphaFold2 prediction library (user-provided or server pre-calculated) is aligned to the initial model; 3) each aligned AlphaFold2 prediction is scored and the top hits subsequently analyzed. To see this figure in color, go online.
The next question will be how many AlphaFold2 models to generate for the screening library. The answer depends on how well the user can estimate the length of the target protein. For a segmented map generated by an experienced EM person, the user may have a very reasonable estimate of the total length of the protein. Therefore, a user-defined search range (e.g., 150–250 residues in a given organism) will significantly speed up the calculations. Otherwise, it is also possible to search against the AlphaFold2 predictions of the entire organism’s proteome, which takes a longer time of course.
3D alignment of AlphaFold2 predictions to map-traced model
We use three different approaches, PyMOL-align (21), PyMOL-cealign (22), and FATCAT (23), to align the AlphaFold2 predicted structures to the backbone model traced directly from the cryo-EM map (Fig. 1). The aligned AlphaFold2 predictions are saved separately for subsequent root-mean-squared deviation (RMSD) analysis. Three different alignment algorithms are used, as we expect they have different advantages and disadvantages depending on the input cryo-EM map. PyMOL-align considers both similarities in sequence and structure, and this is the default alignment option because of its overall most robust performance (see below). Specifically, PyMOL-align performs a sequence alignment first followed by a structural superposition, after which the alignment is refined in more cycles to reject structural outliers found during the fit. PyMOL-cealign aligns two proteins using the combinational extension algorithm (22). It has been reported as helpful for proteins with little to no sequence similarity. Therefore, PyMOL-cealign may be more useful when the protein side-chain densities are not well resolved in the cryo-EM map. FATCAT is an approach for flexible protein structure comparison. It achieves two features during the structure alignment: both optimizing the alignment and minimizing the number of rigid-body movements around pivot points introduced in the reference structure. Using FATCAT could help minimize some errors introduced by the AlphaFold2 predictions, and it may be more useful for smaller proteins or proteins that rely on the local environment to form an ordered 3D structure.
Protein identification scoring matrix
Each AlphaFold2 prediction is scored with two factors: 1) the RMSD to the DeepTracer model and 2) the percentage of residues in the DeepTracer model that can be aligned to the corresponding AlphaFold2 prediction (Eq. 1). The RMSD calculation between the AlphaFold2 prediction and the DeepTracer model is carried out using the methods previously implemented in DeepTracer (17). The length of the DeepTracer model is , and the length of matching residues in the superposed AlphaFold2 prediction is . The AlphaFold2 predictions are then listed from lowest to highest based on this score, and the correct protein is expected to be detected within the AlphaFold2 predictions with lowest score:
| (1) |
Using this approach, we expect to not only detect the correct protein but also other isoforms or homologs that are structurally similar to the correct protein. Therefore, after scoring all AlphaFold2 predictions, we use the DaliLite v5 (27) to perform an all-to-all analysis on the top hits.
Results
Protein identification from segmented cryo-EM maps
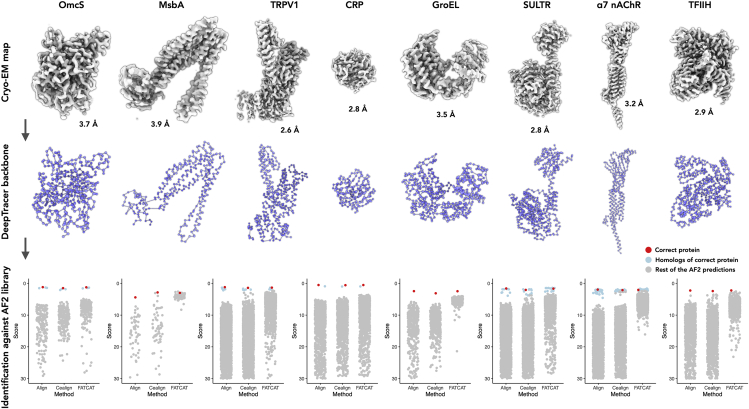
We tested our method on a benchmark set of eight segmented cryo-EM maps from published literature, where we assumed the authors assigned the correct protein sequences to the EM maps. These proteins range in length from 224 to 838 residues, and the maps have a resolution ranging from 2.6 to 3.9 Å. Initially, five cryo-EM maps were selected as test sets to optimize our approach. The selection criteria considered the most likely pitfalls for this approach, and we tried to include cases where: 1) the map lacks secondary structure and has multiple ligands bound; 2) only a portion of the map reaches high resolution; 3) the corresponding protein has multiple domains, and their relative orientation is unlikely to be accurately predicted by AlphaFold2. In all five cases, despite these existing possible pitfalls, DeepTracer-ID was able to identify the correct protein as the top hit based on score (Eq. 1 and Fig. 2). We then moved on to test three more EM maps from eukaryotes with larger genomes, two from H. sapiens and one from A. thaliana. DeepTracer-ID could also identify the correct protein as the top hit, except for one case (α7 nAChR from H. sapiens). In that case the correct protein was ranked third, while the top 32 hits are all structurally related homologs (Figs. 2 and 3).
Figure 2.
Protein identification from eight benchmark cryo-EM maps. Top: cryo-EM maps used for de novo protein identification. Their reported resolution is labeled. Middle: the Cα backbone of the model generated by DeepTracer, from the maps on top. Bottom: the scores of corresponding AF2 predictions are displayed in scatterplots. The results from all three different alignment algorithms are shown. The correct protein is shown as a red dot, the proteins with significant structural similarity to the correct protein are shown in blue dots, and the rest of the AF2 predictions are shown as gray dots. The size of the AF2 library and the corresponding organism for the eight benchmark data sets are OmcS (Geobacter sulfurreducens PCA, N = 226), MsbA (E. coli BL21-DE3, N = 65), TRPV1 (Rattus norvegicus, N = 3679), CRP (H. sapiens, N = 1419), GroEL (E. coli K12, N = 413), SULTR (A. thaliana, N = 3850), α7 nAChR (H. sapiens, N = 2847), and TFIIH (H. sapiens, N = 1347). To see this figure in color, go online.
Figure 3.
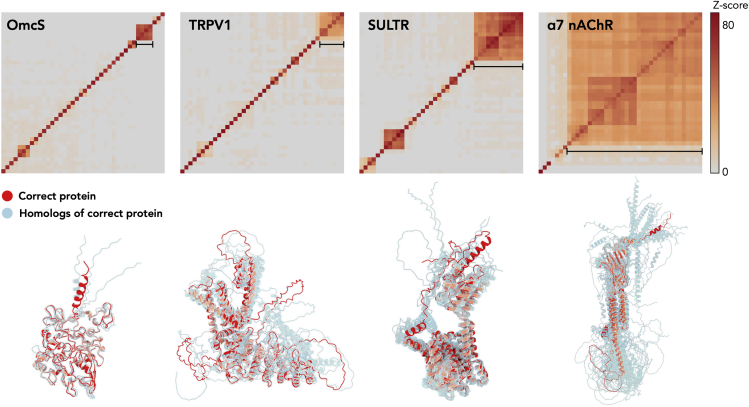
DALI all-to-all analysis of top 40 scoring AF2 predictions. Top: DALI all-to-all analysis of four representative benchmark data sets shown in Fig. 2. The matrix is based on the pairwise Z-score comparisons calculated using the DALI server. The color scale on the right indicates the corresponding Z scores. Bottom: the structures clustered (indicated by black line) in the top matrix are shown. The correct protein is colored red. The remaining proteins are colored transparent cyan and aligned to the correct protein. To see this figure in color, go online.
Among the proteins in the benchmark set, OmcS (14) has six hemes as ligands, and TFIIH (28) has a bound double-stranded DNA. Interestingly, our method detected the correct protein using cryo-EM maps in the presence or absence of ligand/DNA densities (Fig. S1), suggesting that a small portion of non-protein density does not diminish the accuracy of our method. Importantly, although OmcS lacks pronounced secondary structure elements, DeepTracer correctly placed Cα atoms into the 3.7 Å map. In fact, the backbone tracing of DeepTracer is normally robust when the map is better than 4.2 Å. Therefore, when the map is 4.2 Å or better, a successful DeepTracer-ID protein identification will then primarily rely on the accuracy of AlphaFold2 predictions.
The 4.2 Å resolution is normally the cutoff when β-sheets can be clearly resolved in cryo-EM map and strands can be traced unambiguously. Of course, DeepTracer-ID may still work satisfactorily for a helix-rich map at even lower resolution. To obtain a better idea of the resolution required for a successful protein identification, we low-pass filtered the existing 2.6 Å TRVP1 volume shown in Fig. 2 to 3.4 Å, 4.2 Å, 5.0 Å, 5.8 Å, and 6.6 Å, then ran them through DeepTracer-ID (Fig. S2). Interestingly, at 4.2 Å the correct protein and its homologs can be detected as well as in the 2.6 Å map. At 5.0 Å, the correct protein still has the best score using the PyMOL-align method while its homologs start to fall off the radar. This suggests that the quality of DeepTracer backbone tracing decreases when the resolution worsens, leading to poor subsequent alignments with the AlphaFold2 predictions.
Closely related structural homologs of the correct protein
If the correct protein has structural homologs/isoforms, we expect both the correct protein and its homologs/isoforms to be detected using our method. This is inevitable because the correct protein and its homologs/isoforms are structurally similar. In addition, model errors are expected in both Cα tracing in DeepTracer and AlphaFold2 prediction. As a result, the correct protein and its homologs might not be distinguished at this step. Therefore, a subsequent full-length model building for all top hits is required to determine which protein best fits into the cryo-EM map. This is now a routine protocol for de novo model building of an unknown protein (14,15,29). To visualize similarity among possible structural homologs of the top hits, we implemented DALI all-to-all analysis (27) into this pipeline. DALI outputs a protein all-to-all matrix. This additional DALI analysis will not only detect potential sequence ambiguity, it will also reveal possible biologically interesting similarities that were not apparent from scores (Eq. 1). As expected, in several cases our method detected a number of structural homologs of the correct protein (Fig. 3). These can be confidently distinguished by subsequent full-length modeling without the need for additional sequence information.
De novo protein identification from cryo-EM maps of unknown proteins
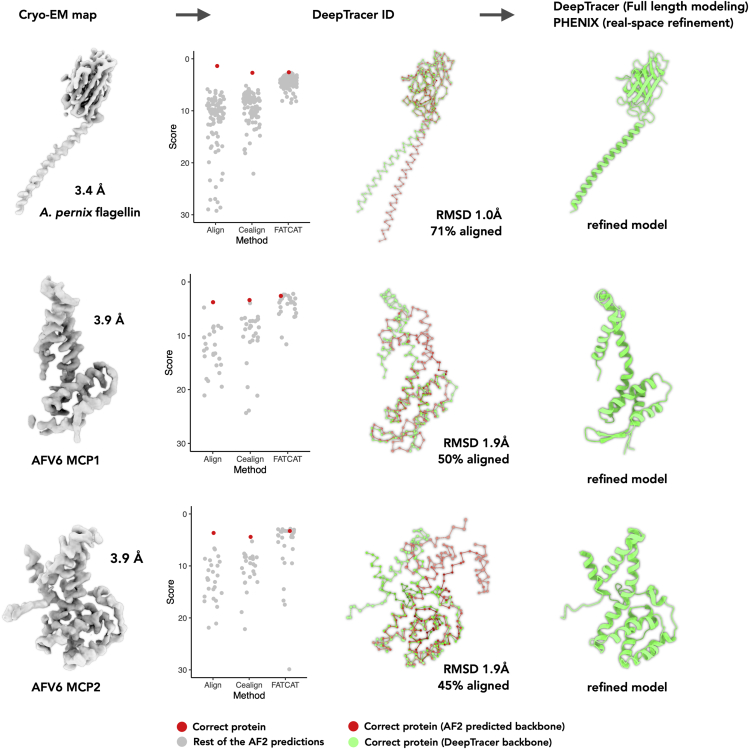
As a proof of principle, we then applied our method to three segmented cryo-EM maps with moderate resolutions from 3.4 to 3.9 Å of unknown proteins. The first protein is an archaeal flagellin from Aeropyrum pernix, with a long N-terminal helix and a C-terminal globular domain (30). Without additional information on how this flagellin packs into a filament, it is challenging for AlphaFold2 to accurately predict how the two domains are oriented with respect to each other. The other two proteins are the two major capsid proteins (MCP) of Acidianus filamentous virus 6 (AFV6), a virus infecting a hyperthermophilic and acidophilic archaeon (31). Similar to Sulfolobus islandicus filamentous virus (SIFV) (32), the two capsid proteins form a heterodimer that wraps around A-form DNA. Without the information about both the helical packing and protein-DNA interactions, AlphaFold2 might not be able to predict the entire structure accurately. Also, archaeal and archaeal virus genomes are generally much more sparsely sampled than those from bacteria and eukaryotes and the viruses that infect them. Therefore, fewer homologous sequences are available for AlphaFold2 to use in the prediction.
With all those potential pitfalls, our method still successfully determined the correct sequences (Fig. 4). In all three cases, only 45%–71% of the AlphaFold2 predicted models could be satisfactorily aligned to the backbone traced from the cryo-EM maps. Nevertheless, such coverage is sufficient for our method to detect the correct protein. We then validated those results with prior information. The detected MCPs in AFV6 are consistent with the annotations, as they are close homologs of MCPs in SIFV. For A. pernix flagellar filaments, proteins that are putative flagellins are easily narrowed down by bioinformatic approaches, and the detected protein is the best fit to the map among all those putative flagellins. The full-length models were subsequently built by DeepTracer and real-space refined by PHENIX (33). The statistics of the deposited models are listed in Table S1.
Figure 4.
Protein identification from A. pernix and archaeal virus AFV6. Far left: cryo-EM maps used for de novo protein identification. Left: the scores of corresponding AF2 predictions are displayed in scatterplots. The correct protein is shown as a red dot and the remaining AF2 predictions are shown as gray dots. The size of the AF2 library and the corresponding organism for the three unpublished data sets are A. pernix flagellin (N = 104), AFV6 MCP1 (N = 28), and AFV6 MCP2 (N = 28). Right: the alignment between DeepTracer model (green) and AF2 prediction of the correct protein (red). The alignment RMSD and percentage of aligned area are also labeled. Far right: the final model after DeepTracer full-length modeling and PHENIX real-space refinement. To see this figure in color, go online.
Structural knowledge mined from large and complicated maps
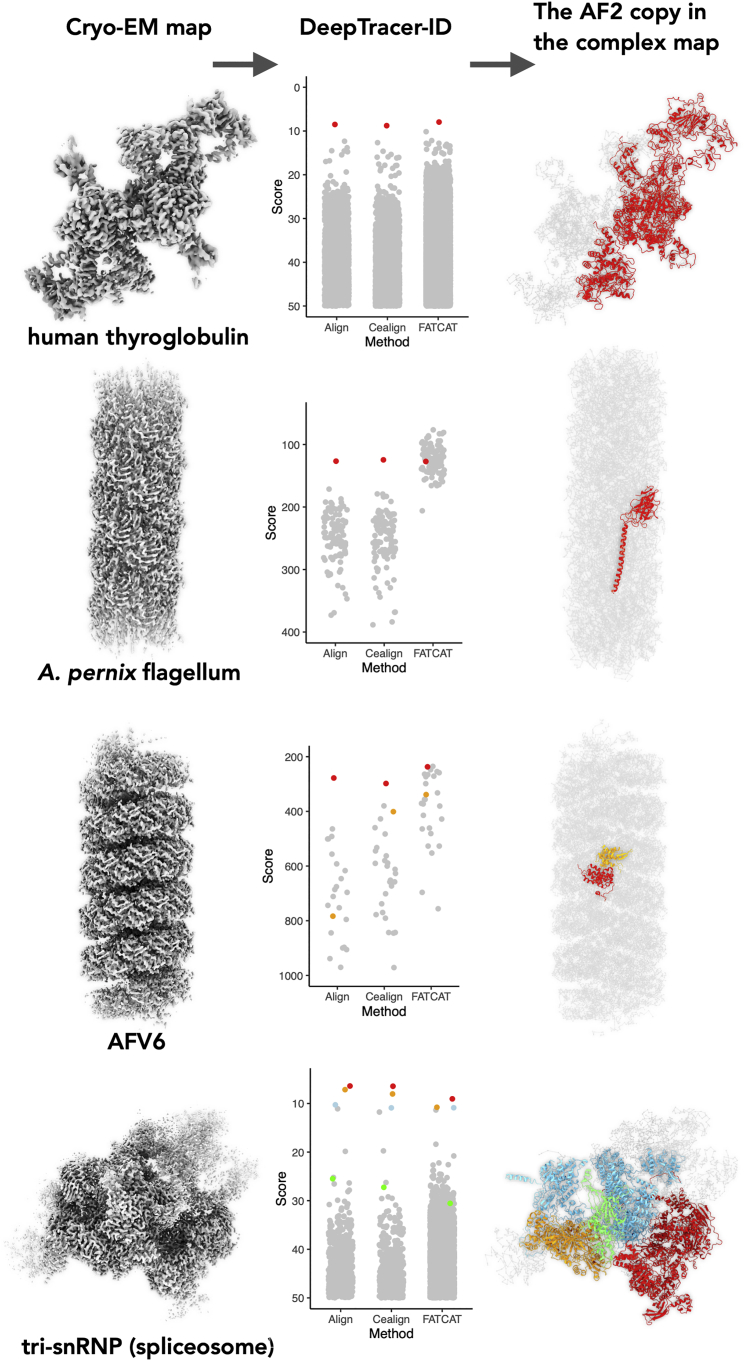
The next question was whether the approach could extract useful information from a more complicated map, such as multimeric protein complexes. We arbitrarily created two tiers for the maps based on their complexity: the “easy” tier contains maps of homomeric protein complexes; the “difficult” tier contains maps of heteromeric protein complexes. We tested our method with two very different maps in the easy tier: the dimeric map of human thyroglobulin with two protein copies and the filament map of an A. pernix flagellum containing ≈50 flagellin subunits. Our method successfully determined the correct protein in both cases (Fig. 5), suggesting that DeepTracer-ID also works for maps of homomeric protein complexes. Interestingly, the gap between the correct protein and the protein with the second best score is comparable with the results from segmented maps. This is presumably because the alignment between AlphaFold2 predictions and DeepTracer models is insensitive to the number of protein copies present in the cryo-EM map. In addition, DeepTracer assigns residues to every corner of the map, thus the subsequent alignment is not affected much by whether the backbone corresponding to a single subunit is successfully traced or not. In such cases, the only benefit of map segmentation is speeding up the calculation process.
Figure 5.
Protein identification from protein complex maps. Left: cryo-EM maps used for de novo protein identification. Middle: the scores of corresponding AF2 predictions are displayed in scatterplots. The correct protein is shown as colored dots and the remaining AF2 predictions as gray dots. The size of the AF2 library and the corresponding organism for the four data sets are H. sapiens thyroglobulin (N = 22,742), A. pernix flagellin (N = 104), archaeal virus AFV6 (N = 28), and H. sapiens tri-snRNP complex (N = 23,391). Right: the identified protein(s) are colored the same as in the scheme shown in the middle; the rest of the complex is colored gray. To see this figure in color, go online.
Next, we were curious as to whether DeepTracer-ID could extract any useful information from complicated cryo-EM maps of heteromeric protein complexes. To this end, we tested two maps (Fig. 5). The first map is that of the filamentous virus AFV6, with ≈50 copies of MCP1, ≈50 copies of MCP2, and ≈600 bp of double-stranded A-form DNA. Interestingly, our method could still detect one of the capsid proteins, MCP2. However, the other capsid protein did not rank high using the default PyMOL-align algorithm. It ranked third using PyMOL-cealign, suggesting that sequence information may not be helpful in a highly complicated map. The second map tested is the tri-snRNP part of the spliceosome complex composed of ≈30 different components. Not surprisingly, our method detected the largest four proteins in the complex, as they are expected to have a higher in Eq. 1, leading to a better score. This suggests that DeepTracer-ID can identify the major components from large complex maps, thereby rendering the subsequent map segmentation and smaller protein identification much easier. For the smaller components, on the other hand, map segmentation will be required prior to DeepTracer-ID protein identification.
Discussion
We present here a server-based pipeline, DeepTracer-ID, which can robustly identify the protein components directly from cryo-EM maps of a given organism without the need for other experimental data. Of course, the detected protein has to be fully modeled into the cryo-EM map and compared thoroughly against its homologs to be confident of the best fit. Furthermore, this approach does not require very high accuracy from AlphaFold2 predicted libraries. As long as ≈45% of the AlphaFold2 prediction reasonably matches the cryo-EM structure, this pipeline can identify the correct protein. As a proof of principle, we successfully identified three proteins within large filamentous complexes directly isolated from natural sources using this pipeline.
The remaining challenge for this pipeline is working with a map of tiny protein that reaches to 4.2 Å or better resolution. Owing to the extremely high signal/noise level in cryo-EM, it is currently almost impossible to reach a near-atomic resolution for a small monomeric protein (<100 amino acids). However, such small proteins frequently exist in larger complexes, such as conjugation pili (34,35) and ciliary doublets (29). In such cases, the DeepTracer model can probably be structurally aligned to many different AlphaFold2 predictions reasonably well. At the same time, sequence information may no longer be beneficial because of the limited length. For example, we tested a virus capsid protein with only ≈90 amino acids in a segmented cryo-EM map (36) (Fig. S3). The protein has the N-terminal ≈60 residues cleaved, a helix-turn-helix structure, glycosylation in the middle, and no cleavage site for trypsin. As a result, we found that this set of features rendered the initial sequence-based alignment method unreliable, while the structure-based PyMOL-cealign remained valid. FATCAT can increase the score of the correct protein by enabling flexible fitting, but it improves the scores for all other proteins at the same time. We will consider introducing other initial alignment methods in the future, such as Chimera matchmaker (37), TM-align (38), DALI (27), or different flexible fitting algorithms.
This approach could be broadly applicable to many scenarios other than identifying an unknown protein in a cryo-EM map resulting from contaminants. Using this method, it is possible to investigate large complexes enriched in a cell extract. The protein of interest can be directly identified when the resolution is better than 4.2 Å, regardless of post-translational modifications and the lack of proteomics data. It may also be utilized to perform a cryo-EM “pull-down” assay where, instead of a long list of candidates from mass spectrometry, the outputs now are protein 3D structures fitted to the maps.
Author contributions
F.W., E.H.E., and D.S. initiated and developed the project. F.W. performed microscopy and image analysis. V.C.-K. and M.K. cultured the viruses and archaeal cells and performed sample preparation. L.C. and K.C. wrote the code, built the web services, and performed the web server testing. H.M. prepared the web server production. F.W. prepared the benchmark data sets for algorithm testing. F.W. and Z.S. analyzed data and developed graphical representations. F.W., E.H.E., Z.S., and D.S. wrote the manuscript with input from all authors.
Acknowledgments
The cryo-EM imaging was done at the Molecular Electron Microscopy Core Facility at the University of Virginia, which is supported by the School of Medicine and built with NIH grant G20-RR31199. This work was supported by NIH grant GM122510 (E.H.E.), K99GM138756 (F.W.), K99CA259526 (Z.S.), NSF grant 2030381 (D.S.), the SRCP Seed Grant at the University of Washington Bothell (D.S.), and l’Agence Nationale de la Recherche grant ANR-21-CE11-0001-01 (M.K.).
Declaration of interests
The authors declare no competing interests.
Editor: Bridget Carragher.
Footnotes
Luca Chang and Fengbin Wang contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2022.06.025.
Contributor Information
Fengbin Wang, Email: jerrywang@virginia.edu.
Edward H. Egelman, Email: egelman@virginia.edu.
Dong Si, Email: dongsi@uw.edu.
Supporting material
Data availability
The DeepTracer-ID described here is free for academic use, available at https://deeptracer.uw.edu/home. The DeepTracer-ID is not open-source software at the moment. The atomic models and cryo-EM volumes have been deposited in the PDB and the Electron Microscopy Data Bank (A. pernix flagellum, PDB: 7TXI and EMD: 26158; AFV6, PDB: 7TXJ and EMD: 26159). The raw cryo-EM micrographs of AFV6 have been deposited to the Electron Microscopy Public Image Archive (EMPIAR: 11040). Other data are available from the corresponding authors upon reasonable request.
References
- 1.Bai X.C., McMullan G., Scheres S.H.W. How cryo-EM is revolutionizing structural biology. Trends Biochem. Sci. 2015;40:49–57. doi: 10.1016/j.tibs.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Kühlbrandt W. Biochemistry. The resolution revolution. Science. 2014;343:1443–1444. doi: 10.1126/science.1251652. [DOI] [PubMed] [Google Scholar]
- 3.Egelman E.H., Wang F. Cryo-EM is a powerful tool, but helical applications can have pitfalls. Soft Matter. 2021;17:3291–3293. doi: 10.1039/d1sm00282a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callaway E. The revolution will not be crystallized: a new method sweeps through structural biology. Nature. 2015;525:172–174. doi: 10.1038/525172a. [DOI] [PubMed] [Google Scholar]
- 5.Fitzpatrick A.W.P., Falcon B., et al. Scheres S.H.W. Cryo-EM structures of tau filaments from Alzheimer's disease. Nature. 2017;547:185–190. doi: 10.1038/nature23002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skalidis I., Kyrilis F.L., et al. Kastritis P.L. Cryo-EM and artificial intelligence visualize endogenous protein community members. Structure. 2022:575–589.e6. doi: 10.1016/j.str.2022.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Punjani A., Rubinstein J.L., et al. Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 8.Kimanius D., Dong L., et al. Scheres S.H.W. New tools for automated cryo-EM single-particle analysis in RELION-4.0. Biochem. J. 2021;478:4169–4185. doi: 10.1042/BCJ20210708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong E.D., Bepler T., et al. Davis J.H. CryoDRGN: reconstruction of heterogeneous cryo-EM structures using neural networks. Nat. Methods. 2021;18:176–185. doi: 10.1038/s41592-020-01049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spaulding C.N., Schreiber H.L., et al. Egelman E.H. Functional role of the type 1 pilus rod structure in mediating host-pathogen interactions. Elife. 2018;7:e31662. doi: 10.7554/elife.31662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F., Baquero D.P., et al. Egelman E.H. The structures of two archaeal type IV pili illuminate evolutionary relationships. Nat. Commun. 2020;11:3424. doi: 10.1038/s41467-020-17268-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ho C.M., Li X., et al. Zhou Z.H. Bottom-up structural proteomics: cryoEM of protein complexes enriched from the cellular milieu. Nat. Methods. 2020;17:79–85. doi: 10.1038/s41592-019-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X., Fu Y., et al. Zhang R. Cryo-EM structure of cortical microtubules from human parasite Toxoplasma gondii identifies their microtubule inner proteins. Nat. Commun. 2021;12:3065. doi: 10.1038/s41467-021-23351-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F., Gu Y., et al. Malvankar N.S. Structure of microbial nanowires reveals stacked hemes that transport electrons over micrometers. Cell. 2019;177:361–369.e10. doi: 10.1016/j.cell.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F., Cvirkaite-Krupovic V., et al. Egelman E.H. An extensively glycosylated archaeal pilus survives extreme conditions. Nat. Microbiol. 2019;4:1401–1410. doi: 10.1038/s41564-019-0458-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lubec G., Afjehi-Sadat L. Limitations and pitfalls in protein identification by mass spectrometry. Chem. Rev. 2007;107:3568–3584. doi: 10.1021/cr068213f. [DOI] [PubMed] [Google Scholar]
- 17.Pfab J., Phan N.M., Si D. DeepTracer for fast de novo cryo-EM protein structure modeling and special studies on CoV-related complexes. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2017525118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Si D., Nakamura A., et al. Zhao M. Artificial intelligence advances for de novo molecular structure modeling in cryo-electron microscopy. Wires Comput. Mol. Sci. 2021;12 doi: 10.1002/wcms.1542. [DOI] [Google Scholar]
- 19.Chojnowski G., Simpkin A.J., et al. Rigden D.J. findMySequence: a neural-network-based approach for identification of unknown proteins in X-ray crystallography and cryo-EM. IUCrJ. 2022;9:86–97. doi: 10.1107/S2052252521011088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jumper J., Evans R., et al. Hassabis D. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeLano W.L., Lam J.W. PyMOL: a communications tool for computational models. Abstr. Pap. Am. Chem. Soc. 2005;230:U1371–U1372. [Google Scholar]
- 22.Shindyalov I.N., Bourne P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- 23.Li Z., Jaroszewski L., et al. Godzik A. Fatcat 2.0: towards a better understanding of the structural diversity of proteins. Nucleic Acids Res. 2020;48:W60–W64. doi: 10.1093/nar/gkaa443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang F., Gnewou O., et al. Egelman E.H. Cryo-EM of helical polymers. Chem. Rev. 2022 doi: 10.1021/acs.chemrev.1c00753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varadi M., Anyango S., et al. Velankar S. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022;50:D439–D444. doi: 10.1093/nar/gkab1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mirdita M., Schuetze K., et al. Steinegger M. ColabFold - making protein folding accessible to all. bioRxiv. 2022 doi: 10.1101/2021.08.15.456425. Preprint at. 2021.2008.2015.456425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm L. Using Dali for protein structure comparison. Methods Mol. Biol. 2020;2112:29–42. doi: 10.1007/978-1-0716-0270-6_3. [DOI] [PubMed] [Google Scholar]
- 28.Aibara S., Schilbach S., Cramer P. Structures of mammalian RNA polymerase II pre-initiation complexes. Nature. 2021;594:124–128. doi: 10.1038/s41586-021-03554-8. [DOI] [PubMed] [Google Scholar]
- 29.Ma M., Stoyanova M., et al. Zhang R. Structure of the decorated ciliary doublet microtubule. Cell. 2019;179:909–922.e12. doi: 10.1016/j.cell.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Egelman E.H. Cryo-EM of bacterial pili and archaeal flagellar filaments. Curr. Opin. Struct. Biol. 2017;46:31–37. doi: 10.1016/j.sbi.2017.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vestergaard G., Aramayo R., et al. Prangishvili D. Structure of the acidianus filamentous virus 3 and comparative genomics of related archaeal lipothrixviruses. J. Virol. 2008;82:371–381. doi: 10.1128/JVI.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang F., Baquero D.P., et al. Egelman E.H. Structures of filamentous viruses infecting hyperthermophilic archaea explain DNA stabilization in extreme environments. Proc. Natl. Acad. Sci. USA. 2020;117:19643–19652. doi: 10.1073/pnas.2011125117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Afonine P.V., Poon B.K., et al. Adams P.D. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D Struct. Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng W., Pena A., et al. Egelman E.H. Cryoelectron-microscopic structure of the pKpQIL conjugative pili from carbapenem-resistant Klebsiella pneumoniae. Structure. 2020;28:1321–1328.e2. doi: 10.1016/j.str.2020.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Costa T.R.D., Ilangovan A., et al. Waksman G. Structure of the bacterial sex F pilus reveals an assembly of a stoichiometric protein-phospholipid complex. Cell. 2016;166:1436–1444.e10. doi: 10.1016/j.cell.2016.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang F., Cvirkaite-Krupovic V., et al. Egelman E.H. Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome. Cell. 2022;185:1297–1307.e11. doi: 10.1016/j.cell.2022.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meng E.C., Pettersen E.F., et al. Ferrin T.E. Tools for integrated sequence-structure analysis with UCSF Chimera. BMC Bioinf. 2006;7:339. doi: 10.1186/1471-2105-7-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y., Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The DeepTracer-ID described here is free for academic use, available at https://deeptracer.uw.edu/home. The DeepTracer-ID is not open-source software at the moment. The atomic models and cryo-EM volumes have been deposited in the PDB and the Electron Microscopy Data Bank (A. pernix flagellum, PDB: 7TXI and EMD: 26158; AFV6, PDB: 7TXJ and EMD: 26159). The raw cryo-EM micrographs of AFV6 have been deposited to the Electron Microscopy Public Image Archive (EMPIAR: 11040). Other data are available from the corresponding authors upon reasonable request.