Summary
Transcriptome-wide association studies (TWASs) are a powerful approach to identify genes whose expression is associated with complex disease risk. However, non-causal genes can exhibit association signals due to confounding by linkage disequilibrium (LD) patterns and eQTL pleiotropy at genomic risk regions, which necessitates fine-mapping of TWAS signals. Here, we present MA-FOCUS, a multi-ancestry framework for the improved identification of genes underlying traits of interest. We demonstrate that by leveraging differences in ancestry-specific patterns of LD and eQTL signals, MA-FOCUS consistently outperforms single-ancestry fine-mapping approaches with equivalent total sample sizes across multiple metrics. We perform TWASs for 15 blood traits using genome-wide summary statistics (average nEA = 511 k, nAA = 13 k) and lymphoblastoid cell line eQTL data from cohorts of primarily European and African continental ancestries. We recapitulate evidence demonstrating shared genetic architectures for eQTL and blood traits between the two ancestry groups and observe that gene-level effects correlate 20% more strongly across ancestries than SNP-level effects. Lastly, we perform fine-mapping using MA-FOCUS and find evidence that genes at TWAS risk regions are more likely to be shared across ancestries than they are to be ancestry specific. Using multiple lines of evidence to validate our findings, we find that gene sets produced by MA-FOCUS are more enriched in hematopoietic categories than alternative approaches (p = 2.36 × 10−15). Our work demonstrates that including and appropriately accounting for genetic diversity can drive more profound insights into the genetic architecture of complex traits.
Keywords: statistical genetics, GWAS, TWAS, gene fine-mapping, multi-ancestry
MA-FOCUS leverages the heterogeneity in linkage disequilibrium and eQTL patterns across multiple ancestries to improve the identification of the putative causal genes of complex traits. Our work demonstrates that including and appropriately accounting for genetic diversity can drive more profound insights into the genetic architecture of complex traits.
Introduction
Genome-wide association studies (GWASs) have identified genomic risk regions for numerous complex traits and diseases but leave unclear the underlying causal mechanisms responsible for risk. Multiple lines of evidence have suggested that genomic risk is imparted through perturbed regulation of nearby target genes, which predicts that the steady-state abundance of expression levels at target genes is associated with disease risk.1, 2, 3, 4, 5, 6 Transcriptome-wide association studies (TWASs),1,2 which explicitly test this hypothesis, have successfully identified novel genomic risk regions and specific genes that influence complex diseases.7, 8, 9 Much of the recent success of TWASs is due to the use of genetically predicted, rather than directly assayed, gene expression, which enables their application to existing large-scale GWASs, thus significantly increasing statistical power. Recently, we and others have demonstrated that TWASs also suffer from confounding due to expression quantitative trait loci (eQTL) pleiotropy and linkage disequilibrium (LD), which can induce correlation in test statistics between causal and non-causal genes analogously to causal and tagging variants in GWASs.10, 11, 12, 13, 14, 15, 16
Despite these recent breakthroughs, our understanding of the genetic architecture of complex traits has been limited by a lack of diversity in human genetics studies: individuals with primarily European genetic ancestry comprise 79% of all GWAS participants, despite representing only 16% of the global population.17 Although risk loci frequently replicate across ancestries,18, 19, 20, 21, 22 the LD patterns, minor allele frequencies (MAFs), and number of causal variants with their effect sizes can vary across genetic ancestries.21 This heterogeneity in genetic architecture hinders clinical applications of GWASs such as polygenic risk scores (PRSs), an issue that has been highlighted by the poor portability of PRS models across ancestries.23,24 On the other hand, the trans-ancestry design of recent GWASs has highlighted the benefits of taking an integrative multi-ancestry approach to study complex disease biology, both by leveraging genetic heterogeneity across human groups to aid in fine-mapping and by enabling the discovery of ancestry-specific disease etiologies.20,21,25, 26, 27 As with GWASs, we expect the integration of genetically diverse datasets into TWAS methodologies will improve our understanding of trait architectures that are both shared and unique to particular genetic ancestries.28, 29, 30
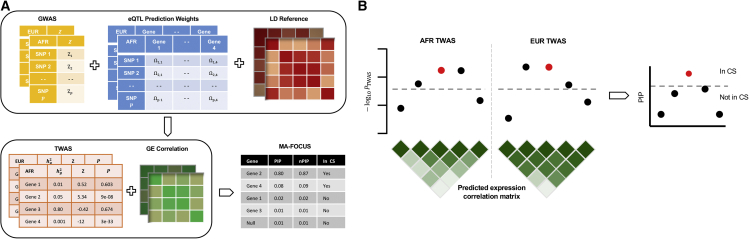
In this work, we present MA-FOCUS (multi-ancestry fine-mapping of causal gene sets), an approach that integrates GWASs, eQTL, and LD data from multiple ancestries to assign a posterior inclusion probability (PIP) that a given gene explains the TWAS signals at a risk region.31, 32, 33 It uses inferred PIPs to compute credible sets of causal genes at a predefined confidence level (Figure 1). A key feature of MA-FOCUS is that it does not assume that the eQTL architecture underlying gene expression is shared across ancestries.34,35 Instead, MA-FOCUS assumes only that the causal genes for a focal trait or disease are shared across ancestries without restrictions on their effect sizes. It is expected that gene-level effects are likely more transferable across ancestry groups than single-nucleotide polymorphism (SNP)-level effects as genes are inherently a more meaningful biological unit.36 As a result, MA-FOCUS leverages cross-ancestry heterogeneity in LD patterns and eQTL associations to identify causal genes with improved precision and accuracy when compared with alternative approaches.
Figure 1.
MA-FOCUS disentangles the causal gene from tagging genes by leveraging GWAS, eQTL, and LD data from multiple ancestries
(A) MA-FOCUS consists of two steps. First, it requires genome-wide association study (GWAS), expression quantitative trait locus (eQTL), and linkage disequilibrium (LD) data from multiple ancestries to calculate the transcriptome-wide association study (TWAS) and gene expression (GE) correlation matrix; second, it outputs a credible gene set (CS) with posterior inclusion probability (PIP) and locus-normalized PIP (nPIP).
(B) Toy example of TWAS Manhattan plots for European (EUR) and African (AFR) ancestries illustrating association signals at a locus for the causal gene (in red) and tagging genes (in black). The correlation among association signals is a combined result of eQTL signals and LD (see material and methods). By accounting for heterogeneity in eQTL effect sizes and LD across different ancestries, MA-FOCUS produces a smaller gene credible set with a more posterior probability assigned to the causal gene. The gray dashed line indicates transcriptome-wide significance.
By performing extensive simulations, we demonstrate that MA-FOCUS consistently outperforms the analogous single-ancestry method with equivalent total sample sizes and a “baseline” approach based on meta-analyzed GWAS statistics from different ancestries. In addition, we show that MA-FOCUS is robust in simulations where the trait-relevant tissue is missing and a proxy tissue is used instead. To illustrate its applicability to real multi-ancestry data, we conduct multiple TWAS and fine-mapping analyses with MA-FOCUS for 15 blood traits in European- and African-ancestry cohorts using large-scale GWAS summary statistics18 (average nEA = 511 k, nAA = 13 k) and eQTL weights calculated from the Genetic Epidemiology Network of Arteriopathy (GENOA) study37 (nEA = 373, nAA = 441). We recapitulate results demonstrating the shared genetic architecture for gene expression and blood traits between the two ancestries. We also find evidence that gene-level effects inferred from TWASs correlate 20% more strongly across ancestries than SNP-level effects. Fine-mapping 23 genomic regions that exhibit TWAS signals for both ancestries, we find that MA-FOCUS identifies genes relevant to hematopoietic and cardiovascular disease etiology missed by the baseline approach. Using multiple validation strategies,38 we show that genes in MA-FOCUS credible sets are more strongly enriched for hematological measurements compared with the baseline approach. Overall, our analyses using MA-FOCUS emphasize the importance of incorporating genetic information from diverse genetic ancestries to drive new insights into the genetic architecture of complex traits.
Material and methods
Multi-ancestry FOCUS model
For the of k total ancestries, we model a centered and standardized complex trait from individuals as a linear combination of gene expression levels at genes as
where are the causal effects of gene expression on the complex trait, and is random environmental noise with and . Additionally, we model ancestry-specific gene expression as a linear combination of genotypes as
where is the centered and standardized genotype matrix at SNPs, is the ancestry-specific eQTL effect-size matrix, and is random environmental noise.
Performing a TWAS using predicted gene expression requires the latent ancestry-matched eQTL weights , which are unknown. In practice, we use expression weights estimated from an independent, ancestry-matched eQTL reference panel using penalized linear models (or Bayesian counterparts).1,2 We model the ith ancestry’s marginal TWAS summary statistics for the gene with a complex trait as , where is the predicted expression imputed by the eQTL panel. By algebraic expansion for genes, we have
where we re-parameterize the causal effects of gene expression as and the ancestry-matched LD at SNPs as . Assuming that expression weights and causal effects are fixed, we can compute the sampling distribution of as
and as sample size increases, asymptotically approaches .
Next, we model a prior distribution for the causal effects as , where , is an causal configuration binary vector (where if the gene at the region is causal and 0 otherwise), denotes the number of non-zero elements of , and denotes the sample-size scaled causal effect prior variance.10 We marginalize out to obtain the TWAS sampling distribution conditioned on a causal gene set as
where is the estimated expression correlation matrix. Therefore, downstream fine-mapping inference will not be affected by ancestry-specific causal effect sizes of gene expression. We assume that the causal genes underlying a complex trait are shared across ancestries, which we model by sharing the vector. Since we do not know the causal genes indicated by beforehand, we adopt a Bayesian approach and compute the posterior for a given causal configuration as
where for some prior causal probability , and is the space of causal gene configurations. In practice, we set to be where denotes the number of known but not necessarily tested genes at the region. Intuitively, this reflects the naive expectation that a given risk locus contains a single causal gene. For computational tractability, we can constrain the space defined by to exclude complex configurations with for some reasonable threshold (e.g., 3–5). In addition, our likelihood, and thus posterior, depends on , which governs the variance of scaled causal gene effects . Previously, we recommended using a genome-wide mean as a heuristic, which works well under polygenic architectures10 but may perform poorly in sparser situations. Motivated by Shi et al. (2016),39 here we describe a local heuristic that estimates as
which is an unbiased estimator of causal effect variance (see supplemental information). In the case of negative estimates, we instead use .
Computing posterior inclusion probabilities and -credible sets
Our model describes the posterior probability for a given causal configuration across ancestries; however, we are more interested in the probability that a specific gene is causal across ancestries. We define the PIP for the gene by marginalizing over all causal configurations where as:
To capture the probability that none of the genes included in our analysis explain the observed TWAS Z scores at a risk region, we include the null model as a possible outcome in the credible set, . To compute a -credible set,31, 32, 33 where reflects the desired confidence that a gene set contains a causal gene, we take a greedy approach that traverses genes ordered decreasingly by their locus-normalized PIPs until the cumulative sum reaches at least .
Overview of the simulation pipeline
Here we provide a high-level summary of our multi-ancestry TWAS simulation pipeline described in five main steps (Figure S1), with details for each step described in the following sections. First, we computed approximately independent LD blocks and sampled genotypes for GWAS and eQTL reference panels in two ancestry groups. Second, we simulated ancestry-matched eQTL data using simulated eQTL reference genotypes from the first step, sampled eQTL effects under a sparse architecture, and simulated gene expression at causal and non-causal genes. Third, we simulated a complex trait in the ancestry-matched GWAS data as a linear function of eQTL effects of the causal gene from the second step and simulated GWAS genotypes from the first step. Fourth, we performed ancestry-matched TWAS using penalized models fitted in the respective eQTL reference panels. Fifth, we performed fine-mapping using single-ancestry FOCUS and MA-FOCUS. We provide details for each step below.
Computing independent LD blocks and simulating reference eQTL panels
We performed simulations using genotype data from phase three of the 1000 Genomes Project (1000G) for individuals of European (EUR; n = 490) and African (AFR; n = 639) ancestries (see Table S1).40 We restricted genotypes to high-quality HapMap SNPs and removed for missingness (>1%), MAF (<1%), and violations of Hardy-Weinberg equilibrium (HWE mid-adjusted p < 1 × 10−5). To identify approximately independent regions consistent with both EUR and AFR ancestries, we used a recently described extension of LDetect that considers LD information from multiple ancestries.21,41 Briefly, we constructed chromosome-wide ancestry-matched LD matrices and computed a chromosome-wide trans-ancestry LD matrix such that it incorporates shared recombination loci across ancestries (see Shi et al., 202021). Applying LDetect21,41 to resulted in 1,278 approximately independent LD blocks. We sampled 100 blocks that carried between five and eight annotated genes (based on hg19 RefSeq release 63) as risk regions. Additionally, we extended each LD block 500 kb upstream of the first gene’s transcription start site (TSS) and 500 kb downstream of the last gene’s transcription end site (TES) and updated accordingly.
At each risk region, we simulated ten genes whose expression is under partial genetic control by first sampling the number of eQTLs for the gene, . Next, we assigned SNPs uniformly at random to be eQTLs (out of total for a given locus) and simulated effect-sizes vector at the causal eQTLs and 0 at the non-causal SNPs where is the proportion of variance in gene expression attributable to cis-eQTLs (i.e., SNP heritability of gene expression).5,42
In addition, we simulated eQTLs as either independent or shared across ancestries; in the former case, SNPs and their effect sizes were chosen for each ancestry individually (under shared and parameters) as described above; in the latter, these were chosen once and then fixed for all ancestries.34,35 Then, we simulated an centered and standardized continuous genotype matrix using a multivariate normal distribution where is the ancestry-matched eQTL panel sample size. For gene , we calculated expression according to , where is random environmental noise for expression , and . To estimate ancestry-matched expression weights , we regressed on using least absolute shrinkage and selection operator (LASSO) regularization. To simulate eQTL effects when only a genetically correlated proxy tissue is available, we sampled proxy eQTL effects under a bivariate normal distribution as
where are the causal tissue eQTLs and is the genetic covariance between two tissues.
Simulating complex traits and statistical fine-mapping of TWASs
To reflect the practical reality that participants in GWASs and eQTL panels are usually different, we re-simulated genotypes at the risk region to compute GWAS summary statistics while keeping eQTLs of the ten simulated genes from the previous step. Then, we randomly sampled one gene as causal and used its eQTLs to simulate a complex trait as
where is the causal gene expression effect, is random environmental noise for (where ), and is the proportion of complex trait variation explained by the genetic component of gene expression. Next, to compute ancestry-matched GWAS summary statistics, we performed linear regression on marginally for each SNP in and calculated GWAS Z scores using the resulting Wald test statistic. We then performed an ancestry-matched summary-based TWAS using the predicted expression for each gene with .
Lastly, we performed TWAS fine-mapping using single-ancestry FOCUS10 and MA-FOCUS on to generate 90% credible sets for each ancestry and under the joint model, respectively. To determine whether the improvement of MA-FOCUS is solely due to increased sample sizes, we also evaluated a “baseline” approach. Specifically, the baseline approach consists of computing meta-analyzed GWAS statistics as , where is the inverse variance weight. Rather than constructing meta-analysis expression weights, is then computed by using the EUR expression weights . Finally, we conducted fine-mapping on using single-ancestry FOCUS and computed 90% credible sets. In all, we ran four methods (EUR FOCUS, AFR FOCUS, baseline, and MA-FOCUS) on 100 LD blocks to output one credible set per LD block per method. To test whether including information from additional ancestries of diverse genetic ancestries increases the performance of MA-FOCUS, we evaluated scenarios including individuals simulated using 1000G East Asian (EAS; n = 481) ancestry data40 (Table S1) and performed MA-FOCUS on three ancestries by fixing per-ancestry eQTL sample size, , and and allowing the total GWAS sample size to vary.
Simulating ancestry-specific genetic architectures and data-missing cases
To characterize the performance of MA-FOCUS when the mediating gene-trait heritability is ancestry specific, we simulated cases with varied values in one ancestry group while keeping it fixed in the other to represent heterogeneity in genetic architectures. Additionally, in practice, eQTL panels for a particular tissue of interest may be either unavailable or underpowered due to the small sample size. To evaluate the performance of MA-FOCUS in cases where relevant eQTL data are unavailable,43 we tested two scenarios that used different types of “proxy” data.1,8,10,34,35,44 First, we simulated cases where the trait-relevant tissue was unavailable in AFR, and a proxy tissue from the same ancestry with correlated gene expression was substituted. Second, we simulated cases where eQTL weights for AFR were entirely unavailable, and weights from EUR were used instead. The latter differs from the baseline approach in that the TWASs and FOCUS were conducted with ancestry-matched, not meta-analyzed, GWAS results.
Description of simulation parameters and fine-mapping performance metrics
We compared MA-FOCUS results to single-ancestry FOCUS results for EUR and AFR and the baseline approach across multiple simulations, which varied according to whether or not eQTLs were shared. We also varied four additional parameters: GWAS sample sizes, eQTL panel sample sizes, cis-SNP heritability of gene expression (cis-), and the proportion of trait variance explained by genetically regulated gene expression (). Unless stated otherwise, the simulation parameters were set to defaults of 100,000 for the per-ancestry GWAS sample size, 200 for the per-ancestry eQTL panel size, expression cis-, and trait . We evaluated fine-mapping performance based on three metrics: mean PIP of the causal genes, mean 90% credible set size, and frequency with which the causal genes are included in 90% credible sets per simulation (sensitivity). In addition, we fit linear regression adjusted for corresponding parameters to report one-sided Wald test p values.
Fitting SNP-based prediction models of LCL expression in the GENOA study
To calculate ancestry-specific gene expression weights in real data, we used genotype and lymphoblastoid cell line (LCL) derived gene expression data from European ancestry (EA) and admixed African American (AA) individuals from the GENOA study.37 Genotype data were generated using Affymetrix and Illumina genotyping arrays; in total, 1,384 EA and 1,263 AA individuals were assayed on the Affymetrix 6.0 array, 20 EA and 269 AA on the Illumina 1M array, and 103 EA on the Illumina 660 k array. All genotype data analyses were conducted using PLINK 1.9, vcftools, and bcftools.45, 46, 47, 48 We imputed genotype data using the TOPMed server, implementing Minimac4 v1.0.2 and eagle v2.4 phasing based on GRCh38.49 Each ancestry dataset was imputed separately, except for EA individuals assayed on Illumina arrays, which we merged prior to imputation. We retained biallelic SNPs with good imputation quality () for both EA and AA cohorts AND removed for MAF <1% and for HWE p < 1 × 10−6, resulting in 1,160,917 and 1,330,340 quality-controlled (QC) SNPs for EA and AA, respectively. We used GCTA50 to compute genotype principal components (PCs) and genetic relatedness matrices within the EA and AA cohorts after further filtering for SNPs with imputation and low pairwise LD (using --indep-pairwise 200 1 0.3 in PLINK46). We filtered out individuals such that no pair exhibited a relatedness coefficient greater than 0.05, resulting in 373 EA and 441 AA individuals. For downstream eQTL model fitting, we used only HapMap v3 SNPs.51
Expression data for the EA and AA cohorts were assayed at 16,944 and 32,881 genes (overlap of 14,797) on the Affymetrix Human Exon 1.0 and Affymetrix Human Transcriptome 2.0 arrays, respectively, and processed by Shang et al.37 After lifting over the expression data to GRCh38, for each gene in its respective ancestry, we ran FUSION1,7,9 to estimate cis-, and to calculate ancestry-specific eQTL weights, limiting the analysis to SNPs falling within a window including 500 kb upstream and downstream of each gene’s TSS and TES, respectively. We included 30 gene expression PCs, five genotype PCs, age, sex, and genotyping platform as covariates in building SNP models.1,7,9 We identified 3,680 and 4,291 genes in EA and AA, respectively, with an estimated cis- of at least 0.01 (nominal p < 0.01) of which 2,496 genes overlapped both ancestries. We limited our downstream analyses to 4,646 unique genes that had evidence for significant cis-, as defined above, in either ancestry and non-zero weights in both ancestries.1,7,9
Global ancestry estimates of GENOA participants
To estimate global ancestry proportions for African American individuals in the GENOA study, we ran ADMIXTURE52 on genotype data. First, we merged the imputed and filtered GENOA genotypes with 1000G phase three genotype data40 of 1,436 individuals from additional ancestries in Africa, Europe, and the Americas (Table S2). We removed the combined genotype data for missingness >1%, MAF <1%, and HWE p < 1 × 10−6. We also removed all palindromic variants and filtered for pairwise LD (using --indep-pairwise 200 1 0.3 in PLINK46). This resulted in 479,763 SNPs being retained for analysis. Next, we ran 30 replicates of ADMIXTURE for using a random seed and the default parameters.52 Finally, we used pong53 to identify a single mode across all replicates and to visualize the results.
Validation of LCL prediction models in GEUVADIS
To validate our estimated ancestry-specific gene expression weights derived from the GENOA study,37 we obtained paired genotypes and LCL-derived mRNA expression data at 22,721 genes for 373 EUR participants and 89 Yoruba in Ibadan (YRI) participants from the GEUVADIS study.54 First, we performed the same relatedness- and variant-based filtering described above, resulting in 358 EUR and 89 YRI participants and 8,403,216 and 14,855,241 SNPs, respectively. Next, focusing on the 4,581 genes that overlapped with GENOA, we performed FUSION1,7,9 to estimate cis- of LCL gene expression in GEUVADIS and compute ancestry-specific eQTL weights analogously to the GENOA described above adjusted for participants’ sex and three genotype PCs.50 Finally, we predicted LCL expression for GEUVADIS and GENOA participants using GENOA-based and GEUVADIS-based expression weights, respectively, and calculated the coefficients of determination () between corresponding predicted and measured expression levels.
TWAS and fine-mapping of 15 blood traits from GWAS summary data
We obtained published GWAS summary statistics for 15 blood traits (Table S3) from Chen et al.18 After lifting SNPs over to GRCh38 and updating their identifiers to dbSNP v153, we used LDSC munge55 to perform quality control by keeping summary statistics based on imputation INFO scores >0.9, MAF >0.01, and chi-squared statistics <80 to limit the influence of outlier SNPs. We flipped alleles as necessary for consistent orientation across European-ancestry and African-ancestry GWAS statistics. The average GWAS sample size was 511,471 for European and 13,298 for African ancestries across all SNPs and 15 blood traits, reflecting an approximately 40-fold difference in sample sizes. As in our simulations, we calculated TWAS Z scores of EA, AA, and the baseline approach for each trait by leveraging corresponding GWAS summary statistics in Chen et al.,18 FUSION-fitted LCL eQTL reference weights in GENOA,1,37 and reference LD estimated from 1000G individuals.40 To validate our TWAS results, we re-performed TWAS analyses using LCL eQTL weights fitted from the GEUVADIS reference data.
To quantify the extent to which LD blocks that contained TWAS significant signals (maximum p < 0.05/4,579, the number of genes with TWAS statistics) but did not exhibit GWAS significant signals (minimum p > 5 × 10−8, the genome-wide threshold) had increased GWAS signals on average, we performed a permutation test that first sampled from all LD blocks that did not exhibit GWAS signals, agnostic to TWAS signal, then computed the average GWAS chi-squared statistics at the sampled region, and last computed an empirical value that counted how frequently these sampled regions exhibited stronger signals than the original observed statistics.
To shed light on ancestry similarity in genetic architecture, we first computed the cross-ancestry correlations and their corresponding standard errors of GWAS and TWAS normalized Z scores for each trait using a blocked jackknife approach. We normalized Z scores by dividing original Z scores by the square root of GWAS sample sizes (for TWAS, it is GWAS sample sizes of the most significant eQTL in the gene) to account for sample size differences. To compute an average across all 15 blood traits, we meta-analyzed individual correlations across 15 blood traits and tested the difference with pooled standard errors. Second, we estimated TWAS effect sizes from the original Z scores as and tested for a difference in means across ancestries using a two-sample t test.8
Next, we fine-mapped the original resulting TWAS Z scores using MA-FOCUS, single-ancestry FOCUS, and the baseline approach, focusing on independent genomic regions (e.g., LD blocks computed by LDetect and lifted over to GRCh38)41 that exhibited transcriptome-wide significant signals in both EA-specific and AA-specific TWAS. We annotated genes based on their inclusion in the 90% credible set, as described above. To validate our fine-mapping results, we re-ran MA-FOCUS on the GEUVADIS-based TWAS Z scores. Given that not all genes tested for association in GENOA data have corresponding weights computed in GEUVADIS, we restricted on overlap genes and calculated how well inferred PIPs correlate across genes assayed in either dataset, and how often the rank of lead genes in GENOA (i.e., highest PIP in a credible set) changed.
To provide evidence of the causal genes being shared rather than ancestry specific, we performed a Bayesian model comparison. Specifically, we used PIPs computed from MA-FOCUS and individual ancestry FOCUS to calculate log-Bayes factors (logBF) for each gene in an MA-FOCUS credible set as
Here, genes with large positive logBF values have better statistical support for shared causal roles than ancestry-specific genes.
Validation of blood trait fine-mapping results
To determine if the genes prioritized by MA-FOCUS are more biologically meaningful than those prioritized by other methods, we validated credible sets using three different approaches. First, we performed a gene set enrichment analysis for genes identified in credible sets (i.e., aggregating genes identified across all loci) for a given fine-mapping method and blood trait using the R package enrichR.56,57 We manually selected 20 trait categories related to hematological measurement in DisGeNET, a database of curated gene-trait associations,38 based on the most relevant body system using MeSH (see web resources) and The Experimental Factor Ontology (EFO)58 hierarchies (Table S4). We counted the number of significantly enriched categories with Bonferroni correction (p < 0.05/n, where n is the number of enrichment testing) for each method and performed meta-analyses on these categories using Fisher’s method. Second, we performed enrichment analyses directly comparing fine-mapped gene sets of blood traits in Chen et al.18 with the counterparts in DisGeNET.38 Third, we evaluated gene sets using a previously published “silver standard” (see web resources) to determine whether they better predict causal genes of 159 blood-related Mendelian and rare diseases (Table S5). Since these diseases are monogenic or oligogenic, their causal genes are affirmative in high confidence and are likely to have moderate effects on blood-related complex traits. Leveraging the database from Online Mendelian Inheritance in Man (OMIM) and Orphanet, we performed logistic regression to calculate the area under the receiver operating characteristic (AUROC) within each method and each blood-related trait in Chen et al.18
Results
MA-FOCUS improves power to identify causal genes in simulations
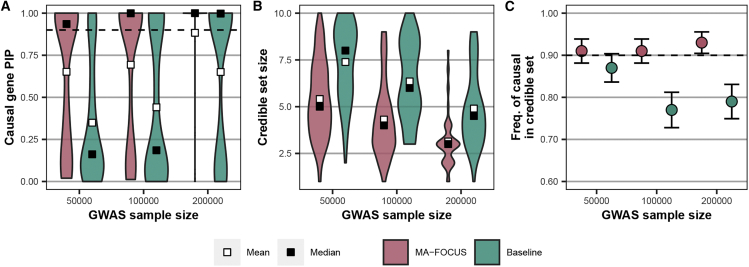
We first evaluated the performance of MA-FOCUS in simulations and compared it with the baseline approach, which consists of GWAS meta-analysis across ancestries followed by TWASs and fine-mapping with a single ancestry’s weights (see material and methods). Briefly, we simulated a complex trait as a function of genetically regulated gene expression for both ancestries when the causal tissue was known (see material and methods) while varying GWAS and eQTL sample sizes and features of the underlying genetic architecture. Across all simulation scenarios where causal eQTLs were independent across ancestries, we found MA-FOCUS reported higher PIPs for causal genes than the baseline approach (0.62 compared with 0.45; p = 9.05 × 10−40), smaller credible sets (4.89 compared to 6.62; p = 2.13 × 10−131), and higher sensitivity (88.30% compared to 81.30%; p = 9.35 × 10−9). Specifically, consistent with previous TWASs and TWAS fine-mapping simulation studies,1,10 performance improved as GWAS and eQTL sample sizes increased, likely reflecting increased statistical power (Figures 2 and S2). In addition, we found that increasing eQTL panel size affected MA-FOCUS sensitivity more dramatically than increasing GWAS sample size. For instance, increasing the eQTL panel size 2-fold, from 200 to 400, improved sensitivity from 91% to 97%, whereas the same proportionate increase in the GWAS sample size, from 100,000 to 200,000, increased sensitivity from 91% to 93% (Figures 2 and S2). Furthermore, we re-performed these simulations assuming that the causal eQTLs were shared across ancestries and observed that MA-FOCUS consistently outperformed the baseline (Figure S3). However, this performance advantage was slightly attenuated compared to the independent eQTL setting, highlighting the ability of MA-FOCUS to improve performance while being agnostic to eQTL architecture. Hereafter, we focused on presenting results where eQTLs were simulated independently in each ancestry to highlight the potential advantage of MA-FOCUS in real-world applications where eQTLs exhibit heterogeneity across ancestries.37
Figure 2.
MA-FOCUS outperforms the baseline approach in all three metrics as GWAS sample sizes vary when eQTLs are independent across ancestries
(A–C) Posterior inclusion probabilities (PIPs) for 100 simulated causal genes (A), the distribution of 90% credible set sizes for 100 simulated gene regions (B), and the sensitivity (C) from MA-FOCUS, and baseline approach, varying genome-wide association study (GWAS) sample sizes across multi-ancestry ancestries. See the material and methods for default parameters. The black dashed lines indicate 90%. Error bars are constructed using a 95% confidence interval.
Next, we sought to quantify the increases in fine-mapping power that could be gained by including individuals from diverse genetic ancestries rather than increasing the sample size of a single-ancestry GWAS. Specifically, we assumed an existing eQTL panel of 200 individuals for AFR and EUR ancestries and compared the performance of MA-FOCUS with single-ancestry fine-mapping, given a fixed number of total GWAS participants. We found that MA-FOCUS estimated higher PIPs at causal genes (mean of 0.67 compared to 0.57; p = 0.01) and produced credible sets containing fewer genes (mean of 4.86 compared to 5.33; p = 0.03) with better sensitivity (0.91 compared to 0.83; p = 0.01) when compared with those computed from FOCUS applied to equivalently powered EUR-only TWAS data (Figure S4). This relative performance advantage held when we compared two-ancestry to three-ancestry scenarios (Figure S5). Consistent with previous multi-ancestry SNP-based fine-mapping approaches,20,27 our results suggest that incorporating additional ancestry genetic diversity in GWASs drives more significant payoffs in fine-mapping performance than simply increasing the sample sizes of GWASs on previously studied ancestries.
To evaluate the performance of MA-FOCUS as a function of the underlying genetic architecture, we next performed simulations varying the cis-SNP heritability of gene expression (cis-) and the proportion of trait heritability attributable to a causal gene (). Across architectures, MA-FOCUS significantly outperformed the baseline (p = 2.52 × 10−14 for PIP metric, p = 7.45 × 10−49 for credible set metric, and p = 3.61 × 10−4 for sensitivity; Figures S6 and S7). Moreover, when there is no causal gene effect (i.e., ), we found that MA-FOCUS returned larger PIPs for the null model (p = 2.88 × 10−5) and smaller credible sets (p = 1.64 × 10−25) on average compared with the baseline (Figure S7). Our results show that MA-FOCUS is better powered than the baseline to identify the true causal model, including the null model, across a range of heritabilities for gene expression and the overall trait.
Multi-Ancestry FOCUS is robust to genetic-architectural and data-dependent assumptions
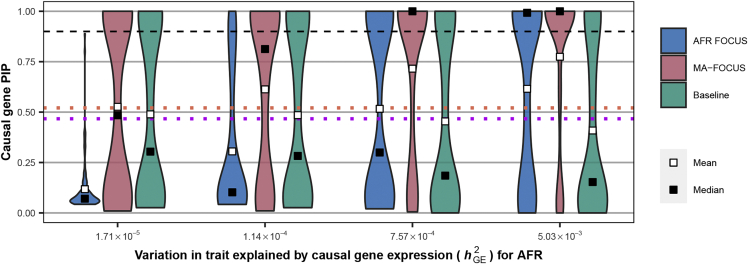
Next, we sought to characterize the performance of MA-FOCUS when assumptions of the underlying model were partially violated. First, we simulated a complex trait where the mediating gene-trait effects differed across ancestries by setting ancestry-specific values (i.e., fixed for EUR and varying for AFR across a range; see material and methods). Again, we found that MA-FOCUS consistently reported higher PIPs for causal genes (p = 3.42 × 10−11) and smaller 90% credible sets (p = 6.80 × 10−33) compared with the baseline (Figures 3 and S8). Furthermore, the sensitivity of gene sets reported by MA-FOCUS was robust to up to 7-fold differences in ancestry-specific (i.e., for EUR compared to for AFR). Only when the AFR was ∼2% of the EUR ( for EUR compared to for AFR) did we find MA-FOCUS performance to degrade, which was consistent with reduced statistical power under a fixed sample size. Together, these results show that MA-FOCUS is generally robust to ancestry-specific architectures.
Figure 3.
MA-FOCUS remains robust in having higher causal gene PIPs when trait heritability mediated by gene expression differs across ancestries
Distribution of inferred PIPs at the causal gene when the trait architecture varies across ancestries. We fixed trait variation explained by causal gene expression to for simulated European (EUR) individuals while varying its amount in African (AFR) individuals. The orange and purple dotted lines indicate the mean and the median of PIPs using EUR FOCUS. The black dashed lines indicate 90%.
To investigate the impact of imbalanced GWAS sample sizes, we performed simulations matching the sample sizes of a recent multi-ancestry blood trait GWAS18 ( and ; see material and methods). In this setting, MA-FOCUS computed credible sets that were smaller compared to the baseline (p = 3.54 × 10−6; Figure S9B) with similar mean PIPs at the causal genes (p = 0.13; Figure S9A) and sensitivity (p = 0.17; Figure S9C). This demonstrates that, even when GWAS sample sizes vary by an order of magnitude across ancestries, MA-FOCUS provides improved fine-mapping performance.
Next, we performed simulations where the trait-relevant tissue for AFR was unavailable and was substituted with eQTL data quantified in a proxy tissue with correlated genetic effects (see material and methods). The performance of MA-FOCUS was highly dependent on the underlying correlation between proxy and causal tissues and increased with increasing inter-tissue genetic covariance, as expected (Figure S10). We again observed that MA-FOCUS outperformed the baseline approach and AFR FOCUS across all metrics (p < 1 × 10−7 for all PIP and credible set metrics, p = 0.02 with MA-FOCUS/baseline comparison, and p = 0.09 with MA-FOCUS/AFR FOCUS comparison for sensitivity; Figure S10).
Finally, we performed simulations where eQTL reference panels for AFR were unavailable and EUR weights were used instead for TWASs and fine-mapping. We found that the relative performance of MA-FOCUS was mixed across different metrics, estimating similar causal PIPs and sensitivity (p = 0.32 and 0.34; Figures S11A and S11C) and smaller credible set sizes (p = 3.73 × 10−9; Figure S11B). In all, this highlights the importance of a multi-ancestry study design collecting gene expression data from different ancestries when possible.
Multi-ancestry TWAS identifies shared architecture in blood traits
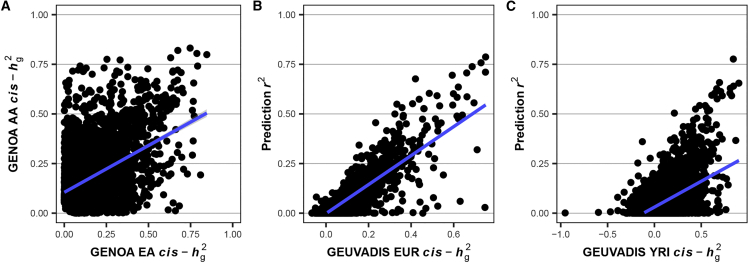
After confirming that MA-FOCUS outperforms other methods of TWAS fine-mapping, we next sought to apply it to real data from cohorts of European (EA) and African (AA) ancestries. We performed ancestry-matched TWASs for 15 blood traits using GWAS summary statistics18 (Tables S3 and S6; nEA = 511,471, nAA = 13,298) together with an eQTL reference panel of LCLs from the GENOA study37 (eQTL: nEA = 373, nAA = 441; see material and methods). First, we estimated SNP heritability (cis-) for expression at 14,797 genes assayed in EA and AA GENOA cohorts (see material and methods). We observed that, across all genes, cis- was significantly non-zero with an average of 0.057 for EA compared to 0.072 for AA (p < 1 × 10−100 for both tests). Furthermore, focusing on the 4,646 genes whose expression was significantly heritable in at least one of the cohorts, cis- estimates were positively correlated across ancestries with r = 0.45 (p < 1 × 10−100 for both tests against 0 and 1; Figure 4A), which is consistent with previous results suggesting that the genetic architecture of gene expression is significantly shared across ancestries.37 Next, we trained prediction models using the FUSION pipeline and performed in-sample validation with 5-fold cross-validation (CV; see material and methods). We found that CV was significantly non-zero (EA CV r2 = 0.105; AA CV r2 = 0.110; p < 1 × 10−100 for both), which were strongly correlated with cis- estimates (r = 0.93 with p < 1 × 10−100 for both; Figure S12), suggesting that in-sample prediction models perform well and are consistent with the theory that heritability provides a predictive upper bound.37,42,59
Figure 4.
Heritability and correlation analysis reveal evidence for shared genetic architecture for expression in LCLs
(A) The scatter plot for the SNP heritability (cis-) of lymphoblastoid cell line (LCL) gene expression for African American (AA) and European American (EA) ancestry in the GENOA study.
(B and C) The scatterplots where the y axis is a squared correlation () between measured LCL gene expression in GEUVADIS and predicted by eQTL panels from GENOA, and the x axis is cis-. Each point represents a gene. The blue line is estimated using ordinary linear regression.
Next, we further validated the predictive performance of LCL expression models by evaluating their out-of-sample performance in the European- and Yoruba-ancestry cohorts (EUR and YRI, compared with GENOA EA and GENOA AA, respectively) of the independent GEUVADIS study (see material and methods).54 While YRI is not an ideal ancestry proxy for admixed African Americans, we expect a significant degree of genetic similarity between the two given that 441 GENOA AA individuals had an average West African ancestry proportion of 83% (Figures S13 and S14), which YRI is commonly used to represent.37 Focusing on 4,581 genes that overlapped with GENOA, we calculated out-of-sample between measured LCL gene expression from GEUVADIS individuals and predicted expression using inferred GENOA-based weights. We found that estimates between measured expression from GEUVADIS individuals and expression predicted using GENOA-based weights were significantly correlated with estimates of GEUVADIS cis-, with r = 0.85 and 0.56 for EUR and YRI (p < 1 × 10−100 for both tests against 0; p < 1 × 10−40 for testing correlation difference; Figures 4B and 4C).60 The comparatively poorer performance of our AA expression weights in the GEUVADIS YRI was not unexpected, given the ancestry differences between the Yoruba people and African Americans discussed above, which likely impacted the genetic regulation of gene expression. Indeed, we found that cis- for AA and YRI were less correlated than EA and EUR (r = 0.27 and 0.49 with p < 1 × 10−75 for both tests against 0; p < 1 × 10−40 for testing correlation difference).60
Next, we evaluated across-ancestry prediction performance by predicting LCL gene expression levels for GEUVADIS EUR individuals using GENOA AA weights (similarly for GEUVADIS YRI and GENOA EA) and estimated an average of r2 = 0.040 and 0.033 for EUR and YRI (p < 1 × 10−100 for both tests; Figure S15). Consistent with the previous work,59 we found a decrease in accuracy for GEUVADIS YRI individuals compared to within-ancestry results (p = 1.75 × 10−31) and similar levels of accuracy for GEUVADIS EUR (p = 0.09). In addition, we observed that GENOA data had a higher estimate of LCL cis-, and its corresponding weight produced higher prediction accuracy for both ancestries than GEUVADIS data (p < 6.33 × 10−15 for all tests; see supplementary note). Together, these results demonstrate that prediction models using the GENOA dataset accurately capture the heritable component of gene expression within ancestry groups and recapitulate previous findings on the limited transportability of cross-ancestry prediction models for gene expression.37,59,61,62
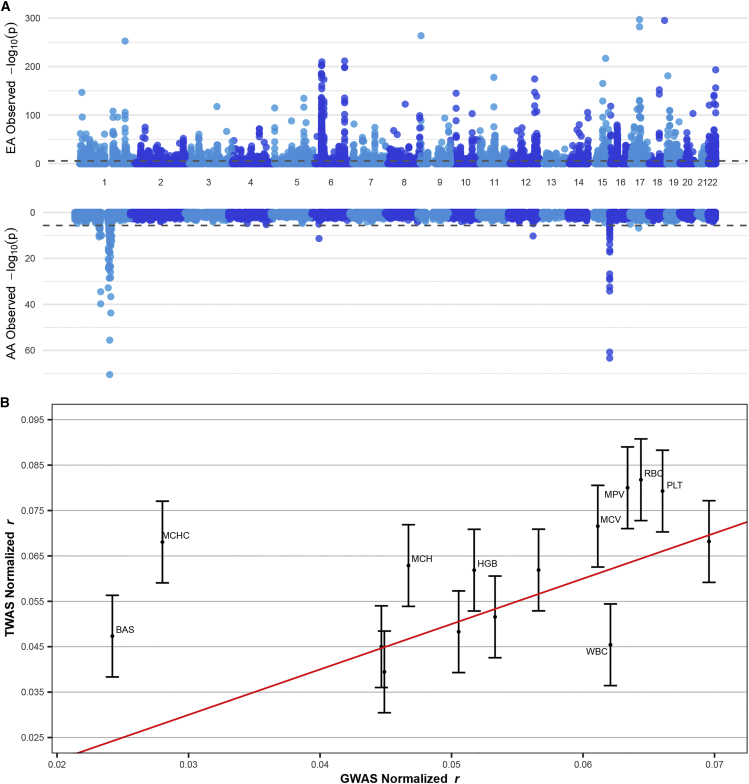
Having validated our SNP-based LCL expression prediction models, we conducted multi-ancestry TWASs for each of the 15 blood traits on 4,579 genes in 989 unique independent regions (see material and methods). Across all traits, we identified a total of 6,236 (2,009 unique) and 116 (57 unique) genome-wide TWAS significant genes in EA and AA, respectively, in 3,032 (622 unique) regions (p < 0.05/4,579, the number of genes with TWAS statistics; Figure 5A; Table S7; see data and code availability for the full results). We observed 28 (17 unique) genes significantly associated in both ancestry groups across 23 (11 unique) regions. Of the 8,243 trait-matched LD blocks that contained genome-wide significant signals (p < 5 × 10−8) in either ancestry or the meta-analysis, 2,940 also exhibited transcriptome-wide significant signals in either ancestry. Conversely, 115 trait-matched LD blocks contained transcriptome-wide significant signals that did not exhibit genome-wide significant signals, which we considered as putative novel risk regions. We observed that these 115 regions exhibited greater GWAS signals on average when compared to their trait-matched genomic background (p = 0.001, 0.02, and 0.001 for EA, AA, and meta-analysis; see material and methods). Of the 3,032 (622 unique) LD blocks containing TWAS hits, 1,329 (315 unique) contained multiple TWAS significant associations (average 3.60 genes per region), thus motivating the use of gene fine-mapping.
Figure 5.
The TWAS Manhattan plot indicates highly correlated genes in certain regions
(A) The upper plot is the Manhattan plot for European American (EA) TWAS and the lower is for African American (AA) TWAS across all 15 blood traits. Colors differentiate adjacent chromosomes.
(B) Cross-ancestry correlations () of normalized TWAS and GWAS effect sizes (see material and methods). Each point represents a trait (see Table S3 for each trait’s full name). The red line is the identity line. Error bars are constructed using a 95% confidence interval.
To validate our multi-ancestry TWAS associations, we re-performed TWASs using GEUVADIS-derived prediction models (see material and methods). Of the 6,352 significantly associated genes from GENOA across 15 traits and two ancestries, 4,315 were assayed in GEUVADIS, and 2,265 exhibited transcriptome-wide significance (p < 0.05/4,579). Overall, we observed stronger TWAS signals using GENOA-derived weights (mean chi-squared statistics of 8.74 and 1.24 for EA and AA) compared with GEUVADIS-derived models (mean chi-squared statistics of 8.11 and 1.13 for EUR and YRI; p = 0.002 and 7.16 × 10−7, respectively), suggesting that the larger sample size in GENOA LCL data has improved prediction accuracy and TWAS performance when compared with the smaller GEUVADIS LCL dataset.
Lastly, we investigated how gene- and SNP-level effect sizes differ across ancestry groups. Both normalized GWAS and TWAS Z score correlations between EA and AA were significantly non-zero for all traits (Table S8; Figure S16; see material and methods). Interestingly, we found that across-ancestry correlations were 20% higher on average for TWAS-based gene effects than GWAS-based SNP effects (r = 0.061 and 0.052, respectively; p = 0.028; Figure 5B; Table S8), which is consistent with previous findings demonstrating that predicted transcriptomic risk scores better correlate across ancestry groups59,63,64 and suggests that gene-level effects on average better reflect shared biology compared with SNP-level effects.36 In addition, we observed little support that TWAS-based gene effects sizes differ across EA and AA (p = 0.57), shedding light on ancestry similarity in the genetic architecture of LCL.
Multi-ancestry fine-mapping prioritizes likely causal genes in blood traits
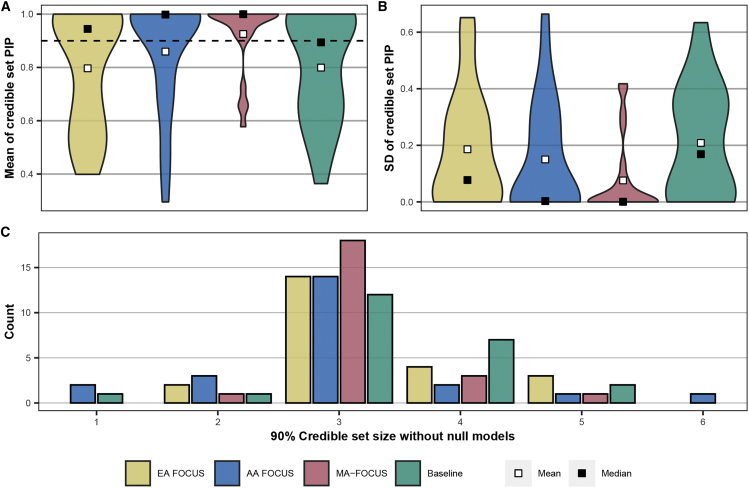
Next, we applied MA-FOCUS to TWAS results for blood traits focusing on 163 genes overlapping the 11 unique regions that contained TWAS signals for both EA and AA ancestry for a given trait (see material and methods). Across these 23 trait-specific regions, each contained an average of 6.13 TWAS significant associations across ancestries and 3.17 genes in the 90%-credible gene set, none of which included the null model. We estimated an average of 2.88 causal genes per region by summing over local PIPs in the credible sets, with 19 out of 23 credible sets containing three or fewer genes (Table S10; see data and code availability). The average maximum PIP across credible sets was 0.99 (SD = 0.02) and retained similar PIPs for the second and the third rank (Figure S17). Then, we compared credible gene sets across different approaches. Although estimated PIPs correlated between MA-FOCUS and the baseline (Figure S18A), we observed MA-FOCUS output higher means and smaller SDs of PIPs (P < 0.05 for all tests; Figure 6). Despite this, MA-FOCUS obtained a smaller credible gene set on average (3.17) compared to the baseline (3.35); however, this result was not significant due to low statistical power (p = 0.22; Figure 6). In addition, EA FOCUS did not prioritize 30 out of 73 trait-gene pairs in MA-FOCUS credible gene sets and missed 7 out of 23 lead genes, suggesting that incorporating non-European data in well-powered loci can prioritize additional putative causal genes (Figure S19A). We observed little support for a difference in the percentage of genes co-prioritized by AA FOCUS/MA-FOCUS (40.2%) compared with EA FOCUS/MA-FOCUS (49.5%; two-sample proportion test p = 0.24; Figure S19B), suggesting that contributing ancestry groups do not disproportionately influence prioritized genes. To determine the extent to which prioritized genes are likely to be shared or ancestry specific, we performed a model comparison using Bayes factors computed from MA-FOCUS and FOCUS PIPs (see material and methods). We observed an average log-scale BF of 1.44 (SD = 3.76), suggesting that credible-set genes underlying these blood traits are much more likely to be shared across ancestries than ancestry-specific genes (Figure S20). For instance, NPRL3 in the trait mean corpuscular volume (MCV) had a logBF of 17.1, which we discuss below (Figure S20).
Figure 6.
Credible sets output by MA-FOCUS have higher mean PIPs and lower standard deviation while exhibiting similar credible set size of EA FOCUS and the baseline approach
(A) Violin plot of the mean of gene PIPs in credible sets.
(B) Violin plot of the standard deviation of gene PIPs in the credible sets.
(C) The bar plot of the count for each credible set size. Calculations do not include null models. The black dashed lines indicate 90%. The methods include European American (EA) FOCUS, African American (AA) FOCUS, MA-FOCUS, and the baseline.
To investigate the stability of MA-FOCUS results, we re-performed fine-mapping, varying the maximum number of causal genes allowed in a configuration (see material and methods), and found that although inferred PIPs were relatively stable (p = 5.07 × 10−51), credible gene set sizes were sensitive to the upper bound on causal genes (see supplemental information; Figure S21). Moreover, to validate our results using eQTL weights in GEUVADIS, of the 49 genes in GENOA-based credible sets, 17 had GEUVADIS-weight-derived results with a PIP correlation estimate of 0.84 (p = 2.05 × 10−5). In addition, 9 out of 17 genes were the lead genes from GENOA, and among these 9 genes, 8 remained lead genes from GEUVADIS, suggesting that our findings are robust when integrating different expression data from a similar context (LCL).
Next, we investigated genes to which MA-FOCUS assigned a high PIP () and which were included in a credible set but not identified by the baseline approach. We refer to these genes hereafter as the “MA-FOCUS-specific genes.” We also examined the converse situation: genes for which the baseline approach found strong support but that were not prioritized by MA-FOCUS, referred to as the “baseline-specific genes.” Importantly, we found that all 22 baseline-specific genes had low PIPs () from ancestry-specific fine-mapping in at least one ancestry, while 11 of these genes had a low PIP in both ancestries. On the other hand, only 1 out of 31 total MA-FOCUS-specific genes had PIPs below 0.1 in both AA and EA. We found that 6 out of 31 total MA-FOCUS-specific genes achieved a moderate PIP of at least 0.25 in both EA and AA ancestry-specific fine-mapping (ARNT, BAK1, MRPL28, NPRL3, PHTF1, and TARS2; Figure S22), suggesting that MA-FOCUS is better able to identify genes that have evidence of causality in at least one ancestry, while the baseline approach identifies genes that have weak or no evidence of causality in either ancestry. A literature search for these six MA-FOCUS-specific genes uncovered additional evidence for roles in cardiovascular system disease and development (specifically, blood cell and vasculature formation, diabetes, leukemia, coronary artery disease, and cardiomyopathy; Figure S23).65, 66, 67, 68, 69, 70, 71, 72, 73, 74, 75 Overall, this result suggests that by appropriately modeling across-ancestry heterogeneity, MA-FOCUS can prioritize disease-relevant genes that naive meta-analyses would otherwise miss.
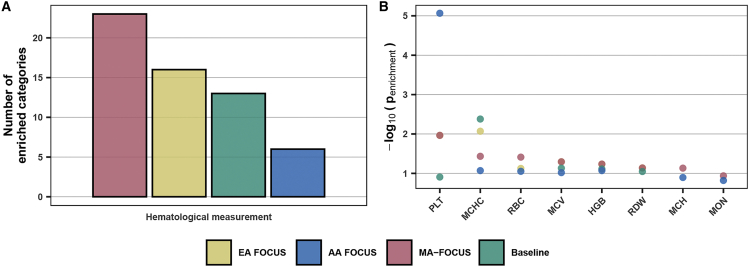
Lastly, to validate genes prioritized by MA-FOCUS and the baseline approach, we performed a series of tests comparing the credible sets (see material and methods). First, we performed gene set enrichment analysis on the credible-set genes using the DisGeNET dataset across all 15 blood traits. We found that MA-FOCUS’s credible sets were enriched more in hematological measurement categories than the baseline approach (23 and 13 categories, meta-analysis p value of compared to ; Figure 7; Table S11). Second, by restricting our focus to trait-matched DisGeNET enrichment categories, we observed that MA-FOCUS output more significantly enriched credible gene sets than the baseline approach (meta-analysis p value of compared to ; Figure 7; Table S12). Third, using curated “silver standard” databases consisting of OMIM and Orphanet for 159 blood-related diseases (see web resources and material and methods), we observed MA-FOCUS output a higher average AUROC with 0.57 compared to 0.43, suggesting improved performance in predicting causal genes of monogenic and oligogenic blood-related Mendelian and rare diseases (Table S13). Altogether, we find that credible set genes computed using MA-FOCUS reflect relevant disease biology better than single-ancestry and alternative approaches.
Figure 7.
Genes prioritized by MA-FOCUS are enriched in hematological categories more often than other methods
(A) The bar plot shows the number of enriched categories in DisGeNET identified by each method within the hematological-measurement-related category. The enriched category is defined as Bonferroni-corrected p value less than 0.05.
(B) The dot plot shows enrichment by categories in DisGeNET corresponding to eight blood traits. See Table S3 for each trait’s full name. EA represents European American ancestry and AA represents African American ancestry.
Case study of white blood cell count credible set genes
To further characterize the performance of MA-FOCUS, we narrowed our focus to the results of white blood cell counts (WBCs), which contributed the largest number of fine-mapped regions () from all analyzed blood traits. First, we found fewer genes in the MA-FOCUS credible sets on average (3.3) compared to the baseline approach (3.8); however, similar to the credible set size difference across all blood traits discussed in the previous section, this result was not significant due to low statistical power (p = 0.15). The fewer genes in MA-FOCUS credible sets were likely due to their significantly higher PIPs (mean = 0.86) compared with genes in the baseline approach credible sets (mean = 0.63; p = 0.01; Table S9). Next, we observed that MA-FOCUS credible gene sets resulted in a greater AUROC curve in the “silver standard” validation (0.63) compared with baseline credible gene sets (0.36; Table S13).
Next, we discussed the role of three genes (UBAP2L, HDGF, and FCGR2B; lead genes in their respective credible sets) in regulating WBCs. First, focusing on fine-mapped genes at 1q21.3, MA-FOCUS attributed the largest PIP to UBAP2L. Experimental manipulation of UBAP2L has confirmed its role in regulating the activity of hematopoietic stem cells in mice via interaction with BMI1.76 Additionally, this study found that UBAP2L mRNA levels were associated with leukemic stem cell frequency in patient-derived samples. Therefore, this gene is a plausible candidate for regulating the white blood cell trait in human ancestries. Furthermore, MA-FOCUS identifies HDGF at 1q23.1 and FCGR2B at 1q23.3, which previous studies have linked to angiogenesis and blood-related diseases, respectively.77, 78, 79, 80 Functional studies have confirmed the direct role of HDGF in promoting the development of blood vessels in cancers.77,78 Furthermore, association studies have linked FCGR2B, a B cell receptor that plays an important role in immune function, with multiple blood-related diseases, such as thrombocytopenia and systemic lupus erythematosus.79,80 Specifically, thrombocytopenia is a disorder characterized by abnormally low platelet counts. Although not directly related to the WBC, numerous studies have found that these blood traits are correlated, particularly in smokers and in disease contexts.81, 82, 83 Similarly, systemic lupus erythematosus is an autoimmune disorder that frequently results in low WBCs and is 2- to 4-fold more common in Asian and African ancestries than in European ancestry.80 A SNP in FCGR2B that has been found to impede this receptor’s normal signaling function is also associated with increased susceptibility to systemic lupus erythematosus.80,84 However, this SNP has also been found to confer protection against severe malaria, potentially explaining its higher frequency in ancestries where malaria is endemic. FCGR2B is therefore an interesting example of a gene that exhibits both significant ancestry-specific variation and a similar functional effect across genetic ancestry backgrounds. These are features that our multi-ancestry fine-mapping method is uniquely equipped to leverage in order to prioritize this gene as a strong candidate for regulating WBC.
Discussion
In this work, we present MA-FOCUS, a Bayesian fine-mapping method that incorporates GWAS and eQTL data together with LD reference panels from multiple ancestries of diverse genetic ancestries to estimate credible sets of causal genes for complex traits. Our method is unique in that it explicitly accounts for, and takes advantage of, heterogeneity in LD and the genetic architecture of gene expression to improve TWAS fine-mapping performance. Importantly, our method assumes only that the causal genes for complex traits are shared across ancestries and makes no assumptions on underlying eQTL architectures. This is an essential feature of our method considering recent findings that SNP-level replication across genetic ancestries is weaker than gene-level replication36 and that only ∼30% of SNP-gene expression associations are shared between European and African American ancestry.37 Through extensive simulations, we demonstrate that the ability of MA-FOCUS to identify causal genes is superior to baseline approaches and robust to data-dependent limitations (see material and methods).
We perform ancestry-specific TWASs and apply MA-FOCUS to 15 blood traits using GWAS statistics in Chen et al. and LCL eQTL data in GENOA from cohorts of primarily European and African continental ancestry. We report 6,236 and 116 TWAS significant genes for EA and AA in 622 unique regions across all blood traits. The cross-ancestry heritability analysis on LCL gene expression data, together with correlation analysis on blood traits of GWAS and TWAS statistics, recapitulate evidence for the shared genetic architecture of blood traits between the two ancestries and provide evidence for gene-level effects correlating better across ancestries than SNP-level effects. Next, in 23 regions that contain TWAS signals for both ancestries, MA-FOCUS reports 3.17 genes in the credible sets and estimates 2.88 putative causal genes per region across all blood traits. Finally, we validate the MA-FOCUS credible sets by performing enrichment analyses and referencing the results of functional studies. We show that MA-FOCUS’s credible sets are more strongly enriched for relevant genes associated with hematological traits in the DisGeNET platform, a database of genotype-trait associations compiled from various sources (Figure 7). Importantly, MA-FOCUS identifies genes that are known to have functional relevance for cardiovascular system disease and development but are not identified by the baseline approach.
Despite MA-FOCUS’s advantages in performance, as demonstrated through extensive simulations, we note several limitations to our analysis of blood traits. First, MA-FOCUS’s performance advantage is attenuated when the EA sample size is approximately 40 times greater than the AA sample size (Figure S6). Across the 11 blood traits evaluated for fine-mapping, all methods output similarly sized 90% credible sets (Figure 6), and MA-FOCUS’s PIPs correlate with PIPs computed by other approaches (Figure S18). Despite this, as discussed previously, we find evidence that MA-FOCUS is more successful than other approaches at identifying genes that are functionally associated with blood traits. Secondly, the gene expression data for our eQTL reference panel are derived from immortalized cell lines,37 which differ from complex living organisms in fundamental ways. Therefore, this tissue type may not be the most appropriate tissue for identifying causal relationships with blood traits. When we explore this scenario using simulations, we find that causal gene PIPs and sensitivity both are substantially reduced when poorly correlated tissue is used for one of the ancestries (Figure S10). We expect this effect would be exacerbated if we used a poorly correlated tissue to estimate weights for both ancestries. Thirdly, our eQTL reference panel and GWAS cohort for the AA ancestry represent genetically admixed individuals whose genomes are a combination of (West) African and European ancestry. Therefore, when estimating weights for this ancestry, the local ancestry at any given locus would include some proportion of European-derived genotypes. This likely introduces noise and further reduces the power of our weight estimates. In total, our analysis limitations motivate us to perform large-scale GWAS and eQTL studies on non-European-ancestry and admixed populations with comprehensive types of tissues and cell types.
Here, we describe some general caveats of our multi-ancestry TWAS fine-mapping approach. First, MA-FOCUS assumes that genes causal for complex traits are shared across ancestries, neglecting the possibility of ancestry-specific causal genes. However, because several large-scale multi-ancestry GWASs have shown that most risk signals replicate in ancestries, we believe this to be a relatively minor issue.18, 19, 20, 21, 22 Second, MA-FOCUS models complex traits as a linear combination of steady-state gene expression, neglecting potential gene-environment interaction (GxE) or gene-gene interaction (GxG). While several works have supported linear assumptions for complex traits through large-scale GWAS results,42,85 recent work analyzing large-scale GWAS from multiple ancestries has provided evidence that allelic heterogeneity across ancestries may be due to GxE,19 and we acknowledge this as an interesting potential direction.
Overall, MA-FOCUS provides Bayesian inference on gene causality for complex traits in specific genomic regions, leveraging GWAS, eQTL, and LD data of multiple ancestries. It improves precision in gene fine-mapping by accounting for eQTL and LD heterogeneity across different ancestral groups and sheds light on the genetic architecture of complex traits.
Acknowledgments
We are thankful to Drs. Sharon Kardia and Jennifer Smith for providing resources to enable model fitting in GENOA eQTL data. GENOA genotype and gene expression data were supported by grants from NHLBI (HL054457, HL054464, HL054481, HL119443, and HL087660). In addition, this work was funded in part by the National Institutes of Health (NIH) under awards R01HG012133, R01GM140287, R01HG006399, P01CA196569, and U01CA261339.
Declaration of interests
The authors declare no competing interests.
Published: August 4, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.07.002.
Contributor Information
Zeyun Lu, Email: zeyunlu@usc.edu.
Nicholas Mancuso, Email: nicholas.mancuso@med.usc.edu.
Web resources
ADMIXTURE, https://dalexander.github.io/admixture/index.html
bedtools, https://bedtools.readthedocs.io/en/latest/
EnrichR, https://cran.r-project.org/web/packages/enrichR/index.html
FUSION, http://gusevlab.org/projects/fusion/
GCTA, https://cnsgenomics.com/software/gcta/
ggvenn, https://github.com/yanlinlin82/ggvenn
LDSC, https://github.com/bulik/ldsc
MESH, https://www.nlm.nih.gov/mesh/meshhome.html
PLINK, https://www.cog-genomics.org/plink/
pong, https://github.com/ramachandran-lab/pong
Silver analysis, https://github.com/hakyimlab/silver-standard-performance
Supplemental information
Data and code availability
MA-FOCUS software: https://github.com/mancusolab/ma-focus
LCL prediction models, sample GWAS statistics, and LD reference data: https://www.mancusolab.com/ma-focus
Analysis codes, and complete TWAS fine-mapping results: https://github.com/mancusolab/MA-FOCUS-data-code
GEUVADIS data: https://www.internationalgenome.org/data-portal/data-collection/geuvadis
The dbGaP accession number for GENOA genotype data: phs001238.v2.p1
The GEO accession numbers for GENOA gene expression data: GSE138914 for AA and GSE49531 for EA
We complied with the data use agreements for the GEUVADIS and GENOA datasets.
References
- 1.Gusev A., Ko A., Shi H., Bhatia G., Chung W., Penninx B.W.J.H., Jansen R., de Geus E.J.C., Boomsma D.I., Wright F.A., et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamazon E.R., Wheeler H.E., Shah K.P., Mozaffari S.V., Aquino-Michaels K., Carroll R.J., Eyler A.E., Denny J.C., Cox N.J., Im H.K., GTEx Consortium A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu Z., Zhang F., Hu H., Bakshi A., Robinson M.R., Powell J.E., Montgomery G.W., Goddard M.E., Wray N.R., Visscher P.M., Yang J. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat. Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 4.Maurano M.T., Humbert R., Rynes E., Thurman R.E., Haugen E., Wang H., Reynolds A.P., Sandstrom R., Qu H., Brody J., et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gusev A., Lee S.H., Trynka G., Finucane H., Vilhjálmsson B.J., Xu H., Zang C., Ripke S., Bulik-Sullivan B., Stahl E., et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am. J. Hum. Genet. 2014;95:535–552. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roadmap Epigenomics Consortium. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gusev A., Mancuso N., Won H., Kousi M., Finucane H.K., Reshef Y., Song L., Safi A., Neale B.M., McCarroll S., et al. Schizophrenia Working Group of the Psychiatric Genomics Consortium Transcriptome-wide association study of schizophrenia and chromatin activity yields mechanistic disease insights. Nat. Genet. 2018;50:538–548. doi: 10.1038/s41588-018-0092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mancuso N., Shi H., Goddard P., Kichaev G., Gusev A., Pasaniuc B. Integrating gene expression with summary association statistics to identify genes associated with 30 complex traits. Am. J. Hum. Genet. 2017;100:473–487. doi: 10.1016/j.ajhg.2017.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancuso N., Gayther S., Gusev A., Zheng W., Penney K.L., Kote-Jarai Z., Eeles R., Freedman M., Haiman C., Pasaniuc B., PRACTICAL consortium Large-scale transcriptome-wide association study identifies new prostate cancer risk regions. Nat. Commun. 2018;9:4079. doi: 10.1038/s41467-018-06302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mancuso N., Freund M.K., Johnson R., Shi H., Kichaev G., Gusev A., Pasaniuc B. Probabilistic fine-mapping of transcriptome-wide association studies. Nat. Genet. 2019;51:675–682. doi: 10.1038/s41588-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wainberg M., Sinnott-Armstrong N., Mancuso N., Barbeira A.N., Knowles D.A., Golan D., Ermel R., Ruusalepp A., Quertermous T., Hao K., et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawlor D.A., Harbord R.M., Sterne J.A.C., Timpson N., Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 13.Barfield R., Feng H., Gusev A., Wu L., Zheng W., Pasaniuc B., Kraft P. Transcriptome-wide association studies accounting for colocalization using Egger regression. Genet. Epidemiol. 2018;42:418–433. doi: 10.1002/gepi.22131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davey Smith G., Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 2014;23:R89–R98. doi: 10.1093/hmg/ddu328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowden J., Davey Smith G., Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pierce B.L., Burgess S. Efficient design for Mendelian randomization studies: subsample and 2-sample instrumental variable estimators. Am. J. Epidemiol. 2013;178:1177–1184. doi: 10.1093/aje/kwt084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen M.-H., Raffield L.M., Mousas A., Sakaue S., Huffman J.E., Moscati A., Trivedi B., Jiang T., Akbari P., Vuckovic D., et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746, 667 individuals from 5 global populations. Cell. 2020;182:1198–1213.e14. doi: 10.1016/j.cell.2020.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi H., Gazal S., Kanai M., Koch E.M., Schoech A.P., Siewert K.M., Kim S.S., Luo Y., Amariuta T., Huang H., et al. Population-specific causal disease effect sizes in functionally important regions impacted by selection. Nat. Commun. 2021;12:1098. doi: 10.1038/s41467-021-21286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wojcik G.L., Graff M., Nishimura K.K., Tao R., Haessler J., Gignoux C.R., Highland H.M., Patel Y.M., Sorokin E.P., Avery C.L., et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi H., Burch K.S., Johnson R., Freund M.K., Kichaev G., Mancuso N., Manuel A.M., Dong N., Pasaniuc B. Localizing components of shared transethnic genetic architecture of complex traits from GWAS summary data. Am. J. Hum. Genet. 2020;106:805–817. doi: 10.1016/j.ajhg.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakaue S., Kanai M., Tanigawa Y., Karjalainen J., Kurki M., Koshiba S., Narita A., Konuma T., Yamamoto K., Akiyama M., et al. A cross-population atlas of genetic associations for 220 human phenotypes. Nat. Genet. 2021;53:1415–1424. doi: 10.1038/s41588-021-00931-x. [DOI] [PubMed] [Google Scholar]
- 23.Martin A.R., Gignoux C.R., Walters R.K., Wojcik G.L., Neale B.M., Gravel S., Daly M.J., Bustamante C.D., Kenny E.E. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 2017;100:635–649. doi: 10.1016/j.ajhg.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncan L., Shen H., Gelaye B., Meijsen J., Ressler K., Feldman M., Peterson R., Domingue B. Analysis of polygenic risk score usage and performance in diverse human populations. Nat. Commun. 2019;10:3328. doi: 10.1038/s41467-019-11112-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown B.C., Ye C.J., Price A.L., Zaitlen N., Asian Genetic Epidemiology Network Type 2 Diabetes Consortium Transethnic genetic-correlation estimates from summary statistics. Am. J. Hum. Genet. 2016;99:76–88. doi: 10.1016/j.ajhg.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wyss A.B., Sofer T., Lee M.K., Terzikhan N., Nguyen J.N., Lahousse L., Latourelle J.C., Smith A.V., Bartz T.M., Feitosa M.F., et al. Multiethnic meta-analysis identifies ancestry-specific and cross-ancestry loci for pulmonary function. Nat. Commun. 2018;9:2976. doi: 10.1038/s41467-018-05369-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kichaev G., Pasaniuc B. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 2015;97:260–271. doi: 10.1016/j.ajhg.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiorica P.N., Schubert R., Morris J.D., Abdul Sami M., Wheeler H.E. Multi-ethnic transcriptome-wide association study of prostate cancer. PLoS One. 2020;15:e0236209. doi: 10.1371/journal.pone.0236209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhattacharya A., García-Closas M., Olshan A.F., Perou C.M., Troester M.A., Love M.I. A framework for transcriptome-wide association studies in breast cancer in diverse study populations. Genome Biol. 2020;21:42. doi: 10.1186/s13059-020-1942-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharya A., Hirbo J.B., Zhou D., Zhou W., Zheng J., Kanai M., Pasaniuc B., Gamazon E.R., Cox N.J., the Global Biobank Meta-analysis Initiative Best practices for multi-ancestry, meta-analytic transcriptome-wide association studies: lessons from the Global Biobank Meta-analysis Initiative. 2021. [DOI] [PMC free article] [PubMed]
- 31.Wellcome Trust Case Control Consortium. Maller J.B., McVean G., Byrnes J., Vukcevic D., Palin K., Su Z., Howson J.M.M., Auton A., Myers S., et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat. Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kichaev G., Yang W.-Y., Lindstrom S., Hormozdiari F., Eskin E., Price A.L., Kraft P., Pasaniuc B. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hormozdiari F., Kichaev G., Yang W.-Y., Pasaniuc B., Eskin E. Identification of causal genes for complex traits. Bioinformatics. 2015;31:i206–i213. doi: 10.1093/bioinformatics/btv240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith S.P., Shahamatdar S., Cheng W., Zhang S., Paik J., Graff M., Haiman C., Matise T.C., North K.E., Peters U., et al. Enrichment analyses identify shared associations for 25 quantitative traits in over 600, 000 individuals from seven diverse ancestries. Am. J. Hum. Genet. 2022;109:871–884. doi: 10.1016/j.ajhg.2022.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shang L., Smith J.A., Zhao W., Kho M., Turner S.T., Mosley T.H., Kardia S.L.R., Zhou X. Genetic architecture of gene expression in European and african Americans: an eQTL mapping study in GENOA. Am. J. Hum. Genet. 2020;106:496–512. doi: 10.1016/j.ajhg.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi H., Kichaev G., Pasaniuc B. Contrasting the genetic architecture of 30 complex traits from summary association data. Am. J. Hum. Genet. 2016;99:139–153. doi: 10.1016/j.ajhg.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berisa T., Pickrell J.K. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32:283–285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wray N.R., Yang J., Hayes B.J., Price A.L., Goddard M.E., Visscher P.M. Pitfalls of predicting complex traits from SNPs. Nat. Rev. Genet. 2013;14:507–515. doi: 10.1038/nrg3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ongen H., Brown A.A., Delaneau O., Panousis N.I., Nica A.C., GTEx Consortium. Dermitzakis E.T. Estimating the causal tissues for complex traits and diseases. Nat. Genet. 2017;49:1676–1683. doi: 10.1038/ng.3981. [DOI] [PubMed] [Google Scholar]
- 44.Liu X., Finucane H.K., Gusev A., Bhatia G., Gazal S., O’Connor L., Bulik-Sullivan B., Wright F.A., Sullivan P.F., Neale B.M., Price A.L. Functional architectures of local and distal regulation of gene expression in multiple human tissues. Am. J. Hum. Genet. 2017;100:605–616. doi: 10.1016/j.ajhg.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup Genome project data processing subgroup (2009). The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Loh P.-R., Danecek P., Palamara P.F., Fuchsberger C., A Reshef Y., K Finucane H., Schoenherr S., Forer L., McCarthy S., Abecasis G.R., et al. Reference-based phasing using the haplotype reference consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J., Lee S.H., Goddard M.E., Visscher P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.HapMap Consortium International. The international HapMap project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 52.Alexander D.H., Novembre J., Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Behr A.A., Liu K.Z., Liu-Fang G., Nakka P., Ramachandran S. Pong: fast analysis and visualization of latent clusters in population genetic data. Bioinformatics. 2016;32:2817–2823. doi: 10.1093/bioinformatics/btw327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lappalainen T., Sammeth M., Friedländer M.R., ’t Hoen P.A.C., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G., et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bulik-Sullivan B.K., Loh P.-R., Finucane H.K., Ripke S., Yang J., Patterson N., Daly M.J., Price A.L., Neale B.M., Schizophrenia Working Group of the Psychiatric Genomics Consortium LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malone J., Holloway E., Adamusiak T., Kapushesky M., Zheng J., Kolesnikov N., Zhukova A., Brazma A., Parkinson H. Modeling sample variables with an experimental factor ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keys K.L., Mak A.C.Y., White M.J., Eckalbar W.L., Dahl A.W., Mefford J., Mikhaylova A.V., Contreras M.G., Elhawary J.R., Eng C., et al. On the cross-population generalizability of gene expression prediction models. PLoS Genet. 2020;16:e1008927. doi: 10.1371/journal.pgen.1008927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diedenhofen B., Musch J. cocor: a comprehensive solution for the statistical comparison of correlations. PLoS One. 2015;10:e0121945. doi: 10.1371/journal.pone.0121945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mogil L.S., Andaleon A., Badalamenti A., Dickinson S.P., Guo X., Rotter J.I., Johnson W.C., Im H.K., Liu Y., Wheeler H.E. Genetic architecture of gene expression traits across diverse populations. PLoS Genet. 2018;14:e1007586. doi: 10.1371/journal.pgen.1007586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mikhaylova A.V., Thornton T.A. Accuracy of gene expression prediction from Genotype data with PrediXcan varies across and within continental populations. Front. Genet. 2019;10:261. doi: 10.3389/fgene.2019.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liang Y., Pividori M., Manichaikul A., Palmer A.A., Cox N.J., Wheeler H.E., Im H.K. Polygenic transcriptome risk scores (PTRS) can improve portability of polygenic risk scores across ancestries. Genome Biol. 2022;23 doi: 10.1186/s13059-021-02591-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hu X., Qiao D., Kim W., Moll M., Balte P.P., Lange L.A., Bartz T.M., Kumar R., Li X., Yu B., et al. Polygenic transcriptome risk scores for COPD and lung function improve cross-ethnic portability of prediction in the NHLBI TOPMed program. Am. J. Hum. Genet. 2022;109:857–870. doi: 10.1016/j.ajhg.2022.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wondimu A., Weir L., Robertson D., Mezentsev A., Kalachikov S., Panteleyev A.A. Loss of Arnt (Hif1β) in mouse epidermis triggers dermal angiogenesis, blood vessel dilation and clotting defects. Lab. Invest. 2012;92:110–124. doi: 10.1038/labinvest.2011.134. [DOI] [PubMed] [Google Scholar]
- 66.Slager S.L., Skibola C.F., Di Bernardo M.C., Conde L., Broderick P., McDonnell S.K., Goldin L.R., Croft N., Holroyd A., Harris S., et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood. 2012;120:843–846. doi: 10.1182/blood-2012-03-413591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Q., Wu Y., Zhang Y., Sun H., Lu Z., Du K., Fang S., Li W. miR-125b regulates cell progression in chronic myeloid leukemia via targeting BAK1. Am. J. Transl. Res. 2016;8:447–459. [PMC free article] [PubMed] [Google Scholar]
- 68.Li Y., Zhuang J. miR-345-3p serves a protective role during gestational diabetes mellitus by targeting BAK1. Exp. Ther. Med. 2021;21:2. doi: 10.3892/etm.2020.9434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kowalczyk M.S., Hughes J.R., Babbs C., Sanchez-Pulido L., Szumska D., Sharpe J.A., Sloane-Stanley J.A., Morriss-Kay G.M., Smoot L.B., Roberts A.E., et al. Nprl3 is required for normal development of the cardiovascular system. Mamm. Genome. 2012;23:404–415. doi: 10.1007/s00335-012-9398-y. [DOI] [PubMed] [Google Scholar]
- 70.Miyata M., Gillemans N., Hockman D., Demmers J.A.A., Cheng J.-F., Hou J., Salminen M., Fisher C.A., Taylor S., Gibbons R.J., et al. An evolutionarily ancient mechanism for regulation of hemoglobin expression in vertebrate red cells. Blood. 2020;136:269–278. doi: 10.1182/blood.2020004826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sapkota Y., Qin N., Ehrhardt M.J., Wang Z., Wilson C.L., Estepp J., Rai P., Hankins J.E., Burridge P., Jefferies J.L., et al. Cardiomyopathy risk among childhood cancer survivors of African ancestry and its molecular mechanisms. J. Clin. Oncol. 2020;38:10514. [Google Scholar]
- 72.Douroudis K., Kisand K., Nemvalts V., Rajasalu T., Uibo R. Allelic variants in the PHTF1-PTPN22, C12orf30 and CD226 regions as candidate susceptibility factors for the type 1 diabetes in the Estonian population. BMC Med. Genet. 2010;11:11. doi: 10.1186/1471-2350-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang X., Geng S., Weng J., Lu Z., Zeng L., Li M., Deng C., Wu X., Li Y., Du X. Analysis of the expression of PHTF1 and related genes in acute lymphoblastic leukemia. Cancer Cell Int. 2015;15:93. doi: 10.1186/s12935-015-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reiling E., van Vliet-Ostaptchouk J.V., van ’t Riet E., van Haeften T.W., Arp P.A., Hansen T., Kremer D., Groenewoud M.J., van Hove E.C., Romijn J.A., et al. Genetic association analysis of 13 nuclear-encoded mitochondrial candidate genes with type II diabetes mellitus: the DAMAGE study. Eur. J. Hum. Genet. 2009;17:1056–1062. doi: 10.1038/ejhg.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Talukdar H.A., Foroughi Asl H., Jain R.K., Ermel R., Ruusalepp A., Franzén O., Kidd B.A., Readhead B., Giannarelli C., Kovacic J.C., et al. Cross-tissue regulatory gene networks in coronary artery disease. Cell Syst. 2016;2:196–208. doi: 10.1016/j.cels.2016.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bordeleau M.-E., Aucagne R., Chagraoui J., Girard S., Mayotte N., Bonneil E., Thibault P., Pabst C., Bergeron A., Barabé F., et al. UBAP2L is a novel BMI1-interacting protein essential for hematopoietic stem cell activity. Blood. 2014;124:2362–2369. doi: 10.1182/blood-2014-01-548651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhao W.-Y., Wang Y., An Z.-J., Shi C.-G., Zhu G.-A., Wang B., Lu M.-Y., Pan C.-K., Chen P. Downregulation of miR-497 promotes tumor growth and angiogenesis by targeting HDGF in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2013;435:466–471. doi: 10.1016/j.bbrc.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 78.Thirant C., Galan-Moya E.-M., Dubois L.G., Pinte S., Chafey P., Broussard C., Varlet P., Devaux B., Soncin F., Gavard J., et al. Differential proteomic analysis of human glioblastoma and neural stem cells reveals HDGF as a novel angiogenic secreted factor. Stem Cell. 2012;30:845–853. doi: 10.1002/stem.1062. [DOI] [PubMed] [Google Scholar]
- 79.Bruin M., Bierings M., Uiterwaal C., Révész T., Bode L., Wiesman M.-E., Kuijpers T., Tamminga R., de Haas M. Platelet count, previous infection and FCGR2B genotype predict development of chronic disease in newly diagnosed idiopathic thrombocytopenia in childhood: results of a prospective study. Br. J. Haematol. 2004;127:561–567. doi: 10.1111/j.1365-2141.2004.05235.x. [DOI] [PubMed] [Google Scholar]
- 80.Willcocks L.C., Carr E.J., Niederer H.A., Rayner T.F., Williams T.N., Yang W., Scott J.A.G., Urban B.C., Peshu N., Vyse T.J., et al. A defunctioning polymorphism in FCGR2B is associated with protection against malaria but susceptibility to systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA. 2010;107:7881–7885. doi: 10.1073/pnas.0915133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tell G.S., Grimm R.H., Jr., Vellar O.D., Theodorsen L. The relationship of white cell count, platelet count, and hematocrit to cigarette smoking in adolescents: the Oslo Youth Study. Circulation. 1985;72:971–974. doi: 10.1161/01.cir.72.5.971. [DOI] [PubMed] [Google Scholar]
- 82.Jesri A., Okonofua E.C., Egan B.M. Platelet and white blood cell counts are elevated in patients with the metabolic syndrome. J. Clin. Hypertens. 2005;7:705–711. doi: 10.1111/j.1524-6175.2005.04809.x. quiz 712-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Santimone I., Di Castelnuovo A., De Curtis A., Spinelli M., Cugino D., Gianfagna F., Zito F., Donati M.B., Cerletti C., de Gaetano G., et al. White blood cell count, sex and age are major determinants of heterogeneity of platelet indices in an adult general population: results from the MOLI-SANI project. Haematologica. 2011;96:1180–1188. doi: 10.3324/haematol.2011.043042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Floto R.A., Clatworthy M.R., Heilbronn K.R., Rosner D.R., MacAry P.A., Rankin A., Lehner P.J., Ouwehand W.H., Allen J.M., Watkins N.A., Smith K.G.C. Loss of function of a lupus-associated FcgammaRIIb polymorphism through exclusion from lipid rafts. Nat. Med. 2005;11:1056–1058. doi: 10.1038/nm1288. [DOI] [PubMed] [Google Scholar]
- 85.Barbeira A.N., Bonazzola R., Gamazon E.R., Liang Y., Park Y., Kim-Hellmuth S., Wang G., Jiang Z., Zhou D., Hormozdiari F., et al. Exploiting the GTEx resources to decipher the mechanisms at GWAS loci. Genome Biol. 2021;22:49. doi: 10.1186/s13059-020-02252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
MA-FOCUS software: https://github.com/mancusolab/ma-focus
LCL prediction models, sample GWAS statistics, and LD reference data: https://www.mancusolab.com/ma-focus
Analysis codes, and complete TWAS fine-mapping results: https://github.com/mancusolab/MA-FOCUS-data-code
GEUVADIS data: https://www.internationalgenome.org/data-portal/data-collection/geuvadis
The dbGaP accession number for GENOA genotype data: phs001238.v2.p1
The GEO accession numbers for GENOA gene expression data: GSE138914 for AA and GSE49531 for EA
We complied with the data use agreements for the GEUVADIS and GENOA datasets.